Abstract
The Graffi murine leukemia virus is a nondefective ecotropic retrovirus that was originally reported to induce myeloid leukemia in some strains of mice (A. Graffi, Ann. N.Y. Acad. Sci. 68:540-558, 1957). Using provirus-flanking sequences as DNA probes, we identified a new common retroviral integration site called Gris1 (for Graffi integration site 1). Viral integrations in Gris1 were detected in 13% of the tumors analyzed. The Gris1 locus was mapped to the distal region of mouse chromosome 6, 85 kb upstream of the cyclin D2 gene. Such viral integration in Gris1 causes overexpression of the normal 6.5-kb major transcript of cyclin D2 but also induces the expression of a new, alternatively spliced 1.1-kb transcript from the cyclin D2 gene that encodes a truncated cyclin D2 of 17 kDa. The expression of this 1.1-kb transcript is specific to tumors in which Gris1 is rearranged but is also detected at low levels in normal tissue.
Insertional mutagenesis is the mechanism by which nondefective murine retroviruses can perturb normal hematopoiesis and cause leukemia. The replication cycle of retroviruses results in the integration of the reverse-transcribed genome into the host genomic DNA. When an integration occurs in the vicinity of a cellular proto-oncogene, the strong regulatory elements present in the long terminal repeat (LTR) of the retrovirus can override the expression of that proto-oncogene, a major step toward tumor formation. Therefore, the proviruses present in the tumors can be used as molecular tags to identify the altered genes. Over the years, the use of these retroviruses has proven to be very successful in the identification of many proto-oncogenes and tumor suppressor genes (for reviews, see references 16 and 44).
The Graffi viral complex was first isolated from a cell-free filtrate of Ehrlich sarcoma and induced almost exclusively myeloid leukemias when inoculated in newborns of some strains of mice (10). Two infectious molecular clones (GV-1.2 and GV-1.4) have been isolated from that viral complex and characterized (30). These two molecular clones are very similar in structure, except that clone GV-1.2 induces the disease with a shorter latency period and shows a perfect 60-bp duplication in the U3 enhancer region of the long terminal repeat (LTR) (30). Newborn BALB/c and NFS mice inoculated with one of the two molecular clones or with the parental mixture develop, to different degrees, hepatosplenomegaly, thymic enlargement, generalized lymphadenopathy, and anemia in the leukemic stage. Therefore, the Graffi murine leukemia virus (MuLV) was considered an excellent model to study the mechanisms of myeloid leukemia induction and progression and to identify novel proto-oncogenes.
The exact mechanism by which the Graffi virus induces leukemia is not well understood. However, activation of cellular proto-oncogenes by insertional mutagenesis is a key step in the leukemic process. We have previously reported activation of the c-myc, Fli1, Pim1, and Spi1/PU.1 genes by Graffi virus integration in 20, 10, 3.3, and 3.3%, respectively, of the tumors tested (6). In an attempt to identify new cellular proto-oncogenes, we have cloned and characterized a novel viral integration site from a Graffi MuLV-induced tumor.
In this study, we report the identification and characterization of Gris1 (for Graffi integration site 1), a new common site of integration in Graffi MuLV-induced tumors. Viral integrations in Gris1 were detected in 13% of the tumors analyzed. The Gris1 locus was mapped to the distal region of mouse chromosome 6 and is located 85 kbp upstream of the cyclin D2 gene. The viral integration in Gris1 increases the expression of the 6.5-kb major transcript of the cyclin D2 gene and activates the expression of a new, yet-uncharacterized alternative 1.1-kb transcript from the same gene.
MATERIALS AND METHODS
Primary tumors.
NFS/N and BALB/c mice were purchased from Frederick Cancer Research Facility, Frederick, Md., and from Charles River Inc., St. Constant, Quebec, Canada, respectively. Newborn mice (<48 h old) were inoculated intraperitoneally with approximately 105 PFU in 0.1 to 0.2 ml of filtered viral supernatant. The inoculated mice were observed daily and sacrificed when they showed signs of advanced disease, and the tumors were frozen immediately on dry ice.
Genomic-DNA and -RNA analysis.
High-molecular-weight genomic DNAs were extracted from frozen normal and leukemic tissues by standard procedures as described previously (31). For Southern analysis, 15 μg of genomic DNA from tumors or control organs was digested with a suitable restriction endonuclease, separated on a 0.7% agarose gel, and transferred to a nylon membrane (Osmonics).
Total RNA was extracted from frozen normal and leukemic tissues with the TRIzol reagent (Life Technologies) according to the instructions of the manufacturer. For Northern analysis, 20 μg of total RNA was separated on a 1.2% formaldehyde agarose gel as described previously (34) and transferred to a nylon membrane (Osmonics).
All the blots were hybridized in 50% formamide-10% dextran sulfate-0.5% sodium dodecyl sulfate (SDS)-1 M NaCl at 42°C with the appropriate probe. Labeling of the probe (gel-purified fragments) was done by the random primer extension method using oligohexamers (Pharmacia P-L Biochemicals, Montreal, Canada). The membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl-0.015 M sodium citrate) for 30 min at 42°C and in 0.2× SSC-0.1% SDS for 30 min at 60°C.
Viral insertion site amplification.
