Abstract
Epstein-Barr virus (EBV) is associated with the development of malignant lymphomas and lymphoproliferative disorders in immunocompromised individuals. The LMP2A protein of EBV is thought to play a central role in this process by allowing the virus to persist in latently infected B lymphocytes. We have demonstrated that LMP2A, when expressed in B cells of transgenic mice, allows normal B-cell developmental checkpoints to be bypassed. To identify cellular genes targeted by LMP2A that are involved in this process, we have utilized DNA microarrays to compare gene transcription in B cells from wild-type versus LMP2A transgenic mice. In B cells from LMP2A transgenic mice, we observed decreased expression of many genes associated with normal B-cell development as well as reduced levels of the transcription factors that regulate their expression. In particular, expression of the transcription factor E2A was down-regulated in bone marrow and splenic B cells. Furthermore, E2A activity was inhibited in these cells as determined by decreased DNA binding and reduced expression of its target genes, including the transcription factors early B-cell factor and Pax-5. Expression of two E2A inhibitors, Id2 and SCL, was up-regulated in splenic B cells expressing LMP2A, suggesting a possible mechanism for E2A inhibition. These results indicate that LMP2A deregulates transcription factor expression and activity in developing B cells, and this likely allows for a bypass of normal signaling events required for proper B-cell development. The ability of LMP2A to interfere with B-cell transcription factor regulation has important implications regarding its role in EBV latency.
Epstein-Barr virus (EBV) is the etiological agent of infectious mononucleosis, a self-limiting lymphoproliferative disease occurring in adolescents and young adults upon primary infection (for reviews, see references 18, 38, 41, and 60). Most infections are uncomplicated, resulting in the establishment of viral latency in B lymphocytes following primary infection. Virus-related pathologies can occur, however, and are of particular concern in immunocompromised individuals (4, 5, 48). EBV is associated with the development of several malignancies, including Burkitt's lymphoma, Hodgkin's lymphoma, nasopharyngeal carcinoma, and various lymphoproliferative disorders arising in immunocompromised patients (2, 3, 4, 15, 37, 74).
The LMP2A protein of EBV is the only viral protein consistently identified in latently infected B cells in vivo, suggesting that LMP2A plays an important role in viral persistence and in the development of EBV-associated diseases (16, 58, 70, 71). In latently infected lymphocytes, LMP2A localizes to small glycolipid-enriched microdomains in the plasma membrane (21). By localizing to membrane microdomains, LMP2A may mimic an activated B-cell receptor (BCR). Studies have demonstrated that BCR activation in LMP2A-expressing B cells fails to activate the downstream signaling molecules Lyn, Syk, phosphatidylinositol 3-kinase (PI3-K), phospholipase C-γ2, Vav, Shc, and mitogen-activated protein kinase (MAPK). Instead, Syk, PI3-K, phospholipase C-γ2, and Vav are constitutively phosphorylated in LMP2A-expressing cells (45, 46, 47). In these cells, the amino-terminal domain of LMP2A is tyrosine phosphorylated and associates with Src family protein tyrosine kinases as well as Syk (11, 45). Mutational analyses indicate that phosphotyrosines at positions 74 and 85 (an ITAM motif) in LMP2A bind Syk, while tyrosine 112 binds Lyn. All three residues are essential for the LMP2A-mediated block in BCR signal transduction (25, 26). It is likely that LMP2A provides a constitutive positive signal and, by sequestering Lyn and Syk, prevents normal BCR signal transduction. By preventing B-cell activation, LMP2A may prevent the induction of lytic EBV replication and subsequent immune recognition (42, 46).
