Abstract
The zinc finger C36-X1-C38-X7-C46-X6-H53 of the nuclearly localized C2 protein of Tomato yellow leaf curl virus China is involved in pathogenicity and suppression of posttranscriptional gene silencing (PTGS). Here, we demonstrate that the zinc finger is indispensable for the C2 protein to bind zinc and DNA. Mutation of cysteine residue C36, C38, or C46 reduced the zinc and DNA binding capacity of C2 protein. When expressed from potato virus X, all three mutants, C2-C36R, C2-C38N, and C2-C46I, tagged with a green fluorescent protein (GFP) were still capable of transporting GFP into but aggregated abnormally in nuclei. Our data establish that zinc- and DNA-binding activity correlates with C2-mediated pathogenesis and PTGS suppression.
Posttranscriptional gene silencing (PTGS) in plants, RNA interference in animals, and gene quelling in fungi share a common molecular mechanism in which a target RNA is transinactivated by homology-dependent RNA degradation, representing a conserved cellular defense system for controlling foreign gene expression across kingdoms (3, 8, 14, 31, 39, 42). In plants, PTGS protects the host against virus infection, down-regulates transgene expression, and may also be an important component in the control of development (39, 42). Consistent with the active role of PTGS in antiviral defense, plant viruses have evolved counterattack functions by encoding proteins that are capable of suppressing PTGS. PTGS suppressors often enhance viral pathogenicity, and a number of them have been characterized, including 2b, HC-Pro, P1, and P19 proteins from cucumoviruses, potyviruses, sobemoviruses, and tombusviruses; p25 movement protein of Potato virus X (PVX); and AC2 and C2 proteins of African cassava mosaic virus and Tomato yellow leaf curl virus China (TYLCV-C) (6, 37, 39). There is much variation in the extent of PTGS suppression by different virus-encoded suppressors that may be mediated by targeting distinct steps in the PTGS pathway (2, 40, 41).
Geminiviruses are a family of unique small circular single-stranded (ss) DNA viruses that replicate via double-stranded (ds) DNA intermediates by a rolling circle mechanism in plant cell nuclei (15). TYLCV-C, a distinct species of the genus Begomovirus in the family Geminiviridae, has a monopartite genome resembling the DNA A component of bipartite begomoviruses (43). We have previously demonstrated that TYLCV-C C2, a nuclearly localized protein, induces necrosis and suppresses PTGS when expressed from a PVX vector and that a cysteine-rich motif (C36-X1-C38-X7-C46-X6-H53) is required for C2 protein-mediated pathogenesis and PTGS suppression (37, 38). However, whether the C2 protein binds zinc and indeed whether the cysteine-rich domain represents an authentic zinc-finger motif remained unclear.
To address these issues, we first used the Bac-to-Bac system (Invitrogen Life Technologies) to express wild-type and mutant C2 proteins, all of which have a six-histidine affinity tag. Baculovirus containing the C2 gene or its derivatives was recovered after homologous recombination between the baculovirus expression vector pBac/C2, pBac/C2-C36R, pBac/C2-C38N, or pBac/C2-C46I (Table 1) and the viral DNA according to the manufacturer's protocol. Sf9 cells were then infected with recombinant baculoviruses, and after 72 h of incubation at 28°C, Sf9 cells were harvested, resuspended in extraction buffer (EB; 50 mM Tris-HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride) containing 50 mM NaCl, and lysed by sonication. Insoluble pellets were discarded after centrifugation, and high-speed clarified supernatants were collected. Aliquots of Sf9 extracts were adjusted to either 50, 100, or 200 mM NaCl. The amount of C2 protein in Sf9 extracts was normalized using competitive enzyme-linked immunosorbent assay (19) and an antihistidine monoclonal antibody (Amersham Pharmacia Biotech).
TABLE 1.
