Abstract
Except for ribavirin, no other antiviral drugs for treating hantaviral diseases have been identified. It is well established that ribavirin will inhibit the production of infectious Hantaan virus (HTNV); however, its mechanism of action is unknown. To characterize the inhibitory effect of ribavirin on HTNV, the levels of viral RNAs, proteins, and infectious particles were measured for 3 days posttreatment of HTNV-infected Vero E6 cells. HTNV-infected cells treated with ribavirin showed a slight reduction in the levels of cRNA, viral RNA, and mRNA populations on the first day postinfection. The amount of cRNA and viral RNA increased to that observed for untreated HTNV-infected cells on day 2, whereas mRNA levels were more greatly reduced on days 2 and 3. Despite the finding of S-segment mRNA, albeit low, three of the viral proteins—nucleocapsid (N) protein and glycoproteins G1 and G2—could not be detected by immunohistochemistry in ribavirin-treated cells. To test the hypothesis that these effects were caused by incorporation of ribavirin into nascent RNA and a resultant “error catastrophe” was occurring, we cloned and sequenced the S-segment cRNA/mRNA from ribavirin-treated or untreated cells from day 3. We found a high mutation frequency (9.5/1,000 nucleotides) in viral RNA synthesized in the presence of ribavirin. Hence, the transcripts produced in the presence of the drug were not functional. These results suggest that ribavirin's mechanism of action lies in challenging the fidelity of the hantavirus polymerase, which causes error catastrophe.
Hantaviruses, which are endemic in most regions of the world, persistently infect murid rodents and are shed through rodent excreta (20). Transmission of hantaviruses from rodent hosts to humans causes two illnesses, hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome. Hantaan virus (HTNV), carried by Apodemus agrarius, produces one of the more severe HFRS illnesses caused by the Old World hantaviruses, causing death in 5 to 15% of the cases (14, 15). HTNV infections cause a renal dysfunction with fever, hemorrhaging, cardiovascular instability, and shock. Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) is the only antiviral drug shown to have efficacy against HFRS in clinical trials (11). Ribavirin has been tested for its effectiveness in clinical trials with patients suspected to have hantavirus pulmonary syndrome; however, its therapeutic benefits are still not known (2).
Ribavirin has a broad spectrum of antiviral activity against both RNA and DNA viruses. Its mechanism of action, however, has been difficult to elucidate, primarily because of its pleiotropic effects (10, 17). Ribavirin 5′-monophosphate resembles GMP and can decrease cellular GTP pools due to the inhibition of the enzyme inosine monophosphate dehydrogenase dehydrogenase; however, this decrease does not completely account for the observed antiviral activity. Inhibitory effects have also been noted on the capping (9) and translation efficiency (23) of viral mRNA, as well as a direct suppressive effect on the viral polymerase activity (7, 8, 25). Crotty et al. showed that ribavirin acts as a potent RNA virus mutagen in poliovirus-infected cells (4). They showed that genomes synthesized in the presence of ribavirin had a substantial increase in C-to-U transition mutations. The incorporation of ribavirin triphosphate as a GTP analogue resulted in templates that are copied to yield C or U with equal frequency at the sites of incorporation during negative-strand synthesis (4). Further, Lanford et al. showed that treating GB virus B-infected hepatocytes with ribavirin resulted in a fourfold reduction in viral RNA (vRNA) levels (13). These authors suggest that the incorporation of ribavirin induced error-prone replication, resulting in a reduction in viral titer (13).
The hantavirus genome consists of three segments of negative-sense single-strand RNA, designated S, M, and L, encoding the nucleocapsid (N) protein, the envelope glycoproteins (G1 and G2), and the RNA-dependent RNA polymerase (RdRp), respectively (19). The N protein encapsidates the three vRNAs within the virion particle, and these serve as templates for transcription and replication in the cytoplasm of an infected host cell. Hantavirus replication proceeds by synthesis of an antigenomic, cRNA, intermediate transcribed from the vRNA by the RdRp, which is then used to produce the vRNA. Transcription of viral mRNAs initiates with the endonucleolytic cleavage of host mRNAs to yield capped oligonucleotides that prime mRNA synthesis from the vRNA templates, and termination occurs shortly after the coding information (19).
