Abstract
Dobrava virus (DOBV) occurs in two different rodent species, Apodemus flavicollis (DOBV-Af) and A. agrarius (DOBV-Aa). We sequenced the S and M genomic segments from sympatric DOBV-Af and DOBV-Aa strains which fell into two distinct genetic lineages. Molecular phylogenetic analyses gave evidence for genetic reassortment between S and M segments of DOBV-Af and DOBV-Aa and indicated homologous recombination events in DOBV evolution. DOBV-Af and DOBV-Aa are distinct but also subject to genetic exchanges that affect their evolutionary trajectories.
Hantaviruses are emerging viruses which cause two human zoonoses, hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome (8, 11, 19). In contrast to other genera of the Bunyaviridae family, they are transmitted to humans from rodents of the family Muridae, their natural reservoirs. The virus genome consists of three segments of negative-stranded RNA: the large (L) segment encodes the viral RNA polymerase, the medium (M) segment encodes the envelope glycoproteins G1 and G2, and the small (S) segment encodes the nucleocapsid (N) protein.
Prominent examples of hantaviruses that cause human disease are the Hantaan virus (HTNV) and Seoul virus (SEOV) in Asia, Puumala virus (PUUV) and Dobrava virus (DOBV) in Europe, Sin Nombre virus (SNV) in North America, and Andes virus in South America. The close phylogenetic correspondence between hantaviruses and their reservoir hosts has been interpreted as evidence for coevolution (cospeciation) of virus and host (16).
Interestingly, DOBV is hosted by two different rodent species, Apodemus flavicollis (yellow-necked mice) and A. agrarius (striped field mice). In southeastern Europe, A. flavicollis is the natural reservoir for DOBV. A virus isolate from Slovenia (1, 2) has been taken as the prototypical DOBV lineage from A. flavicollis (DOBV-Af). In contrast, in eastern, northeastern, and central Europe, DOBV is carried by A. agrarius (14). A virus of the DOBV-Aa group, Saaremaa virus, was isolated in Estonia, northeastern Europe (12), and was later suggested to represent the prototype for a unique virus species (13).
Although a large overlap between the distribution of A. agrarius and A. flavicollis exists in Europe, so far the two host-specific subtypes have been detected sympatrically only in the border region between central and southeastern Europe (3, 23). The coexistence of two different virus lineages in the same geographic region indicates some genetic stability and evolutionary distinctness.
We have determined the total nucleotide sequences of the S and M segments of DOBV-Af (DOBV/Esl/400Af/98) and DOBV-Aa (DOBV/Esl/856Aa/97 and DOBV/Esl/862Aa/97) strains which were found to occur only 13 km apart in Slovakia, central Europe. This discovery offered an opportunity to remove the effects of geographic isolation on the supposedly host-specific genetic determinants that distinguish DOBV-Af from DOBV-Aa and to examine whether the two virus types are subject to genetic interactions with one another that could influence their evolutionary trajectories.
In 1997 and 1998, rodents were trapped in eight localities in eastern Slovakia by using bridge-metal traps and blood and tissue samples were collected as previously described (23). Rodents were tested for the presence of hantavirus-specific immunoglobulin G antibodies by enzyme-linked immunosorbent assay as described by Sibold et al. (22, 23). The RNA of seropositive rodents was extracted from homogenized lung tissues by using Trizol reagent (Gibco BRL Invitrogen, Karlsruhe, Germany) according to a standard protocol (4). Nested reverse transcription-PCR was carried out with the Robust reverse transcription-PCR system (Finnzymes, Espoo, Finland). A variety of oligonucleotide primers (available from the authors upon request) were used to generate six overlapping cDNA fragments for the entire M genome RNA segment. Amplification of the whole S segment was undertaken by use of a single genus-specific primer, RT-DOB (5′-ttctgcag TAG TAG TAK RCT CCC TAA ARA G), as described earlier (23).