Amplification of the provirus-cellular DNA junctions was performed by PCR (37). Tumor DNAs (100 ng) were subjected to a first round of amplification using a biotinylated primer specific for the Graffi U3 region of the LTR (5′ AGGCGCAAGGTCGTTTCAGGTCTTTGGG 3′) and one of five partially degenerated primers (FP) described previously (36). The amplified biotinylated fragments were isolated on streptavidin-coated magnetic particles (Boehringer Mannheim). A second PCR was then performed on the recovered biotinylated amplicon using a nested Graffi virus-specific primer (5′ ATGGGTCTCTTGAAACTGCTGAGGG 3′) and a primer corresponding to the nondegenerated portion of the FP primer used in the first PCR (5′ CAGTTCAAGCTTGTCCAGGAATTC 3′). For tumors 6 and 8, viral integrations in the region between Gris1 and Ccnd2 were analyzed by PCR using a series of 10 primers specific for the genomic region between Gris1 and Ccnd2 (one primer every 10 kbp) in combination with one of the two primers specific for the Graffi LTR. The primers specific for the Graffi LTR were Graffi 254-229 (5′ GGGGCAACCTGGAAACATCTGATGGG 3′) and Graffi 10-34 (5′ CCCCACCATAAGGCTTAGCAAGCTAG 3′). The genomic specific primers were designed according to the sequence obtained at the Ensembl mouse genome server (http://www.ensembl.org/Mus_musculus/) for the chromosome 6 region 128.05 to 128.15 Mb. The primers were as follows: GRIS1F, 5′ GTCTCTTTTGTTGCTTTGAGGGGC 3′; GRIS2F, 5′ GAGAAGGTGAATGGTTTTTATGCCC 3′; GRIS3F, 5′ CAGAGTTAGAACCTGGGTGTGTGC 3′; GRIS4F, 5′ CTGACAAGCCGCCCTAGGTG 3′; GRIS5F, 5′ GTCCATTTTCCAGGCAGGACTGAG 3′; GRIS6F, 5′ CTGAGCCCTCCATATCAGTCATCA 3′; GRIS7F, 5′ TTGATTTTTGCCTGCTTGTCCTCAC 3′; GRIS8F, 5′ TCTTCCACTTCCTGGTTTGGTATC 3′; GRIS9F, 5′ CAGGAGTAACGGGAGAGAGCTA 3′; and GRIS10F, 5′ TGCGTTAAGAGGACTGAGACCC 3′.
Genetic mapping.
Gris1 was mapped by Southern blot analysis of two sets of genetic crosses: (NFS/N or C58/J × Mus mus musculus)F1 × M. m. musculus (20) and (NFS/N × Mus spretus) × M. spretus or C58/J (1). Progeny of these crosses have been typed for over 1,200 markers distributed over all 19 autosomes and the X chromosome. Linkage distances were determined from the number of recombinants, and loci were ordered by minimizing the number of recombinations.
Genomic-clone isolation.
BAC genomic clones containing the Gris1 locus were isolated by screening the RPCI-129S6 segment 2 mouse BAC library (BACPAC resources; CHORI, Oakland, Calif.) with Gris1 probe B.
Shotgun sequencing.
The shotgun library was constructed as described previously (43). Briefly, BAC DNA was prepared by large-scale alkaline lysis with the Large Construct kit (Qiagen). DNA (20 μg) was sonicated in mung bean nuclease repair buffer (30 mM sodium acetate, 50 mM NaCl, 1 mM ZnCl2, 5% glycerol). Fragment ends were repaired at 30°C for 30 min with 40 U of mung bean nuclease (Amersham-Pharmacia Biotech). Samples were electrophoresed on a 1% agarose gel, and 800- to 1,200-bp fragments were gel purified and ligated into the EcoRV cloning site of pBlueScript II KS(+) (Stratagene). The ligation products were transformed into chemically competent Dh5α cells. For sequencing, plasmid DNA clones were prepared by small-scale alkaline lysis with the Miniprep kit from Qiagen. The sequencing reactions were performed with the Big-Dye Terminator Cycle Sequencing kit version 2.0 (Applied Biosystems, Foster City, Calif.). The sequencing reactions were run on a model 3700 DNA analyzer (Applied Biosystems).
RACE.
5′ and 3′ rapid amplification of cDNA ends (RACE) experiments were performed using the GeneRacer kit (Invitrogen). The RACE-ready cDNA pool was obtained using the GeneRacer oligo(dT) primer on tumor F6 total RNA. The amplification of the 5′ end of expressed sequence tag (EST) AW060277 was accomplished using the GeneRacer 5′ primer and the EST-specific primer 5′ GTCTTGGTTAGTGTGGCGGCCTTA 3′. The GeneRacer 3′ primer and the EST-specific primer 5′ TCACACTAAGGCCGCCACACACTAA 3′ were used to obtain the 3′ end.
Cell culture conditions, plasmid construction, and transfections.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% calf serum (Life Technologies). The pCMV-D2trc construct was made by cloning the complete coding sequence of the truncated cyclin D2 obtained by PCR with the following primers: 5′ TTTCACCCTCTAGAATTTCCATG 3′ and 5′ AGGAGGTAAGGGAAGCTTTCC 3′. The fragment was cloned in the HindIII-XbaI site of the pRcCMV vector (Invitrogen). Transfections were made with Polyfect (Qiagen) according to the manufacturer's instructions.
Antibody preparation, protein extraction, and Western blotting.