We have utilized a transgenic mouse model to further define the function of LMP2A in B cells in vivo. Expression of LMP2A interferes with normal B-cell development, allowing BCR-negative cells to exit the bone marrow and colonize peripheral organs (12, 13). In normal bone marrow, appropriate immunoglobulin (Ig) heavy-chain gene rearrangement is required for transition from the CD19+ CD43+ pre-B stage to the CD19+ CD43− pre-B stage. Subsequent rearrangement of Ig light-chain genes and expression of both heavy and light chains at the cell surface allow for transition to the CD19+ IgM+ immature B-cell stage, which is required for exit from the bone marrow (Fig. 1B) (24, 28). The TgE LMP2A transgenic line contains significantly reduced numbers of CD19+ B cells in the bone marrow and spleen. Additionally, the majority of bone marrow and splenic CD19+ B cells do not express surface IgM. Interestingly, these cells are CD43 negative and interleukin-7 (IL-7) responsive (13). The presence of CD43-negative cells also lacking IgM suggests a defect at the pre-B stage of development. Bone marrow B cells from these mice also undergo Ig light-chain, but not heavy-chain, gene rearrangement (13). This indicates that LMP2A signaling bypasses the requirement for Ig recombination and allows IgM-negative cells, which would normally undergo apoptosis, to colonize peripheral lymphoid organs.
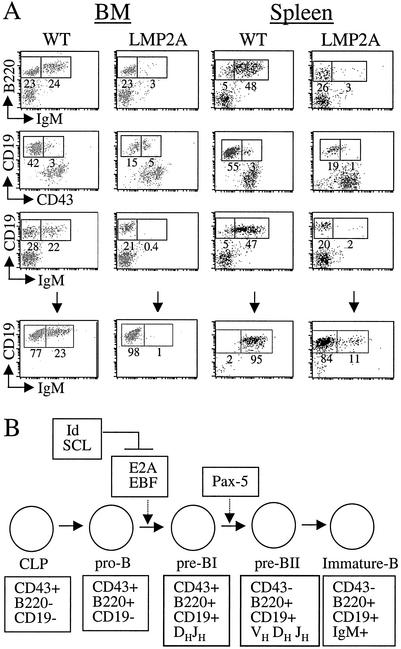
FIG. 1.
LMP2A transgenic mice and B-lymphocyte development. (A) Upper panel, bone marrow (BM) (left) and splenic (right) B cells were purified from wild-type (WT) and LMP2A transgenic mice. Cells were stained with antibodies to CD19, B220, CD43, and IgM to detect cell surface expression. The numbers indicate the percentage of cells positive for expression. Lower panel, CD19+ bone marrow B cells were purified in methylcellulose cultures containing IL-7, and splenic B cells were isolated by using CD19-coated magnetic beads as described in Materials and Methods. (B) Stages of B-cell development in the bone marrow, beginning from common lymphoid progenitor (CLP) cells and progressing to immature B cells, which exit the bone marrow and colonize peripheral lymphoid organs. The transition from the pro-B stage to the pre-B stage of development requires both E2A and EBF transcription factors. Pax-5 is required for B-cell commitment and transition through the pre-B stage. The activity of E2A is inhibited by Id and SCL proteins (for reviews, see references 27, 29, and 61).
Precursor B-cell growth in IL-7 in the absence of bone marrow stromal cells requires progression to the CD19+ CD43− stage, which depends on proper Ig rearrangement (24, 28). B cells from LMP2A transgenic mice form colonies in methylcellulose, and the B cells in these colonies lack IgM expression, indicating that LMP2A bypasses the requirement for Ig rearrangement and allows for IL-7-driven B-cell proliferation (12, 13). LMP2A-mediated effects on B-cell development and survival are abolished in transgenic mice expressing LMP2A with tyrosines 74 and 85 mutated to phenylalanines, indicating that the ITAM and Syk are required for LMP2A-mediated developmental and survival signals in B lymphocytes (43). Additionally, studies utilizing mice deficient in the downstream B-cell signaling component BLNK (SLP-65) or Bruton's tyrosine kinase (Btk) have demonstrated that LMP2A utilizes these molecules for its effects on B-cell development and survival (23, 44).