The amplified products were cloned into the pCR 2.1 vector (TA cloning kit; Invitrogen, Leek, The Netherlands). Three recombinant plasmids were sequenced in both directions, and the consensus sequence from them was used. Dideoxy sequencing was performed on a LICOR sequencer with the Autoread kit (Pharmacia-Biotech, Freiburg, Germany) as described by the manufacturer. Nucleic acid and amino acid sequence data were analyzed with the BioEdit software package (6). Sequences were aligned with Clustal W (25) with default parameters and checked manually.
Comparison of S segment sequences.
We have established the total nucleotide sequence of the S segment of DOBV/Esl/400Af/98 (Esl/400Af). At 1,673 nucleotides (nt), this DOBV-Af S segment is shorter than the 1,704-nt DOBV/Esl/856Aa/97 (Esl/856Aa) and DOBV/Esl/862Aa/97 (Esl/862Aa) S segments (23).
Table 1 shows the nucleotide and amino acid percent identities among the N-encoding open reading frame (ORF) sequences and deduced N proteins. Esl/400Af exhibited high nucleotide and amino acid sequence similarities to DOBV/Slovenia-Af (Slo/Af) but lower similarity to all DOBV-Aa strains. Virus strains Esl/856Aa and Esl/862Aa exhibited the lowest nucleotide sequence similarities (about 85 to 87%) to the two DOBV-Af strains as well as to the two DOBV/Saaremaa-Aa strains. At the amino acid level, the similarity of Esl/856Aa and Esl/862Aa to Saaremaa-Aa is lower than that to the DOBV-Af strains.
TABLE 1.
S segment nucleotide and amino acid percent identities of DOBV strains, HTNV, and SEOVa
| Segment portion | Strain | % Identity with strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Esl/856Aa | Esl/862Aa | Kur/53Aa | Kur/44Aa | Saa/160v | Saa/90Aa | Esl/400Af | Slo/Af | HTNV | SEOV | ||
| ORF | Esl/856Aa | 99.6 | 90.5 | 90.2 | 87.0 | 87.4 | 85.0 | 86.7 | 74.0 | 73.5 | |
| Esl/862Aa | 99.5 | 90.4 | 90.2 | 86.9 | 87.3 | 85.0 | 86.7 | 74.0 | 73.6 | ||
| Kur/53Aa | 98.6 | 98.6 | 99.7 | 87.8 | 87.7 | 87.6 | 88.1 | 74.2 | 74.2 | ||
| Kur/44Aa | 98.1 | 98.1 | 99.5 | 87.5 | 87.4 | 87.4 | 87.8 | 74.0 | 74.1 | ||
| Saa/160v | 96.7 | 96.7 | 96.2 | 95.8 | 98.3 | 87.5 | 87.9 | 73.3 | 72.5 | ||
| Saa/90Aa | 96.5 | 96.5 | 96.0 | 95.5 | 99.7 | 87.2 | 88.0 | 73.1 | 72.8 | ||
| Esl/400Af | 97.6 | 97.6 | 97.2 | 96.7 | 97.2 | 96.9 | 95.3 | 73.8 | 72.5 | ||
| Slo/Af | 97.9 | 97.9 | 97.2 | 96.7 | 97.2 | 96.9 | 99.0 | 74.2 | 73.6 | ||
| HTNV | 82.9 | 82.9 | 83.4 | 83.2 | 82.5 | 82.2 | 82.7 | 82.7 | 73.9 | ||
| SEOV | 80.8 | 80.8 | 81.3 | 80.8 | 79.9 | 79.9 | 80.1 | 80.1 | 82.2 | ||
| 3′ NCR | Esl/856Aa | 100 | 85.3 | 85.3 | 83.6 | 84.1 | 84.4 | 83.6 | 51.3 | 40.7 | |
| Esl/862Aa | 85.3 | 85.3 | 83.6 | 84.1 | 84.4 | 83.6 | 51.3 | 40.7 | |||
| Kur/53Aa | 100 | 89.0 | 90.0 | 91.8 | 90.3 | 53.7 | 40.6 | ||||
| Kur/44Aa | 89.0 | 90.0 | 91.8 | 90.3 | 53.7 | 40.6 | |||||
| Saa/160v | 98.7 | 89.6 | 89.9 | 53.6 | 40.6 | ||||||
| Saa/90Aa | 90.2 | 89.9 | 53.8 | 40.6 | |||||||
| Esl/400Af | 96.6 | 54.4 | 40.1 | ||||||||
| Slo/Af | 54.7 | 39.7 | |||||||||
| HTNV | 46.3 | ||||||||||
| SEOV | |||||||||||
The identity values were calculated by the Clustal method. The percentage differences for nucleotide (above the diagonal) and amino acid (below the diagonal) sequences are presented. Strain name abbreviations: Esl/856Aa, DOBV/Esl/856Aa/97; Esl/862Aa, DOBV/Esl/862Aa/97; Kur/53Aa, DOBV/Kurkino/53Aa/97; Kur/44Aa, DOBV/Kurkino/44Aa/97; Saa/160v, DOBV/Saaremaa/160v; Saa/90Aa, DOBV/Saaremaa/90Aa/97; Esl/400Af, DOBV/Esl/400Af/98; Slo/Af, DOBV prototype strain. HTNV strain 76-118 and SEOV strain SR11 were used. For sequence references see the legend to Fig. 1.