In Vero E6 cells infected with HTNV, ribavirin acts as an inhibitor of the life cycle with an 50% effective dose (ED50) of 14.8 μg/ml (12). The mechanism of action for the observed sensitivity of HTNV to ribavirin at the molecular level is not known. We examined here the effect of ribavirin on synthesis of each of the vRNAs, as well as three of the viral protein products. After HTNV-infected cells were treated with ribavirin, we monitored replication and transcription of the cell-associated HTNV S-segment vRNA, cRNA, and mRNA populations; viral protein production; and infectious viral particle formation during infection.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides were synthesized on an Applied Biosystems model 380B DNA synthesizer or purchased from Integrated DNA Technologies (Coralville, Iowa). Oligonucleotides were 5′-end labeled with [γ-32P]ATP (6,000 Ci/mmol; Dupont/New England Nuclear, Boston, Mass.) and T4 polynucleotide kinase reagent (Gibco-BRL, Grand Island, N.Y.). The labeled products were separated from unincorporated nucleotides on G50 Sephadex columns.
Ribavirin treatments and RNA isolation.
Six-well cell culture plates containing Vero E6 cells (ATCC CRL 1586) were infected with HTNV, strain 76-118, at a multiplicity of infection (MOI) of 0.01 or 0.1 and then incubated at 37°C. After 1 h of adsorption, the monolayers were overlaid with 2 ml of Eagle minimum essential medium with 10% fetal bovine serum containing 0 or 24 μg of ribavirin (ICN Pharmaceuticals, Costa Mesa, Calif.) and returned to 37°C. Seven six-well plates were established for each treatment (0 or 24 μg of ribavirin). For each of 3 days after infection, total intracellular RNA was isolated with Trizol LS reagent (Gibco-BRL) from each well from one plate, resulting in six separate samples per treatment per day. RNA was stored at −80°C.
Probe design.
Three species of viral S-segment RNA were measured, vRNA, cRNA, and mRNA (Table 1). The vRNA and cRNA are complementary; therefore, probes could easily distinguish between these two species. However, the probe designed for detecting the mRNA can also detect the cRNA, although the cRNA probe cannot hybridize to mRNA, because mRNA is truncated at the 3′ end relative to cRNA, and the cRNA probe binds to the deleted region. Therefore, we estimated the amount of mRNA by subtracting the levels measured for the cRNA from the total RNA detected with the mRNA probe.
TABLE 1.
Oligonucleotides used to measure HTNV S-segment vRNA, cRNA, and mRNA populations
RNA analysis.
To follow the production of the different populations of vRNAs relative to replication and transcription, we designed an RNA slot blot assay and used oligonucleotide probes to detect HTNV S-segment vRNA, mRNA, and cRNA, as well as host actin levels (Table 1). A total of 8 μg of RNA was bound to nitrocellulose membranes (Schleicher & Schuell, Inc., Keene, N.H.) as described by the manufacturer (Bio-Rad Laboratories, Richmond, Calif.) and probed sequentially with S-vRNA, S-mRNA, S-cRNA, and actin-specific oligonucleotides. Nitrocellulose filters were prehybridized in 50 mM PIPES (pH 7.4), 100 mM NaCl, 50 mM sodium phosphate, 1 mM EDTA, and 5.0% sodium dodecyl sulfate for 60 min at the appropriate temperature (54°C for S-vRNA, 48°C for S-mRNA, 50°C for S-cRNA, and 72°C for actin) (24). The prehybridization buffer was discarded and replaced with fresh hybridization buffer (same as above) containing 106 cpm of radiolabeled oligonucleotide per ml of buffer. Hybridization was carried out in a water bath for 16 h at the prehybridization temperatures. Blots were washed once at room temperature and twice at the hybridization temperature in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% sodium dodecyl sulfate. Dried membranes were visualized with a Storm 860 Molecular Dynamics PhosphorImager, and signals were measured by using ImageQuant version 4.2 software (Sunnyvale, Calif.). The probe for actin RNA levels to control for RNA loading and to normalize RNA levels between experiments was designed from Macaca fascicularis actin coding sequence (GenBank accession no. U20576). Thus, the pixels obtained by phosphorimager analysis for a sample were divided by the pixels obtained by using the actin probe for that sample. In addition, each filter contained serial dilutions (0.05, 0.5, and 5 ng) of unlabeled RNA transcript of the HTNV S coding region and the HTNV S-segment vRNA. Transcripts were produced from linearized plasmids by using the MaxiScript SP6/T7 RNA transcription kit (Ambion, Austin, Tex.). These acted as standards to adjust for probe specific activities and thus allow comparisons among different experiments.
Assay for infectious particles.
For each of 7 days, the supernatants from six wells were pooled and stored at −80°C. Once all samples were collected, the supernatants were assayed for infectious viral particles as described previously (21).