Polyclonal antibodies against the entire truncated cyclin D2 protein and also against the last 20 amino acids from the C-terminal end were generated. Briefly, the corresponding cDNA sequences were PCR amplified and cloned in frame with the glutathione S-tranferase protein into the pGEX-4T-1 vector (Amersham Biosciences). After induction, the bacterial extracts were passed on a glutathione-Sepharose 4B column (Amersham Biosciences), and the purified proteins were injected subcutaneously into New Zealand White rabbits. Serum was collected following clot retraction and stored at −70°C. NIH 3T3 fibroblasts transfected with the pCMV-D2trc plasmid were washed twice with ice-cold PBS and lysed in RIPA buffer (0.15 M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.05 M Tris-HCl [pH 8]) and complete protease inhibitor cocktail (Roche Applied Science) for 30 min at 4°C. Protein extracts from tumors and tissues were prepared in SDS-urea buffer (0.5% SDS, 8 M urea). The protein concentrations in the different extracts were measured using the Bio-Rad protein assay. Equal amounts of lysate proteins (30 to 80 μg) were diluted in 2× sample buffer (30% glycerol, 4% SDS, 160 mM Tris-HCl [pH 6.8], 10% β-mercaptoethanol, and 0.02% bromophenol blue) and boiled for 3 min. The proteins were separated on 12% SDS-polyacrylamide gel electrophoresis and electrophoretically transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). The membranes were blocked in PBS containing 0.2% Tween 20 and 4% skim milk powder (PBS-Tween-milk) for 1 h at room temperature. Incubation with the purified anti-truncated cyclin D2 peptide antibody (1:100 dilution in PBS-Tween-milk) or the anti-truncated cyclin D2 serum (1:5,000 dilution in PBS-Tween-milk) was performed for 1 h at room temperature. After incubation, the blots were washed in PBS containing 0.2% Tween 20 (PBS-Tween) and incubated for 1 h with an anti-rabbit immunoglobulin G peroxidase conjugate (1:5,000 in PBS-Tween-milk) (BD Pharmingen). The immune complexes were revealed using the ECL Plus chemiluminescence reagents (Amersham Biosciences).
RESULTS
Identification of Gris1, a common site of integration in Graffi MuLV-induced tumors.
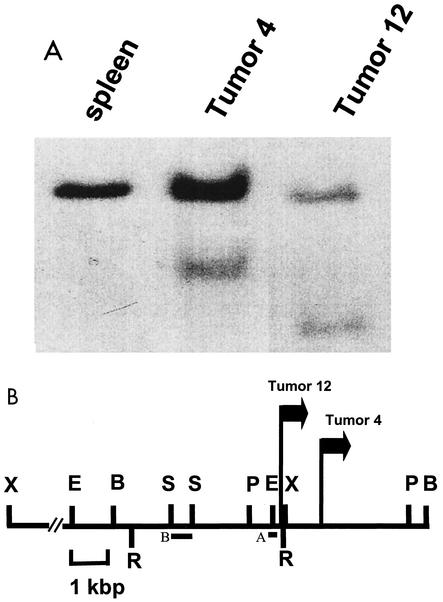
To identify new proto-oncogenes activated in Graffi MuLV-induced tumors, a PCR approach described by Sorensen and colleagues (37) was applied to obtain the cellular DNA flanking the 5′ end of the Graffi proviruses from tumor 12. A unique genomic PCR probe of ∼400 bp was generated from that tumor and used to screen a panel of 30 other tumor DNAs. On Southern blots, this probe (probe A [Fig. 1B]) could detect the rearranged allele in tumor 12 DNA and also in one other tumor (tumor 4) (Fig. 1A). The lower intensity of the rearranged allele compared with the normal allele in tumor 4 suggests that the viral integration at this site for that tumor is partially clonal. The two proviruses in that locus are both in the same orientation and are located 1 kbp apart from each other. Two out of 30 tumors tested contained a proviral integration in this novel locus that we have called Gris1.
FIG. 1.
Southern blot analysis of Graffi MuLV-induced leukemias. (A) Normal spleen and tumor DNAs digested with BamHI and hybridized with probe A. (B) Restriction map of the Gris1 locus. The positions and orientations of the proviruses are indicated by arrows. The probes used to detect rearrangements and for genetic mapping are shown as solid boxes (A and B, respectively). Probe A was obtained by PCR, and probe B was derived from the genomic DNA lambda clone. Abbreviations for restriction endonucleases: E, EcoRI; B, BamHI; R, EcoRV; S, SacI; P, PstI; X, XbaI. The XbaI fragment is 13 kbp in length.
The Gris1 probe A obtained by PCR was used to screen a genomic lambda phage library from mouse 129/Sv to obtain clones representing the germ line Gris1 locus. One 8-kb clone was recovered and used to construct a restriction map of the locus and to derive a longer unique probe (probe B) that was used subsequently for genetic mapping (Fig. 1B).
Genetic mapping of Gris1.
To determine the genetic-map location of Gris1, we examined the inheritance of this marker in two sets of genetic crosses. In the first set of crosses, Southern blot analysis identified 6.8-kb EcoRI fragments in M. m. musculus and 5.2-kb fragments in NFS/N mice. In the second set of crosses, SacII digestion produced 3.0-kb fragments in M. spretus and 1.2-kb fragments in NFS/N mice. The patterns of inheritance of the variant fragments in the two sets of crosses were compared with those of the 1,200 markers previously typed and mapped. Linkage was observed with markers on distal chromosome 6. The gene order and recombinational distances are as follows: Raf1 (3.0 ± 1.1)-Tnfr1 Ltbr (0.4 ± 0.4)-Scnn1a (0.8 ± 0.8)-Gris1 Ccnd2 (5.2 ± 1.5)-Kras2.
Identification of genes activated by viral integration in Gris1.