The single ORF (nt 36 to 1325) for the Esl/400Af N protein encodes a putative protein of 429 amino acids (aa) in length. No putative ORF2 was found. In general, the N-terminal half of the predicted N protein (approximately 200 aa) is more conserved than the C-terminal half. Cysteine residues at amino acid positions 219, 244, 293, 315, and 319 were highly conserved.
The 3′ noncoding region (3′ NCR) of the S segment of strain Esl/400Af is 348 nt long and appears highly similar to Slo/Af but most different from Esl/856Aa and Esl/862Aa (Table 1). Interestingly, the 3′ NCR of Esl/856Aa and Esl/862Aa is longer than that of Esl/400Af, due to a 32-nt insert that is an imperfect repeat of a downstream sequence, beginning 28 nt downstream of the termination codon of the N gene. No other DOBV S segment sequences contain this insertion.
Altogether, S segments of the two DOBV-Af strains were found to be very similar, whereas the DOBV-Aa strains exhibited higher diversity. The central and eastern European DOBV-Aa strains were found to be rather dissimilar from the Saaremaa-Aa strains.
Comparison of M segment sequences.
Total M segment nucleotide sequences were determined for the sympatrically occurring strains Esl/862Aa and Esl/400Af. Their lengths were 3,643 and 3,648 nt, respectively. Both virus types encode a predicted glycoprotein precursor (GPC) of 1,135 aa. Table 2 shows the degree of nucleotide and deduced amino acid sequence identity between the glycoprotein-encoding ORFs of DOBV strains sequenced so far. Strain Esl/400Af resembles the DOBV/Slovenia-Af prototype sequence, whereas greater differences were found between Esl/862Aa and Saa/160v and, even more pronounced, between Esl/862Aa and the DOBV-Af strains. Similar relationships could be deduced from the comparative analysis of the 3′ NCR of the M segment (Table 2). The M segment 3′ NCR exhibits a higher variability than does the S segment 3′ NCR (compare Tables 1 and 2).
TABLE 2.
M segment nucleotide and amino acid percent identities of DOBV strains, HTNV, and SEOVa
| Segment portion | Strain | % Identity with strain:
|
|||||
|---|---|---|---|---|---|---|---|
| Esl/862Aa | Saa/160v | Esl/400Af | Slo/Af | HTNV | SEOV | ||
| ORF | Esl/862Aa | 87.3 | 82.6 | 82.5 | 70.5 | 70.8 | |
| Saa/160v | 96.1 | 82.2 | 82.4 | 71.2 | 70.6 | ||
| Esl/400Af | 94.1 | 94.3 | 93.0 | 71.0 | 70.6 | ||
| Slo/Af | 94.1 | 94.5 | 99.2 | 70.8 | 70.7 | ||
| HTNV | 77.0 | 76.9 | 77.7 | 77.5 | 72.3 | ||
| SEOV | 76.8 | 76.4 | 77.3 | 77.5 | 76.8 | ||
| 3′ NCR | Esl/862Aa | 79.7 | 62.5 | 63.1 | 37.8 | 44.2 | |
| Saa/160v | 62.0 | 63.6 | 41.3 | 44.2 | |||
| Esl/400Af | 91.9 | 40.5 | 43.2 | ||||
| Slo/Af | 42.3 | 44.5 | |||||
| HTNV | 39.1 | ||||||
| SEOV | |||||||
Whereas the central European DOBV-Aa strains exhibited a similar, limited level of identity with both Saaremaa-Aa and DOBV-Af strains, the M segment of the central European DOBV-Aa strain showed higher similarity to the Saaremaa strains than to the DOBV-Af strains. In addition, the M segments of the DOBV-Af strains appeared more similar to each other than did the DOBV-Aa M segments to one another.