Antibodies.
Three monoclonal antibodies specific to HTNV N (ECO2-BDO1) (22), G1 (H13-16D2-1-1) (1), and G2 (EBO6-AAO2) (1) were used. The reactivity patterns of these antibodies are described elsewhere (1, 18). All monoclonal antibodies were used at a 1:100 dilution. Rabbit anti-laminin (Sigma, St. Louis, Mo.) was used at a 1:25 dilution.
Immunohistochemical analysis and confocal microscopy.
Vero E6 cells were removed from one well of the six-well plate by trypsin. Then, 4 ml of medium was added, and the resuspended cells were placed dropwise onto 10-well HTC-coated microscope slides. After 24 h at 37°C, the cells were washed in phosphate-buffered saline (PBS) for 3 min and fixed for 10 min in ice-cold acetone. Slides were incubated with 30 μl of the appropriate HTNV monoclonal antibody in PBS for 30 min in a humid chamber at 37°C. After two consecutive 3-min washes in PBS, cells were incubated for 30 min with 20 μl of fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG) (H+L) F(ab′)2 (Kirkegaard & Perry Laboratories, Gaithersberg, Md.) secondary antibody at a 1:100 dilution in a humid chamber at 37°C. Slides were then washed twice (3 min/wash) in PBS and incubated with 30 μl of anti-laminin antibody (Sigma) in PBS for 1.5 h in a humid chamber at 37°C. After two consecutive 3-min washes in PBS, cells were incubated with 20 μl of tetramethylrhodamine isothiocyanate (TRITC)-conjugated anti-rabbit IgG (H+L) F(ab′)2 (Calbiochem, La Jolla, Calif.) as a secondary antibody at a 1:40 dilution for 2.5 h in a humid chamber at 37°C. Slides were then washed twice (3 min/wash) in PBS and once with distilled water and mounted in Vector shield. The antiviral effects of ribavirin were recorded and stored by using a Zeiss axioscope connected to an Orca 100 camera interfaced to a Macintosh computer with Improvision software. Confocal images were recorded by a LSM 510 META microscope (Zeiss).
Sequencing.
The S-segment coding region for the HTNV N protein was sequenced to monitor mutation frequency. For sequence analysis, HTNV-infected, ribavirin-treated RNA was isolated by Trizol LS reagent (Gibco-BRL) from 106 cells (treated as previously described). Reverse transcription (RT)-PCRs were performed by using SuperScript One-Step RT-PCR (Gibco-BRL). cDNA was synthesized from 10 μg of total RNA, and HTNV S-segment sequence was amplified by PCR with HTNV S-segment-specific primers (HTNV S-ORF forward [CGATGGCAACTATGGAGGAATTAC] and HTNV S-ORF reverse [GAGTTTCAAAGGCTCTTGGTTGG]). The 1,289-bp PCR product was ligated into pGEM-T vector (Promega, Madison, Wis.) and transformed, and plasmid DNA was prepared from independent bacterial colonies. Plasmid DNA was sequenced from positions 37 to 1326 (cRNA) of the HTNV N protein coding region by using BigDye terminator cycle sequencing and analyzed with Vector NTI software (Informax, Bethesda, Md.). RT-PCRs were tested without RT-Taq polymerases as a control to eliminate false-positive results.
RESULTS
Inhibition of HTNV antigenomic synthesis by ribavirin.
The effect of ribavirin on the synthesis of cRNA was monitored with the cRNA probe indicated in Table 1. In untreated Vero E6 cells, infected at MOIs of 0.01 and 0.1, HTNV S-segment cRNA levels peaked on days 1 and 3 postinfection (p.i.) (Fig. 1A and B). In both the low and high MOIs (Fig. 1A and B), adding ribavirin reduced S-cRNA levels on day 1 p.i. The decrease in cRNA levels was approximately threefold in the 0.01 MOI sample (Fig. 1A). In the case of cells infected at an MOI of 0.1, the cRNA levels were 6.8-fold reduced compared to untreated cells (Fig. 1B). Thereafter, the levels of cRNA in the ribavirin-treated cells increased over the remaining days examined and then showed slightly lower levels that, for the most part, did not differ from the levels in the untreated cells. Particularly in the case of the higher HTNV MOI, there was little difference between the mock ribavirin treatment (no drug) and the ribavirin treatments after day 1 p.i.
FIG. 1.