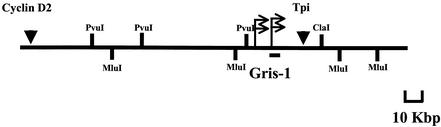
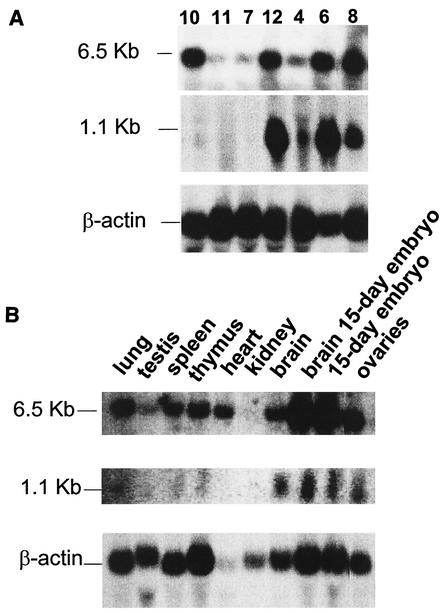
In order to identify potential candidate genes either activated or inactivated by the viral integration in Gris1, we first screened the 8-kb Gris1 genomic lambda clone for expressed sequences. However, no transcriptional unit could be detected by Northern blot analysis (data not shown). It is well known that gene activation by viral integration can occur over large distances (12, 21, 35). Therefore, we looked for candidate genes in a region of about 200 kbp surrounding the Gris1 insertion site. A 210-kbp contig of the Gris1 region was obtained by screening a mouse 129/Sv genomic BAC library. Two overlapping BAC clones were recovered and used to create a restriction map of the region (Fig. 2). The longest clone recovered was used in sample shotgun sequencing designed to obtain a 2.5-fold sequence redundancy of the 200-kbp region of Gris1. Each sequence was then analyzed individually against the NR, dbEST, and HTGS nucleotide databases using the BLAST algorithm. We found sequence homology in the databases with two known genes and with several ESTs unrelated to known genes. The two known genes were those for triose phosphate isomerase (Tpi) and cyclin D2 (Ccnd2). The positions of the two genes relative to Gris1 were determined by restriction mapping (Fig. 2). The Tpi gene was positioned approximately 30 kb upstream of Gris1, whereas the Ccnd2 gene was positioned about 85 kbp downstream (Fig. 2). To determine if expression of the Tpi and Ccnd2 genes was altered by the viral integrations in Gris1, genomic fragments that presented homologies with either Tpi or Ccnd2 were used in Northern blot analysis. Even though we positioned the Gris1 locus only 30 kbp from Tpi, no alterations in the expression profile of the 1.5-kb transcript of that gene could be detected in the tumors with viral integration in Gris1 compared with other Graffi tumors and control organs (data not shown). However, when the cyclin D2 expression was verified, the two tumors in which Gris1 is rearranged (tumors 4 and 12) showed the normal cyclin D2 message at the expected size of 6.5 kb, but surprisingly, the two tumors presented an additional intense 1.1-kb band (Fig. 3A). This new cyclin D2-specific band is absent in the tumors in which Gris1 is not rearranged (tumors 7, 10, and 11). Tumors 6 and 8 also presented an intense 1.1-kb band, even though we could not detect a DNA rearrangement in the Gris1 locus with probes A and B. Since the integration site is located about 85 kbp away from the cyclin D2 gene, we speculated that tumors 6 and 8 most probably harbored a viral integration somewhere between Gris1 and the cyclin D2 gene, outside the limit of probe A and B detection. To determine if a viral integration had occurred in the Gris1 locus in these two tumors, we PCR amplified tumors 6 and 8 with different pairs of primers, one specific for the Graffi MuLV U3 LTR and the other derived from every 10 kbp between the cyclin D2 gene and the Gris1 locus. This process was also performed with U3 and genomic primer pairs corresponding to the other retroviral orientation. The PCR products were hybridized with a U3 LTR-specific probe. As expected, we could detect a unique U3-specific fragment for each tumor with only one combination of primers (results not shown), confirming the presence of a provirus in the Gris1 locus in tumors 6 and 8 at 4 and 5.5 kbp, respectively, upstream of the XbaI site which was the 5′ limit of detection in the first analysis. The presence of provirus in the Gris1 locus in tumors 6 and 8 was also documented by Southern blot analysis using three different derived genomic probes (results not shown). As determined by PCR, the proviruses from tumors 6 and 8 were in the same orientation as those in tumors 4 and 12. Therefore, 4 tumors out of 30 (13%) presented a Graffi MuLV integration in the Gris1 locus and expressed a new 1.1-kb cyclin D2-specific mRNA.
FIG. 2.
Predicted physical map of the Gris1 genomic region. BAC clones recovered with the Gris1 probe were used for restriction mapping. The positions of the two genes found by random sequencing are indicated by inverted triangles above the map. Each gene was positioned on the map by hybridization to blots of MluI-, ClaI-, and PvuI-digested BAC clones. The small arrows above the map indicate the positions and orientations of the viral integrations for tumors 6 and 8 and tumors 4 and 12. The Gris1 probe used is shown and is not drawn to scale.
FIG. 3.
Northern blot analysis showing expression of the 6.5- and 1.1-kb transcripts of cyclin D2. Ten micrograms of RNA extracted from different Graffi MuLV-induced tumors (A) or from normal tissues (B) as indicated above the lanes was transferred to nylon membranes and hybridized with a cyclin D2 cDNA probe to reveal the 6.5-kb transcript and a genomic fragment derived from the cyclin D2 intron 2 to reveal the 1.1-kb transcript. Note that the cyclin D2 cDNA probe also reveals the 1.1-kb transcript. Size markers are indicated to the left of the panels. The same blots were also hybridized with a β-actin probe. (A) Tumors 7, 10, and 11 were obtained from BALB/c mice, and tumors 4, 6, 8, and 12 were obtained from NFS/N mice. Tumors 7, 8 and 12 were generated by the parental Graffi virus, and tumors 4, 6, 10, and 11 were generated by the GV-1.2 variant. The latency varied between 3 and 4 months.
