Abstract
The two major envelope proteins of arteriviruses, the membrane protein (M) and the major glycoprotein (GP5), associate into a disulfide-linked heterodimer that is incorporated into the virion and has been assumed to be a prerequisite for virus assembly. Using an equine arteritis virus (EAV) infectious cDNA clone, we have analyzed the requirement for GP5-M heterodimerization and have identified the Cys residues involved in the formation of the GP5-M disulfide bond. The single Cys residue (Cys-8) in the M ectodomain was crucial for heterodimerization and virus infectivity. Mutagenesis of any of the five Cys residues in the GP5 ectodomain or removal of the single GP5 N-glycosylation site also rendered the full-length clone noninfectious. However, an analysis of revertants yielded an exceptional pseudorevertant in which residues 52 to 79 of the GP5 ectodomain had been deleted and the original Cys-80→Ser mutation had been maintained. Consequently, this revertant lacked the GP5 N-glycosyation site (Asn-56) and retained only a single cysteine residue (Cys-34). By using this GP5 deletion, we confirmed that Cys-34 of GP5 and Cys-8 of M are essential for GP5-M heterodimerization, a key event in the assembly of the EAV envelope.
Arteriviridae (order Nidovirales) are positive-sense, single-stranded RNA viruses with a polycistronic genome of 12 to 16 kb (for reviews, see references 43 and 44). In addition to the prototype equine arteritis virus (EAV) (19), the family contains the lactate dehydrogenase-elevating virus (LDV) of mice, porcine reproductive and respiratory syndrome virus (PRRSV), and simian hemorrhagic fever virus (SHFV).
Arterivirus particles are enveloped and have a diameter of 40 to 60 nm. The virion probably contains a set of seven structural proteins, a number that is unusually large compared to other positive-stranded RNA viruses. The isometric arterivirus nucleocapsid is composed of the RNA genome and a small nucleocapsid protein (N; 110 to 128 amino acids [aa]), whereas the virion envelope contains six envelope proteins. Major envelope components are the nonglycosylated integral membrane protein (M) and the “major” glycoprotein (15, 20, 34). Different numbers of “minor” envelope proteins have been identified for different arteriviruses (for reviews, see references 12, 43, 44, and 49). Also, the nomenclature of arterivirus envelope glycoproteins has not yet been standardized. For example, the major glycoprotein has been designated GL, VP-3P, GP5, and p54 for EAV, LDV, PRRSV, and SHFV, respectively. In this paper, we refer to the glycoproteins as GP x, where x indicates the number of the open reading frame (ORF) in the genome from which the protein is expressed.
The arterivirus genome contains a large 5′-proximal replicase gene (ORF1a plus ORF1b) that is translated from the genome RNA. The downstream structural protein genes are expressed from subgenomic mRNAs (44). The three major structural proteins of EAV, GP5, M, and N, are encoded by the three most 3′-proximal ORFs of the genome. The M protein (162 to 173 aa), the most highly conserved envelope protein of arteriviruses, is assumed to span the membrane three times (15, 20). M contains a short ectodomain of only 10 to 18 residues (Fig. 1A) and forms disulfide-linked heterodimers with the major glycoprotein (GP5) (17, 20). GP5 contains an N-terminal signal sequence that is cleaved from a relatively small ectodomain (Fig. 1B). In EAV, this ectodomain is 95 residues long and possesses one or two N-linked polylactosamine side chains (5, 15, 22). In other arteriviruses (LDV and PRRSV), the ectodomain can be as short as 30 residues and carries one to three N-linked glycans (20, 32, 36). The internal hydrophobic region of GP5 (Fig. 1B) probably spans the membrane three times and is followed by a cytoplasmic domain of 50 to 72 amino acids.
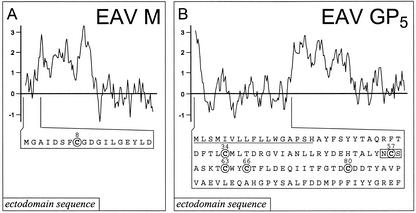
FIG. 1.
(A) Hydrophobicity plot and ectodomain sequence of the EAV M protein. The hydrophobicity plot was generated using the method of Kyte and Doolittle (26) and a moving window of 11; above the axis is hydrophobic. The position of the single, conserved Cys residue (Cys-8) in the M protein ectodomain sequence is indicated. (B) Hydrophobicity plot and ectodomain sequence of EAV GP5. See panel A for further details. In the GP5 ectodomain sequence, the (predicted) N-terminal signal sequence is underlined, the five Cys residues targeted in this study are indicated, and the single N-glycosylation site (Asn-56) is boxed.
Neutralizing antibodies from animals infected with arteriviruses are directed predominantly against GP5 (6, 10, 23, 38). Furthermore, all published LDV- and EAV-neutralizing monoclonal antibodies (MAbs) are GP5 specific (4, 11, 14, 24, 25), and also MAbs recognizing the PRRSV ORF5 protein were reported to neutralize virus infectivity (38, 40). EAV- neutralizing horse antibodies and MAbs bind to the putative ectodomain (aa 19 to 115) of GP5 (3, 10, 14, 37). Recently, Balasuriya et al. reported that the simultaneous expression of both GP5 and M, and probably their heterodimerization, is required for the induction of neutralizing immunity in horses (1). Nevertheless, neutralizing antibodies against EAV in horses could also be induced by using peptides derived from only the GP5 ectodomain (7, 10).
