Abstract
The crystal structure of recombinant hepatitis B virus (HBV) capsids formed by 240 core proteins has recently been published. We wanted to map sites on the surface of the icosahedral 35-nm particle that are important for nucleocapsid envelopment by HBV surface proteins during virion morphogenesis. For this purpose, we individually mutated 52 amino acids (aa) within the N-terminal 140 aa of the 185-aa long core protein displaying their side chains to the external surface of the capsid to alanine residues. The phenotype of the mutations with respect to virion formation was tested by transcomplementation of a core gene-negative HBV genome in transiently cotransfected cells, immunoprecipitation of nucleocapsids from cells and secreted virions from culture media, and detection of the particles by radioactive endogenous polymerase reactions. Thirteen point mutations impeded nucleocapsid detection by endogenous polymerase reactions. Twenty-seven mutations were compatible with virion formation. Among these were all capsid-forming mutations in the upper half of the spike protruding from the particle shell and two additional triple mutations at tip of the spike. Eleven mutations (S17, F18, L60, L95, K96, F122, I126, R127, N136, A137, and I139) allowed nucleocapsid formation but blocked particle envelopment and virion formation to undetectable levels. These mutations map to a ring-like groove around the base of the spike and to a small area at the capsid surface close to the pores in the capsid shell. These residues are candidate sites for the interaction with envelope proteins during virion morphogenesis.
Different mechanisms have evolved by which enveloped viruses acquire their outer cover (7). For togaviruses (22) and hepadnaviruses (3) it is postulated that preformed cytosolic nucleocapsids bind directly to cytosolic endodomains of transmembrane envelope proteins and that this binding is an essential step during the budding of nucleocapsids through a cellular membrane and the concomitant envelopment of the capsid. For the human hepatitis B virus (HBV), the prototype of the family Hepadnaviridae comprising small hepatotrophic DNA viruses replicating by reverse transcription (6), the main evidence supporting this model is deduced from the phenotypic characterization of mutations in structural genes causing defects in nucleocapsid envelopment (19).
HBV has three envelope proteins (large [L], middle [M], and small [S]), which are expressed from three staggered start sites of a single 1,200-bp open reading frame (9) of the viral 3.2-kb DNA genome. They traverse the membrane multiple times and form homo- and heterodimers with each other (27). The envelope proteins not only are incorporated into the outer shell of virions but also form spherical and filamentous subviral lipoprotein particles half the diameter of the virion (20-nm particles) which are released from virus-producing cells in large excess over virions. The L and S proteins are essential for virion formation (4). Two regions in endodomains of these proteins have been identified in which mutations block nucleocapsid envelopment but still allow the secretion of subviral 20-nm particles: one linear stretch of 22 amino acids (aa) between residues 103 and 124 in L (3, 16) (a domain which is not present in M or S) and a second region between residue 35 and 46 in the S protein (17). Although the latter sequence is also present in the C-terminal part of L, the mutations affected virion morphogenesis mainly in the context of S.
The formation of the nucleocapsid is initiated by binding of the viral P protein to a terminal redundant 3.5-kb viral RNA molecule (pregenome) containing all the viral genetic information. This complex, together with host factors, is packaged by 90 or 120 homodimers of the 21-kDa core protein, forming the icosahedral shell of the nucleocapsid with a diameter of approximately 34 nm (19). This newly formed particle is not ready for envelopment. Rather, a maturation step is necessary which is linked to the reverse transcription of the encapsidated RNA molecule to DNA by the P protein (8, 23, 25). The assumed structural change of the capsid accompanying this maturation step is still obscure.
The core protein (185 aa residues in the genotype A used for this work) forms capsids by self-assembly when expressed in bacteria. Icosahedral shells of two sizes are observed, 32-nm particles with a T=3 symmetry containing 90 homodimers and 36-nm particles with a T=4 symmetry consisting of 120 homodimers (26, 30). The identity of the particle species present in virions in patients is not clear (11). Deletion of an arginine-rich, 30-aa domain at the C terminus, which is thought to bind to the viral nucleic acid during particle morphogenesis, allows efficient expression of the protein in bacteria and favors the formation of T=4 over T=3 capsids.