Phylogenetic trees and reassortment.
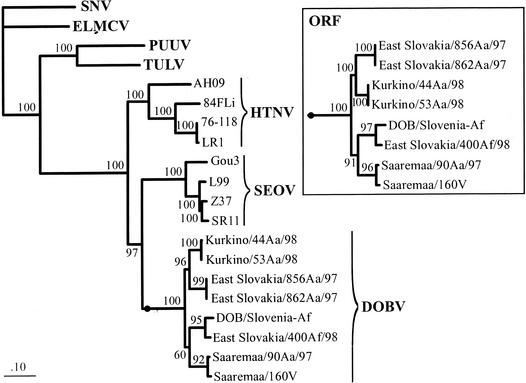
To reconstruct maximum likelihood (ML) phylogenetic trees, we applied quartet puzzling by using the TREE-PUZZLE package (20, 24). We used the Tamura-Nei model; missing parameters were reconstructed from the data sets. The values at the tree branches represent the PUZZLE support values (Fig. 1). The phylogeny of complete S segment nucleotide sequences from the DOBV strains indicates that central European DOBV-Aa strains form a well-supported monophyletic group with the Russian Kurkino-Aa strains whereas the strain Esl/400Af forms a monophyletic group together with Slo/Af. With use of the complete S segment sequence for analysis, the PUZZLE support for position of the Saaremaa-Aa strains within the tree was slightly below the 70% threshold limit. Therefore, we repeated the analysis on the basis of ORF nucleotide sequences (Fig. 1, upper right) and amino acid sequences of the N proteins (data not shown). These analyses unambiguously group Saaremaa-Aa sequences together with the DOBV-Af strains and well apart from the central European and Russian DOBV-Aa lineages. This shows that the S segment of Saaremaa-Aa strains is related to the S segment of A. flavicollis-derived DOBV strains. There are also five amino acid variable sites in the N protein wherein the Saaremaa-Aa strains encode the same amino acid as do Esl/400Af and/or Slo/Af, in contrast to the central European and Russian DOBV-Aa lineage (data not shown).
FIG. 1.
Phylogenetic ML trees from the available complete S segment nucleotide sequences of different members of the DOBV lineage and further hantaviruses, computed with TREE-PUZZLE. The main tree is based on sequences of the complete S segments. The insert shows the subtree of the complete ORF sequences for the DOBV strains. The S segment sequences analyzed included DOBV/East Slovakia/856Aa/97 and DOBV/East Slovakia/862Aa/97 (accession numbers Aj269549 and Aj269550, respectively), DOBV/Kurkino/44Aa/98 and DOBV/Kurkino/53Aa/98 (Aj131672 and Aj131673, respectively), DOBV/Saaremaa/160v and DOBV/Saaremaa/90Aa/97 (Aj009773 and Aj009775, respectively), HTNV strain 76-118 (M14626), HTNV strain 84FLi (AY017064), HTNV strain LR1 (AF288294), HTNV strain AH09 (AF285264), SEOV strain SR11 (M34881), SEOV strain L99 (AF288299), SEOV strain Z37 (AF187082), SEOV strain Gou3 (AB027522), PUUV strain Vranica-Hällnäs (U14137), TULV strain Moravia/5302v/95 (Z69991), SNV strain NM H10 (L25784), and El Moro Canyon hantavirus (ELMCV) strain RM-97 (U11427). The S segment sequence of DOBV/Slovenia (2) (L41916) was proven by sequencing of DOBV obtained from cell culture supernatant of Vero E6 cells, revealing inversion of two nucleotides at position 1035/1036 (GT instead of TG), resulting in an amino acid exchange (Val instead of Cys).