Arteriviruses acquire their envelope by budding into the lumen of smooth membranes of the exocytic pathway, probably including those of the Golgi complex. The specific roles of the various envelope proteins in virus assembly and infectivity have not yet been established. However, the recent development of infectious cDNA clones for arteriviruses (16, 31, 47) has opened the possibility of studying arterivirus assembly by modifying the expression and properties of structural proteins. It was shown that the proteins encoded by all seven genes in the 3′-terminal region of the EAV genome (ORF2a to ORF7) are required for the production of infectious progeny virus (35). Using the reverse genetics approach, we have now addressed the only well-established interaction between two EAV structural proteins, the heterodimerization of the GP5 and M proteins. We have identified the interacting Cys residues, obtained novel data concerning the properties of the GP5 ectodomain, and shown that the formation of the disulfide bond between the GP5 and M proteins is indeed crucial for virus infectivity.
MATERIALS AND METHODS
Cells, virus, and antisera.
Baby hamster kidney (BHK-21) cells were used for infection experiments with the EAV Bucyrus strain (19) and transfection with in vitro-generated full-length transcripts (47). The EAV nsp3-specific rabbit antiserum was described previously (39). Rabbit antiserum SP06 (15), recognizing the C-terminal domain of the EAV M protein, was obtained from A. A. F. de Vries and P. J. M. Rottier (Utrecht University). Mouse MAb 6D10 and 10H4 (3) against EAV GP5 were kindly provided by U. Balasuriya and N. J. MacLachlan (University of California at Davis).
Construction of mutant EAV full-length cDNA clones.
EAV infectious cDNA clone pEAV030 (47) was the backbone for all mutant constructs used in this study. The previously designed construct pA45 (18), in which the small overlap between EAV ORF4 and ORF5 had been removed (16), was used for site-directed mutagenesis of the GP5 ectodomain and residue Cys-8 in the M protein (Tables 1 and 2). The small insertion made to functionally separate ORF4 and ORF5 did not significantly impair virus replication or infectivity and was stable on repeated virus passaging (16). Site-directed PCR mutagenesis was performed as described by Landt et al. (27). Restriction fragments carrying the desired mutations were cloned into pA45 as AflII-EcoRI (GP5 ectodomain mutants) or EcoRI-XbaI (Cys-8-to-Ser mutation in M) fragments. Subsequently, constructs were sequenced using an ABI PRISM sequencing kit (Applied Biosystems) and an ABI PRISM 310 Genetic analyzer (Perkin-Elmer). The 5′ half of ORF5 (encoding the GP5 ectodomain) of pseudorevertant 80.4 was amplified from intracellular RNA (passage 4) by reverse transcription-PCR (RT-PCR), cloned into the pA45 backbone as an AflII-EcoRI fragment, and sequenced completely to produce clone pEAN80.4.
TABLE 1.
Primers used in this study for RT-PCR and site-directed mutagenesis
| Primer | Sequence (5′ → 3′)a | Location in the genomeb | Polarity | Purpose |
|---|---|---|---|---|
| E272 | ATGAAGATCTACGGCTGC | 10701-10718 | + | ORF5 PCR primer |
| E280 | GCGTAGCATAGGGTAGTACTG | 11525-11545 | − | ORF5 PCR primer |
| E301 | CCGTCAGCATGCTCAAGGTGAAG | 11234-11257 | − | Cys-34 → Ser mutation in GP5 |
| E302 | CAACAGGTTTTGCTAGCGGAAGAATTGTACAAAG | 11303-11336 | − | Cys-57 → Ser mutation in GP5 |
| E303 | CAATACCAACTAGTTTTACTGG | 11321-11342 | − | Cys-63 → Ser mutation in GP5 |
| E304 | CGTCCAGGAAAGTACTATACCAACAG | 11331-11356 | − | Cys-66 → Ser mutation in GP5 |
| E305 | CACGTTTGGTACCGATTCTGATGACACC | 11367-11394 | + | Cys-80 → Ser mutation in GP5 |
| E263 | CATGCCCCCTTTTATTTACT | 11460-11479 | + | ORF6 PCR primer |
| E163 | CCACCAGTTGGCGATGGTTG | 12185-12204 | − | ORF6 PCR primer |
| E306 | GATTCATTTTCCGGAGACGGGATTTTAG | 11913-11940 | + | Cys-8 → Ser mutation in M |
| E336 | GGCGGAACACTGGTACAAAGC | 11302-11322 | − | Asn-56 → Gln mutation in GP5 |
| E363a | GGTTTTACTGGCGCTGCAGCCGTACAAAGCAGT | 11299-11332 | − | Asn-56 → Gly mutation in GP5 |
| E363a | GGTTTTACTGGCGCTGCAGCTGTACAAAGCAGT | 11299-11332 | − | Asn-56 → Ser mutation in GP5 |
| E364 | GGTTTTACTGGCGCCGCAATTGTACAAAGC | 11302-11332 | − | Ser-58 → Gly mutation in GP5 |
| E123 | GCCCATGGCCAAGTAGGCCCCG | 11625-11646 | − | RT primer |
Mutated nucleotides are depicted in bold, and (translationally silent) restriction sites engineered to select and identify mutants are underlined.
Genome positions are based on the sequence of EAV full-length cDNA clone pEAV030 (EMBL database accession number Y07862).
TABLE 2.
Composition of the GP5 and M proteins of the EAV constructs
| Construct | Mutated protein | Mutation(s) | Phenotype |
|---|---|---|---|
| pA45 | NAa | NA | Wild type |
| pA45/C8S | M | Cys-8 → Ser | No infectivity |
| pA45/C34S | GP5 | Cys-34 → Ser | No infectivity |
| pA45/C57S | GP5 | Cys-57 → Ser | No infectivity |
| pA45/C63S | GP5 | Cys-63 → Ser | No infectivity |
| pA45/C66S | GP5 | Cys-66 → Ser | No infectivity |
| pA45/C80S | GP5 | Cys-80 → Ser | No infectivity |
| pA45/QCS | GP5 | Asn-56 → Gln | No infectivity |
| pA45/GCS | GP5 | Asn-56 → Gly | No infectivity |
| pA45/SCS | GP5 | Asn-56 → Ser | No infectivity |
| pA45/NCG | GP5 | Ser-58 → Gly | No infectivity |
| pA45-80.4 | GP5 | Thr-52 to Asp-79 replaced by Asn; Cys-80 → Ser | Attenuated |
| pA45-80.4/C34S | GP5 | Thr-52 to Asp-79 replaced by Asn; Cys-80 → Ser; Cys-34 → Ser | No infectivity |
NA, not applicable.