A few years ago, a model for the folding of a C-terminally deleted core protein in T=4 capsids was proposed based on cryoelectron micrographs of particles (2, 5). Mutations in the core protein allowing capsid formation and viral DNA genome synthesis but blocking nucleocapsid envelopment mapped to the surface of the particle (12), and it was suggested that these sites might interact with viral surface proteins during virion morphogenesis.
Recently, the crystal structure of recombinant capsids was published (28). This allowed a refinement of the analysis of capsid surface structures involved in virion formation. Based on the crystal structure, we selected 52-aa residues exposed at the exterior of the capsid for mutation to alanine residues (or, for two positions carrying alanine residues in the wild-type [WT] protein, to glycine residues) and expressed these variants together with a core-gene negative HBV genome in transiently transfected cells. Testing the presence of viral DNA-containing capsids in the cells and virions in the culture medium allowed us to map several amino acids on the capsid surface which were essential for virion but not nucleocapsid formation. These residues are potential binding sites for viral envelope protein domains contacting the capsid during virion morphogenesis. They cluster in a relatively small region on the capsid surface.
MATERIALS AND METHODS
Plasmids.
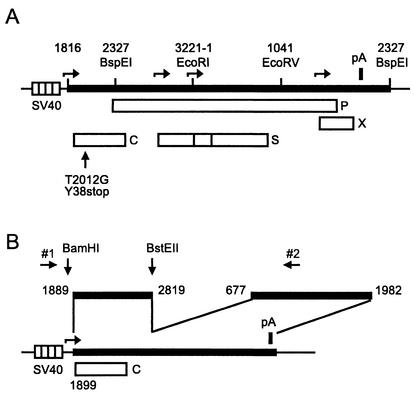
For plasmid pSVHBV1.5, a fusion between a 280-bp simian virus 40 (SV40) fragment carrying the SV40 early promoter and the HBV sequence was constructed by PCR. The fusion site has the sequence ggccgaggccgcctcAACTTTTTCACCTC, where the HBV sequence is in capital letters and starts with HBV nucleotide (nt) 1816. Numbering of the plus strand of the HBV DNA used in this work (24) (genotype A; GenEMBL data bank accession no. X02763) starts with the deoxycytidine residue of the unique EcoRI site in the pre-S2 region. The HBV sequence in pSVHBV1.5 ends with HBV nt 2327. The SV40-HBV construct was inserted between the HindIII and SacI sites of vector pBluescript KS(+) (Stratagene). pSVHBV1.5core− (Fig. 1A) is identical to pSVHBV1.5 with the exception of HBV nt 2012 in the 5′ part of the terminally redundant HBV genome. This nucleotide was changed from dT to dG, creating a stop codon at triplet 38 of the core gene. The construction of plasmid pSVcore (Fig. 1B) has been described previously (12).
FIG. 1.
Maps of plasmids. (A) pSVHBV1.5core− contains an overlength copy of the HBV genome (thick bar) inserted downstream of the SV40 early promoter (striped box). Open boxes indicate genes of the HBV genome: C, core protein; S, envelope proteins; P, P protein; X, X protein. The C gene carries the point mutation T2012G, introducing a stop codon at triplet 38. Some restriction enzyme sites are shown. Numbers indicating the locations of restriction sites and the 5′ start of the HBV DNA insertion correspond to the numbering of the HBV genome. Horizontal arrows point to transcription start sites. The thick vertical bar (pA) indicates the polyadenylation site of the HBV genome. (B) pSVcore contains the HBV DNA fragment from nt 1889 to 2819 carrying the HBV core gene (C) starting at HBV nt 1899 under the transcriptional control of a SV40 early promoter (striped box). The HBV DNA fragment from nt 677 to 1982 containing a posttranscriptional regulatory element for mRNA nuclear export (10) and the polyadenylation signal were fused downstream of HBV nt 2819. Restriction enzyme sites and binding sites for primers 1 and 2 used for the construction of core gene mutations are indicated.
Site-directed mutagenesis.