The database searches also identified sequence similarity in the 200-kb region around the viral integrations with several mouse and human ESTs that presented no homology with any known genes (Table 1). Only one EST among those listed in Table 1 was found by Northern blot analysis to be modulated in the tumors in which Gris1 is rearranged compared with tumors in which Gris1 is not rearranged. Interestingly, this EST showed sequence homology with the 3′ end of Ccnd2 (exon 2 and intron 2 of the gene). A specific probe derived from the intron 2 region revealed a 1.1-kb transcript strongly overexpressed in one of three tumors in which Gris1 is rearranged (tumors 6, 8, and 12) (Fig. 3A) compared to tumors in which Gris1 is not rearranged, which do not express the 1.1-kb message. The lower level of expression detected in tumor 4 is due to the fact that this tumor shows a nonequimolar DNA rearrangement for Gris1 (Fig. 1A). The tumors in which Gris1 is rearranged (except tumor 4) also show high levels of the normal 6.5-kb cyclin D2 transcript. The 1.1-kb transcript corresponding to EST AW060277 was also detected at much lower levels in normal tissues (Fig. 3B). The highest expression was seen in tissues from the brain, the ovaries, 15-day-old embryo brain, and 15 day-old whole embryos (Fig. 3B).
TABLE 1.
Human and mouse ESTs identified by BLAST searches
| ESTa | Gene name or homology |
|---|---|
| AW813396 | None |
| BB028209 | None |
| AV360664 | None |
| AI853374 | None |
| BB272521 | None |
| AI523988 | None |
| AW060277 | Cyclin D2 exon 2 and intron 2 |
GenBank accession number.
The 1-kb transcript of EST AW060277 encodes a truncated cyclin D2.
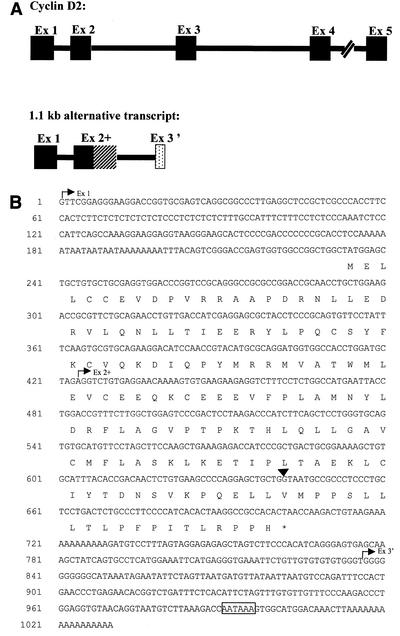
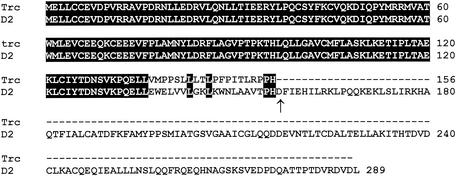
A 1,023-bp cDNA clone representing the full-length EST AW060277 cDNA was obtained by performing 5′ and 3′ RACE on mRNA extracted from the tumor (Fig. 4). Analysis of the sequences of the RACE products revealed that this 1.1-kbp cDNA is in fact an alternative transcript of the cyclin D2 gene. It contains the sequence of cyclin D2 exon 1 spliced to a longer exon 2 (exon 2+) resulting from the utilization of an alternative splice donor site located in the intron region of the gene (Fig. 4A). The transcript contains an alternative 3′ exon which harbors a new consensus poly(A) signal (AATAAA) (Fig. 4B). The open reading frame of this cDNA predicts a 156-amino-acid protein representing a cyclin D2 with a truncated cyclin box. The use of an alternative splice site for exon 2 results in the addition of 20 novel amino acids to the first 136 amino acids of cyclin D2 and the premature termination of the protein due to the presence of a TAA stop codon just after residue 156 (Fig. 4B). The truncated cyclin D2 includes the LXCXE motif known to be implicated in binding to pRb and other pocket proteins (7, 8) and the highly conserved cyclin box lacking the last 23 amino acids (Fig. 5). Interestingly, sequence comparison between the full-length and the truncated cyclin D2 proteins showed sequence conservation of residues L143, L146, P155, and H156 in the region where the two proteins diverge at the C-terminal end of the cyclin box (Fig. 5).
FIG. 4.
Schematic representation of the normal and alternatively spliced cyclin D2 cDNAs. (A) The solid boxes represent the five exons (Ex 1 to -5) of the normal cyclin D2 gene. In the alternatively spliced 1.1-kb transcript, exons 1 and 2 are the same as those of the normal cyclin D2, and the hatched box represents the readtrough region which constitutes exon 2+. The stippled box represents the alternative exon 3′. (B) Nucleotide sequence of the 1.1-kb cDNA and the deduced amino acid sequence. The start points of the three exons are indicated by arrows. The arrowhead above the sequence indicates the end of the 6.5-kb transcript exon 2. The polyadenylation consensus signal is boxed.
FIG. 5.
Comparison of the amino acid sequences of the truncated (Trc) and the full-length (D2) cyclin D2. The first 136 residues are identical and encoded by the same sequences. The end of the cyclin box is indicated by an arrow. The solid boxes indicate identical amino acids. The dashes represent missing amino acids.
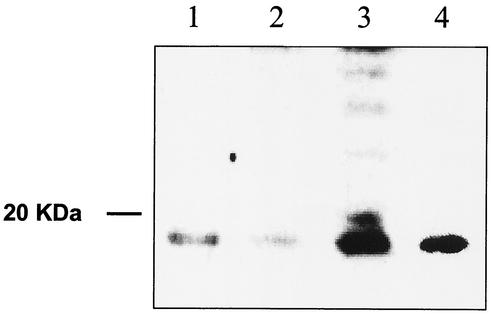
To verify if the truncated protein was present in normal and tumor tissues, specific antibodies were raised against the entire truncated protein and also against the last 20 amino acids from the C-terminal end. These specific antibodies were used to analyze protein extracts from tumors in which Gris1 is rearranged (Fig. 6). The truncated cyclin D2 protein is clearly detected at the expected mass of 17 kDa by the polyclonal serum raised against the 20-amino-acid sequence specific for this protein. Protein extracts from brain and ovary tissues, which were the major producers of the 1.1-kb transcript (Fig. 3B), were also analyzed and revealed the presence of the truncated protein as well. A control, protein extracts from NIH 3T3 fibroblasts transfected with pCMV-D2trc, an expression vector containing the cDNA encoding the truncated protein, also contained detectable amounts of the truncated protein (Fig. 6). Similar results were also obtained with the polyclonal serum raised against the complete truncated cyclin D2 protein (data not shown). This serum also detected the cyclin D2 protein at 34 kDa (data not shown).