Infection and transfection experiments.
Infection experiments with EAV were performed with BHK-21 cells by the method of de Vries et al. (15). BHK-21 cells were also used for transfection experiments with in vitro-generated transcripts from the EAV infectious cDNA clone pEAV030 and derivatives thereof by using the previously described electroporation protocol (47). For plaque assays, subconfluent monolayers of BHK-21 cells were infected with 10-fold serial dilutions of wild-type (wt) or recombinant EAV. Following a 1-h incubation, a 1.5% agarose overlay was applied and cells were incubated at 39.5°C. Plaques were visible between 2 and 3 days after infection.
Immunofluorescence microscopy.
Cells were prepared for immunofluorescence assays (IFAs) essentially as described by van der Meer et al. (46). As secondary antibodies, a Cy3-conjugated donkey anti-rabbit immunoglobulin G antibody and a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G antibody (both from Jackson ImmunoResearch Laboratories) were used. Samples were examined using an Olympus microscope equipped with a digital camera and Qfluoro software (Leica).
Protein labeling and immunoprecipitation analysis.
Transfected cells were starved for 15 min in methionine- and cysteine-free medium, and proteins were metabolically labeled from 10 to 13 h posttransfection with ∼500 μCi of [35S]methionine-[35S]cysteine (Promix; Amersham) per ml. Cell lysis and immunoprecipitation of labeled EAV GP5 and M proteins (under both reducing and nonreducing conditions) were performed essentially as described by de Vries et al. (15, 17). To prevent the formation of disulfide bridges during or after cell lysis, 20 mM N-ethylmaleimide was included in the lysis buffer. After immunoprecipitation, immune complexes were eluted from the immunoadsorbent in Laemmli sample buffer without dithiothreitol (DTT). Samples were divided into two aliquots; one was analyzed directly (nonreducing conditions), whereas 100 mM DTT was added to the other sample for analysis under reducing conditions. Proteins were separated in sodium dodecyl sulfate (SDS)-containing 12.5% polyacrylamide gels and visualized by autoradiography.
RESULTS
GP5-M heterodimerization is required for transport to the Golgi complex.
Previously, biochemical studies by de Vries et al. (17) established the formation of a disulfide-linked heterodimer between the EAV GP5 and M proteins. In EAV-infected cells, heterodimerization coincided with transport of the two proteins to the Golgi complex, as evidenced by the acquisition of endoglycosidase H resistance by the N-linked oligosaccharide of GP5. Interestingly, the two partners were recruited into the GP5-M heterodimer with different kinetics, which may be explained by the presence of an excess of M protein in the endoplasmic reticulum (ER), where heterodimerization is assumed to occur (18). In addition to heterodimerizing with GP5, M was found to homodimerize, but the functional implications of this observation are unclear since the M-M homodimer was not detected in extracellular virions (17). More recently, we visualized the heterodimerization and transport of the EAV GP5 and M proteins to the Golgi complex by using double labeling in an immunofluorescence assay (IFA) (Fig. 2A1 and A2) (18). In experiments with chimeric GP5 proteins that were defective in heterodimerization with M, it was shown that both proteins were retained in the ER when complex formation was blocked and that they continued to be recognized by their respective antibodies. Likewise, we established that when the expression of ORF5 or ORF6 is completely blocked, the product of the other gene is retained in the ER and virus (or virus-like) particles are not released into the medium (data not shown). The latter experiments were carried out with previously described mutant EAV full-length cDNA clones in which either ORF5 or ORF6 had been disrupted (35).
FIG. 2.
Localization of EAV M and GP5 in BHK-21 cells transfected with wt EAV and a selection of mutants. Cells were fixed at 8 to 10 h posttransfection and processed for a double IFA using an anti-M rabbit serum, an anti-GP5 mouse MAb, and appropriate fluorescent conjugates. (A1 to E1) Labeling for M; (A2 to E2) signal for GP5. Constructs: A, pA45 (wt); B, pA45/C8S (M mutant); C, pA45/C34S (GP5 mutant); D, pA45-80.4 (pseudorevertant); E, pA45-80.4/C34S (pseudorevertant with the Cys-34- to-Ser mutation in GP5). (A and D) Transport of both M and GP5 to the Golgi complex and the accumulation of an excess of M in the ER (18). Transport of pA45-80.4 GP5 (D2) seems to be less efficient than for wt EAV (A2); see the text for details. For the Cys mutants in panels B, C, and E, transport to the Golgi complex was not observed.
Together, the data summarized above had firmly established the formation of intermolecular disulfide bonds by Cys residues in the ectodomains of both M and GP5 of EAV. As shown in Fig. 1, the (predicted) M ectodomain contains a single Cys residue at position 8, which is also conserved in other arteriviruses (18). In contrast, the putative GP5 ectodomain contains five Cys residues (at positions 34, 57, 63, 66, and 80), opening the possibility that in addition to the intermolecular interaction with M, intramolecular disulfide bonds may be formed.
Site-directed mutagenesis of Cys residues in GP5 and M.