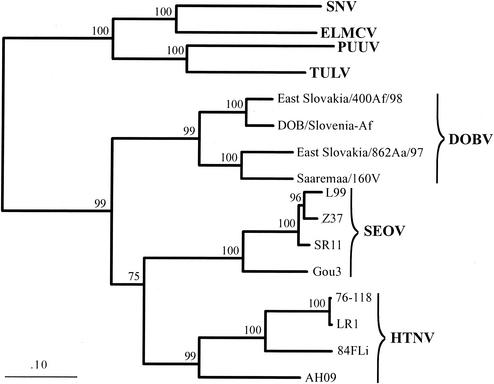
Figure 2 shows the phylogenetic tree based on the available complete M segment sequences of DOBV strains. In addition to our two newly determined complete sequences, Esl/862Aa and Esl/400Af, only two other complete M segment sequences were available for analysis. However, the results clearly showed that the Esl/862Aa strain formed one well-supported group with Saa/160v whereas the strain Esl/400Af demonstrated, as expected, a close relationship to the Slo/Af prototype.
FIG. 2.
Phylogenetic ML tree of the available complete M segment nucleotide sequences of different members of the DOBV lineage and further hantaviruses computed with TREE-PUZZLE. The M segment sequences included HTNV strain 76-118 (M14627), HTNV strain 84FLi (AF366569), HTNV strain LR1 (AF288293), HTNV strain AH09 (AF285265), SEOV strain SR11 (M34882), SEOV strain L99 (AF035833), SEOV strain Z37 (AF187081), SEOV strain Gou3 (AB027521), PUUV strain Vranica-Hällnäs (U14136), TULV strain Moravia/5302v/95 (Z69993), SNV strain NM H10 (L25783), and El Moro Canyon virus (ELMCV) strain RM-97 (U26828). The M segment sequence of DOBV/Saaremaa/160v (12) (Aj009774) was corrected by nucleotide exchanges found by resequencing of plasmids M1, M2, and M3, provided by those authors (12); we found two frameshift mutations at nucleotide positions 2630 (ΔC; −1) and 2638/2639 (+A; +1) and nucleotide exchanges resulting in amino acid exchanges (nt 471, C instead of T [C/T], Thr instead of Met [Thr/Met]; position 2121, C/T, Pro/Leu; position 2675, A/C, Lys/Gln; position 2679, A/T, Asp/Val; position 2712, A/C, His/Pro; and position 2954, G/A, Val/Ile). The M segment sequence of DOBV/Slovenia (2) (L33685) was corrected according to sequencing data obtained from DOBV/Slovenia from cell culture supernatant of Vero E6 cells; we have found several nucleotide exchanges causing amino acid exchanges (nucleotide position 2777, G/A, Gly/Arg; position 2861: T/C, Trp/Arg; position 3181, T/A, His/Gln) and an insertion of TTT (Phe) between nt 3381 and 3382.
The difference in the phylogenetic placement of Saaremaa-Aa in respect to S and M segments shows that this virus was involved in reassortment processes during its evolution; its S segment is more closely related to DOBV-Af whereas its M segment resembles the central European strain Esl/862Aa. To ascertain whether this contradiction of S and M segment-derived trees could have been caused simply by the absence of additional sequences (e.g., Kurkino-Aa strains) in the M but not S segment ML analysis, we repeated the S segment analysis without involving sequences of Kurkino-Aa strains; however, the phylogenetic grouping of the S segments of Saaremaa-Aa with DOBV-Af remained unchanged (data not shown).
Natural genetic reassortment of genomic RNA segments has been also found within the SNV species (7, 9). Moreover, genetic reassortants involving S and M segments were detected after mixed infections in tissue culture by using two distinguishable strains of SNV. One virus reassortant was observed also after mixed infections between SNV and genetically more distant Black Creek Canal virus (17). This together with our own results indicates that genetic reassortment is not an uncommon process in hantavirus evolution.
Recombination analysis.