FIG. 6.
Detection of the expressed product of the mouse truncated cyclin D2 in NIH 3T3 fibroblasts and the endogenous truncated cyclin D2 protein. Western blot analysis of normal tissues and Graffi MuLV-induced tumors is shown. Proteins were extracted from adult mouse brain (lane 1); ovaries (lane 2); tumor F6, in which Gris1 is rearranged (lane 3); and NIH 3T3 fibroblasts transfected with pCMV-D2trc plasmid (lane 4). Proteins (30 to 80 μg) were loaded on SDS-12% polyacrylamide gel electrophoresis gels and electrophoretically transferred to polyvinylidene difluoride membranes, and immunodetection was performed with the antibody raised against the last 20 amino acids of the truncated cyclin D2 protein.
Sequence analysis of the human cyclin D2 genomic locus and analysis of EST data bases revealed a number of human ESTs representing cDNA clones with a similar readthrough transcript for exon 2. One of these human ESTs was obtained (IMAGE:1703052) and sequenced. The open reading frame found on this human cDNA predicts, as for the mouse, an isoform of cyclin D2 with a truncated cyclin box (Fig. 7). Others have reported the isolation of a Xenopus laevis cyclin D2 cDNA that is predicted to encode a similar truncated cyclin D2 (38) (Fig. 7). However, these authors did not elucidate the possible role of such a transcript, and no further studies of this putative truncated cyclin D2 have been reported. Taken together, these results show that alternative transcripts encoding a cyclin D2 with a truncated cyclin box are conserved among human, mouse, and Xenopus and suggest an important role for this protein.
FIG. 7.
Comparison of the predicted amino acid sequences of the truncated cyclin D2 from mouse, Xenopus (accession no. Y10075), and human (IMAGE:1703052). The homology is indicated by solid boxes. Note that all three forms originate from a readtrough at the end of normal exon 2. The alignment was performed with CLUSTAL W (40).
DISCUSSION
In this study, proviral tagging was used as a strategy to identify new proto-oncogenes in a model of leukemia induced by the Graffi MuLV. We have identified a new locus, Gris1, which appears to be important in the development of leukemias, since it is rearranged in 4 of 30 tumors. Our results demonstrate that retroviral integrations in Gris1 have the ability to deregulate the expression of the cyclin D2 gene located 85 kbp downstream. Indeed, retroviral integration in Gris1 leads to increased expression of two transcripts of the cyclin D2 gene: the already-characterized 6.5-kb transcript encoding the known 289-amino-acid full-length protein (19) and a 1.1-kb transcript derived from alternative splicing which encodes a 156-amino-acid isoform of the cyclin D2 protein. Deregulated expression of the cyclin D2 gene has also been reported in T-cell leukemias induced by the BL/VL3 radiation leukemia virus (11, 41). In that case, two tumors were reported to be rearranged, but the BL/VL3 MuLV integrations were located in the 5′ noncoding region of the first cyclin D2 exon and in the same orientation as the gene. This resulted in the overexpression of the major 6.5-kb transcript. Interestingly, an abundant 1.1-kb band was also detected but not further characterized (41). We analyzed this transcript present in the BL/VL3-induced tumor 133-1 and found that it had the same mobility as the transcripts in tumors in which Gris1 is rearranged. More importantly, the transcript from tumor 133-1 hybridized strongly with the cyclin D2 intron 2 probe (result not shown). Therefore, retroviral integrations in the 5′ region of the cyclin D2 gene seem to induce the expression of an alternatively spliced transcript and a truncated protein as well. In our case, the Graffi proviruses are located approximately 85 kbp upstream of the cyclin D2 gene, suggesting activation through an enhancer insertion mechanism (29, 42). Enhanced expression of this gene by retroviral insertion also seems to alter its splicing regulation by a mechanism not yet investigated. Shorter transcripts (2.9 and 1.3 kb) of the cyclin D2 gene have also been reported in murine erythroleukemia cells (19) and in various human tumor cell lines (15, 24), indicating that the splicing regulation of this gene could be very complex. It is known that cyclin D2 act as a potent proto-oncogene, as it can transform primary rat embryo fibroblasts when overexpressed in conjunction with activated Ha-Ras (17). In addition, overexpression of cyclin D2 is often seen in various human malignancies, such as T-cell lymphoma, testicular carcinoma, and ovarian tumors (2, 33, 39). The data presented here show that the 1.1-kb transcript overexpressed in the Graffi tumors encodes a C-terminally truncated cyclin D2 protein. The transcript is unlikely to be an aberrant, nontranslated message, since the truncated protein at the expected mass of 17 kDa is clearly detectable in tumors in which Gris1 is rearranged and in normal tissues that express the 1.1-kb transcript. Thus, it is conceivable that this new isoform may also act as a proto-oncogene alone or in collaboration with the full-length isoform or with other oncogenes.