To assess the importance of the six Cys residues (one in M and five in GP5) described above, we used our reverse-genetics system to generate six EAV mutants in which these residues were individually mutated to Ser. These mutants were initially screened by using the double IFA described above (18) to study the subcellular localization of M and GP5 in transfected cells. As expected, the Cys-8-to-Ser mutation in M abrogated the transport of both M and GP5 to the Golgi complex (Fig. 2B1 and B2) and resulted in the accumulation of both proteins in the ER. Remarkably, the same result was obtained for each of the five GP5 mutants (Fig. 2, C1 and C2 and data not shown), suggesting that the five Cys residues in the GP5 ectodomain were all critical for folding and/or heterodimerization of this glycoprotein (Table 2). Whereas the wt control virus had infected all initially untransfected cells by 24 h, the six mutant viruses were unable to spread from cell to cell (data not shown) unless reversion occurred (see below). At 24 h posttransfection, only some small clusters of infected cells were observed for some mutants and infection of all cells required at least 48 h (i.e., four cycles of EAV replication). Plaque assays with the cell culture supernatant harvested at 24 h posttransfection confirmed that the virus titers of the mutants were reduced at least 10,000-fold compared to the titer obtained for the wt control (data not shown).
Analysis of revertant viruses.
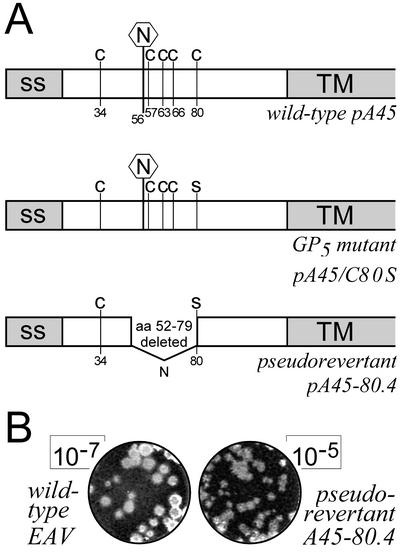
Since all Cys-to-Ser mutants required just a single nucleotide change to convert the mutant Ser codon to the original Cys codon, we expected that the low virus titers observed at 24 h posttransfection were due to rapid reversion of the mutants to the wt genotype. Thus, single plaques were picked from the plaque assay mixtures described above, cloned viruses were grown in BHK-21 cells, and the ORF5 or ORF6 region of their genome was amplified by RT-PCR. Direct sequence analysis of these amplicons indeed confirmed reversion to the wt Cys codon for all of these viruses, with one exception. One of the four revertants analyzed for the construct pA45/C80S had retained the mutant Ser codon at position 80 in GP5. In addition, its ORF5 contained an in-frame deletion, replacing the codons for residues 52 to 79 with a single AAU codon (for Asn). The junction generating this deletion in ORF5 was CACA to AUUCU, with the CAC codon in the first sequence encoding the wt His-51 and the UCU codon in the second sequence specifying the Cys-80-to-Ser mutation. Strikingly, the deletion in this revertant, which was named 80.4, resulted in the loss of the codons for the single GP5 N-glycosylation site (Asn-56) and residues Cys-57, Cys-63, and Cys-66. Thus, only a single Cys residue (Cys-34) was left in the GP5 ectodomain, which was now predicted to be unglycosylated, since the novel Asn codon that was created at the site of the deletion (see above) was not in the right sequence context to yield an alternative N-glycosylation site (Fig. 3A).
FIG. 3.
(A) Schematic representation of the structure of the GP5 ectodomain for wt EAV (pA45), the GP5 Cys-80-to-Ser mutant (pA45/C80S), and pseudorevertant 80.4 (pA45-80.4). The putative GP5 signal sequence (ss) and transmembrane domain (TM) are depicted in gray. Cys residues and the single GP5 N-glycosylation site (Asn-56) are indicated. (B) Example of a plaque assay of the virus harvested (24 h postinfection) from BHK-21 cells infected with wt EAV (left; dilution, 10−7) or pseudorevertant A45-80.4 (right; dilution, 10−5). The latter virus consistently produced 20- to 100-fold-reduced infectivity titers and somewhat smaller plaques.
Characterization of pseudorevertant 80.4.
To ascertain that a virus carrying the ORF5 deletion detected in pseudorevertant 80.4 was indeed viable, an RT-PCR product specifying the 80.4 deletion was cloned into the backbone of the original pA45 full-length cDNA clone, yielding construct pA45-80.4. Thus, we could rule out the possibility that the pseudoreversion of the 80.4 virus was based on the synergistic effect of the ORF5 deletion and additional mutations that might have been present in other viral genes. Transfection of full-length RNA transcribed from clone pA45-80.4 indeed yielded an infectious virus. Although GP5 of the A45-80.4 virus was no longer N glycosylated, the protein continued to be recognized by MAb 6D10, which recognizes an epitope formed by residues 99 to 104 (2, 3), just downstream of the deletion in GP5 (Fig. 2 and data not shown; also see Fig. 4). The GP5 and M proteins of the A45-80.4 virus were transported to the Golgi complex (Fig. 2D1 and D2), although a somewhat stronger labeling of the ER was observed for GP5 (Fig. 2D2), suggesting slower or incomplete export from the ER (see also below). The A45-80.4 virus was found to be substantially attenuated, since it grew to titers that were 20- to 100-fold lower than those of the wt virus control and produced somewhat smaller plaques (Fig. 3B). Nevertheless, the ORF5 deletion was found to be genetically stable on passaging. Nucleotide changes were not detected when the ORF5 sequence of the A45-80.4 virus was amplified by RT-PCR and sequenced after five consecutive 24-h passages in BHK-21 cells (comprising approximately 10 replication cycles).
