Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with three types of human tumor: Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. The virus encodes a number of proteins that participate in disrupting the immune response, one of which was predicted by sequence analysis to be encoded by open reading frame 4 (ORF4). The predicted ORF4 protein shares homology with cellular proteins referred to as regulators of complement activation. In the present study, the transcription profile of the ORF4 gene was characterized, revealing that it encodes at least three transcripts, by alternative splicing mechanisms, and three protein isoforms. Functional studies revealed that each ORF4 protein isoform inhibits complement and retains a C-terminal transmembrane domain. Consistent with the complement-regulating activity, we propose to name the proteins encoded by the ORF4 gene collectively as KSHV complement control protein (KCP). KSHV ORF4 is the most complex alternatively spliced gene encoding a viral complement regulator described to date. KCP inhibits the complement component of the innate immune response, thereby possibly contributing to the in vivo persistence and pathogenesis of this virus.
Viral mechanisms of immune system evasion are numerous. They include active strategies, in which viruses have acquired immunomodulatory genes, and passive strategies, where error-prone replication facilitates rapid antigenic evolution and consequent evasion of the adaptive immune response (10). Like other large DNA viruses, herpesviruses encode proteins that specifically subvert the immune response and thus enable persistent infection and facilitate pathogenesis in the host. One example is Kaposi's sarcoma-associated herpesvirus (KSHV), which is associated with three types of tumor: Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma (PEL) (9; reviewed in references 26 and 27). This virus encodes a number of proteins that potentially participate in disrupting the immune response. One of these proteins was predicted by sequence analysis to be encoded by open reading frame 4 (ORF4), since it shares homology with cellular proteins referred to as regulators of complement activation (RCA) (20, 23).
Complement is part of the innate immune defense system composed of serum proteins that interact in an amplification cascade (36). Invading pathogens activate complement either spontaneously due to differences in envelope or membrane composition compared to the host (alternative pathway) or through antipathogen antibody binding (classical pathway). Central to complement activation is the formation of the C3 and C5 complement convertase enzymatic complexes following proenzyme cleavage and release of smaller chemoattractant and anaphylatoxin fragments. Host soluble and membrane-bound regulators of these convertases that protect autologous cells from complement-mediated damage are encoded on chromosome 1 and are referred to as RCA (19). These proteins are composed of 4 to 35 short consensus repeat (SCR) modules that have four conserved cysteine residues and are rich in proline and glycine residues. Complement convertases are negatively regulated through increased dissociation of the enzyme complex (i.e., decay-accelerating activity) or through promoting further enzymatic cleavage of either C3b or C4b by factor I (i.e., cofactor activity).
In the present study, we report the first characterization of KSHV ORF4 transcription and protein function. We identified the transcription initiation site of the gene and the transcripts that it encodes. All three predicted ORF4 protein isoforms retained the region of high homology to a human complement regulator, decay-accelerating factor (DAF), and a putative transmembrane region, and each was found to inhibit C3b deposition on the surface of cells under complement-activating conditions. Consistent with the complement-regulating activity, we propose to name the proteins encoded by the ORF4 gene collectively as KSHV complement control protein (KCP). KSHV ORF4 is the most complex alternatively spliced gene encoding a viral complement regulator described to date. KCP inhibits the complement component of the innate immune response, and it is unique in that all three isoforms retained both membrane attachment and complement regulation. This activity could provide a defense mechanism for KSHV-infected cells and KSHV virions, either by protecting them from elimination by complement or by reducing the recruitment of antiviral inflammatory cells.
MATERIALS AND METHODS
Cell culture.
The KSHV-infected PEL cell lines were JSC-1 (8), provided by R. Ambinder (Johns Hopkins Medical School) and BCBL-1 (21) and HBL-6, provided by T. Schulz (Hannover Medical School). They were cultured as described previously (8, 21). CHO cells were obtained from the European Collection of Animal Cell Cultures (Salisbury, United Kingdom) and cultured as directed. Most PEL cells (>95%) are latently infected, but lytic replication can be induced by treating the PEL cells with the phorbol ester phorbol 12-myristate 13-acetate (PMA) (20 ng/ml; Sigma, Poole, United Kingdom).
ORF4 cloning and RT-PCR.
The ORF4 gene was cloned from the BCBL-1 PEL cell line by reverse transcription-PCR (RT-PCR) with primers designed to flank the predicted ORF (23). The ORF4 5′ primer was GCG CTC TAG AGC TAG CAT GGC CTT TTT AAG ACA AAC, and the ORF4 3′ primer was GCG CGA TAT CCT AAC GAA AGA ACA GAT AG. RNA was prepared with Trizol (Invitrogen, Paisley, Scotland), and cDNA synthesis was performed on 800 ng of whole-cell RNA as described previously (5). Primers specific to p53 provided the positive control to ensure the integrity of the RT reaction (5′ primer, CTG AGG TTG GCT CTG ACT GTA CCA CCA TCC; 3′ primer, CTC ATT CAG CTC TCG GAA CAT CTC GAA GCG). To avoid contamination of reaction mixtures with product, PCRs were established in a laminar flow PCR workstation (Labcair) located in a laboratory that was remote from the site of amplification and analysis. ORF4 PCR products were TA cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced.
5′-RACE PCR.