The development of mature B cells from multipotent progenitors requires the coordinated action of a number of transcriptional regulators, including E2A, early B-cell factor (EBF), and Pax-5 (BSAP) (for reviews, see references 27 and 61). E2A proteins (E proteins) encode two alternatively spliced gene products, E47 and E12, which contain a basic DNA binding domain and a helix-loop-helix (bHLH) dimerization motif. These proteins bind DNA as homo- or heterodimers and function as transcriptional activators (27, 35, 66). Mice deficient in E2A demonstrate a block in the early pro-B-cell stage of development and are defective for Ig heavy-chain gene rearrangement (7, 75). E2A induces expression of EBF, and the two proteins function together for induction of gene transcription from target promoters necessary for the pro-B to pre-B stage transition. These genes include those for components of the pre-BCR complex (Ig-α [mb-1], Ig-β [B29], λ5, and Vpre-B) and factors involved in recombination (terminal deoxynucleotidyl transferase [TdT], RAG-1, and RAG-2) (35, 55, 67). E2A and EBF, together with IL-7 receptor signaling, are necessary to activate Pax-5 expression. Pax-5 functions later than E2A and EBF during the pre-B stage of development and is necessary for the generation of fully rearranged V(D)J heavy-chain genes. Pax-5 mutant pre-B cells can respond to IL-7 in the presence of stromal cells but fail to differentiate into more mature B-cell stages (27, 52, 61, 62). Pax-5 has been shown to repress J-chain gene expression and induce germ line IgH, Ig-α (mb-1), CD19, N-myc, and lymphoid enhancer factor-1 (LEF-1) (31, 51, 62). In summary, E2A, EBF, and Pax-5 function together to regulate the expression of components of the pre-BCR complex and are required for the pro-B to pre-B-cell transition when Ig heavy-chain recombination is beginning.
E2A activity is negatively regulated by Id proteins, which also contain a helix-loop-helix domain but lack a DNA binding domain and thereby inhibit E2A family members from forming functional dimers (22, 29, 69). Another E2A inhibitor is SCL (TAL1), a bHLH factor that can also form heterodimers with E proteins. Constitutive expression of SCL in the bone marrow has been shown to induce a dose-dependent block at the B-cell commitment step (10, 32). Not surprisingly, B lymphocytes from mice in which Id proteins or SCL is overexpressed resemble those from E2A-deficient mice (22, 30, 68).
Expression of LMP2A in transgenic mice alters B-cell development by inhibiting Ig heavy-chain gene rearrangement and expression of a functional BCR. This suggests that LMP2A may alter the expression and/or activity of transcription factors or signaling molecules required for proper B-cell development. In this study, we have utilized DNA microarray technology to identify alterations in gene expression in B cells from mice expressing the LMP2A transgene. We have identified not only decreased expression of genes associated with normal B-cell development but also reduced levels of the transcription factors that regulate expression of those genes. These studies demonstrate that LMP2A, when expressed during B-cell development, can repress the expression and activities of critical transcription factors necessary for proper B-cell maturation.
MATERIALS AND METHODS
Mice.
Construction and characterization of the Eμ TgE LMP2A transgenic mice has been described previously (13). All animals were housed at the Northwestern University Center for Experimental Animal Resources in accordance with university animal welfare guidelines.
Isolation of primary B cells.
Bone marrow cells were flushed from femurs and tibias with using 1× phosphate-buffered saline containing 1% penicillin-streptomycin. Red blood cells were lysed in 155 mM ammonium chloride, and 2 × 106 cells were placed in 3 ml of Methocult M3630 methylcellulose cultures containing 10 ng of recombinant mouse IL-7 (Stemcell Technologies, Vancouver, British Colombia, Canada) per ml. Colonies formed in 7 to 10 days were routinely >95% CD19+ B cells as demonstrated by flow cytometry as previously described (13), and these cells were utilized for microarray analysis (Fig. 1A, bottom panels). Spleens were dissociated between frosted slides in RPMI to prepare single cell suspensions. Red blood cells were lysed, and B cells were purified at 4°C on magnetic cell sorting columns with magnetic beads coated with CD19 antibodies (Miltenyi Biotec, Auburn, Calif.). B cells were tested for purity by fluorescence-activated cell sorter analysis, and cells shown to be >95% CD19+ B cells by flow cytometry were utilized for microarray analysis (Fig. 1A, bottom panels).