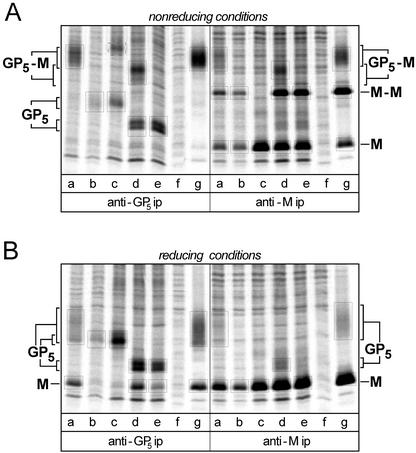
FIG. 4.
Analysis of GP5-M heterodimerization in BHK-21 cells transfected with wt EAV and a selection of mutants. Protein synthesis in transfected cells was35S labeled from 10 to 13 posttransfection, and immunoprecipitations were carried out with antisera recognizing GP5 (left side of panels) or M (right side of panels) of EAV. The immunoprecipitates were divided into two aliquots, which were analyzed under nonreducing conditions (A) or under reducing conditions (B), which have been described to disrupt the GP5-M disulfide bond. In each lane, the relevant bands (see the text) are boxed. The sizes of GP5 and the GP5-M complex in the assay under nonreducing conditions are variable, depending on the presence of the 80.4 deletion in GP5 (see the text). Constructs: lanes a, pA45 (wt); lanes b, pA45/C34S (GP5 mutant); lanes c, pA45/C8S (M protein mutant); lanes d, pA45-80.4 (pseudorevertant); lanes e, pA45-80.4/C34S (pseudorevertant with Cys-34-to-Ser mutation in GP5); lanes f, mock transfection; lanes g, control infection with wt EAV (MOI 10).
The EAV GP5 glycosylation site can be deleted, but not mutated, without loss of infectivity.
The properties of pseudorevertant 80.4 revealed that Asn-56, the single target for N-linked glycosylation of GP5 in the Bucyrus strain of EAV, and its flanking sequences can be deleted without complete loss of virus infectivity. This unexpected finding prompted us to also inactivate this glycosylation site by site-directed mutagenesis. Four mutants were engineered in which the Asn-Cys-Ser N-linked glycosylation motif at position 56-58 of GP5 was changed to Gln-Cys-Ser, Gly-Cys-Ser, Ser-Cys-Ser, or Asn-Cys-Gly, each of which should render the protein unsuitable as a target for N-linked glycosylation. On transfection, each of these mutants was replication and transcription competent, but infectious progeny was not produced. For all mutants, GP5 accumulated in the ER and neither GP5 nor M protein were transported to the Golgi complex (data not shown).
Thus, the N-linked glycosylation site of EAV GP5 could apparently be removed in the context of the 80.4 deletion, but its inactivation by single point mutations completely blocked GP5 function. Together with the observation that the replacement of each of the five Cys residues in the GP5 ectodomain also abrogated virus infectivity (see above), these data suggested that the internal organization of the EAV GP5 ectodomain is complex and that its disruption readily affected transport and processing of the glycoprotein.
Cys-8 of the M protein interacts with Cys-34 of GP5.
The properties of GP5 of the A45-80.4 virus strongly suggested that it is Cys-34 that interacts with Cys-8 of the M protein to form the GP5-M heterodimer. To test this hypothesis, the mutation specifying the Cys-34-to-Ser mutation in GP5 was introduced into pA45-80.4 (pA45-80.4/C34S). Cells were transfected with RNA transcribed from a number of full-length cDNA clones, including mutants pA45-C34S, pA45-C8S, pA45-80.4, and pA45-80.4/C34S (Table 2), and protein synthesis was 35S labeled from 10 to 13 h posttransfection. Transfection efficiencies were determined by IFA and were found to range from 30 to 50% (data not shown). Subsequently, immunoprecipitations were carried out with antisera recognizing the M protein or GP5. The immunoprecipitates were divided into two aliquots that were analyzed either under reducing conditions (with 100 mM DTT in the sample buffer) or under nonreducing conditions, which have been shown to leave the disulfide bond between GP5 and M intact (17).
The SDS-polyacrylamide gel electrophoresis (PAGE) results of this biochemical analysis are shown in Fig. 4. Because at least 50% of the cells had not been transfected, the background in the immunoprecipitation analysis of transfected cell cultures was relatively high compared to that in the lanes of the control infection with wt virus at a multiplicity of infection (MOI) of 10 (Fig. 4, lanes g). Nevertheless, when using nonreducing conditions we readily detected the GP5-M heterodimer, which was precipitated by the anti-GP5 MAb and the anti-M rabbit serum from lysates of cells transfected with the wt pA45 transcript (lanes a). In addition to the GP5-M complex, the anti-M serum precipitated the M-M homodimer and M monomers. Under reducing conditions, the GP5-M and M-M complexes disintegrated and only M and GP5 monomers were observed, the latter migrating as the usual broad 30 to 44-kDa band due to its heterogeneous glycosylation state (15). The mutant carrying the Cys-34-to-Ser mutation in GP5 (A45/C34S) did not produce the GP5-M heterodimer (lanes b); only a quite homogeneous GP5 monomer was observed, both under reducing and nonreducing SDS-PAGE conditions, suggesting that the molecule accumulated in the ER. The Cys-8-to-Ser mutation in M (construct A45/C8S; Fig. 4, lanes c) abrogated the formation of both the GP5-M heterodimer and the M-M homodimer. Under nonreducing conditions, the anti-GP5 immunoprecipitate contained an additional high-molecular-weight band, which may represent a GP5-GP5 complex that was not observed previously.