Effect of ribavirin on HTNV S-segment RNA replication and transcription. Six-well cell culture plates containing Vero E6 cells per well were infected with HTNV at an MOI of 0.01 (A, C, and E) or an MOI of 0.1 (B, D, and F). After 1 h, the monolayers were overlaid with 2 ml of Eagle minimum essential medium containing 0 (▪) or 24 (▩) μg of ribavirin. For each of the following 3 days, total RNA was isolated and probed for cRNA (A and B), vRNA (C and D), and mRNA (E and F) synthesis as described in Materials and Methods. An actin probe was used to control the proportional quantity of cellular RNA (Table 1). The graph shows the area obtained from PhosphorImager analysis of the hybridization signals from two separate experiments of the RNA levels in which the RNA levels were analyzed in triplicate (± the standard deviation). ✽, Up to 4-fold differences from wild-type levels (0 μg of ribavirin); ✽✽, up to 8-fold differences from wild-type levels (0 μg of ribavirin); ✽✽✽, a >8-fold difference from wild-type levels (0 μg of ribavirin).
Effect of ribavirin on HTNS vRNA replication differs at low and high MOIs.
To investigate the influence of MOI on S-vRNA, we examined ribavirin's antiviral effect on HTNV-infected Vero E6 cells at a low MOI (MOI = 0.01; Fig. 1C) and at a higher MOI (MOI = 0.1; Fig. 1D). In untreated cells infected at an MOI of 0.01, HTNV S-segment vRNA levels were fairly constant (Fig. 1C). However, in the ribavirin-treated cells, there was an increase in the level of vRNA of 2.6-fold on day 3 p.i. In the cells infected at an MOI of 0.1, the highest vRNA levels were observed on days 1 and 3 p.i. (Fig. 1D). We observed initial reductions in S-vRNA levels of ∼2.3-fold on day 1 p.i. and of 2.9-fold on day 3 p.i. in the ribavirin-treated cells compared to the untreated virus-infected cells. At the other time points, there was no difference between the untreated and ribavirin-treated HTNV-infected cells. In summary, at the higher MOI, we noted a greater effect on vRNA synthesis for the ribavirin-treated cells, which had lower vRNA levels than levels in the untreated HTNV-infected cells on days 1 and 3 (Fig. 1D).
Ribavirin inhibits HTNV S mRNA levels.
The cRNA and mRNA are synthesized with the same positive-sense polarity. The mRNA is truncated at the 3′-end, however, relative to the cRNA. Thus, the probe designed for detecting the mRNA will also detect the cRNA, although the cRNA probe will not hybridize to mRNA because of the 3′ truncation. Therefore, we estimated the amount of HTNV S mRNA by subtracting the levels measured for the cRNA from the total RNA detected with the mRNA probe. In HTNV-infected cells (MOI = 0.01, no drug), the amount of S mRNA remained fairly high and constant from days 1 through 3. In HTNV-infected cells treated with ribavirin, S-mRNA levels were substantially lower than levels in untreated HTNV-infected cells from days 1 through 3 p.i. (Fig. 1E). In the cells treated with ribavirin, S mRNA levels were reduced 1.9-, 6.3-, and 6.7-fold on days 1, 2, and 3 p.i., respectively, compared to levels in untreated HTNV-infected cells.
In untreated, HTNV-infected cells (MOI = 0.1), the level of S-segment mRNA increased over the 3 days (Fig. 1F). In the ribavirin-treated cells, reduced levels of S mRNA were detected for the 3 days. There was a decrease in S mRNA levels on day 1 of 2.8-fold, followed by a more dramatic drop in mRNA level on day 2, in which the levels were reduced 22-fold compared to the levels in wild-type cells. In summary, the fold decrease observed for mRNA levels at the lower MOI on day 1 was similar to that observed for the higher MOI. On day 2, however, the reduction in mRNA levels in the higher-MOI experiments was much greater than that observed for the lower MOI. The levels of mRNA remained low on day 3 in the low-MOI experiments but recovered substantially in the higher-MOI experiments.
Effect of ribavirin on infectious HTN virus production.
A plaque assay was used to monitor the yield of infectious HTNV in pooled supernatants collected during the experiment. In both the low and high HTNV infections, adding ribavirin affected the number of HTNV PFU compared to the untreated HTNV-infected Vero E6 cells (Fig. 2). No plaques were observed on days 1 through 3 p.i. when the MOI was 0.01 (Fig. 2A). Infectious virus was produced on days 4, 5, and 6, although the levels were decreased by 550-, 311-, and 17-fold, respectively, versus the levels in untreated HTNV-infected cells. In the ribavirin-treated, virus-infected cells (MOI = 0.1), there were no plaques observed on days 1 or 2 p.i. (Fig. 2B). On day 3 p.i., infectious viral particles were produced; however, the level was 379-fold lower than the levels in untreated HTNV-infected Vero E6 cells.