Point mutations were introduced using antisense (or sense) oligodeoxynucleotides (primers) if the mutation was introduced into the 5′ half (or 3′ half, for sense oligodeoxynucleotides) of the core gene. The primers were between 18 and 35 bases in length and carried the point mutation in the middle of the sequence. Ten nanograms of plasmid pSVcore, 10 pmol of the primer carrying the mutation, 10 pmol of sense primer 1 (5′CCCATTCTCCGCCCCATGGC) annealing at bp 146 to 126 upstream of the core gene start codon in the SV40 early promoter of pSVcore (Fig. 1B) (or antisense primer 2 [5′GAAAGTGAAAGCC] annealing between bp 1096 and 1084 of the HBV genome downstream of the core gene), 10 nmol of each of the four deoxynucleoside triphosphates, and 2 to 5 U of Taq (Roche Diagnostics GmbH, Mannheim, Germany) or Pfu (Stratagene) DNA polymerase were mixed in a total volume of 50 μl. A fragment between 160 and 410 bp in length was amplified by 30 cycles of 96°C for 30 s, 50°C for 30 s, and 72°C for 10 min. The DNA product was purified using an PCR purification kit (Qiagen GmbH, Hilden, Germany) and dissolved in 50 μl of water. Then 1/10 to 1/50 of the purified product was mixed with 10 ng of pSVcore, 10 pmol of primer 2 (or primer 1), 10 nmol of each of the four deoxynucleoside triphosphates, and 2 to 5 U of Taq or Pfu DNA polymerase in 50 μl (total volume) and amplified by 40 cycles of 96°C for 30 s, 50°C for 1 min, and 72°C for 1 min. The 1,493-bp product was purified by agarose gel electrophoresis and cut with BamHI upstream of the core gene and BstEII at nt 2819 in the HBV sequence. The resulting 932-bp fragment was purified and inserted into BamHI-BstEII-cut pSVcore. After molecular cloning, the sequence between the BamHI and BstEII sites of the resulting plasmid was determined and plasmids carrying unintentional mutations were discarded.
Cell culture, transfection, and detection of viral particles.
Transient transfection of HuH7 cells by the calcium phosphate precipitation method, immunoprecipitation of cytoplasmic nucleocapsids with anti-HBc and of secreted virions from the culture medium with anti-HBs, labeling of the viral genome using the endogenous polymerase reaction, isolation, and separation of the DNA by agarose gel electrophoresis have been described previously (12). The radioactive signal was detected in some experiments by autoradiography using X-ray films and in other experiments by a phosphorimager.
Three-dimensional view of the core protein fold.
The data for the crystal structure of the HBV capsid (28) were retrieved from http://pdb.gmd.de (accession number 1QGT). The data were visualized using the program Swiss-PdbView version 3.7b2 retrieved from www.expasy.ch/spdbv/mainpage.html. For refined three-dimensional inspection, we used the program InsightII running on a Silicon Graphics Octane computer and three-dimensional-viewing spectacles.
RESULTS
Selection of amino acid residues externally exposed on the capsid structure.
The crystal structure of the capsid was visualized as a three-dimensional spatial model with the help of a computer, and 52 aa residues were chosen by eye for the mutational analysis (Table 1). The criteria for the selection were that the amino acid side chains should be accessible from outside for the export machinery and that the residues should cover almost the whole surface of the capsid. Several residues were omitted just to limit the number of mutants for the analysis.
TABLE 1.