Our analysis of the cells transfected with the A45-80.4 virus (Fig. 4, lanes d) revealed that its GP5 migrated as a double band of 21 and 24 kDa. The calculated size of the A45-80.4 GP5, after removal of the predicted 18-residue signal sequence, is 23.6 kDa. The origin of the second band is unclear, but both GP5 species seem to participate in the formation of the GP5-M complex, since the intensities of both bands increased equally upon disruption of the complex under reducing conditions. Under nonreducing conditions, the GP5-M complex of A45-80.4 migrated as a smaller and more homogeneous complex than its counterpart from the wt A45 virus, an observation that matched the 27-residue deletion in GP5 and the lack of N glycosylation. Remarkably, a substantial amount of A45-80.4 GP5 showed up as monomers under nonreducing conditions, indicating that the mutant GP5 was probably impaired in heterodimerization (see also Fig. 2D2). On introduction of the Cys-34-to-Ser mutation into A45-80.4 (construct pA45-80.4/C34S; Fig. 4, lanes e), the GP5-M complex could no longer be detected, although some M monomers still coprecipitated with the GP5 monomers. Together, these data firmly established that heterodimerization of M and GP5 of deletion mutant A45-80.4, and most probably also of the wt A45 virus, depends on the presence of Cys-34 in the GP5 ectodomain. Furthermore, the formation of both the GP5-M heterodimer and the M-M homodimer depends on the presence of Cys-8 in the M ectodomain.
DISCUSSION
The recent development of reverse-genetics systems (16, 31, 47) has created new avenues to explore the properties and functions of the unique but poorly characterized set of structural proteins that is used by arteriviruses. It has now been established that the products of all seven genes in the 3′-proximal region of the genomes of EAV and the swine arterivirus PRRSV can be detected in virus particles (15, 33, 34, 45, 48, 49, 50). Moreover, using reverse genetics, we have previously demonstrated that in EAV, each of these seven proteins (E-GP2b-GP3-GP4-GP5-M-N) is required to produce infectious progeny (35). These proteins clearly fall into two categories, the major structural proteins N, M, and GP5 and the minor structural proteins E, GP2b, GP3, and GP4. Although contradictory reports have been published on the structural nature of some of these proteins, in particular GP3 of PRRSV and both GP3 and E of LDV (21, 30, 41), it seems unlikely that there would be fundamental differences between arteriviruses in this respect. Possibly, technical complications explain some of these observations, in particular the lack of (suitable) antibodies for these proteins and/or the low abundance of the minor structural proteins in the virion.
How the RNA genome, the N protein, and the six envelope polypeptides of EAV interact with each other during virus assembly remains an open question. In particular, the presence of subsets of major and minor structural proteins is intriguing. Clearly, the GP5-M heterodimer is the major protein component of the viral envelope (15, 20, 34). Its formation triggers a series of important events, including transport of the two proteins to the Golgi complex, the extensive but variable modification of the GP5 sugar moiety into a polylactosaminoglycan (15), and probably also the incorporation of the GP5-M complex into the budding virion. The PRRSV GP5-M heterodimer was recently implicated in attachment to a heparinlike receptor on the surface of porcine alveolar macrophages (13), although recent studies with EAV chimeras make it clear that another factor than the GP5 ectodomain sequence may determine the host specificity of arteriviruses (18).
In this study we have shown that the critical interaction between the luminal domains of the two major envelope proteins of EAV depends on the formation of a disulfide bridge between two Cys residues, Cys-8 of M and Cys-34 of GP5. Replacement of either Cys residue completely blocked heterodimerization of the two proteins (Fig. 4), resulting in their accumulation in the ER (Fig. 2), a block in maturation of the GP5 sugar moiety (Fig. 4), and a complete block in the production of infectious progeny (unless reversion occurred). An initial complication was the fact that replacement of each of the five Cys residues in the GP5 ectodomain abolished virus infectivity. However, the fortuitous isolation of pseudorevertant 80.4 (Fig. 3), containing a large deletion and just a single Cys residue in its GP5 ectodomain, allowed us to identify Cys-34 of GP5 as the partner of Cys-8 in the M ectodomain. Obviously, it can be argued that one of the other GP5 Cys residues may play this role in the full-length GP5 of the wt virus, but it is remarkable that the sole Cys residue in the much shorter ectodomain of PRRSV and LDV GP5 occupies exactly the same position as Cys-34 in EAV GP5, 16 to 17 residues downstream of the (predicted) N terminus of the protein after signal sequence cleavage (18). The deleterious effect of the replacement of the GP5 Cys residues at positions 57, 63, 66, and 80 may be explained by assuming that intramolecular disulfide bridges are formed between these residues, which may be critical for proper folding and function of the glycoprotein. Disruption of one of these bonds would leave an unpaired Cys residue, which could interfere with the formation of other intramolecular disulfide bonds or the intermolecular bridge with the M protein. This may explain why the deletion of this entire region of GP5 in pseudorevertant 80.4 largely neutralized the adverse effects of the Cys-80-to-Ser mutation.
Similar GP5 deletions in response to a specific selective pressure were observed previously by Balasuriya et al. (3). Using a panel of anti-GP5 monoclonal antibodies, they isolated two different neutralization escape mutants that contained large deletions in their GP5 ectodomain (residues 66 to 112 and 62 to 101, respectively). In both escape mutants, the deletion affected the region around residues 99 and 100, which constitutes a major neutralization site of EAV (2, 22). In pseudorevertant 80.4, this region was maintained, but surprisingly the single conserved GP5 glycosylation site at Asn-56 was deleted. Most nonlaboratory strains of EAV contain a second GP5 glycosylation site at position 81, and it has been noticed that this site has been lost in all cell culture-adapted strains and the ARVAC vaccine virus (3, 22). However, an infectious EAV (or arterivirus) variant without a single GP5 sugar moiety has not been described before. As with the individual replacements of the Cys residues at positions 57, 63, 66, and 80, GP5 function was completely lost when Asn-56 was “simply” removed by site-directed mutagenesis (to either Gln, Gly, or Ser) or when glycosylation was abrogated by mutating the downstream Ser-58, which is part of the Asn-X-Ser N-linked glycosylation motif. Thus, the isolation of pseudorevertant 80.4 once again underlines the capacity of RNA viruses to rapidly adapt to changing circumstances and “engineer” a protein structure that will ensure their survival.