FIG. 2.
Effect of ribavirin on the yield of infectious HTNV. Supernatants were harvested from HTNV-infected cells at MOIs of 0.01 (A) and 0.1 (B) for the various treatments, i.e., 0 (•) or 24 (▪) μg of ribavirin, and then assayed for plaque formation on monolayers of Vero E6 cells.
HTNV N protein and glycoprotein expression is inhibited by ribavirin.
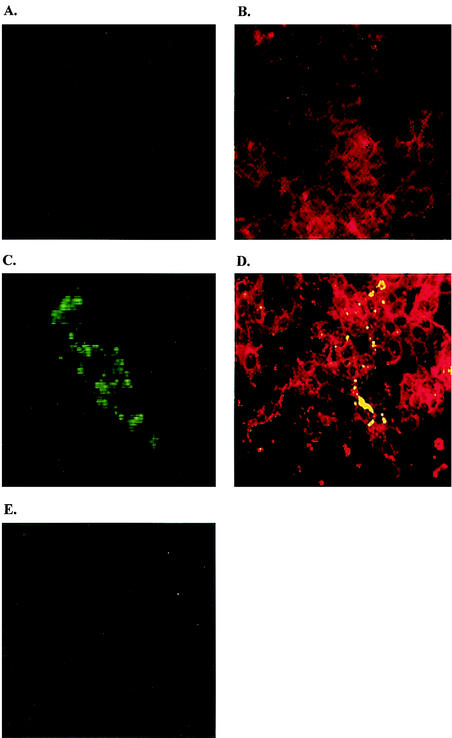
The level of HTNV N protein synthesis was measured for each of the HTNV-infected ribavirin-treated Vero cells by immunofluorescence with monoclonal antibody ECO2-BDOl (22). We failed to detect HTNV N protein on days 1 and 2 p.i. However, on day 3 p.i., HTNV N protein was visible in the cytoplasm of control cells lacking ribavirin (Fig. 3C and D), whereas viral proteins were not detected in the presence of ribavirin (Fig. 3E). These results suggest that a reduction in the N protein blocked the assembly of nucleocapsids.
FIG. 3.
Inhibition of HTNV N protein synthesis by ribavirin. (A) Monolayers of Vero E6 cells. (B) Monolayers of Vero cells stained with anti-laminin antibody, followed by TRITC (tetramethyl rhodamine isothiocyanate)-conjugated anti-rabbit immunoglobulin. (C) Vero cells infected with HTNV 76-118 and stained with N mouse monoclonal antibody (EC02-BD01) (22), followed by FITC-conjugated anti-mouse immunoglobulin. (D) Confocal image (overlay) of Vero cells infected with HTNV, stained with anti-laminin TRITC and N mouse monoclonal antibody FITC and observed by using an LSM 510 META microscope (Zeiss). (E) Vero cells infected with HTNV, treated with 24 μg of ribavirin/ml, and stained with N mouse monoclonal antibody. Cells were fixed 72 h after infection and analyzed by immunofluorescence for the presence of HTNV N protein by using a Zeiss axioscope connected to an Orca 100 camera interfaced to a Macintosh computer with Improvision software.
HTNV-infected ribavirin-treated Vero cells were analyzed by immunofluorescence for the presence of HTNV G1 protein with monoclonal antibody H13-16D2-1-1 (1) and HTNV G2 protein with monoclonal antibody EBO6-AAO2 (1). We did not detect expression of viral glycoproteins G1 and G2. This suggests that protein synthesis was inhibited on day 3 p.i. with 24 μg of ribavirin/ml (Fig. 4).
FIG. 4.
Inhibition of HTNV glycoprotein synthesis by ribavirin. (A) Monolayers of Vero cells infected with HTNV 76-118 and stained with monoclonal antibody for G1 (H13-16D2-1-1) (1) as described in Material and Methods. (B) Vero cells infected with HTNV and stained with monoclonal antibody for G2 (EBO6-AAO2) (1). (C) Vero cells infected with HTNV, treated with 24 μg of ribavirin/ml, and stained with G1 mouse monoclonal antibody. Cells were fixed 72 h after infection and analyzed by immunofluorescence for the presence of HTNV G1 and G2 proteins with a Zeiss axioscope connected to an Orca 100 camera interfaced to a Macintosh computer with Improvision software.