The 5′ transcriptional start site was mapped for total RNA prepared from phorbol ester-treated HBL-6 cells. cDNA was prepared with the SMART RACE (rapid amplification of cDNA ends) cDNA amplification kit (Clontech Laboratories, Basingstoke, United Kingdom) according to the manufacturer's instructions. In this protocol, first-strand cDNA is synthesized from an oligo(dT) primer, extending to the 5′ end of the mRNA (i.e., the transcription start site), and a stretch of cytosine residues is added by the intrinsic terminal transferase activity of the enzyme. A custom SMART oligonucleotide (Clontech) then hybridizes to the cytosine stretch, providing a further template for the reverse transcriptase, such that the SMART oligonucleotide sequence is incorporated into the 3′ end of the cDNA. The region spanning the transcription start site was then PCR amplified with a forward primer complementary to the SMART oligonucleotide and a reverse primer mapping within ORF4 (TTT AAG GCA TCT TGT CCA CCA TGA). The PCR product thus encompassing the transcription start site was TA cloned into pCR2.1-TOPO vector (Invitrogen) and sequenced.
Northern blotting.
Northern blotting was performed by standard methods with polyadenylated RNA. Essentially, total RNA was prepared from PEL cells as described above, and polyadenylated RNA was then isolated by using oligo(dT) cellulose (Sigma). RNA (3 μg/lane) was electrophoretically separated on denaturing formaldehyde agarose gels and immobilized on a nylon membrane (Hybond N; Amersham Pharmacia Biotech, Little Chalfont, England) by capillary transfer. Radiolabeled ORF4 probe was synthesized by the random priming reaction (Megaprime; Amersham Pharmacia Biotech) with the full-length ORF4 cDNA as a template, and hybridization was performed overnight in RapidHyb (Amersham Pharmacia Biotech). The results were visualized with a phosphorimager (Molecular Imager Fx; Bio-Rad). The transcript sizes were assigned through comparison with RNA molecular size markers (Invitrogen).
Derivation of anti-KCP antibody.
The full-length ORF4 PCR product was subcloned from pCR2.1 into the pDR2ΔEF1α eukaryotic expression vector, as described previously for human DAF (14). Recombinant ORF4 genes encoding soluble KCP chimeric proteins of different sizes were also cloned into this vector. They were created by truncating the ORF4 gene at different positions and replacing (in frame) the eliminated portion with part of the human immunoglobulin G1 (IgG1) gene encoding the C-terminal domain of human IgG1 Fc. Three primers were used in combination with the 5′ ORF4 primer described above: AGC TGC TGT ATG GGT GTC TTC A (truncation immediately 3′ of the SCR region), TGG CTG GGA TGT AGT TTT CTC AT (truncation immediately 3′ of the additional dicysteine region), or AAT GGG AGG GAG TGT TGG TTC T (truncation immediately 5′ of the domain encoding the transmembrane region) (Fig. 1). All constructs were transfected into CHO cells and selected in hygromycin B as previously described (31). Supernatants from stable transfectant cells were stored at 4°C until the volume exceeded 1 liter. The Fc fusion proteins were then isolated by using protein A-Sepharose as previously described for other Fc fusion proteins (14). Anti-KCP rabbit polyclonal antibodies were raised against the shortest KCP-Fc fusion protein. Contaminating anti-human Fc rabbit polyclonal antibodies were removed with an affinity column containing human coxsackievirus and adenovirus receptor-Fc fusion protein. A specific oligoclonal anti-KCP antibody was then isolated by binding to an affinity column containing soluble recombinant KCP and was subsequently eluted.
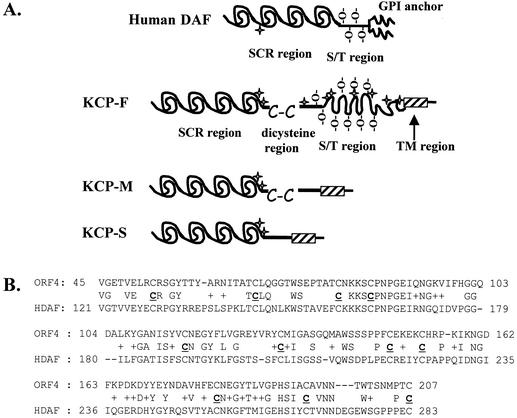
FIG. 1.
(A) Diagram of human DAF and KSHV KCP. Both human DAF and KSHV KCP contain SCR motifs (60 aa) and have serine/threonine (S/T) regions that are predicted to be highly O glycosylated (-O-). Putative N-glycosylation sites are marked with star symbols. Full-length KCP (KCP-F) has an additional dicysteine motif (C-C). The region demarcated by the hatched box is composed of hydrophobic and neutrally charged amino acids and is a potential transmembrane (TM) domain. (B) Best-fit amino acid alignment of SCR motifs in KCP and human DAF. Essential conserved cysteine residues are indicated (C), as are identical and similar (+) residues.
Western blotting.
Cell lysates were prepared in lysis buffer (100 mM Tris-HCl [pH 8], 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% NP-40, 0.5% sodium deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride), and 5 × 105 cell equivalents per lane were resolved on a 10% polyacrylamide gel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under nonreducing conditions. Proteins were immobilized on nylon membranes (Immobilon-P; Millipore) by electroblotting, and KCP proteins were detected with the affinity-purified rabbit anti-KCP antibody, which in turn was detected with a peroxidase-conjugated donkey anti-rabbit immunoglobulin antibody (Diagnostics Scotland). Specific anti-KCP reactive bands were visualized with the ECL detection system (Amersham Pharmacia Biotech).
IFA.
Immunofluorescence assays (IFA) were performed on cells that were air dried on glass slides (eight-well multitest slides [ICN]; 2 μl of cells per well at 3 × 106 cells/ml) and fixed by incubation for 5 min in a 1:1 solution of acetone and methanol at −20°C. IFA slides were used within 2 weeks of preparation. Anti-KCP antibody staining was detected with fluorescein isothiocyanate-conjugated anti-rabbit IgG (Diagnostics Scotland), and stained cells were visualized with a laser scanning microscope (Zeiss).