Construction of C2 expression cassettesa
| Construct | Vector | Primer pair (5′ region/ 3′ region) | Cloning sites | Mutation |
|---|---|---|---|---|
| pBac/C2 | pFastBacHTa | PP141/PP142 | NcoI, EcoRI | Wild type |
| pBac/C2-C36R | pFastBacHTa | PP141/PP142 | NcoI, EcoRI | Cys36→Arg |
| pBac/C2-C38N | pFastBacHTa | PP141/PP143 | NcoI, BamHI | Cys38→Asn |
| pBac/C2-C46I | pFastBacHTa | PP141/PP143 | NcoI, BamHI | Cys46→Ile |
| pEHT/C2 | pEHISTEV | PP141/PP142 | NcoI, EcoRI | Wild type |
| pEHT/C2-C36R | pEHISTEV | PP141/PP142 | NcoI, EcoRI | Cys36→Arg |
| pEHT/C2-C38N | pEHISTEV | PP141/PP143 | NcoI, BamHI | Cys38→Asn |
| pEHT/C2-C46I | pEHISTEV | PP141/PP143 | NcoI, BamHI | Cys46→Ile |
The TYLCV-C C2 gene and mutant derivatives were PCR amplified using PVX/C2, PVX/C2-C36R, PVX/C2-C38N, and PVX/C2-C46I (37) as DNA templates together with primer PP141 (cctgtatcATGaGATCTTCGTCTCCCTC) and either PP142 (ccgaatTcAAATACTCTTAAGAAATGCGAGGTC) or PP143 (ccggatccTAAATACTCTTAAGAAATGCGAGGTC) (introduced restriction endonuclease sites are underlined and modified nucleotides are lowercase). PCR products were digested with BspHI plus EcoRI or BamHI and were cloned into the NcoI and EcoRI or NcoI and BamHI sites of pEHISTEV (a modified version of pET28a [unpublished data]) and pFastBacHTa (Invitrogen Life Technologies) to produce vectors for C2 protein expression in E. coli and insect cells, respectively.
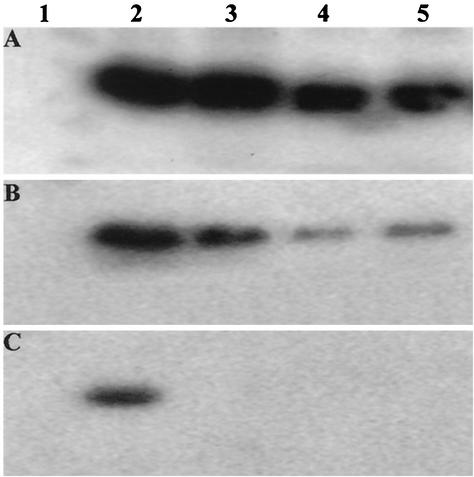
We performed a zinc-affinity pull-down assay, for which extracts from wild-type or recombinant baculovirus-infected Sf9 cells containing equal amounts of wild-type or mutant C2 protein in either 50, 100, or 200 mM NaCl were incubated with 50-μl aliquots of zinc chelate affinity resins (iminodiacetic acid-Sepharose 6B; Sigma) preequilibrated with EB containing either 50, 100, or 200 mM NaCl, as appropriate. Resins were then washed three times with the same buffer, resuspended in 100 μl of 1× gel loading buffer (21), and boiled for 3 min before loading samples onto a sodium dodecyl sulfate-15% polyacrylamide gel. After electrophoresis, proteins were immobilized on nitrocellulose membranes, immunodetected with the ECL detection system by use of a monoclonal antibody raised against the histidine tag (Amersham Pharmacia Biotech), and analyzed using a PhosphorImager. In a low-salt (50 mM NaCl) buffer, C2 protein and all three mutants remained bound to the resin. There was little difference in binding affinity between the wild-type protein and mutant C2-C46I at this salt concentration, although mutants C2-C36R and C2-C38N bound less well (Fig. 1A). A reduction in the ability of all three mutants to bind to the resin in comparison with the wild-type protein became more evident at 100 mM NaCl and was particularly marked for mutants C2-C36R and C2-C38N (Fig. 1B). Some wild-type protein remained bound at 200 mM NaCl, although none of the three mutants was capable of binding at this salt concentration (Fig. 1C). Our data indicate that histidine tags made little contribution to the differential capabilities of the wild-type and mutant C2 proteins to bind zinc. Indeed, the affinity of the wild-type and mutant proteins for zinc chelate resin indicates that C2 protein is capable of binding zinc, as reported for Tomato golden mosaic virus TrAP (transcriptional activator protein; also known as AL2 or AC2) (16), and that the cysteine-rich C36-X1-C38-X7-C46-X6-H53 domain represents a bona fide zinc finger required for this purpose. We previously demonstrated that mutation of C36, C38, and C46 eliminated C2 protein-mediated induction of necrosis and PTGS suppression (37) and that these mutations also significantly reduce the ability of C2 protein to bind zinc, suggesting that the three cysteines are additionally responsible for this activity. Interestingly, mutation of C38N and C36R had a greater adverse effect on zinc binding activity than mutation of C46I.