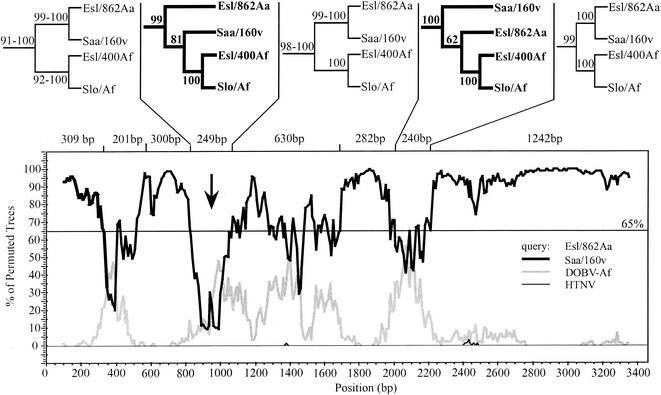
To study more precisely the evolutionary history of the two genomic segments, we performed bootscan analyses on the alignments of the S and M segment ORFs for the viruses Esl/400Af, Esl/856Aa, and Esl/862Aa. Bootscan analysis (18) was performed with Stuart Ray's SimPlot 2.5 (10) in combination with the PHYLIP package (version 3.5c; J. Felsenstein, Seattle, Wash., 1993). As the underlying phylogeny reconstruction algorithm we used neighbor joining (NEIGHBOR, DNADIST) under the Kimura model with a Ts/Tv ratio of 2.0 for the distances. For subsequent phylogenetic analysis the alignments were split according to the bootscan diagrams at cross points of a 65% threshold, even if only one curve dropped down while another one did not come up. The subalignments were tested for phylogenetic information with likelihood mapping and then used to reconstruct ML phylogenies by using quartet puzzling (20, 24).
In the analysis of the M segment (Fig. 3), the main DOBV topology divided into Esl/862Aa with Saa/160v on the one hand and Slo/Af with Esl/400Af on the other yielding high support (92 to 100%). But in two regions of the ORF this topology changed. While this change had moderate support (62%) in the area of nt 1971 to 2211, the 81% support in the area of nt 810 to 1059 was high. Homologous recombination events in the natural history of DOBV could explain the creation of this significant nucleotide sequence exchange. However, the phylogenetic analysis of this recombination event might be influenced by the short extent (249 bp) of the region. Hence, this issue needs to be further evaluated, taking into account biological properties of M segment gene products which are currently not very well characterized.
FIG. 3.
Recombination analysis for the M segment ORFs. The lower part shows the bootscan diagram with Esl/862Aa as query and the other viruses grouped into DOBV-Af, Saaremaa-Aa (Saa/160v), and HTNV. For clarity we removed the curves of SEOV, PUUV, TULV, El Moro Canyon virus, and SNV, which never exceeded values of 2.5%. The upper part shows the DOBV subtrees obtained from ML-based phylogenetic analysis with TREE-PUZZLE from the subalignments split at a 65% threshold in the bootscan diagram. At the branches we give the PUZZLE support values for the subtrees. Where neighboring subalignment resulted in the same tree topology, only one, with ranges of support values, is shown. The subalignments were tested for phylogenetic information by likelihood mapping (24). A window size of 200 nt, a step size of 10 nt, and 500 bootstrap samples per window were used.
The availability of more sequenced virus strains would be helpful to further elucidate the scenario of recombination processes in the phylogenetic development of strains of DOBV. Nevertheless, the recombination event in DOBV evolution, after similar findings for Tula virus (TULV) in its natural evolution (21) and under in vitro conditions (15), offers another example of natural homologous recombination in the evolution of hantaviruses.
Evolution of DOBV and the problem of hantavirus species definition.
Since DOBV-Af and DOBV-Aa seem to exhibit stable association with two different host species, it has been proposed elsewhere to consider them as different virus species and to take the only available virus isolate of the DOBV-Aa group, Saa/160v, as the prototype of the new virus species, called Saaremaa virus (13, 16). According to the species demarcation criteria in the genus Hantavirus, as defined by the International Committee on Taxonomy of Viruses (5), hantavirus species have to meet the following essentials.