Mutation rate of HTNV suggests error catastrophe.
To evaluate the mutations induced by ribavirin, we analyzed 62 sequences derived from independently cloned cDNAs of HTNV S-segment coding region from virus grown in the presence of 24 μg of ribavirin/ml. Ten sequences were obtained from untreated HTNV-infected cells. A total of 95,386 nucleotides (nt) were sequenced to obtain a representative population of mutations. In control experiments measuring mutations of HTNV S-segment in the absence of ribavirin, we calculated a mutation frequency of 1.1/1,000 nt. In contrast, the HTNV-infected ribavirin-treated cells, had a mutation frequency of 9.5/1,000 nt (Table 2). Ribavirin incorporation increased the mutation rate ca. 8.6-fold. There were on average 1.8 transitions and 2.0 transversions per S-segment sequence in the presence of ribavirin. There were about eight insertions per sequence, with a notable increase in the insertions of guanines and cytosines.
TABLE 2.
Summary of mutations noted in sequence analysis of wild-type and ribavirin-treated HTNV S-segment genomes
| Treatment | Mutation category |
||||||
|---|---|---|---|---|---|---|---|
| Transitions | Transversions | Insertions |
Totala | ||||
| G | C | A | T | ||||
| No ribavirin | 0.3 | 0.3 | 0.5 | 0.2 | 0.1 | 0 | 1.4 |
| Ribavirin | 1.8 | 2.0 | 2.4 | 2.4 | 1.6 | 2.0 | 12.2 |
Total mutations are the sum of all transitions, transversions, and insertions per HTNV S-segment coding region (1,289 bp) divided by the total number of sequences analyzed. A total of 95,386 nt were sequenced.
DISCUSSION
Several members of the Bunyaviridae family are sensitive to the drug ribavirin; of these, HTNV is one of the most sensitive (10, 12). Previous studies examined the effect of ribavirin on the yield of infectious HTNV and determined an ED50 of 15 μg/ml; however, its mechanism of action is still unknown (12). We show here that ribavirin can cause a reduction in infectious HTNV, as measured by plaque assay. Further, Kirsi et al. and our studies found that as the MOI was increased, the antiviral effect was decreased. We hypothesize that the inhibitory effects of ribavirin observed by plaque reduction may reflect a suppression of replication and transcription activities by the viral RdRp. To elucidate the target of the antiviral activity shown by ribavirin, we examined the levels of HTNV S-segment vRNA, cRNA, and mRNA over 3 days after Vero E6 cells were infected at two different MOIs. Since the 2.4-μg/ml concentration showed a non-dose-response effect on RNA synthesis (data not shown), even when a low MOI was used, we focused our attention on the effect of 24 μg of ribavirin/ml. Studies with the 24 μg of ribavirin/ml showed a greater decrease in viral S-segment mRNA compared to vRNA or cRNA synthesis. Interestingly, the level of vRNA in the untreated and treated HTNV-infected cells decreased on day 2 p.i. This may be explained by the release of infectious virus; hence, untreated vRNA was encapsidated and secreted, resulting in low vRNA levels. In contrast, we speculated that the ribavirin-treated RNA was less efficiently encapsidated. Therefore, there was more intracellular vRNA present, and subsequently no infectious virus was produced. Previously, Cassidy et al. showed that ribavirin affected the replication of LaCrosse virus at the level of mRNA transcription (3). In accordance with our studies, their results show that when the ribavirin concentration increased from 10 to 30 μg/ml, there was a fivefold decrease in mRNA levels. The effect of ribavirin on infectious virus yield was not examined in that study.