FIG. 1.
Zinc-binding activity of C2 protein. Zinc-affinity pull-down assays were performed in buffers containing 50 mM NaCl (A), 100 mM NaCl (B), and 200 mM NaCl (C) with protein extracts from Sf9 cells infected with wild-type baculovirus (lane 1) and recombinant baculovirus expressing His-tagged C2 (lane 2), C2-C46I (lane 3), C2-C38N (lane 4), and C2-C36R (lane 5). His-tagged C2 protein with the predicted size was detected using a monoclonal antibody raised against the histidine tag.
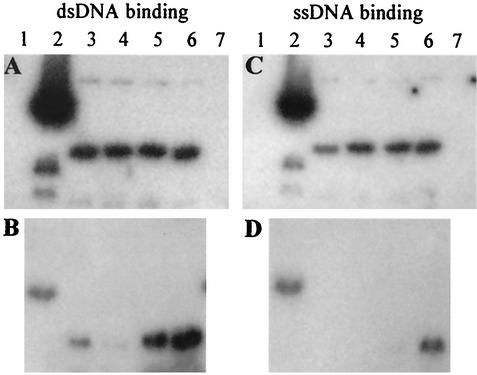
Zinc-finger proteins frequently possess DNA-binding activity. We therefore investigated whether C2 protein bound ssDNA and/or dsDNA and the role of the zinc finger in DNA binding. To achieve this, Escherichia coli strain BL21 (DE3; Invitrogen Life Technologies) was transformed with pEHT/C2, pEHT/C2-C36R, pEHT/C2-C38N, or pEHT/C2-C46I (Table 1), and expression of C2, C2-C36R, C2-C38N, and C2-C46I was induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Viral and nonviral ssDNA and dsDNA fragments were 32P labeled using a Ready-to-Go labeling kit (Amersham Pharmacia Biotech), and Southwestern assays were performed to determine interactions between C2 protein and ssDNA and dsDNA probes with equal amounts of radioactivity as described by Liu et al. (24). The C2 protein bound to TYLCV-C C2 gene ssDNA (405 nucleotides) and dsDNA (405 bp) probes (Fig. 2A and C). At 300 mM KCl, all three mutants bound to ssDNA and dsDNA, although C2-C46I bound less efficiently to ssDNA at this salt concentration. At 600 mM KCl, C2 protein bound dsDNA more efficiently than ssDNA (Fig. 2B and D). At this salt concentration, however, all three mutants failed to bind ssDNA, and the ability of C2-C38N and C2-C46I to bind dsDNA was significantly reduced in comparison with the wild-type protein. Moreover, in a parallel experiment, the C2 protein was found to preferably bind dsDNA probes (1,161 bp) prepared from a TYLCV-C unrelated plant gene, the LFY-like gene involved in flower development (23; H. Liu and Y. Hong, unpublished data). The results indicate that C2 protein binds DNA in a sequence-nonspecific manner with a preference for dsDNA, in contrast to other TrAPs that bound ssDNA with higher efficiency (16, 27), and that the cysteine-rich motif participates in this binding activity. Although C2 protein is basic, its interaction with DNA is not necessarily due to a net positive charge since the basic protein lysozyme (pI, ∼9.6) bound neither ssDNA nor dsDNA under the prevailing conditions. Clearly, DNA binding by C2 protein was affected by mutation of the three zinc-finger cysteine residues. C2-C36R, C2-C38N and C2-C46I mutants were unable to bind ssDNA in high salt (600 mM KCl), and both C2-C38N and C2-C36R were significantly impaired in their ability to bind dsDNA under these conditions, although C2-C46I maintained substantial binding activity, reflecting the effect of these mutations on zinc binding.
FIG. 2.