Flow cytometry and GPI anchor analysis.
Expression of KCP in CHO cells stably transfected with the full-length ORF4 gene was measured by flow cytometry with the affinity-purified rabbit polyclonal anti-KCP antibody. Cells were disaggregated in flow cytometry medium (FCM) (phosphate-buffered saline [PBS] containing 15 mM EDTA, 30 mM sodium azide, and 1% bovine serum albumin), counted, and adjusted to a concentration of ∼106 per ml. Aliquots (100 μl) were incubated with an empirically determined saturating dilution of anti-KCP antibody. Bound primary antibody was detected with phycoerythrin-conjugated goat anti-rabbit immunoglobulin (Sigma). Cells were washed three times in FCM and analyzed on a fluorescence-activated cell sorter (FACScalibur; Becton-Dickinson, Oxford, United Kingdom). All studies were carried out in triplicate.
Glycophosphatidylinositol (GPI) anchor analysis was performed by subculturing cells in six-well dishes (Invitrogen) and incubating them with 0.01 U of phosphoinositol-specific phospholipase C (PIPLC) (Funikoshi Ltd., Osaka, Japan) in 0.5 ml of culture medium for 30 min at 37°C prior to disaggregation with FCM and flow cytometry analysis. CHO cells stably transfected with human DAF were included as positive controls. Human DAF was detected by flow cytometry with rabbit polyclonal anti-DAF antiserum. Trypsin sensitivity was also investigated for both KCP- and DAF-expressing CHO cells, where cells were disaggregated with 0.05 mg of trypsin (Invitrogen) in 0.5 ml of PBS prior to being washed once in cell medium and then in FCM before processing for flow cytometry as detailed above.
Construction of GPI-anchored, truncated KCP proteins.
Cell surface expression of the three truncated KCP-Fc fusion proteins was accomplished by exchanging the in-frame human Ig Fc portions of the chimeric proteins described above for the serine/threonine region and GPI anchor of human DAF (Fig. 1). This DAF region was obtained by PCR amplification (5′ primer, ATC TCT AAC TTC CAA GGT C; 3′ primer, CTA GTA ACC ATG GGC TTG CTG ACT TAG) of cDNA obtained from I. Anegon (INSERM U437, Nantes, France). The resultant, truncated ORF4 genes encoding GPI-anchored proteins were transfected into CHO cells, and stable transfectants were generated by hygromycin B selection as detailed above. Transfected cells expressing high levels of stable KCP proteins (GPI-anchored, truncated forms and full-length forms) were obtained by incubation with rabbit polyclonal anti-KCP antibody, followed by selection of positive cells by using goat anti-rabbit immunoglobulin-conjugated magnetic beads according to the manufacturer's instructions (Dynabeads; Dynal, Oslo, Norway). Cell surface expression was confirmed by flow cytometry.
C3 deposition assay.
C3 convertase regulation was measured as described previously (31). Briefly, CHO cells expressing KCP or DAF, or cells transfected with empty vector, were disaggregated in FCM. Cells (5 × 105) were sensitized by incubation (15 min, room temperature) with an equal volume of different dilutions of heat-inactivated polyclonal rabbit anti-CHO antiserum (raised in house). Cells were washed in PBS (800 × g, 3 min, 4°C) followed by complement fixation diluent (Oxoid, Basingstoke, United Kingdom) and then resuspended in 10% normal human serum diluted in complement fixation diluent before incubation (15 min, 37°C) and washing with FCM. Cell-bound C3b/iC3b was detected with mouse monoclonal C3/30 (anti-human C3c) (14) as a primary antibody, which was in turn detected with phycoerythrin-conjugated goat anti-mouse immunoglobulin (DAKO Ltd., High Wycombe, United Kingdom). Samples were analyzed on a FACScalibur, as described above. Background levels were set by using controls that consisted of cells incubated with anti-C3 antibody but not exposed to complement and cells exposed to complement but not incubated with primary antibody. Single-variable analysis of variance was performed on the data with the Bonferroni multiple-comparisons test.
RESULTS
Cloning of ORF4.
RT-PCR analyses confirmed the expression of ORF4 RNA in virus-infected BCBL-1 cells, and the transcript was much more abundant following phorbol ester treatment, suggesting that it is a lytic cycle gene (data not shown). The identity of the PCR product was confirmed by sequence analysis and differed by only five nucleotides from the previously published BC-1 virus sequence (accession number U75698) (23). Relative to the ATG initiation codon, these differences were as follows: T51C (silent), A474G (Lys to Glu), G628A (Ala to Thr), T1055C (Ile to Thr), and G1155A (silent).
Northern blot analysis of ORF4 expression.