We examined untreated and treated HTNV-infected cells for N, G1, or G2 protein from day 3. We were unable to detect any N, G1, or G2 protein in the ribavirin-treated cells. These results suggested that the mRNA produced from cRNA templates was defective, perhaps in its ability to be translated. These findings are similar to those reported by Murphy et al., who showed that ribavirin inhibits the synthesis of viral proteins and RNA and viral shedding of another hantavirus, Seoul virus, strain SR-11 (16). However, the mechanism of inhibition was not elucidated. We hypothesize that ribavirin may be acting as a mutagen, as reported for poliovirus by Crotty et al. In that study the authors showed that ribavirin triphosphate is incorporated into nascent RNA by the poliovirus RNA polymerase (3Dpol), forcing the virus into “error catastrophe” (4). That study showed that extended reaction times for 3Dpol synthesis allows multiple cycles of ribavirin incorporation, which does not terminate elongation of nascent RNA (4). Also, ribavirin did not decrease RNA synthesis since viral replication, measured by RNA accumulation in ribavirin-treated poliovirus-infected cells, reached nearly wild-type levels. Crotty et al. suggested that ribavirin, upon incorporation by the 3Dpol, acts as a mutagenic ribonucleoside, thereby generating templates that are copied incorrectly, resulting in a significant increase in the production of defective genomes. In our study, sequencing data revealed an increased mutation frequency in the cRNA and mRNA in ribavirin-treated cells. In contrast, ribavirin decreased mRNA levels throughout the study, whereas vRNA and cRNA levels were similar to wild-type levels. Specifically, in comparing sequences derived from HTNV-infected Vero cells with those from HTNV-infected ribavirin-treated cells, we observed an 8.6-fold increase in the mutation frequency (Table 2). Three sequences had multiple mutations; therefore, they were not considered in the final analysis. In the presence of ribavirin, there were approximately four base substitutions per S-segment. The transitions and transversions were used to determine the mutation frequency (Table 2). Most nucleotide substitutions were located in the first 120 nt or the last 300 nt of the cRNA coding region. Interestingly, two key transversions, T-to-A and A-to-T, occurred in the stop codon for the S-segment open reading frame. Therefore, the stop codon was replaced by an asparagine. This was the only apparent “hot spot” for base substitutions. Replacing the stop codon with asparagine opens another open reading frame immediately after the stop codon (22). There were also approximately eight insertions per sequence, with guanines and cytosines preferred. The insertions were distributed randomly throughout the sequence; however, we noted that the nucleotide inserted was always the same as the preceding nucleotide in the sequence. For example, if a G was inserted, the preceding nucleotide was a G. We postulate that these insertions may cause the polymerase to slip, resulting in deleterious effects. In summary, we hypothesize that ribavirin is acting as a mutagen upon incorporation into the S-segment cRNA. This suggests that in the presence of ribavirin, hantavirus succumbs to error catastrophe, whereby nucleotide sequences of vRNA become essentially random, with a total loss of information (5, 6). We hypothesize that during hantavirus replication the genomic and antigenomic templates, vRNA and cRNA, sustain a lethal accumulation of errors. Further, transcription of viral mRNA from these mutant templates, which can also incorporate additional ribavirin, may substantially reduce the stability of the transcripts within the cell. The mRNAs would have increased genetic variation that could affect their structure and function. In support of this hypothesis, these studies have shown that there was little to no reduction in the amount of the vRNA or cRNA templates even though sequence analysis of the cRNA showed an 8.6-fold increase in mutation frequency. In contrast, viral mRNA levels were reduced substantially, and we were unable to detect virus N, G1, or G2 protein by immunohistochemistry. Further, the HTNV-infected ribavirin-treated cells did not produce infectious virus as measured by the plaque assay. Therefore, the phenomenon of error catastrophe directly affected the fidelity of the replication and transcription of the virus polymerase, which indirectly effected virus protein levels and, hence, virion assembly.
Outbreaks of hantavirus infections in the Americas over the past decade substantiate the need for antiviral agents to treat hantavirus-related syndromes. Continued exploration of the molecular mechanism of action of ribavirin and other drugs that inhibit hantavirus replication may provide insight into enzymatic targets for rational drug development. Future efforts will be directed to the development of in vitro assays for the various enzymatic functions associated with RdRp to explore the effect of ribavirin, as well as other antiviral agents, on polymerase function.
Acknowledgments
We thank Jay Hooper (USAMRIID) for critical reading of the manuscript.
W.E.S. was supported in part by a fellowship from the Coulston Foundation. This work was supported in part by Department of Defense grant DAMD17-00-1-0513 to C.B.J.
REFERENCES
- 1.Arikawa, J., A. L. Schmaljohn, J. M. Dalrymple, and C. S. Schmaljohn. 1989. Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies. J. Gen. Virol. 70:615-624. [DOI] [PubMed] [Google Scholar]
- 2.Butler, J. C., and C. J. Peters. 1994. Hantaviruses and hantavirus pulmonary syndrome. Clin. Infect. Dis. 19:387-394. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy, L. F., and J. L. Patterson. 1989. Mechanism of La Crosse virus inhibition by ribavirin. Antimicrob. Agents Chemother. 33:2009-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. N. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 5.Domingo, E., and J. J. Holland. 1994. Mutation rates and rapid evolution of RNA viruses, p. 161-184. In S. S. Morse (ed.), Evolutionary biology of viruses. Raven Press, New York, N.Y.