Genotypes and phenotypes of 52 single-point mutations in the HBV core genea
| Positionb in sequence | Amino acidc | EPR signald in:
|
Position in fold
|
|||||
|---|---|---|---|---|---|---|---|---|
| Cytoplasmic nucleocapsids | Secreted virions | Virions/nucleocapsids
|
||||||
| Expt 1 | Expt 2 | |||||||
| 2 | D | + | + | 0.27 | 0.25 | |||
| 4 | D | − | − | —f | — | |||
| 13 | V | + | + | 0.35 | 0.26 | α1 | ||
| 14 | E | + | + | 0.3e | 1.2e | α1 | ||
| 17 | S | + | − | <0.02 | <0.02 | α1 | Lateral loop | |
| 18 | F | + | − | <0.02 | <0.02 | |||
| 20 | P | − | − | — | — | |||
| 21 | S | + | + | 0.6e | 1.0e | |||
| 22 | D | + | + | 0.30 | 0.34 | |||
| 26 | S | + | + | 0.43 | 0.53 | |||
| 27 | V | + | + | 0.3e | 1.0e | α2a | ||
| 28 | R | + | + | 0.07 | 0.08 | α2a | ||
| 29 | D | − | − | — | — | α2a | Lateral helix | |
| 31 | L | + | + | 0.14 | 0.33 | α2a | ||
| 32 | D | + | + | 0.24 | 0.25 | α2a | ||
| 59 | I | + | + | 0.3e | 1.0e | α3 | ||
| 60 | L | + | − | <0.02 | <0.02 | α3 | ||
| 62 | W | + | + | 0.43 | 0.24 | α3 | Spike upward | |
| 66 | M | + | + | 0.4e | 1.2e | α3 | ||
| 67 | T | + | + | 0.61 | 0.55 | α3 | ||
| 70 | T | + | + | 0.37 | 0.60 | α3 | ||
| 73 | G | − | − | — | — | α3 | ||
| 74 | N | + | + | 0.59 | 0.73 | |||
| 75 | N | + | + | 0.66 | 0.79 | |||
| 76 | L | + | + | 0.64 | 0.82 | Tip | ||
| 77 | E | + | + | 0.55 | 0.79 | |||
| 78 | D | − | − | — | — | |||
| 79 | P | + | + | 0.09 | 0.06 | α4a | ||
| 83 | D | + | + | 0.3e | 1.5e | α4a | ||
| 84 | L | + | + | 0.32 | 0.56 | α4a | Spike downwards | |
| 87 | N | + | + | 0.47 | 0.36 | α4a | ||
| 91 | T | + | + | 0.15 | 0.46 | |||
| 92 | N | − | − | — | — | α4b | ||
| 95 | L | + | − | <0.02 | <0.02 | α4b | ||
| 96 | K | + | − | <0.02 | <0.02 | α4b | ||
| 98 | R | + | + | 0.05 | 0.11 | α4b | ||
| 118 | Y | − | − | — | — | α5 | ||
| 119 | L | + | + | 0.11 | 0.09 | α5 | ||
| 122 | F | + | − | <0.02 | <0.02 | α5 | Protruding helix | |
| 125 | W | − | − | — | — | α5 | ||
| 126 | I | + | − | <0.02 | <0.02 | α5 | ||
| 127 | R | + | − | <0.02 | <0.02 | α5 | ||
| 129 | P | − | − | — | — | |||
| 130 | P | − | − | — | — | |||
| 131 | A | + | + | 0.74 | 0.39 | Proline-rich loop | ||
| 132 | Y | − | − | — | — | |||
| 133 | R | − | − | — | — | |||
| 135 | P | + | + | 0.22 | 0.22 | |||
| 136 | N | + | − | <0.02 | <0.02 | |||
| 137 | A | + | − | <0.02 | <0.02 | Extended strand | ||
| 139 | I | + | − | <0.02 | <0.02 | |||
| 140 | L | − | − | — | — | |||
| WT | + | + | 0.80 | 0.64 | ||||
Results for mutants of the class c+v− forming nucleocapsids but no virions are shown in bold type.
Position in the primary amino acid sequence of the core protein.
Amino acid residue in the WT sequence changed to alanine (at positions 131 and 137, an alanine residue was changed to a glycine residue).
+, signal detectable; −, no signal detectable; Expt 1, and 2, EPR signal from secreted virions quantified by a PhosphorImager divided by the EPR signal from cytoplasmic nucleocapsids in experiments 1 and 2.
In these experiments, the radioactive signal was detected by X-ray films and the ratio was estimated.
—, the ratio could not be calculated because the EPR signal from cytoplasmic cores was close to background. position: position of the mutation in the core protein fold according to reference 28.
Expression of point mutants in cell culture.
Mutations were introduced into the SV40 early promoter expression vector pSVcore carrying the HBV core gene (Fig. 1B) by a PCR-based method. All DNA fragments generated by PCR and transferred into this vector were sequenced to exclude unintended mutations. The plasmids were cotransfected with an overlength HBV genome carrying a stop codon at triplet 38 of the core gene. This HBV genome was inserted downstream of a SV40 early promoter (Fig. 1A). The 5′ end of transcripts initiating at this promoter corresponds to the 5′ end of pregenomic RNA transcribed from the authenic core promoter of the HBV genome (data not shown). This RNA served as a template for viral DNA synthesis and in addition as mRNA for P protein synthesis. Expression of the HBV envelope proteins was governed by two authentic promoters within the HBV genome (horrizontal arrows in Fig. 1A).