D-type cyclin expression is induced by mitogens during the transition of the G1-to-S phase of the cell cycle (24, 25, 32). These induced D-type cyclins form complexes with cyclin-dependent kinases (Cdk-4 and Cdk-6) via the highly conserved cyclin box (3, 23, 27). The recognized function of the cyclin D-Cdk4/6 complex is to phosphorylate pRb, and recent evidence suggests a role for the complex both in the regulation of the G0-to-G1 phase transition and in the regulation of cellular growth in cell cycle progression (5, 9, 26). The new isoform of cyclin D2 identified here is a C-terminally truncated protein that lacks 133 amino acids of the long isoform, including the last 23 amino acids from the cyclin box, but carries a new stretch of 20 amino acids. We do not know yet if this truncated cyclin D2 still retains its capacity to bind the Cdks. However, the short isoform presents sequence conservation with the long isoform in this region of 20 residues at residues L143, L146, P155, and H156, just at the end of the cyclin box, suggesting that these residues could have an important functional role, perhaps in the formation of a complex with the Cdks or with other interaction partners. Recent evidence points to Cdk-independent activation of D-type cyclins. Indeed, it was shown that cyclin D2 interacts with DMP1, a protein with Myb-like repeats, via its carboxyl-terminal half (residues 142 to 253) and that no ternary cyclin D/Cdk/DMP1 complexes could be detected (13). Most likely the short isoform cannot interact with DMP1, since it does not harbor the region important for binding.
As shown by our Northern blot analysis, the 6.5-kb transcript of the cyclin D2 gene is expressed at higher levels in virus-induced tumors, not only in those in which Gris1 is rearranged, and in a variety of nontransformed tissues. The more restricted pattern of expression of the 1.1-kb transcript indicates that the truncated cyclin could play a role in the development of specific tissues like the brain and ovaries, where it is most often expressed. Interestingly, mice bearing a disrupted cyclin D2 gene (with both the 6.5- and 1.1-kb transcripts disrupted) showed defects only in the proliferation of ovarian granulosa cells and granule and stellate interneurons of the cerebellum (33, 14). Since these tissues contain high levels of the 1.1-kb transcript, it is tempting to speculate that the truncated cyclin D2 could also have a role in these cell types.
Other members of the cyclin family have been shown to have alternatively spliced transcripts predicting truncated proteins. Such transcripts have been reported for cyclin E in tumor cells only (18, 28) and for cyclin D1 in a variety of normal and tumor cell lines (4). However, the alternatively spliced transcripts described for cyclin D1 were not similar to those found for cyclin D2, as they encoded a protein truncated in a very small portion of the C-terminal end. In addition, an alternatively spliced transcript coding for a shorter isoform of cyclin C with a truncated cyclin box was also reported (22). These findings indicate that shorter cyclin isoforms might play important roles in cell regulation, at least in some specific tissues. The functional relevance of a truncated cyclin D2 is supported by the presence of alternatively spliced messages coding for truncated cyclin D2 in EST databases for humans and characterized by Taieb and Jessus (38) in X. laevis.
We do not know the functional role of this truncated isoform of cyclin D2 in normal cells, but we are investigating its expression through the cell cycle and the specific interaction partners of this novel protein.
Acknowledgments
We are grateful to Elsy Edouard for helpful discussions and critical review of the manuscript. We thank Pierre Lepage and Tom Hudson (Montreal Genome Center) for sequencing and help in bioinformatics analysis.
This work was supported by grant 007072 from the National Cancer Institute of Canada and by grant 37994 from the Canadian Institutes of Health Research. C.D. is a recipient of a Cancer Research Society Inc. studentship and an FCAR Ph.D. scholarship.
REFERENCES
- 1.Adamson, M. C., J. Silver, and C. A. Kozak. 1991. The mouse homolog of the gibbon ape leukemia virus receptor: genetic mapping and a possible receptor function in rodents. Virology 183:778-781. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova, J., E. Rajpert-de Meyts, N. E. Skakkebaek, and J. Bartek. 1999. D-type cyclins in adult human testis and testicular cancer: relation to cell type, proliferation, differentiation, and malignancy. J. Pathol. 187:573-581. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., L. Bonetta, D. MacAllan, D. Parry, A. Holder, C. Dickson, and G. Peters. 1994. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9:71-79. [PubMed] [Google Scholar]
- 4.Betticher, D. C., N. Thatcher, H. J. Altermatt, P. Hoban, W. D. Ryder, and J. Heighway. 1995. Alternate splicing produces a novel cyclin D1 transcript. Oncogene 11:1005-1011. [PubMed] [Google Scholar]
- 5.Datar, S. A., H. W. Jacobs, A. F. de la Cruz, C. F. Lehner, and B. A. Edgar. 2000. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 19:4543-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denicourt, C., E. Edouard, and E. Rassart. 1999. Oncogene activation in myeloid leukemias by Graffi murine leukemia virus proviral integration. J. Virol. 73:4439-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowdy, S. F., P. W. Hinds, K. Louie, S. I. Reed, A. Arnold, and R. A. Weinberg. 1993. Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73:499-511. [DOI] [PubMed] [Google Scholar]
- 8.Ewen, M. E., H. K. Sluss, C. J. Sherr, H. Matsushime, J. Kato, and D. M. Livingston. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73:487-497. [DOI] [PubMed] [Google Scholar]
- 9.Ezhevsky, S. A., A. Ho, M. Becker-Hapak, P. K. Davis, and S. F. Dowdy. 2001. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol. Cell. Biol. 21:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graffi, A. 1957. Chloroleukemia of mice. Ann. N. Y. Acad. Sci. 68:540-558. [DOI] [PubMed] [Google Scholar]