Glycosylation of the major EAV glycoprotein probably affects the structure and immunogenicity of GP5 and may also affect other biological features of the virus. In LDV (and PRRSV [42]), the GP5 ectodomain contains one to three N-linked glycans and (as in EAV) glycosylation occurs by the addition of variable numbers of lactosamine repeats (29). For LDV, Plagemann and coworkers have described a relationship between the extent of GP5 glycosylation and the efficiency of virus neutralization (8, 9, 28). It will be interesting to evaluate the immunological and biological properties of the attenuated EAV A45-80.4 in horses. The A45-80.4 GP5 deletion, which was found to be stable on passaging of the virus in cell culture, may be useful in attempts to develop attenuated recombinant vaccine viruses and may also provide a suitable site for the insertion of heterologous epitopes or marker sequences into the GP5 ectodomain.
Acknowledgments
We gratefully acknowledge Twan de Vries, Amy Glaser, and Peter Rottier (Utrecht University) for providing cDNA constructs and antisera. We are grateful to Udeni Balasuriya and James MacLachlan (University of California, Davis) for providing anti-GP5 monoclonal antibodies. We thank Yvonne van der Meer for assistance with fluorescence microscopy and image processing, and we thank Roeland Wieringa and Peter Rottier for sharing unpublished information.
This work was supported by grant PL96-1666 from the FAIR program of the European Union.
REFERENCES
- 1.Balasuriya, U. B., H. W. Heidner, N. L. Davis, H. M. Wagner, P. A. Hullinger, J. F. Hedges, J. C. Williams, R. E. Johnston, W. D. Wilson, I. K. Liu, and N. J. Maclachlan. 2002. Alphavirus replicon particles expressing the two major envelope proteins of equine arteritis virus induce high level protection against challenge with virulent virus in vaccinated horses. Vaccine 20:1609-1617. [DOI] [PubMed] [Google Scholar]
- 2.Balasuriya, U. B., N. J. Maclachlan, A. A. F. de Vries, P. V. Rossitto, and P. J. M. Rottier. 1995. Identification of a neutralization site in the major envelope glycoprotein (GL) of equine arteritis virus. Virology 207:518-527. [DOI] [PubMed] [Google Scholar]
- 3.Balasuriya, U. B., J. F. Patton, P. V. Rossitto, P. J. Timoney, W. H. McCollum, and N. J. Maclachlan. 1997. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the GL envelope glycoprotein. Virology 232:114-128. [DOI] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., P. V. Rossitto, C. D. DeMaula, and N. J. Maclachlan. 1993. A 29K envelope glycoprotein of equine arteritis virus expresses neutralization determinants recognized by murine monoclonal antibodies. J. Gen. Virol. 74:2525-2529. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya, U. B., P. J. Timoney, W. H. McCollum, and N. J. Maclachlan. 1995. Phylogenetic analysis of open reading frame 5 of field isolates of equine arteritis virus and identification of conserved and nonconserved regions in the GL envelope glycoprotein. Virology 214:690-697. [DOI] [PubMed] [Google Scholar]
- 6.Cafruny, W. A., S. P. Chan, J. T. Harty, S. Yousefi, K. Kowalchyk, D. McDonald, B. Foreman, G. Budweg, and P. G. W. Plagemann. 1986. Antibody response of mice to lactate dehydrogenase-elevating virus during infection and immunization with inactivated virus. Virus Res. 5:357-375. [DOI] [PubMed] [Google Scholar]
- 7.Castillo-Olivares, J., A. A. F. de Vries, M. J. B. Raamsman, P. J. M. Rottier, K. Lakhani, D. Westcott, J. P. Tearle, J. L. Wood, J. A. Mumford, D. Hannant, and N. J. Davis-Poynter. 2001. Evaluation of a prototype sub-unit vaccine against equine arteritis virus comprising the entire ectodomain of the virus large envelope glycoprotein (GL): induction of virus-neutralizing antibody and assessment of protection in ponies. J. Gen. Virol. 82:2425-2435. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., K. Li, and P. G. W. Plagemann. 2000. Neuropathogenicity and sensitivity to antibody neutralization of lactate dehydrogenase-elevating virus are determined by polylactosaminoglycan chains on the primary envelope glycoprotein. Virology 266:88-98. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., R. R. R. Rowland, G. W. Anderson, G. A. Palmer, and P. G. W. Plagemann. 1997. Coexistence in lactate dehydrogenase-elevating virus pools of variants that differ in neuropathogenicity and ability to establish a persistent infection. J. Virol. 71:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chirnside, E. D., A. A. F. de Vries, J. A. Mumford, and P. J. M. Rottier. 1995. Equine arteritis virus-neutralizing antibody in the horse is induced by a determinant on the large envelope glycoprotein GL. J. Gen. Virol. 76:1989-1998. [DOI] [PubMed] [Google Scholar]
- 11.Coutelier, J. P., and J. van Snick. 1988. Neutralization and sensitization of lactate dehydrogenase-elevating virus with monoclonal antibodies. J. Gen. Virol. 69:2097-2100. [DOI] [PubMed] [Google Scholar]
- 12.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:658-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deregt, D., A. A. F. de Vries, M. J. B. Raamsman, L. D. Elmgren, and P. J. M. Rottier. 1994. Monoclonal antibodies to equine arteritis virus proteins identify the GL protein as a target for virus neutralization. J. Gen. Virol. 75:2439-2444. [DOI] [PubMed] [Google Scholar]
- 15.de Vries, A. A. F., E. D. Chirnside, M. C. Horzinek, and P. J. M. Rottier. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries, A. A. F., A. L. Glaser, M. J. B. Raamsman, C. A. M. de Haan, S. Sarnataro, G. J. Godeke, and P. J. M. Rottier. 2000. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology 270:84-97. [DOI] [PubMed] [Google Scholar]