Nucleocapsids from cleared cell lysates of transiently cotransfected cells were bound to antibodies of a polyclonal rabbit antiserum (anti-HBc), concentrated by immunoprecipitation, and detected by radioactive labeling of the viral genome by using the endogenous DNA polymerase which fills in the single-stranded gap of the partially double-stranded viral DNA genome when substrates are supplied (endogenous polymerase reaction [EPR]). The viral DNA was then isolated, separated on an agarose gel, and depicted by fluorography. The anti-HBc antiserum contained antibodies to at least two different separated epitopes on the capsid (12, 21). If one epitope on the particle was destroyed by a point mutation, the mutant should still be able to bind during immunoprecipitation due to the remaining unchanged epitope. Since we did not primarily intend in this analysis to identify core protein mutations influencing capsid formation, RNA/P protein packaging, or reverse transcription, we used the EPR as a readout instead of, e.g., capsid detection by sucrose gradient centrifugation. This had the advantage that only replication-competent nucleocapsids which are suitable candidates to investigate nucleocapsid envelopment were scored. Virions from the culture supernatant were immunoprecipitated with an antiserum against viral envelope proteins (goat anti-HBs) and detected by the same technique as was used for nucleocapsids.
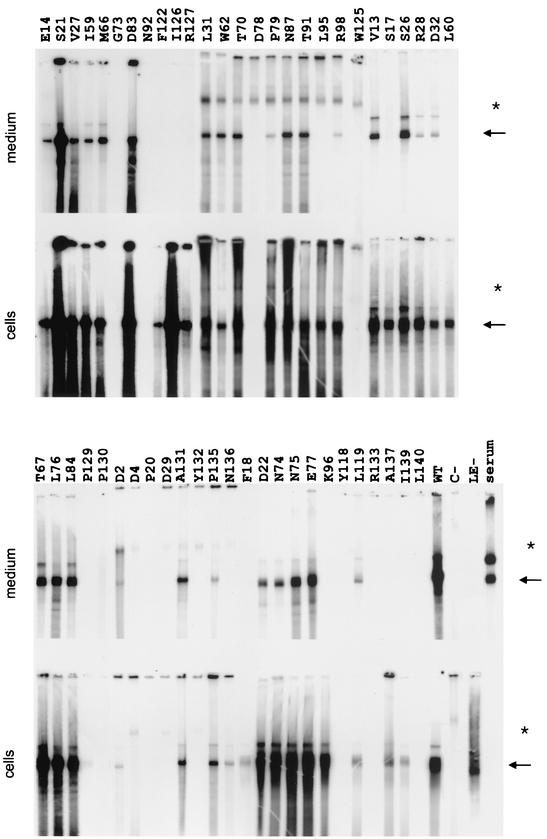
Transfection with the core-negative HBV genome alone did not lead to detectable nucleocapsids in cells or virions in the culture medium, as expected (Fig. 2, lanes C−). However, cotransfection of the genome together with the pSVcore expressing the WT core protein restored nucleocapsid as well as virion formation by transcomplementation (lanes WT). The ratio of the EPR signal from cytoplasmic capsids to secreted virions was around 0.7 (Table 1).
FIG. 2.
Phenotypes of core gene point mutants. HuH7 cells were cotransfected with a core-negative HBV genome and a vector for core protein expression carrying the mutations. Signals from the radioactive endogenous polymerase reactions using virions harvested from the culture medium and capsids harvested from cleared cell lysates are shown. The arrows point to the relevant signal. Signals marked by the asterisks are nonspecific. The duration of the exposure was identical for each pair of lanes (cells and medium), whereas the duration varied for different mutants. The WT amino acid residue changed to alanine (or glycine in two cases) and its position in the core protein is indicated above each pair of lanes. C−, transfection only with the core-negative HBV genome; WT, cotransfection with the core-negative HBV genome and an expression vector carrying the WT core gene; LE-, transfection with an HBV genome carrying two stop codons in the envelope gene, allowing nucleocapsid formation but blocking virion secretion; serum, approximately 2 × 106 virions from a human HBV carrier.