RNA preparation and microarray analysis.
Total RNA was extracted from CD19+ bone marrow and splenic B cells according to the Trizol reagent protocol (Invitrogen, Carlsbad, Calif.). RNA was then subjected to cleanup by using the Rneasy mini kit according to the RNA cleanup protocol (Qiagen, Valencia, Calif.). Aliquots of RNA were then removed and run on 1% agarose-formaldehyde gels to verify that no degradation occurred. Additionally, optical density (OD) readings were taken, and 20 μg of total RNA having OD A260/A280 readings of between 1.9 and 2.1 was utilized for reverse transcription. Double-stranded cDNA was then generated with the Superscript double-stranded cDNA synthesis kit (Invitrogen), using an oligo(dT)-T7 primer [5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′]. Following phenol-chloroform extraction and ethanol precipitation, cRNA was in vitro transcribed and labeled with biotin by using the Enzo Bioarray RNA transcription labeling kit (Affymetrix, Huntsville, Ala.). The cRNA was purified by using CHROMA SPIN 100 columns (Clontech, Palo Alto, Calif.) and ethanol precipitated. cRNAs having OD A260/A280 readings of between 1.9 and 2.1 were again run on agarose-formaldehyde gels to check for transcripts ranging between 35 and 200 bases, and 20 μg of these products was fragmented in fragmentation buffer containing 200 mM Tris-acetate (pH 8.1), 500 mM potassium acetate, and 150 mM magnesium acetate at 95°C for 35 min. Fragmented, biotinylated cRNA was then submitted for Affymetrix microarray analysis by the Children’s Memorial Institute for Education and Research (CMIER) microarray facility at Northwestern University. The murine genome U74A chips were purchased from Affymetrix and utilized for analysis. The results from two separate, identical experiments were compared, and an average differential change in gene expression of twofold or greater in both experiments was considered significant.
RT-PCR Analysis.
Total RNA was extracted from CD19+ B cells by using Trizol reagent (Invitrogen). Five micrograms of RNA was reverse transcribed according to the Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR) protocol (Invitrogen). One-fourth of the RT reaction mixture was utilized in standard PCRs containing 1× PCR buffer (Amersham Pharmacia Biotech, Piscataway, N.J.), a 1 μM concentration of each oligonucleotide primer, 0.2 mM deoxynucleoside triphosphates, and 1 U of Taq polymerase (Amersham Pharmacia Biotech). The amplification cycle (15 s at 94°C, 30 s at 58°C, and 75 s at 72°C) was repeated 16 to 18 times, depending on the primers used, and followed by a single 15-min period at 72°C. Aliquots were removed at the end of cycles 10 through 20 in order to determine the minimum amplification necessary for detection. Oligonucleotides for PCRs were as follows: E47 sense, 5′-GTCCTGGGTGGATGATGAAC-3′; E47 antisense, 5′-CATCCCTGCTGTAGCTGTCA-3′; EBF sense, 5′-CCAACTCACCCTATGCCATT-3′; EBF antisense, 5′-GCAAGGTCGGTGATTTTGTT-3′; Pax5 sense, 5′-CAGCAAAATTCTTGGCAGGT-3′; Pax5 antisense, 5′-TGCTGTGTGAACAGGTCTCC-3′; Id2 sense, 5′-CCAATCTTTTGCAGGCATTT-3′; Id2 antisense, 5′-TCCCCATGGTGGGAATAGTA-3′ SCL sense, 5′-CTGTTTGTGCAGGAGAGCAA-3′; SCL antisense, 5′-CACCACCTGGATTGACACAG-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) sense, 5′-TGGTATCGTGGAAGGACTCATGAC-3′; and GAPDH antisense, 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′. PCR samples were subjected to standard gel electrophoresis in 1% agarose, and the images were scanned with an alphaimager (Alpha Innotech Corporation). The band intensities of each PCR product were normalized to those of GAPDH to determine the relative levels of each transcript.
Protein isolation and Western blots.