- 6.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, B., E. Helgstrand, N. G. Johansson, A. Larsson, A. Misiorny, J. O. Noren, L. Philipson, K. Stenberg, G. Stening, S. Stridh, and B. Oberg. 1977. Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob. Agents Chemother. 11:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Larsson, P., K. O'Connell, E. Koumans, and J. L. Patterson. 1989. Molecular analysis of the inhibitory effect of phosphorylated ribavirin on the vesicular stomatitis virus in vitro polymerase reaction. Antimicrob. Agents Chemother. 33:1668-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goswami, B. B., E. Borek, and O. K. Sharma. 1979. The broad spectrum antiviral agent ribavirin inhibits capping of mRNA. Biophys. Res. Commun. 89:830-836. [DOI] [PubMed] [Google Scholar]
- 10.Huggins, J. W. 1989. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev. Infect. Dis. 11:S759-S761. [DOI] [PubMed] [Google Scholar]
- 11.Huggins, J. W., C. M. Hsiang, T. M. Cosgriff, M. Y. Guang, J. I. Smith, Z. O. Wu, J. W. LeDuc, Z. M. Zheng, J. M. Meegan, Q. N. Wang, et al. 1991. Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 164:1119-1127. [DOI] [PubMed] [Google Scholar]
- 12.Kirsi, J. J., J. A. North, P. A. McKernan, B. K. Murray, G. Canonico, J. W. Huggins, P. C. Srivastava, and R. K. Robins. 1983. Broad-spectrum antiviral activity of 2-β-d-ribofuranosylselanazole-4-carboximide, a new antiviral agent. Antimicrob. Agents Chemother. 24:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanford, R. E., D. Chavez, B. Guerra, J. Y. N. Lau, Z. Hong, K. M. Brasky, and B. Beames. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 75:8074-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, H. W. 1996. Epidemiology and pathogenesis of hemorrhagic fever with renal syndrome, p. 253-267. In R. M. Elliot (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 15.Lee, J. S. 1991. Clinical feature of hemorrhagic fever with renal syndrome in Korea. Kidney Int. Suppl. 35:S88-S93. [PubMed] [Google Scholar]
- 16.Murphy, M. E., H. Kariwa, T. Mizutani, H. Tanabe, K. Yoshimatsu, J. Arikawa, and I. Takashima. 2001. Characterization of in vitro and in vivo antiviral activity of lactoferrin and ribavirin upon hantavirus. J. Vet. Med. Sci. 63:637-645. [DOI] [PubMed] [Google Scholar]
- 17.Patterson, J. L., and P. Fernandez-Larsson. 1990. Molecular mechanisms of action of ribavirin. Rev. Infect. Dis. 12:1139-1146. [DOI] [PubMed] [Google Scholar]
- 18.Ruo, S. L., A. Sanchez, L. H. Elliott, L. S. Brammer, J. B. McCormick, and S. P. Fisher-Hoch. 1991. Monoclonal antibodies to three strains of hantaviruses: Hantaan, R22, and Puumala. Arch. Virol. 119:1-11. [DOI] [PubMed] [Google Scholar]
- 19.Schmaljohn, C. 1996. Bunyaviridae: the viruses and their replication, p. 649-673. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 20.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmaljohn, C. S., S. E. Hasty, S. A. Harrison, and J. M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005-1012. [DOI] [PubMed] [Google Scholar]
- 22.Schmaljohn, C. S., G. B. Jennings, J. Hay, and J. M. Dalrymple. 1986. Coding strategy of the S genome segment of Hantaan virus. Virology 155:633-643. [DOI] [PubMed] [Google Scholar]
- 23.Toltzis, P., and A. S. Huang. 1986. Effect of ribavirin on macromolecular synthesis in vesicular stomatitis virus-infected cells. Antimicrob. Agents Chemother. 29:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virca, G. D., W. Northemann, B. R. Shiels, G. Widera, and S. Broome. 1990. Simplified northern blot hybridization using 5% sodium dodecyl sulfate. BioTechniques 8:370-371. [PubMed] [Google Scholar]
- 25.Wray, S. K., B. E. Gilbert, and V. Knight. 1985. Effect of ribavirin triphosphate on primer generation and elongation during influenza virus transcription in vitro. Antiviral Res. 5:39-48. [DOI] [PubMed] [Google Scholar]