DNA-binding activity of C2 protein. Southwestern assays were performed using dsDNA (A and B) and ssDNA (C and D) in buffers containing 300 mM KCl (A and C) and 600 mM KCl (B and D). Assays contained lysozyme (lanes 1), extracts from E. coli expressing maize streak virus coat protein (lanes 2), C2-C46I (lanes 3), C2-C38N (lanes 4), C2-C36R (lanes 5), and C2 (lanes 6), and extracts from E. coli transformed with expression vector pEHISTEV alone (lanes 7).
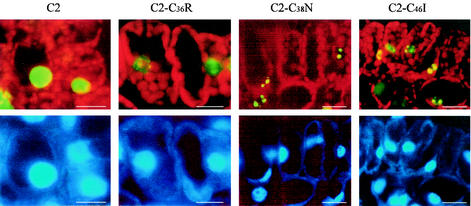
To better understand the relevance of the observed biochemical properties to C2 protein function, the cellular localization of mutants C2-C36R, C2-C38N, and C2-C46I was investigated. PVX RNA transcripts were produced by in vitro transcription and mechanically inoculated onto Nicotiana benthamiana plants as previously described (7). Leaf tissues infected with PVX/C2-GFP (C2), PVX/C2-C36R-GFP (C2-C36R), PVX/C2-C38N-GFP (C2-C38N), and PVX/C2-C46I-GFP (C2-C46I) were collected 7 days postinoculation, cut into 3-mm-wide strips, vacuum infiltrated, and fixed overnight in 4% paraformaldehyde-100 mM phosphate buffer (pH 7.0). Tissues were then infiltrated with 15% sucrose-100 mM phosphate buffer (pH 7.0), embedded in 5% low-melting-point agarose, and sectioned in a cryostat at −20°C (OTS; Bright Instruments). Ten-micrometer sections were mounted in 50% glycerol containing 1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml and examined using a Zeiss Axiophot equipped with a Nikon Digital Camera Coolpix995. When expressed as green fluorescent protein (GFP)-tagged fusion proteins from PVX vectors (PVX/C2-C36R-GFP, PVX/C2-C38N-GFP, and PVX/C2-C46I-GFP), all three mutants localized to the nucleus (Fig. 3, top panels), resembling the behavior of the wild-type protein fused to GFP expressed from PVX/C2-GFP (38). However, fluorescence associated with the mutants was irregularly scattered in the nuclei of leaf mesophyll cells and was distinct from the even distribution of C2-GFP fluorescence. DAPI staining indicated that the nuclei remained intact (Fig. 3, bottom panels), and no nuclear structural alterations were observed by transmission electron microscopy (R. Van Wezel and Y. Hong, unpublished data), suggesting that the zinc-finger mutants aggregate abnormally in nuclei. This remarkable effect may not be due simply to alteration of the primary structure of the protein. Indeed, several C2 protein mutants such as C2-K17D, C2-HR21DV, and C2-KK25DI containing different amino acid substitutions were capable of transporting GFP into nuclei, and the nuclearly localized GFP fluorescence was evenly distributed (R. van Wezel, X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong, Abstr. XII Int. Congr. Virol., p. 148, 2002).
FIG. 3.
Nuclear localization of C2 zinc-finger mutants. Fluorescence was observed with filters for GFP (450- to 490-nm excitation, 520-nm long-pass emission) (top panels) and DAPI (365-nm excitation, 420-nm long-pass emission) (bottom panels). Chloroplast autofluorescence appears red. Bar = 10 μm, indicating that the ratios between the sizes of cells and nuclei are similar.