Bone marrow and splenic B cells were lysed in buffer containing 125 mM HEPES (pH 7.5), 750 mM NaCl, 50 mM MgCl2, 5 mM EDTA, 10% glycerol, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 25 mM NaF, and 1 mM sodium orthovanadate, and protein levels were quantitated in standard Bradford assays (Bio-Rad, Hercules, Calif.). Equivalent amounts of protein were subjected to heat denaturation at 70°C for 10 min. Samples were run on sodium dodecyl sulfate (SDS)-4 to 15% polyacrylamide gels (Bio-Rad) at 100 V for 1.5 h and transferred to Immobilon-P membranes (Millipore, Bedford, Mass.). E2A was detected by using rabbit polyclonal antibodies which recognize both E12 and E47 (Santa Cruz Biotechnology, Santa Cruz, Calif.) diluted 1:500 in TBST (150 mM NaCl, 50 mM Tris-Cl [pH 8], and 0.05% Tween 20), and Pax-5 was detected by using goat polyclonal antibodies (Santa Cruz Biotechnology) diluted 1:500 in TBST. Rabbit polyclonal antibodies to PI3-K were diluted 1:2,000 in TBST and utilized as a loading control. All proteins were detected by using the appropriate secondary antibodies conjugated to horseradish peroxidase at 1:2,000, followed by ECL detection (Amersham Pharmacia Biotech) and autoradiography.
DNA binding assays.
Nuclear extracts were prepared from bone marrow and splenic B cells by lysing cells in hypotonic buffer (20 mM HEPES [pH 7], 10 mM KCl, 1 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Triton X-100, 20% glycerol, 2 μM phenylmethylsulfonyl fluoride, 5 μg of aprotinin per ml, and 5 μg of leupeptin per ml). Extracts were then homogenized and centrifuged, and the pellets containing the nuclear extracts were dissolved in extraction buffer (hypotonic buffer containing 420 mM NaCl). Protein levels in nuclear and cytoplasmic extracts were quantitated in standard Bradford assays (Bio-Rad). Equivalent levels of protein were diluted fourfold in binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, and 0.01% Triton X-100), and 0.3 μg of 5′-biotinylated target DNA was added. The double-stranded oligonucleotide sequence for the κE2 Ig kappa-chain enhancer sequence 5′-TCGAAAGGCAGGTGGCCCAAGCT-3′ (49). Following a 20-min incubation, μMACS streptavidin-conjugated microbeads (Miltenyi Biotec) were added, and samples were incubated for an additional 15 min. Samples were then put through magnetic columns (Miltenyi Biotec) and washed four times with binding buffer. Bound protein was then eluted with elution buffer (binding buffer containing 1 M NaCl) and half of the samples were run on standard SDS-10% polyacrylamide gels as described above. Western blotting was performed with antibodies to E2A as described above, and identical SDS-polyacrylamide gels were silver stained according to the protocol of the manufacturer (Bio-Rad).
RESULTS
LMP2A transgenic mice and B-lymphocyte development.
We have previously reported that LMP2A transgenic mice demonstrate altered B-cell development, with the majority of bone marrow B cells lacking IgM expression (13). The characteristics of these cells are presented in Fig. 1, along with a schematic representation of the stages and transcription factor regulation of normal B-cell development. As shown in Fig. 1A, IgM-negative B cells from LMP2A transgenic mice are B220+ and CD19+, suggesting that the transition from the early pro-B stage of development is not altered by LMP2A (compare columns 1 and 2). Additionally, the majority of CD19+ B cells from LMP2A transgenic mice are CD43 negative and do not rearrange Ig heavy-chain genes (Fig. 1A) (13), which suggests that LMP2A-mediated alterations in development occur sometime during the pre-B stage when Ig is rearranged and a functional BCR is expressed on the cell surface (Fig. 1B). These IgM-negative B cells, which should normally undergo apoptosis, are able to exit the bone marrow and colonize peripheral lymphoid organs such as the spleen (Fig. 1A, compare columns 3 and 4) (13). These observations indicate that LMP2A manipulates cellular components involved in B-cell signaling and survival during B-cell development.