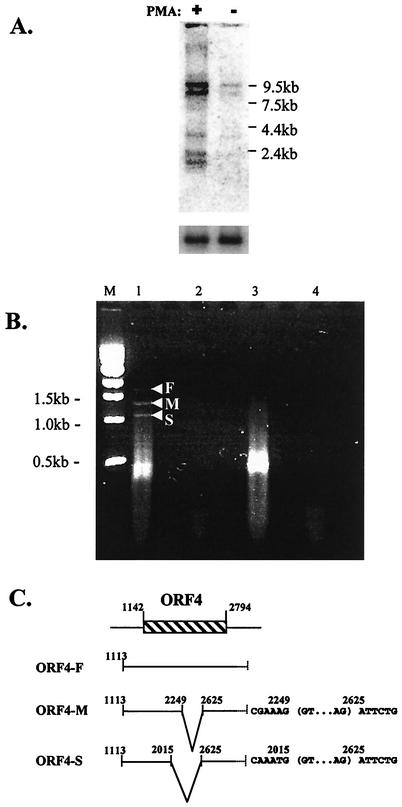
Polyadenylated RNA was prepared from the HBL-6 cell line and analyzed by Northern blotting with a full-length ORF4 gene probe (Fig. 2A). Five prominent transcript species of approximately 9.5, 8.5, 3.5, 2.5, and 2 kb were detected following the induction of lytic virus replication by phorbol ester. These transcripts were expressed at either very low or undetectable levels in untreated cells (Fig. 2A). Temporal analysis of ORF4 transcripts monitored by Northern blotting revealed that the earliest detectable induction occurred at 8 h following phorbol ester treatment (data not shown).
FIG. 2.
(A) ORF4 transcription following induction of lytic cycle replication. Northern blot analysis of mRNA isolated from HBL-6 cells incubated in the presence (+) or absence (−) of PMA for 48 h is shown. The positions of the RNA ladder molecular size markers are given. The upper-panel blot was probed with radiolabeled full-length ORF4 probe. For the lower panel, the blot was stripped of ORF4 probe and hybridized with a radiolabeled cellular gene (glyceraldehyde-3-phosphate dehydrogenase) probe. (B) Splicing of the ORF4 gene. RT-PCR analysis of ORF4 transcripts in BCBL-1 cells treated with phorbol ester revealed the presence of at least three transcript species, ORF-F, ORF4-M, and ORF4-S. Lanes: M, molecular size marker (1-kbp ladder); 1, ORF4 amplification of cDNA from BCBL-1 cells treated with phorbol ester for 48 h; 2, ORF4 amplification of BCBL-1 cells treated as for lane 1, with cDNA synthesis performed in the absence of reverse transcriptase enzyme; 3, p53 amplification of cDNA from BCBL-1 cells treated as for lane 1; 4, p53 amplification of BCBL-1 cells treated as for lane 1, with cDNA synthesis performed in the absence of reverse transcriptase enzyme and template. (C) Transcript map of the ORF4 gene. The locations of the transcript initiation site (identified by 5′-RACE-PCR) and the splice sites (identified by RT-PCR) for each of the transcripts are indicated. Local sequences at the splice sites are shown, with introns bracketed. Five of seven clones yielded position 1113 as the major transcription initiation site, and the two remaining clones indicated transcription start sites at either position 1102 or 1264. Nucleotide positions refer to the published KSHV nucleotide sequence (23). The 3′ ends of the ORF4 transcripts were not mapped.
Transcription initiation site.
The ORF4 transcription initiation site was determined by 5′-RACE PCR. Nucleotide sequence analyses of ORF4 5′-RACE clones indicated that the major transcription initiation site in HBL-6 cells is at position 1113 in the genome sequence (Fig. 2C) (23). This position places the transcription initiation site 31 bp downstream from a putative TATA box (5′ nucleotide at 1081) and 29 bp upstream of the predicted initiation codon (position 1142) (23).
Alternative splicing of the ORF4 gene.
We characterized the posttranscriptional processing of ORF4 mRNA by RT-PCR. Cellular RNA was prepared from phorbol ester-treated BCBL-1 cells and analyzed by RT-PCR with the ORF4 5′ and 3′ PCR primers designed to span the entire ORF (23). In addition to the expected 1,679-bp RT-PCR product (ORF4-F) derived from full-length ORF4 mRNA, at least two other smaller products (ORF4-M and -S) were obtained (Fig. 2B). Sequence analysis of all three RT-PCR products indicated that the ORF4 transcripts are identical for the first 901 nucleotides and last 170 nucleotides, but the smaller spliced forms were missing either a 609-nucleotide intron (ORF4-S) or 375-nucleotide intron (ORF4-M) (Fig. 2C). Both spliced transcripts shared the same 3′ (splice acceptor) sites but differed in the 5′ (splice donor) sites, and each maintained the two exons in frame.
Analysis of predicted KCP products.
The KCP protein encoded by the full-length ORF4-F mRNA was predicted to be 551 amino acid residues (aa) in size. Structural analyses carried out by using the EBI Proteinpredict website (www.ebi.ac.uk) revealed that the first 270 aa form four SCRs (Fig. 1A) exhibiting 24.7% identity to human DAF. BLAST (www.ncbi.nlm.nih.gov/BLAST/) alignments of the SCR regions of KCP and human DAF identified 41% identity when SCR1 to SCR3 of KCP were aligned with SCR2 to SCR4 of human DAF (Fig. 1B). Immediately following the SCR region of KCP is a proline-rich region of about 70 aa ending in a dicysteine motif, followed by a 202-aa S/T region containing 26 serine and 38 threonine residues (32% S+T) that may be heavily O-glycosylated (Fig. 1A). Further analyses with the transmembrane prediction algorithm (www.ch.embnet.org/software/TMPRED_form.html) predicted a 26-aa transmembrane helix followed by the two C-terminal residues in the cytoplasmic domain. The predicted proteins encoded by the two spliced mRNA species (ORF4-M and -S) (Fig. 2C) also contain this putative transmembrane region. However, the S/T region is missing from KCP-M (predicted to be encoded from the ORF4-M transcript), and the dicysteine motif and the S/T region (data not shown) are removed from the KCP-S protein (predicted to be encoded from the ORF4-S transcript) (Fig. 1A).