The three cysteine residues of the zinc finger were changed to either arginine, asparagine, or isoleucine. The choice of mutations was based on the notion that the physicochemical properties of these amino acids are largely unrelated to that of cysteine as discussed previously (37). Thus, the effect of mutations on biological and biochemical functions of the C2 protein would become detectable. Another important issue is that such amino acid replacements should impose a minimum effect on the C2 protein structures. As revealed by Chou-Fasman prediction (9), the wild-type C2 protein and mutant C2-C38N possessed essentially the same secondary structures. Mutant C2-C46I formed a secondary structure similar to that of wild-type C2 and mutant C2-C38N proteins, with the exception of having a β-sheet around the zinc finger instead of an α-helix. C2-C36R had a slight different overall conformation but without local structural alterations in the region of the zinc finger. Surprisingly, C2-mediated zinc- and DNA-binding activities were mostly affected by the mutation of C38N, while the C36R mutation had the least effect on C2 binding to DNA and the C2-C46I mutation had stronger zinc-binding activity than the other two mutants (Fig. 1 and 2). Therefore, the specificity of effects observed in this report is unlikely due to the mutant amino acids globally interfering with C2 folding or conformational stability, which would simultaneously impair all of its activities. Indeed, the modified proteins are soluble in plant cells and at least partially functional since they are still transported to the nucleus. It is likely that the zinc finger specially affects the tertiary and/or quaternary structure of the C2 protein. Single substitution of the three cysteine residues with any amino acid may lead to the formation of unusual disulfide bonds between the two SH groups of the remaining nonmutated cysteine residues of the zinc finger within or between C2 protein molecules. Another possibility is that the zinc finger could control the ability of the C2 protein to form stable structural complexes as has been described for other zinc- and DNA-binding proteins (22). As a consequence of such structural changes, the C2 protein altered its biochemical activities in zinc and DNA binding and its cellular biological property, which led to the deficit of its biological functions in pathogenesis and PTGS suppression in planta.
In summary, we have found that a correlation exists between the biochemical properties of the C2 protein encoded by the monopartite begomovirus TYLCV-C and its biological functions in pathogenesis and PTGS suppression. In plants, PTGS represents an effective defense mechanism against infection caused by RNA and DNA viruses. Initiation, propagation of a systemic silencing signal, and maintenance have been recently proposed to be the three components in the process of PTGS (28, 29) which can be targeted by PTGS suppressors. Notably, potyviral HC-Pro affects a step coincident with, or upstream of, the production of small RNAs that is necessary for PTGS maintenance (25, 26). In contrast, the PVX p25 cell-to-cell movement protein and the 2b protein of Cucumber mosaic virus appear to prevent silencing signal spread (12, 40). While most suppressors that have been identified are from RNA viruses and are involved in virus movement, TrAP and C2 proteins are encoded by ssDNA begomoviruses (1, 4, 20, 37, 41). Collectively, TrAP and its C2 protein homologue, encoded by bipartite and monopartite begomoviruses, respectively, specifically modulate viral gene expression at the transcriptional level, enhance plant susceptibility to viral infection, contribute to viral pathogenicity, and suppress PTGS (5, 10, 11, 13, 16-18, 32-36, 37, 41). TrAP exhibits sequence-nonspecific DNA binding activity and is confined to the nucleus (16, 27, 30, 32, 38). TrAP has also been reported to be phosphorylated and to bind zinc (16). However, the relevance of these diverse molecular and cellular properties of TrAP to pathogenesis and PTGS suppression is less well understood.
Although TrAP and C2 proteins participate in the control of specific viral genes and are probably also involved in host gene expression, these proteins are not canonical transcriptional activators due to their sequence-nonspecific interaction with DNA. It is believed that the zinc- and ssDNA-binding activity is not particularly relevant to TrAP-mediated transcriptional regulation (16, 27), and this may also be the case for the TYLCV-C C2 protein. Clearly, this raises the question of the biological significance of the observed zinc- and DNA-binding activity to the infection process. The results of this study and a previous mutagenesis analysis (37) have indicated that the altered biochemical behavior of the three zinc-finger mutants in zinc and DNA binding correlates with a loss of biological function in inducing necrosis and suppressing PTGS in planta. This suggests that zinc and DNA binding play either a direct or indirect role in C2 protein-mediated pathogenesis and PTG5 suppression.
Acknowledgments
We thank D. Baulcombe for providing the original PVX-based vector and are grateful to T. M. A. Wilson for his encouragement throughout this work.