- 17.de Vries, A. A. F., S. M. Post, M. J. B. Raamsman, M. C. Horzinek, and P. J. M. Rottier. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 69:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobbe, J. C., Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288:283-294. [DOI] [PubMed] [Google Scholar]
- 19.Doll, E. R., J. T. Bryans, W. H. McCollum, and M. E. W. Crowe. 1957. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 47:3-41. [PubMed] [Google Scholar]
- 20.Faaberg, K. S., and P. G. W. Plagemann. 1995. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology 212:512-525. [DOI] [PubMed] [Google Scholar]
- 21.Faaberg, K. S., and P. G. W. Plagemann. 1997. ORF3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology 227:245-251. [DOI] [PubMed] [Google Scholar]
- 22.Glaser, A. L., A. A. F. de Vries, and E. J. Dubovi. 1995. Comparison of equine arteritis virus isolates using neutralizing monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J. Gen. Virol. 76:2223-2233. [DOI] [PubMed] [Google Scholar]
- 23.Gonin, P., B. Pirzadeh, C. A. Gagnon, and S. Dea. 1999. Seroneutralization of porcine reproductive and respiratory syndrome virus correlates with antibody response to the GP5 major envelope glycoprotein. J. Vet. Diagn. Investig. 11:20-26. [DOI] [PubMed] [Google Scholar]
- 24.Harty, J. T., S. P. Chan, and P. G. W. Plagemann. 1987. Characteristics of monoclonal antibodies to the lactate dehydrogenase-elevating virus. Intervirology 27:53-60. [DOI] [PubMed] [Google Scholar]
- 25.Harty, J. T., and P. G. W. Plagemann. 1988. Formalin inactivation of the lactate dehydrogenase-elevating virus reveals a major neutralizing epitope not recognized during natural infection. J. Virol. 62:3210-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 27.Landt, O., H.-P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96:125-128. [DOI] [PubMed] [Google Scholar]
- 28.Li, K., Z. Chen, and P. G. W. Plagemann. 1998. The neutralization epitope of lactate dehydrogenase-elevating virus is located on the short ectodomain of the primary envelope glycoprotein. Virology 242:239-245. [DOI] [PubMed] [Google Scholar]
- 29.Li, K., Z. Chen, and P. G. W. Plagemann. 1998. The neutralization epitope of lactate dehydrogenase-elevating virus is located on the short ectodomain of the primary envelope glycoprotein. Virology 242:239-245. [DOI] [PubMed] [Google Scholar]
- 30.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meulenberg, J. J. M., J. N. A. Bos-de Ruijter, G. Wensvoort, and R. J. M. Moormann. 1998. Infectious transcripts from cloned genome-length cDNA of porcine reproductive respiratory syndrome virus. J. Virol. 72:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meulenberg, J. J. M., M. M. Hulst, E. J. de Meijer, P. L. J. M. Moonen, A. den Besten, E. P. de Kluyver, G. Wensvoort, and R. J. M. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meulenberg, J. J. M., and A. Petersen-den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44-51. [DOI] [PubMed] [Google Scholar]
- 34.Meulenberg, J. J. M., A. Petersen-den Besten, E. P. de Kluyver, R. J. M. Moorman, W. M. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 36.Murtaugh, M. P., M. R. Elam, and L. T. Kakach. 1995. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 140:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nugent, J., R. Sinclair, A. A. F. de Vries, R. Y. Eberhardt, J. Castillo-Olivares, P. N. Davis, P. J. M. Rottier, and J. A. Mumford. 2000. Development and evaluation of ELISA procedures to detect antibodies against the major envelope protein (GL) of equine arteritis virus. J. Virol. Methods 90:167-183. [DOI] [PubMed] [Google Scholar]
- 38.Ostrowski, M., J. A. Galeota, A. M. Jar, K. B. Platt, F. A. Osorio, and O. J. Lopez. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 76:4241-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirzadeh, B., and S. Dea. 1997. Monoclonal antibodies to the ORF5 product of porcine reproductive and respiratory syndrome virus define linear neutralizing determinants. J. Gen. Virol. 78:1867-1873. [DOI] [PubMed] [Google Scholar]
- 41.Plagemann, P. G. W. 2001. An ORF-2a protein is not present at a significant level in virions of the arterivirus lactate dehydrogenase-elevating virus. Virus Res. 74:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland, R. R. R., M. Steffen, T. Ackermann, and D. A. Benfield. 1999. The evolution of porcine reproductive and respiratory syndrome virus: quasispecies and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology 259:262-266. [DOI] [PubMed] [Google Scholar]
- 43.Snijder, E. J., and J. J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 44.Snijder, E. J., and J. J. M. Meulenberg. 2001. Arteriviruses, p. 1205-1220. In D. M. Knipe, and P. M., Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 45.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. B. Raamsman, and A. A. F. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dinten, L. C., J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Nieuwstadt, A. P., J. J. M. Meulenberg, A. van Essen-Zandbergen, A. Petersen-den Besten, R. J. Bende, R. J. M. Moormann, and G. Wensvoort. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wieringa, R., A. A. F. de Vries, M. J. B. Raamsman, and P. J. M. Rottier. 2002. Characterization of two new structural glycoproteins, GP3 and GP4, of equine arteritis virus. J. Virol. 76:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]