Analysis of KCP expressed in transfected cells.
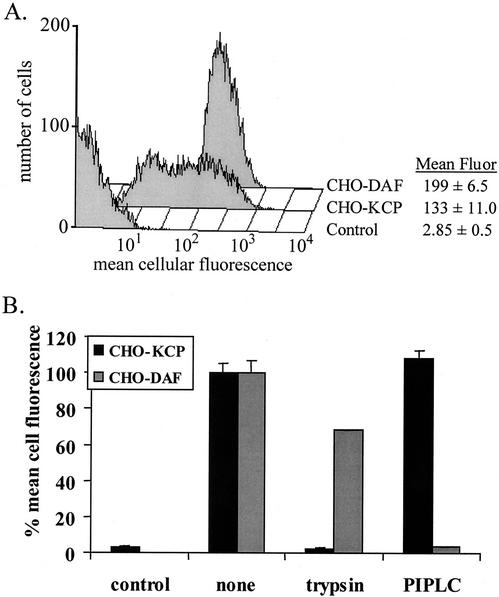
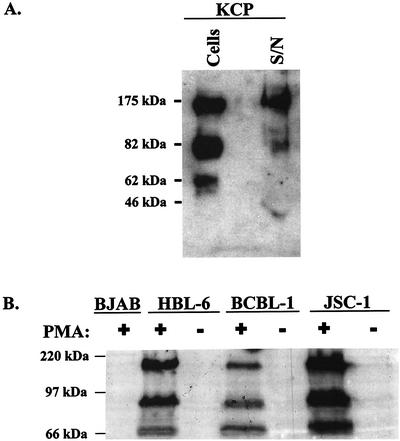
The entire ORF4 coding region, from initiation codon to stop codon, was cloned into a eukaryotic expression vector containing the elongation factor 1α promoter and the hygromycin resistance gene. The resultant plasmid was transfected into CHO cells to make stable transfectants (CHO-KCP cells). KCP proteins were detected on the surface of these cells by flow cytometry (Fig. 3A) with an affinity-purified polyclonal rabbit anti-recombinant KCP antibody. To investigate whether KCP proteins are held in the cell membrane by a GPI anchor (which is often not predicted by analysis algorithms), CHO-KCP cells were incubated with PIPLC. This enzyme removed >95% of cell surface human DAF, but no reduction in KCP expression was detected (Fig. 3B). However, trypsin treatment reduced the surface expression of DAF by only 25% but completely removed cell surface KCP (Fig. 3B). Western blot analysis of CHO-KCP cells identified three protein species of approximately 62, 82, and 175 kDa under nonreducing gel electrophoresis conditions (Fig. 4A). The 175-kDa band remained unchanged in mobility when Western blot analysis was performed under reducing conditions, suggesting that it was not composed of a dimer or complex of the smaller bands (data not shown). Western blot analysis of tissue culture supernatant from CHO-KCP cells identified the 175-kDa and trace amounts of the 82-kDa forms of KCP proteins (Fig. 4A).
FIG. 3.
(A) Flow cytometry analysis of CHO cells stably transfected with ORF4, human DAF cDNA, or empty vector control. Mean cellular fluorescence (fluor) for triplicate assays is given, with standard deviations. (B) Flow cytometry analysis of KCP and DAF expression on CHO-KCP cells following treatment with trypsin or PIPLC. Mean values for three separate analyses on different days are shown, with standard deviations.
FIG. 4.
(A) Western blot analyses of CHO-KCP cells. Blotting was performed with polyclonal anti-KCP or control anti-DAF antibodies on protein extracts prepared from 106 cell equivalents or cell-free supernatant (S/N) from 106 confluent cells as indicated. Molecular size markers are shown to the left. (B) Western blot analyses of KSHV-infected PEL cells. Analyses were performed with affinity-purified anti-KCP antibody lysates on 2.5 × 105 cell equivalents of the negative control BJAB cells (KSHV negative) or KSHV-infected HBL-6, BCBL-1, or JSC-1 PEL cell lines in the presence (+) or absence (−) of PMA. Molecular size markers are shown to the left.
Analysis of expressed KCP in KSHV-infected lymphoma cells.
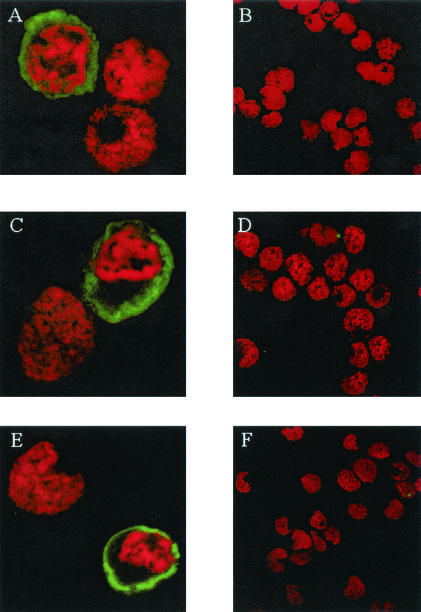
KCP expression in three separate PEL cell lines was analyzed by Western blotting (Fig. 4B). A protein profile similar to that for CHO-KCP cells was observed for all three cell lines. However, significant protein expression was observed only following induction of lytic virus replication by PMA treatment of the cells (Fig. 4B). Expression of the KCP proteins in PEL cells was also examined by confocal microscopy (Fig. 5). Consistent with RT-PCR and Northern blot studies of ORF4 gene expression (Fig. 2) and the Western blot data (Fig. 4B), significant KCP expression was detected only following the induction of lytic virus replication by PMA treatment (Fig. 5A, C, and E). In the absence of PMA treatment, no staining was detected (Fig. 5B, D, and F). The staining pattern for KCP suggested that the protein was most abundant at the plasma membrane, with some expression also in the cytoplasm. This distribution was similar to that in CHO-KCP cells (data not shown). Cell surface expression of KCP was confirmed by flow cytometric analyses of nonpermeabilized PEL cells. In these studies, the level of KCP protein expression at the cell surface was in the order JSC-1 > HBL-6 > BCBL-1, where approximately 50% of JSC-1 cells expressed KCP proteins after phorbol ester treatment (data not shown). Very low or undetectable levels of ORF4 transcripts or KCP proteins were observed in Northern blot, Western blot, flow cytometry, and IFA analyses of PEL cells in the absence of PMA treatment, and we attribute this presence to a small population of the PEL cells undergoing spontaneous lytic KSHV replication.