The coordinated activities of several transcription factors, including E2A, EBF, and Pax-5, are essential for proper B-cell development (Fig. 1B). As shown in Fig. 1B, E2A and EBF are critical for the transition from the pro-B to the pre-B stage of development, while Pax-5 functions later in development at the pre-B stage (for reviews, see references 27, 29, and 61). As mentioned above, the activity of E2A can be blocked by expression of specific inhibitors, such as Id proteins or SCL (Fig. 1B). Based upon the tight regulation of B-cell maturation by specific transcription factors, the developmental defects that we have observed in LMP2A transgenic mice suggest that transcription may be altered in B cells from LMP2A transgenic mice.
DNA microarray analysis of B cells from LMP2A transgenic mice.
In order to identify the molecular basis for the alterations in B-cell development observed in LMP2A transgenic mice, we utilized DNA microarray analysis to directly compare gene transcription in B cells from wild-type and LMP2A transgenic mice. To identify alterations in gene transcription that may be induced by LMP2A during B-cell development, CD19+ bone marrow B cells were selected on methylcellulose containing IL-7 and used for microarray analysis (Fig. 1A, bottom panels). Nonactivated, purified CD19+ splenic B cells from wild-type and LMP2A transgenic mice were also utilized to identify altered transcription maintained by LMP2A in peripheral B cells (Fig. 1A, bottom panels). Identical microarray experiments were performed in duplicate, and transcripts exhibiting an average change in expression of twofold or greater were considered significant. Sequences for over 12,000 genes were contained in the microarrays. In bone marrow B cells from LMP2A transgenic mice, 117 of these transcripts were up-regulated twofold or more compared to those in cells from wild-type mice, and 54 of these transcripts were designated expressed sequence tags (ESTs) or sequences without designation in the genome database. A total of 711 transcripts were down-regulated in LMP2A-expressing bone marrow B cells, and 375 of these were ESTs. In splenic B cells, 285 genes were up-regulated (117 ESTs) and 67 transcripts were down-regulated (20 ESTs). Although expression of many genes was altered by LMP2A, we chose to focus our studies on B-cell-specific transcription factors and other genes important for B-cell development in order to identify the specific transcriptional targets of LMP2A that produce the phenotype we observe in transgenic mice.
Many genes induced during B-cell development and necessary for appropriate maturation were affected by LMP2A expression. The transcription factors E2A, EBF, and Pax-5 were each down-regulated from 2.1- to 3.5-fold in bone marrow and splenic B cells from LMP2A transgenic mice. These results are shown in Table 1, with the average fold change from two identical experiments and standard deviation indicated. Interestingly, expression of two inhibitors of E2A, Id2 and SCL, was up-regulated 3.9- and 2.3-fold, respectively, in splenic B cells. Many other B-cell-specific genes and components of the pre-BCR complex were down-regulated to various degrees, indicating that the activity of their regulating transcription factors is also decreased. As mentioned above, Ig-α (mb-1), Ig-β (B29), RAG-1, RAG-2, Vpre-B, λ5, and TdT are all induced during the early stages of B-cell development, and their expression depends on coordinated activity of E2A, EBF, and Pax-5 (for reviews, see references 27, 29, and 61). In agreement with this, expression of most of these B-cell-specific genes was more strongly down-regulated in bone marrow B cells than in splenic B cells from LMP2A transgenic mice (Table 1). Interestingly, expression of Ig-β (B29) was not significantly affected by LMP2A expression; however, this has also been demonstrated in developing B cells from mice deficient in E2A and suggests an additional regulatory mechanism for Ig-β (B29) expression (55). The TdT gene, which is induced by E2A and involved in Ig recombination, was down-regulated 3.6-fold in bone marrow B cells. Additionally, expression of the Ig J chain, which is normally repressed by Pax-5, was significantly enhanced 4.3-fold in bone marrow and 6.7-fold in splenic B cells expressing LMP2A, further indicating that Pax-5 activity is repressed.