(i) Species are found in a unique ecological niche, i.e., in a different primary rodent reservoir species or subspecies. This is the case for DOBV-Af versus DOBV-Aa. Our results presented here demonstrate that the investigated DOBV strains from A. flavicollis and A. agrarius in Slovakia are clearly different despite their coexistence in the same geographical place. On the other hand, the reassortment origin of Saaremaa-Aa strains indicates the possibility of genetic exchanges between members of the DOBV-Af and DOBV-Aa lineages. Obviously, the precondition for such events was the coinfection of the same host animal.
(ii) Species exhibit at least a 7% difference in amino acid identity on comparison of the complete GPC and N protein sequences. As shown in Table 1, we found amino acid differences in the deduced N protein sequences within the DOBV-Af and DOBV-Aa groups of between only 2.1 and 3.3%. Saaremaa-Aa shows an N protein sequence difference of 2.8% from the DOBV-Af strains investigated. Since the Saaremaa-Aa strain carries a DOBV-Af-like S segment, it is not surprising that it exhibits higher similarities to the DOBV-Af strains than to the other DOBV-Aa strains (Table 1). In the putative GPC the amino acid differences between the DOBV-Af and DOBV-Aa groups have been found to be in the range of 5.5 to 5.9% (Table 2).
(iii) Species show at least a fourfold difference in two-way cross-neutralization tests. By analysis of human serum samples from the Balkans (southeastern Europe) and Estonia (northeastern Europe), Sjölander et al. (3a) have shown that a majority of the sera exhibited this typical titer difference with preference for the local virus (DOBV-Slovenia and Saaremaa, respectively); however, a substantial number of serum samples did not. The authors conclude that DOBV-Af and Saaremaa virus define unique hantavirus serotypes (3a). We have deduced 72 amino acid exchanges from the glycoprotein (M) gene sequences of the virus isolates Slo/Af and Saa/160v (data not shown) which could lead to the distinct serological behavior.
(iv) Species do not naturally form reassortants with other species. However, this was highly probably the case for the Saaremaa-Aa strains (see above). The occurrence of such reassortment events between DOBV-Af and DOBV-Aa should be taken as one argument against the idea that Saaremaa-Aa is a separate species.
The data demonstrate the phylogenetic relationship of DOBV members which are able to sympatrically coexist in different host species. However, the seemingly distinct DOBV clades are interacting in nature and affecting one another's evolution.
Nucleotide sequence accession number.
The sequences of the complete M segments of DOBV strains from A. agrarius and A. flavicollis from eastern Slovakia, DOBV/Esl/862Aa/97 and DOBV/Esl/400Af/98, and of the complete S segment of DOBV/Esl/400Af/98 (the abbreviations stand for virus species/place of occurrence [eastern Slovakia]/number of the sample and animal species/year of detection), have been assigned EMBL accession numbers AY168578, AY168577, and AY168576, respectively.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grants Kr1293/2-2/2-3 and Vi160/3-2), Charité Medical School, Fonds der Chemischen Industrie, United States Public Health Service (grants RO1 AI41692 and E11/CCE620225), and Slovak Scientific Grant Agency (VEGA grant 2/7166). Boris Klempa acknowledges financial support by an EMBO short-term fellowship.
We thank Claus Sibold and Katrin Leitmeyer for their contributions to the early stages of this work. We acknowledge the participation of Brita Haring, Beate Ziaja, Anna Hegele, and Burkhard Jandrig in performing the experiments. We thank Mikka Salminen and Christian Pitra for helpful discussions.