The mutants fell into three classes (Table 1). In 13 mutants, nucleocapsids could not be detected within cells (class c−v−). All mutants of this class also formed no detectable virions, as expected. Twenty-eight mutants were compatible with nucleocapsid as well as virion formation (class c+n+). For these mutants, the absolute level of nucleocapsids and virions varied relative to the WT and the ratio of the signal generated from intracellular nucleocapsids to the signal from secreted virions was between 0.07 (mutant R28) and 1.5 (mutant D83). Three mutations in this class (R28, P79, and R98) led to a reduction of this ratio below 0.1 but still allowed detectable virion secretion. The third class (c+v−) contains 11 mutants forming nucleocapsids within cells but no detectable virions in the culture medium (the ratio of the signal from cytoplasmic nucleocapsids to secreted virions was below 0.02). We conclude that mutants in this class had a defect in late virion morphogenesis and secretion.
Triple mutations at the tip of the spike.
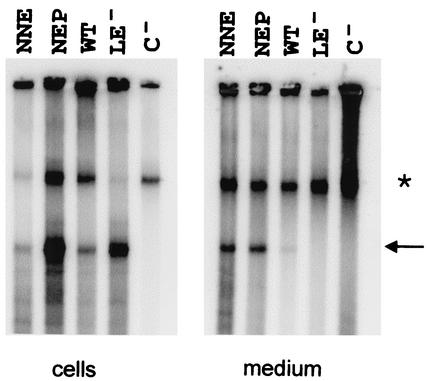
A peptide binding to the tip of the spike which protrudes from the HBV capsid surface was able to inhibit virion formation (1), assuming that this site could be involved directly in the envelopment process. However, none of the four single-amino-acid changes at the tip (N74, N75, L76, and E77) had a marked influence on virion morphogenesis. Only one change (P79), at the N-terminal end of the helix reaching downward from the tip of the spike, reduced virion secretion by a factor of approximately 10. It was possible that the alteration of only one amino acid residue at the tip was not sufficient for a functional block. To exclude this possibility, we generated two mutants (N74A-N75A-E77A and N74A-E77A-P79A) each carrying three amino acid changes in this area and analyzed their phenotypes as described above. Both mutants retained the ability to replicate and secrete virus particles (Fig. 3). We conclude that the tip of the spike most probably does not participate in sequence-specific protein-protein interactions during nucleocapsid envelopment.
FIG. 3.
Phenotypes of triple mutations at the tip of the spike. Two triple mutants (N74A-N75A-E77A [NNE] and N74A-E77A-P79A [NEP]) were phenotyped as described in the legend of Fig. 2. WT, LE-, and C- are as in Fig. 2. ∗, nonspecific band.
Location of the c+v− mutations in the core protein fold.
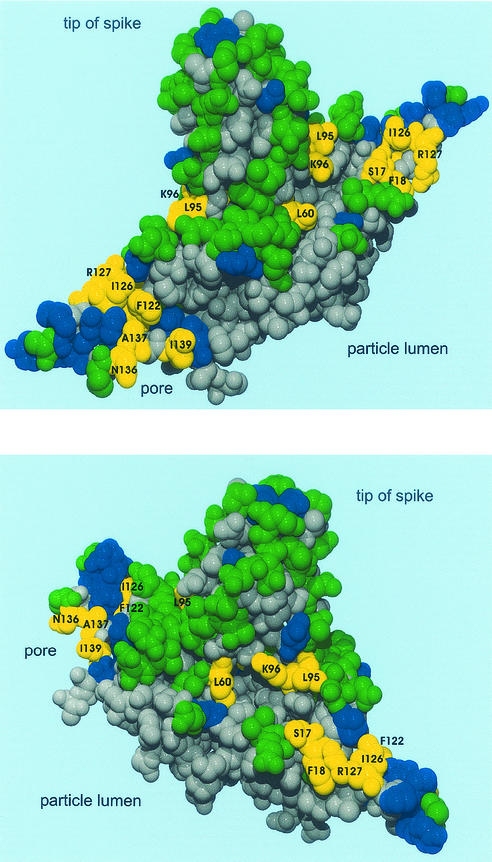
The point mutations of class c+v− causing a block in nucleocapsid envelopment are clustered on the capsid surface (Fig. 4, yellow residues). Residues L60, L95, and K96 form a ring-like groove at the base of the spike, and residues S17, F18, R127, I126, F122, A137, N136, and I139 are located in a relatively small area close to the pores in the capsid shell.