TABLE 1.
Expression of B-cell-specific genes in DNA microarrays
| Gene | Fold changea
|
|
|---|---|---|
| BM | Spleen | |
| Transcription factor genes | ||
| E2A | −2.2 ± 0.2 | −2.3 ± 0.2 |
| EBF | −3.5 ± 1.0 | −2.2 ± 0.3 |
| Pax-5 | −2.1 ± 0.2 | −2.6 ± 0.3 |
| Id2 | 1.7 ± 0.2 | 3.9 ± 0.1 |
| SCL | 1.4 ± 0.3 | 2.3 ± 0.1 |
| PU.1 | −3 ± 0.1 | −1.5 ± 0.3 |
| Spi-B | −1.7 ± 0.2 | −2.3 ± 0.2 |
| LEF-1 | −3.2 ± 0.2 | −1.5 ± 0.2 |
| TCF-1 | 2.6 ± 0.3 | 1.2 ± 0.6 |
| B-cell-specific gene | ||
| Ig-α (mb-1) | −2.1 ± 0.1 | −2.2 ± 0.2 |
| Ig-β (B29) | −1.5 ± 0.3 | −1.6 ± 0.2 |
| RAG-1 | −1.9 ± 0.3 | 1.8 ± 0.2 |
| RAG-2 | −2.8 ± 0.3 | 1.6 ± 0.5 |
| Vpre-B | −2.6 ± 0.2 | −2.4 ± 0.1 |
| Lambda 5 | −2.7 ± 0.5 | −1.3 ± 0.3 |
| TdT | −3.6 ± 0.2 | −1 ± 0.8 |
| J chain | 4.3 ± 0.5 | 6.7 ± 2.1 |
Identical microarray experiments were performed in duplicate with CD19+ bone marrow (BM) and splenic B cells from wild-type and LMP2A transgenic mice. The average fold change in gene expression in cells from LMP2A transgenic mice versus wild-type mice from the two experiments is shown, along with the standard deviation. Negative values indicate decreased expression.
Interestingly, expression of other transcription factors that cooperate with E2A, EBF, and Pax-5 during B-cell development was affected by LMP2A expression. Two members of the Ets family of transcription factors, PU.1 and Spi-B, were down-regulated to various degrees (Table 1). In developing B cells, PU.1 modulates the expression of several B-cell-specific genes, such as those for Ig heavy and light chains, Ig-α (mb-1), Ig-β (B29), Vpre-B, kappa and lambda light chains, Btk, TdT, CD19, and J chain (for reviews, see references 8, 29, 53, and 61). Spi-B, another Ets family member that is closely related to PU.1 in its binding specificity, transcriptional activity, and expression, has been shown to be important for cell proliferation following BCR stimulation (for reviews, see references 8 and 27). In our microarray experiments, PU.1 demonstrated a more significant down-regulation (3-fold) in the bone marrow, while Spi-B was down-regulated 2.3-fold in splenic B cells from LMP2A transgenic mice. One possible explanation for this is that PU.1 functions early during B-cell development and Spi-B is believed to be important for activation of mature B cells.
LEF-1/T-cell factor-1 (TCF-1) factors activate gene transcription during lymphoid development in response to Wnt signaling. LEF-1, which is normally induced by Pax-5, is down-regulated 3.2-fold in bone marrow B cells from LMP2A transgenic mice (Table 1). LEF-1 has been implicated in B-cell development, while the related TCF protein (up-regulated 2.6-fold) is involved in thymocyte development (for reviews, see references 36, 61, 72, and 73). This further indicates that the transcription factor environment in bone marrow cells from LMP2A transgenic mice is not favorable for proper B-cell development. These results, taken together, indicate that the transcription factors critical for proper B-cell-specific transcription, Ig rearrangement, and transition through the pre-B-cell stage of development are down-regulated in LMP2A-expressing B cells.
Down-regulation of transcription factor expression in B cells from LMP2A transgenic mice.