FIG. 5.
Confocal microscopic analysis of KSHV-infected PEL cells. Cells were stained with affinity-purified anti-KCP antibody either following phorbol ester treatment for 48 h (A, C, and E) or without phorbol ester treatment (B, D, and F), (A and B) BCBL-1; (C and D) HBL-6; (E and F) JSC-1. Cell nuclei were identified by propidium iodide staining (red). Fields show a mid-cell section through cells which were induced into the lytic cycle (green and red) as well as cells which were not (red only).
Complement regulation by KCP.
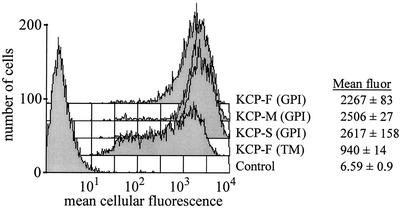
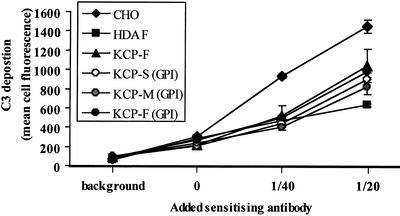
The ability of the three KCP protein isoforms to inhibit complement activation was analyzed in transfected CHO cells expressing recombinant KCP proteins. The predicted transmembrane region in the KCP-F protein was exchanged for the GPI anchor from human DAF. Two truncated forms similar to the KCP-M and KCP-S proteins (Fig. 1A) were also constructed with GPI anchors. The truncated KCP isoforms enabled assessment of the contribution of the SCR, dicysteine, and S/T regions of the recombinant proteins to complement regulation. For comparison, the activity of a recombinant KCP-F protein, in which the putative transmembrane domain was retained, was also evaluated. The results of flow cytometry analysis of the resultant stably transfected CHO cells are shown in Fig. 6. The levels of expression of the GPI-anchored KCP-F, -M, and -S recombinant proteins were similar and were expressed approximately 2.3 times greater than the level of expression of recombinant transmembrane-anchored KCP-F protein (see mean fluorescence data in Fig. 6). All of the KCP isoforms were found to regulate C3 deposition at the cell surface (Fig. 7). The difference in the levels of C3 deposition on the surface of CHO cells lacking heterologous protein expression compared with those expressing KCP-F or GPI-anchored KCP-F, KCP-M, and KCP-S recombinant proteins was statistically significant (P < 0.001 for 1/40 and 1/20 dilutions of sensitizing antibody). Human DAF-expressing CHO cells were included as a positive control and were marginally more efficient at regulating complement than any of the KCP isoforms. This difference between DAF- and KCP-expressing cells was statistically different only when sensitizing antibody was added at a 1/20 dilution. Further analysis showed that all of the recombinant GPI-anchored KCP-F, -M, and -S proteins regulated complement to similar levels and that the presence of the dicysteine region 3′ to the SCR region did not adversely influence complement regulation. The CHO cells expressing the transmembrane-anchored KCP had the lowest complement-regulating ability, but we attribute this finding to lower levels of expression.
FIG. 6.
Flow cytometry analysis of CHO cells stably transfected with recombinant forms of the ORF4 gene. Cells were stained with affinity-purified anti-KCP antibody. The cells were transfected with the ORF4-F gene [KCP-F (TM)], a full-length ORF4 gene in which the transmembrane (TM) region was swapped for the human DAF GPI anchor [KCP-F (GPI)], a medium-length ORF4 gene containing the GPI anchor and with the S/T region removed [KCP-M (GPI)], or a short-length ORF4 gene with the S/T region and the dicysteine motif removed [KCP-S (GPI)]. CHO cells transfected with empty vector (control) were also analyzed. Mean cellular fluorescence (fluor) values are given for triplicate analyses, with standard deviations.
FIG. 7.
KCP regulates complement activation. Flow cytometry analysis of C3 deposition with a human C3b/iC3b-specific monoclonal antibody following incubation of KCP-transfected or control cells with 10% human serum in the presence or absence of CHO-specific activating rabbit polyclonal antibody (as indicated) is shown. Background refers to the addition of sensitizing antibody and complement, but the anti-C3b antibody was omitted to show that neither complement nor the sensitizing antibody were detected nonspecifically by the PE-conjugated secondary antibody in flow cytometry. An antibody dilution of 0 refers to the addition of complement to the cells in the absence of sensitizing antibodies. Mean data are shown for triplicate assays, with standard deviations. These data represent those from one of four replicate experiments, which gave identical results.