FIG. 4.
Location of mutations in the core protein fold. Two views of the core protein homodimer are shown. Amino acid residues analyzed in this study are colored. Green, WT phenotype (c+v+); blue, no detection of nucleocapsids or virions (c−v−); yellow, detection of nucleocapsids but not of virions (c+v−); gray, residues have not been mutated. The c+v− mutations cluster at a ring-like groove around the base of the spike and at a small area on the capsid surface close to the pore.
DISCUSSION
We describe in this paper 11 independent single-amino-acid changes in the HBV core protein (class c+v−) inducing a specific and rather drastic defect: nucleocapsids formed by the slightly altered core proteins are no longer exported from cells as enveloped virions. These mutations are clustered around the base of the spike and in a small area close to the pore. The close proximity of these residues in the fold argues for a common function during virion morphogenesis. The simplest interpretation of this phenotype is that the amino acid side chains of the targeted residues participate in a very specific binding to viral surface proteins during envelopment, as proposed for alphaviruses (22), and that the exchange of one of these residues is sufficient to abolish this interaction. Candidates for binding sites in the envelope proteins are a 22-aa domain in a cytosolic part of the L protein (3, 16) and a 12-aa domain in the S protein (17) mapped by mutational analysis. This view is supported by experiments demonstrating the direct binding of peptides derived from HBV envelope proteins to capsids (20). However, other interpretations are also possible. For example, the maturation signal of the capsid, which is linked to the synthesis of the viral DNA genome by reverse transcription (8, 25), has to be transmitted from the lumen of the capsid to the outer surface in order to be recognizable for the export machinery. This may be a very complex mechanism, involving the displacement of several amino acid residues in the core protein. It is easily seen that such a process is sensitive to single-amino-acid changes and that some of the c+v− mutations identified in this work influence the signaling of maturation. Another possibility is that an envelope protein-independent membrane affinity of mature nucleocapsids, which has been described for duck HBV (18) and which may be a prerequisite for budding, is blocked by c+v− mutations.
The identities of the amino acid residues which have been found to influence nucleocapsid envelopment in studies of naturally occurring HBV core gene mutations are in good agreement with our results. A mutation of core L60 to valine induced a low-level virion secretion phenotype (15). The mutation of L60 to alanine, causing a strong block of nucleocapsid export, has been found independently in our study. The mutation of I97 to leucine enhances the envelopment and secretion of immature nucleocapsids (29). This residue is in close proximity to residues L95 and K96, belonging to class c+v− and identified in our study. The fact that a complementary mutation in the pre-S1 region of the large envelope protein (A119F) restored the WT phenotype of the core I97L mutation (14) supports the interpretation that the L protein contacts the capsid at this area.
One might have assumed that the upper part of the spike is important for nucleocapsid envelopment because these structures protrude prominently from the capsid and are expected to touch the outer cover. A report that a peptide binding to this site inhibited virion morphogenesis supported this view (1). However, all four single-amino-acid changes and two triple changes directly at the tip of the spike, as well as eight additional point mutation at the trunk of the spike, clearly allowed virion formation. The deletion of A80 also had no effect on virion formation (12). However, due to a 10-fold reduction of the level of mutant P79A in virion secretion and due to the lack of detectable nucleocapsids for mutant D78A (class c−v−), the relevance of these two amino acids at the tip to the envelopment process could not be answered with certainty. Possibly, the relatively drastic P79A mutation changing the peptide backbone influences the protein conformation in other relevant parts of the protein.
One potential limitation of our study is that we generated only one mutant for each analyzed amino acid position of the core protein (usually alanine, in some cases glycine). It is likely that introducing another amino acid would cause a different phenotype at some positions. However, the fact that the mutations of the c+v− class cluster in a relatively small region argues for the view that at least the overall picture of the capsid surface areas important for nucleocapsid envelopment and export is sound.