- 11.Hanna, Z., M. Jankowski, P. Tremblay, X. Jiang, A. Milatovich, U. Francke, and P. Jolicoeur. 1993. The Vin-1 gene, identified by provirus insertional mutagenesis, is the cyclin D2. Oncogene 8:1661-1666. [PubMed] [Google Scholar]
- 12.Hansen, G. M., and M. J. Justice. 1999. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B-cell leukemia. Oncogene 18:6531-6539. [DOI] [PubMed] [Google Scholar]
- 13.Hirai, H., and C. J. Sherr. 1996. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol. Cell. Biol. 16:6457-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huard, J. M., C. C. Forster, M. L. Carter, P. Sicinski, and M. E. Ross. 1999. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development 126:1927-1935. [DOI] [PubMed] [Google Scholar]
- 15.Inaba, T., H. Matsushime, M. Valentine, M. F. Roussel, C. J. Sherr, and A. T. Look. 1992. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics 13:565-574. [DOI] [PubMed] [Google Scholar]
- 16.Jonkers, J., and A. Berns. 1996. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim. Biophys. Acta 1287:29-57. [DOI] [PubMed] [Google Scholar]
- 17.Kerkhoff, E., and E. B. Ziff. 1995. Cyclin D2 and Ha-Ras transformed rat embryo fibroblasts exhibit a novel deregulation of cell size control and early S phase arrest in low serum. EMBO J. 14:1892-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keyomarsi, K., D. Conte, Jr., W. Toyofuku, and M. P. Fox. 1995. Deregulation of cyclin E in breast cancer. Oncogene 11:941-950. [PubMed] [Google Scholar]
- 19.Kiyokawa, H., X. Busquets, C. T. Powell, L. Ngo, R. A. Rifkind, and P. A. Marks. 1992. Cloning of a D-type cyclin from murine erythroleukemia cells. Proc. Natl. Acad. Sci. USA 89:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, C. A., L. M. Albritton, and J. Cunningham. 1990. Genetic mapping of a cloned sequence responsible for susceptibility to ecotropic murine leukemia viruses. J. Virol. 64:3119-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammie, G. A., R. Smith, J. Silver, S. Brookes, C. Dickson, and G. Peters. 1992. Proviral insertions near cyclin D1 in mouse lymphomas: a parallel for BCL1 translocations in human B-cell neoplasms. Oncogene 7:2381-2387. [PubMed] [Google Scholar]
- 22.Li, H., J. M. Lahti, and V. J. Kidd. 1996. Alternatively spliced cyclin C mRNA is widely expressed, cell cycle regulated, and encodes a truncated cyclin box. Oncogene 13:705-712. [PubMed] [Google Scholar]
- 23.Matsushime, H., M. E. Ewen, D. K. Strom, J. Y. Kato, S. K. Hanks, M. F. Roussel, and C. J. Sherr. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71:323-334. [DOI] [PubMed] [Google Scholar]
- 24.Matsushime, H., M. F. Roussel, R. A. Ashmun, and C. J. Sherr. 1991. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65:701-713. [DOI] [PubMed] [Google Scholar]
- 25.Matsushime, H., M. F. Roussel, and C. J. Sherr. 1991. Novel mammalian cyclins (CYL genes) expressed during G1. Cold Spring Harbor Symp. Quant. Biol. 56:69-74. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, C. A., H. W. Jacobs, S. A. Datar, W. Du, B. A. Edgar, and C. F. Lehner. 2000. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19:4533-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerson, M., and E. Harlow. 1994. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14:2077-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsubo, M., A. M. Theodoras, J. Schumacher, J. M. Roberts, and M. Pagano. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-586. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 30.Ru, M., C. Shustik, and E. Rassart. 1993. Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J. Virol. 67:4722-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sherr, C. J. 1995. D-type cyclins. Trends Biochem. Sci. 20:187-190. [DOI] [PubMed] [Google Scholar]
- 33.Sicinski, P., J. L. Donaher, Y. Geng, S. B. Parker, H. Gardner, M. Y. Park, R. L. Robker, J. S. Richards, L. K. McGinnis, J. D. Biggers, J. J. Eppig, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1996. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384:470-474. [DOI] [PubMed] [Google Scholar]
- 34.Soares, M. B., and M. de Fatima Bonaldo. 1998. Methods for isolating RNA, p. 66. In B. Birren, E. D. Green, S. Klapholz, R. M. Myers, and J. Roskams (ed.), Genome analysis. A laboratory manual, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 35.Sola, B., D. Simon, M. G. Mattei, S. Fichelson, D. Bordereaux, P. E. Tambourin, J. L. Guenet, and S. Gisselbrecht. 1988. Fim-1, Fim-2/c-fms, and Fim-3, three common integration sites of Friend murine leukemia virus in myeloblastic leukemias, map to mouse chromosomes 13, 18, and 3, respectively. J. Virol. 62:3973-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen, A. B., M. Duch, H. W. Amtoft, P. Jorgensen, and F. S. Pedersen. 1996. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J. Virol. 70:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorensen, A. B., M. Duch, P. Jorgensen, and F. S. Pedersen. 1993. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J. Virol. 67:7118-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taieb, F., and C. Jessus. 1996. Xenopus cyclin D2: cloning and expression in oocytes and during early development. Biol. Cell 88:99-111. [PubMed] [Google Scholar]
- 39.Teramoto, N., K. Pokrovskaja, L. Szekely, A. Polack, T. Yoshino, T. Akagi, and G. Klein. 1999. Expression of cyclin D2 and D3 in lymphoid lesions. Int. J. Cancer 81:543-550. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tremblay, P. J., C. A. Kozak, and P. Jolicoeur. 1992. Identification of a novel gene, Vin-1, in murine leukemia virus-induced T-cell leukemias by provirus insertional mutagenesis. J. Virol. 66:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Lohuizen, M., and A. Berns. 1990. Tumorigenesis by slow-transforming retroviruses—an update. Biochim. Biophys. Acta 1032:213-235. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, R. K., and E. R. Mardis. 1998. Shotgun sequencing, p. 397. In B. Birren, E. D. Green, S. Klapholz, R. M. Myers, and J. Roskams (ed.), Genome analysis. A laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 44.Wolff, L. 1997. Contribution of oncogenes and tumor suppressor genes to myeloid leukemia. Biochim. Biophys. Acta 1332:F67-F104. [DOI] [PubMed] [Google Scholar]