DISCUSSION
Like the majority of virus-encoded complement regulators, KSHV KCP was first described as a complement-regulatory homologue based on its homology to host and other virus-encoded RCA proteins (23). Some members of the Rhadinovirus genus of the Gammaherpesvirinae subfamily, to which KSHV belongs, also carry genes with homology to KSHV ORF4 that encode functional complement regulators. These members include murine gammaherpesvirus 68 (MHV-68), having an ORF4 homologue, and herpesvirus saimiri (HVS) (20, 28, 35). HVS encodes two proteins that control complement activation: a glycosylphosphatidylinositol-anchored CD59 homologue (HVSCD59) (2, 22) that blocks formation of the membrane attack complex and a complement control protein homologue (CCPH) encoded by ORF4 that inhibits C3 convertase (1, 12). The rhesus monkey rhadinovirus is predicted to encode RCA (4, 23), but functional studies have not yet been reported. However, other herpesviruses encode complement regulators that are entirely unique. For example, glycoprotein C (gC) of herpes simplex virus type 1 is an effective complement regulator (13) but does not share significant homology to known host complement regulators (17).
In the present study, we sought to determine empirically the complement-regulatory activity of the KSHV KCP. We found that the KSHV ORF4 gene is expressed as a full-length transcript and as two alternatively spliced transcripts. Three KCP isoforms, predicted to be encoded by the transcripts, contain four conserved SCR domains that are necessary for complement regulation and retain a transmembrane domain. All three isoforms regulate complement activation by inhibiting C3 deposition on the cell surface (Fig. 7).
The SCR region that is conserved in the three KSHV KCP isoforms is homologous to that in human DAF (Fig. 1B), HVS ORF4 (CCPH), and MHV-68 ORF4 (data not shown). Alignment analysis indicates that SCR1 to SCR3 of KCP are most similar to SCR2 to SCR4 of DAF. This alignment is significant, since SCR1 of DAF has been reported to be dispensable for complement regulation (7, 11), which suggests that SCR4 of KSHV KCP may also be dispensable. Brodbeck et al. (6) have reported that the KKK(158-160) and L(180)F(181) motifs are required for complement regulation by DAF. Similar motifs [NKK(81-83) and I(97)F(98)] are present in the KSHV KCP, taking into account the shifted SCR alignment. The predicted S/T regions of these proteins (HVS ORF4 [CCPH], 63 aa; MHV68 ORF4, 90 aa) are similar in length to that in human DAF (70 aa), and the S/T region for the full-length KSHV KCP is predicted from our studies to be 202 aa.
IFA, flow cytometry, and Western blot analyses of KCP expression in PEL cells with an affinity-purified antibody indicated that the protein was expressed at very low levels in the absence of phorbol ester treatment. Such levels are probably due to spontaneous lytic replication of KSHV in a small proportion of cells. However, treatment of PEL cells with phorbol ester resulted in the expression of three KCP species, an expression pattern confirmed in three different PEL cell lines. These studies, taken together with the RT-PCR analyses and temporal Northern blotting, indicate that the ORF4 gene is expressed during lytic cycle replication, approximately 8 h following phorbol ester activation, and that the protein is expressed following reactivation of KSHV replication in cells naturally infected with wild-type virus. These data are consistent with previous DNA microarray and Northern blot studies describing the induction of ORF4 gene expression during lytic cycle replication (15, 25).
Evidence for the significance of complement in antiviral defense is provided by the diversity of strategies that viruses have adopted to avoid this aspect of the immune response. Essentially, viruses either modulate or sequester the expression of RCA proteins, or they encode their own homologues. Human cytomegalovirus, human immunodeficiency virus, human T-cell leukemia virus type 1, and vaccinia virus are examples of viruses that either increase the host cell surface expression of DAF and membrane cofactor protein or acquire DAF, membrane cofactor protein, and the terminal pathway regulator (CD59) in their envelopes on egress from the cell (18, 24, 29, 30, 32, 34). Viruses that encode homologues of RCA proteins include members of the poxvirus (e.g., smallpox virus, vaccinia virus, and cowpox virus) and herpesvirus families (reviewed in references 3 and 33). The ORF4 protein of MHV-68 was predicted to share homology with RCA proteins (35) and has been shown to regulate murine complement activation (16), but no evidence for alternative splicing of the MHV-68 ORF4 mRNA has been reported. HVS ORF4 (CCPH) is encoded by an unspliced (1.7-kb) and by a spliced (1.5-kb) transcript (1). The 1.5-kb species is identical to the 1.7-kb form other than in lacking a 193-bp intron which encodes the transmembrane domain, and it therefore yields a secreted protein with a C terminus different from that of the full-length protein (1). KSHV ORF4 transcription is more complex: in addition to the unspliced mRNA, two spliced transcripts are produced, in both of which removal of the intron maintains two exons in frame (Fig. 2C). KSHV ORF4 therefore encodes three membrane-bound protein isoforms (Fig. 1A), all of which inhibit C3 deposition on the cell surface (Fig. 7). KCP expression at the cell surface, and on the virion itself, may therefore contribute to the evasion of the complement component of the innate immune response and facilitate KSHV infection, persistence, and pathogenesis.
Acknowledgments
We thank Charles Cunningham for performing the 5′-RACE PCR, Maeve MacLeod for the IFA, and Jay A. Levy for critical reading of the manuscript.
This work was supported in part by grants from the Wellcome Trust (059008/Z/99/Z to D.J.B. and 061150/Z/00/Z to O.B.S.), the Cunningham Trust (ACC/KM CT to D.J.B.), The Royal Society (574006.G503/21709/SM to D.J.B.), and Cancer Research UK (C7934 to O.B.S. and D.J.B.).