This project was supported in part by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, C. Mau, A. Mallory, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. [DOI] [PubMed] [Google Scholar]
- 3.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Brough, C. L., G. Sunter, W. E. Gardiner, and D. M. Bisaro. 1992. Kinetics of tomato golden mosaic virus DNA replication and coat protein promoter activity in Nicotiana tabacum protoplasts. Virology 187:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, S., T. Kavanagh, and D. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. [DOI] [PubMed] [Google Scholar]
- 8.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dry, I., L. Krake, P. Mullineaux, and A. Rezaian. 2000. Regulation of tomato leaf curl virus gene expression in host tissues. Mol. Plant-Microbe Interact. 13:529-537. [DOI] [PubMed] [Google Scholar]
- 11.Groning, B. R., R. J. Hayes, and K. W. Buck. 1994. Simultaneous regulation of tomato golden mosaic virus coat protein and AL1 gene expression: expression of the AL4 gene may contribute to suppression of the AL1 gene. J. Gen. Virol. 75:721-726. [DOI] [PubMed] [Google Scholar]
- 12.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haley, A., X. C. Zhan, K. Richardson, K. Head, and B. Morris. 1992. Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2 and AC3 gene products. Virology 188:905-909. [DOI] [PubMed] [Google Scholar]
- 14.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 15.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminivirus: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 16.Hartitz, M. D., G. Sunter, and D. M. Bisaro. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1-14. [DOI] [PubMed] [Google Scholar]
- 17.Hong, Y., K. Saunders, M. R. Hartley, and J. Stanley. 1996. Resistance of geminivirus infection by virus-induced expression of dianthin in transgenic plants. Virology 220:119-127. [DOI] [PubMed] [Google Scholar]
- 18.Hong, Y., K. Saunders, and J. Stanley. 1997. Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology 228:383-387. [DOI] [PubMed] [Google Scholar]
- 19.Hornbeck, P. 1991. Direct competitive ELISA to detect soluble antigens, p. 2.1.6-2.1.11. In J. E. Coligan, A. M. Kruisbeek, D. H. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons Inc., New York. N.Y.
- 20.Kasschau, K. D., and J. C. Carrington. 1998. A counter-defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lewin, B. 1997. Gene VI. Oxford University Press, Oxford, United Kingdom.
- 23.Liu, F.-Q., G.-L. Zhu, D. Luo, X.-Y. Wu, and Z.-H. Xu. 1999. Cloning and analysis of CFL—an LFY-like gene from cucumber. Acta Bot. Sin. 41:813-819. [Google Scholar]
- 24.Liu, H., M. I. Boulton, and J. W. Davies. 1997. Maize streak virus coat protein binds single- and double-stranded DNA in vitro. J. Gen. Virol. 78:1265-1270. [DOI] [PubMed] [Google Scholar]
- 25.Llave, C., K. Kasschau, and J. Carrington. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noris, E., I. Jupin, G. P. Accotto, and B. Gronenborn. 1996. DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217:607-612. [DOI] [PubMed] [Google Scholar]
- 28.Palauqui, J.-C., and H. Vaucheret. 1998. Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95:9675-9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanderfoot, A. A., and S. G. Lazarowitz. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp, P. A. 2001. RNA interference 2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 32.Sung, Y. K., and R. H. A. Coutts. 1996. Potato yellow mosaic geminivirus AC2 protein is a sequence non-specific DNA binding protein. FEBS Lett. 383:51-54. [DOI] [PubMed] [Google Scholar]
- 33.Sunter, G., and D. M. Bisaro. 1991. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416-419. [DOI] [PubMed] [Google Scholar]
- 34.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunter, G., and D. M. Bisaro. 1997. Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232:269-280. [DOI] [PubMed] [Google Scholar]
- 36.Sunter, G., J. L. Sunter, and D. M. Bisaro. 2001. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59-70. [DOI] [PubMed] [Google Scholar]
- 37.van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 38.Van Wezel, R., H. Liu, P. Tien, J. Stanley, and Y. Hong. 2001. Gene C2 of the monopartite geminivirus Tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localised in the nucleus. Mol. Plant-Microbe Interact. 14:1125-1128. [DOI] [PubMed] [Google Scholar]
- 39.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 40.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:57-167. [DOI] [PubMed] [Google Scholar]
- 41.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 43.Yin, Q., H. Yang, Q. Gong, H. Wang, Y. Liu, Y. Hong, and P. Tien. 2001. Tomato yellow leaf curl China virus: monopartite genome organization and agroinfection of plants. Virus Res. 81:69-76. [DOI] [PubMed] [Google Scholar]