Several mutations of classes c+v+ and c+v− reduced the amount of radioactively labeled genomes harvested from cytoplasmic nucleocapsids, and 13 single-amino-acid changes in the core protein totally abolished the detection of nucleocapsids by the endogenous polymerase reaction (class c−v−). We have not yet investigated the reasons for this behavior. Apparently, the mutations blocked one step in the maturation of nucleocapsids, e.g., stable core protein expression, dimer formation, shell assembly, interaction with the pregenomic RNA/P protein complex or with essential host factors, or viral DNA synthesis. Former studies also demonstrated that some regions of the core protein are very sensitive to subtle mutations (13). At least two mutations in class c−v− allowed capsid formation (unpublished results). They map to the rim of the pores in the capsid shell. It is, for example, possible that the introduction of an alanine residue at this site blocked the diffusion of nucleotides required during viral DNA synthesis through this pore into the capsid lumen and therefore prevented the detection of viral particles by EPR.
Acknowledgments
We thank Alexander Wentzel and Andreas Christmann from the Institute for Microbiology and Genetics in Göttingen for their help and for permission to use the Silicon Graphics Octane computer.
This work was supported by the Deutsche Forschungsgemeinschaft, Graduiertenkolleg 521 and Sonderforschungsbereich 402, project C2.
REFERENCES
- 1.Böttcher, B., N. Tsuji, H. Takahashi, M. R. Dyson, S. Zhao, R. A. Crowther, and K. Murray. 1998. Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 17:6839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böttcher, B., S. A. Wynne, and R. A. Crowther. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88-91. [DOI] [PubMed] [Google Scholar]
- 3.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway, J. F., N. Cheng, A. Zlotnick, P. T. Wingfield, S. J. Stahl, and A. C. Steven. 1997. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386:91-94. [DOI] [PubMed] [Google Scholar]
- 6.Ganem, D. 1996. Hepadnaviridae: the viruses and their replication, p. 2703-2737. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 7.Garoff, H., R. Hewson, and D. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerelsaikhan, T., J. E. Tavis, and V. Bruss. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 70:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heermann, K. H., U. Goldmann, W. Schwarz, T. Seyffarth, H. Baumgarten, and W. H. Gerlich. 1984. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 52:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, Z. M., and T. S. Yen. 1995. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 15:3864-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionaly conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 12.Koschel, M., D. Oed, T. Gerelsaikhan, R. Thomssen, and V. Bruss. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J. Virol. 74:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koschel, M., R. Thomssen, and V. Bruss. 1999. Extensive mutagenesis of the hepatitis B virus core gene and mapping of mutations that allow capsid formation. J. Virol. 73:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Pogam, S., and C. Shih. 2002. Influence of a putative intermolecular interaction between core and the pre-S1 domain of the large envelope protein on hepatitis B virus secretion. J. Virol. 76:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Pogam, S., T. T. Yuan, G. K. Sahu, S. Chatterjee, and C. Shih. 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J. Virol. 74:9099-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and R. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löffler-Mary, H., J. Dumortier, C. Klentsch-Zimmer, and R. Prange. 2000. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270:358-367. [DOI] [PubMed] [Google Scholar]
- 18.Mabit, H., and H. Schaller. 2000. Intracellular hepdnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassal, M. 1996. Hepatitis B virus morphogenesis. Curr. Top. Microbiol. Immunol. 214:297-337. [DOI] [PubMed] [Google Scholar]
- 20.Poisson, F., A. Severac, C. Hourioux, A. Goudeau, and P. Roingeard. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228:115-120. [DOI] [PubMed] [Google Scholar]
- 21.Salfeld, J., E. Pfaff, M. Noah, and H. Schaller. 1989. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol. 63:798-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skoging, U., M. Vihinen, L. Nilsson, and P. Liljeström. 1996. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4:519-529. [DOI] [PubMed] [Google Scholar]
- 23.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 24.Valenzuela, P., M. Quiroga, J. Zaldivar, R. Gray, and W. Rutter. 1980. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. UCLA Symp. Mol. Cell. Biol. 18:57-70. [Google Scholar]
- 25.Wei, Y., J. E. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingfield, P. T., S. J. Stahl, R. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 27.Wunderlich, G., and V. Bruss. 1996. Characterization of early hepatitis B virus surface protein oligomers. Arch. Virol. 141:1191-1205. [DOI] [PubMed] [Google Scholar]
- 28.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 29.Yuan, T. T., P. C. Tai, and C. Shih. 1999. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J. Virol. 73:10122-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]