REFERENCES
- 1.Albrecht, J. C., and B. Fleckenstein. 1992. New member of the multigene family of complement control proteins in herpesvirus saimiri. J. Virol. 66:3937-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., J. Nicholas, K. R. Cameron, C. Newman, B. Fleckenstein, and R. W. Honess. 1992. Herpesvirus saimiri has a gene specifying a homologue of the cellular membrane glycoprotein CD59. Virology 190:527-530. [DOI] [PubMed] [Google Scholar]
- 3.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Immunol. Today 21:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackbourn, D. J., L. F. Chuang, S. Sutjipto, K. F. Killam, Jr., P. M. McCready, R. H. Doi, Y. Li, and R. Y. Chuang. 1992. Detection of simian immunodeficiency virus RNA from infected rhesus macaques by the polymerase chain reaction. J. Virol. Methods 37:109-117. [DOI] [PubMed] [Google Scholar]
- 6.Brodbeck, W. G., L. Kuttner-Kondo, C. Mold, and M. E. Medof. 2000. Structure/function studies of human decay-accelerating factor. Immunology 101:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodbeck, W. G., D. Liu, J. Sperry, C. Mold, and M. E. Medof. 1996. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J. Immunol. 156:2528-2533. [PubMed] [Google Scholar]
- 8.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Chaston, T. B., and B. A. Lidbury. 2001. Genetic ′budget' of viruses and the cost to the infected host: a theory on the relationship between the genetic capacity of viruses, immune evasion, persistence and disease. Immunol. Cell. Biol. 79:62-66. [DOI] [PubMed] [Google Scholar]
- 11.Coyne, K. E., S. E. Hall, S. Thompson, M. A. Arce, T. Kinoshita, T. Fujita, D. J. Anstee, W. Rosse, and D. M. Lublin. 1992. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J. Immunol. 149:2906-2913. [PubMed] [Google Scholar]
- 12.Fodor, W. L., S. A. Rollins, S. Bianco-Caron, R. P. Rother, E. R. Guilmette, W. V. Burton, J. C. Albrecht, B. Fleckenstein, and S. P. Squinto. 1995. The complement control protein homolog of herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J. Virol. 69:3889-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries, L. F., H. M. Friedman, G. H. Cohen, R. J. Eisenberg, C. H. Hammer, and M. M. Frank. 1986. Glycoprotein C of herpes simplex virus 1 is an inhibitor of the complement cascade. J. Immunol. 137:1636-1641. [PubMed] [Google Scholar]
- 14.Harris, C. L. 2000. Functional assays for complement regulators. Methods Mol. Biol. 150:83-101. [DOI] [PubMed] [Google Scholar]
- 15.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapadia, S. B., H. Molina, V. van Berkel, S. H. Speck, and H. W. Virgin IV. 1999. Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J. Virol. 73:7658-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNearney, T. A., C. Odell, V. M. Holers, P. G. Spear, and J. P. Atkinson. 1987. Herpes simplex virus glycoproteins gC-1 and gC-2 bind to the third component of complement and provide protection against complement-mediated neutralization of viral infectivity. J. Exp. Med. 166:1525-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montefiori, D. C., R. J. Cornell, J. Y. Zhou, J. T. Zhou, V. M. Hirsch, and P. R. Johnson. 1994. Complement control proteins, CD46, CD55, and CD59, as common surface constituents of human and simian immunodeficiency viruses and possible targets for vaccine protection. Virology 205:82-92. [DOI] [PubMed] [Google Scholar]
- 19.Morgan, B. P., and C. L. Harris. 1999. Complement regulatory proteins. Academic Press, London, United Kingdom.
- 20.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 22.Rother, R. P., S. A. Rollins, W. L. Fodor, J. C. Albrecht, E. Setter, B. Fleckenstein, and S. P. Squinto. 1994. Inhibition of complement-mediated cytolysis by the terminal complement inhibitor of herpesvirus saimiri. J. Virol. 68:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saifuddin, M., M. Ghassemi, C. Patki, C. J. Parker, and G. T. Spear. 1994. Host cell components affect the sensitivity of HIV type 1 to complement-mediated virolysis. AIDS Res. Hum. Retrovir. 10:829-837. [DOI] [PubMed] [Google Scholar]
- 25.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarid, R., S. J. Olsen, and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv. Virus Res. 52:139-232. [DOI] [PubMed] [Google Scholar]
- 27.Schulz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 79:1573-1591. [DOI] [PubMed] [Google Scholar]
- 28.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses, human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381. [PubMed] [Google Scholar]
- 30.Spiller, O. B., S. M. Hanna, D. V. Devine, and F. Tufaro. 1997. Neutralization of cytomegalovirus virions: the role of complement. J. Infect. Dis. 176:339-347. [DOI] [PubMed] [Google Scholar]
- 31.Spiller, O. B., C. L. Harris, and B. P. Morgan. 1999. Efficient generation of monoclonal antibodies against surface-expressed proteins by hyperexpression in rodent cells. J. Immunol. Methods 224:51-60. [DOI] [PubMed] [Google Scholar]
- 32.Spiller, O. B., B. P. Morgan, F. Tufaro, and D. V. Devine. 1996. Altered expression of host-encoded complement regulators on human cytomegalovirus-infected cells. Eur. J. Immunol. 26:1532-1538. [DOI] [PubMed] [Google Scholar]
- 33.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 34.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 95:7544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walport, M. J. 2001. Complement. N. Engl. J. Med. 344:1140-1144. [DOI] [PubMed] [Google Scholar]