REFERENCES
- 1.Avsic-Zupanc, T., S. Y. Xiao, R. Stojanovic, A. Gligic, G. van der Groen, and J. W. LeDuc. 1992. Characterization of Dobrava virus: a Hantavirus from Slovenia, Yugoslavia. J. Med. Virol. 38:132-137. [DOI] [PubMed] [Google Scholar]
- 2.Avsic-Zupanc, T., A. Toney, K. Anderson, Y. K. Chu, and C. Schmaljohn. 1995. Genetic and antigenic properties of Dobrava virus: a unique member of the Hantavirus genus, family Bunyaviridae. J. Gen. Virol. 76:2801-2808. [DOI] [PubMed] [Google Scholar]
- 3.Avsic-Zupanc, T., K. Nemirov, M. Petrovec, T. Trilar, M. Poljak, A. Vaheri, and A. Plyusnin. 2000. Genetic analysis of wild-type Dobrava hantavirus in Slovenia: co-existence of two distinct genetic lineages within the same natural focus. J. Gen. Virol. 81:1747-1755. [DOI] [PubMed] [Google Scholar]
- 3a.Brus Sjölander, K., I. Golovljova, V. Vasilenko, A. Plyusnin, and Å. Lundkvist. 2002. Serological divergence of Dobrova and Saaremaa hantaviruses: evidence for two distinct serotypes. J. Epidemiol. Infect. 128:99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 6.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95./98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 7.Henderson, W. W., M. C. Monroe, S. C. St. Jeor, W. P. Thayer, J. E. Rowe, C. J. Peters, and S. T. Nichol. 1995. Naturally occurring Sin Nombre virus genetic reassortants. Virology 214:602-610. [DOI] [PubMed] [Google Scholar]
- 8.Krüger, D. H., R. Ulrich, and Å. Lundkvist. 2001. Hantavirus infections and their prevention. Microbes Infect. 3:1129-1144. [DOI] [PubMed] [Google Scholar]
- 9.Li, D., A. L. Schmaljohn, K. Anderson, and C. S. Schmaljohn. 1995. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology 206:973-983. [DOI] [PubMed] [Google Scholar]
- 10.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertz, G. J., B. L. Hjelle, and R. T. Bryan. 1997. Hantavirus infection. Adv. Intern. Med. 42:369-421. [PubMed] [Google Scholar]
- 12.Nemirov, K., O. Vapalahti, Å. Lundkvist, V. Vasilenko, I. Golovljova, A. Plyusnina, J. Niemimaa, J. Laakkonen, H. Henttonen, A. Vaheri, and A. Plyusnin. 1999. Isolation and characterization of Dobrava hantavirus carried by the striped field mouse (Apodemus agrarius) in Estonia. J. Gen. Virol. 80:371-379. [DOI] [PubMed] [Google Scholar]
- 13.Plyusnin, A. 2002. Genetics of hantaviruses: implications to taxonomy. Arch. Virol. 147:665-682. [DOI] [PubMed] [Google Scholar]
- 14.Plyusnin, A., D. H. Krüger, and Å. Lundkvist. 2001. Hantavirus infections in Europe. Adv. Virus Res. 57:105-136. [DOI] [PubMed] [Google Scholar]
- 15.Plyusnin, A., S. K. J. Kukkonen, A. Plyusnina, O. Vapalahti, and A. Vaheri. 2002. Transfection-mediated generation of functionally competent Tula hantavirus with recombinant S RNA segment. EMBO J. 21:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plyusnin, A., and S. P. Morzunov. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47-75. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez, L. L., J. H. Owens, C. J. Peters, and S. T. Nichol. 1998. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology 242:99-106. [DOI] [PubMed] [Google Scholar]
- 18.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 19.Schmaljohn, C. S., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 21.Sibold, C., H. Meisel, D. H. Krüger, M. Labuda, J. Lysy, O. Koz̆uch, M. Pejcoch, A. Vaheri, and A. Plyusnin. 1999. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J. Virol. 73:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibold, C., H. Meisel, Å. Lundkvist, A. Schulz, F. Cifire, R. Ulrich, O. Koz̆uch, M. Labuda, and D. H. Krüger. 1999. Simultaneous occurrence of Dobrava, Puumala and Tula hantaviruses in Slovakia. Am. J. Trop. Med. Hyg. 61:409-411. [DOI] [PubMed] [Google Scholar]
- 23.Sibold, C., R. Ulrich, M. Labuda, Å. Lundkvist, H. Martens, M. Schütt, P. Gerke, K. Leitmeyer, H. Meisel, and D. H. Krüger. 2001. Dobrava hantavirus causes hemorrhagic fever with renal syndrome in central Europe and is carried by two different Apodemus mice species. J. Med. Virol. 63:158-167. [PubMed] [Google Scholar]
- 24.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]