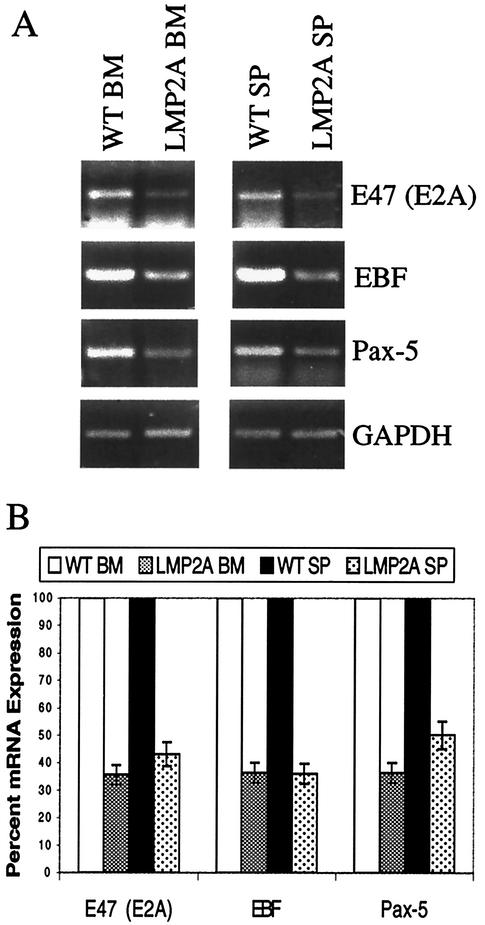
In order to verify that expression of E2A, EBF, and Pax-5 was indeed down-regulated in bone marrow and splenic B cells from LMP2A transgenic mice, semiquantitative RT-PCR was performed (Fig. 2A). Primers specific for E47 were utilized as an indication of E2A expression, since this E protein is predominantly expressed in B cells (66). Experiments were repeated three times, and representative gels are shown (Fig. 2A). The levels of each RT-PCR product were normalized to those of a control transcript, GAPDH, by dividing the intensities of the PCR products by the fraction of GAPDH product present. The mRNA expression in wild-type bone marrow and splenic B cells was then set to a value of 100%, and the relative percentage of E2A, EBF, and Pax-5 expression in B cells from LMP2A transgenic mice is shown graphically (Fig. 2B). In agreement with the microarray data, E2A, EBF, and Pax-5 transcripts were each reduced by two- to threefold in LMP2A-expressing splenic and bone marrow B cells from LMP2A transgenic mice.
FIG. 2.
Decreased expression of B-cell developmental transcription factors in B cells from LMP2A transgenic mice. (A) Semiquantitative RT-PCR was performed with total RNA from CD19+ bone marrow (BM) (left) and splenic (SP) (right) B cells from wild-type (WT) and LMP2A transgenic mice. Primers specific for E47 (E2A), EBF, and Pax-5 were utilized, along with GAPDH as a control for RNA levels. Each experiment was repeated three times, and a representative gel is shown. (B) The levels of expression of E47, EBF, and Pax-5 were normalized to the levels of GAPDH, and the amount of expression in wild-type B cells was set to 100%. Error bars indicate standard deviations.
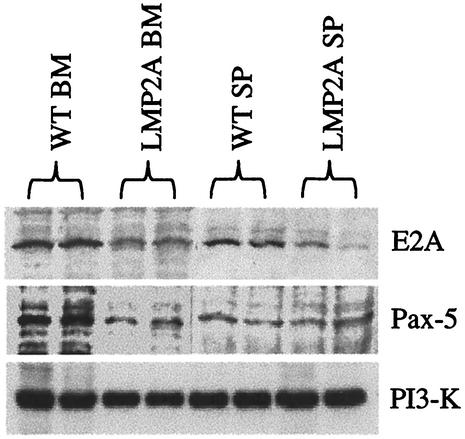
The protein levels of E2A and Pax-5 were also determined by Western blot analysis (Fig. 3). EBF levels were not measured due to the lack of a commercially available antibody against this protein. Experiments were repeated three times, and representative gels are shown. As shown in Fig. 3, E2A protein levels were reduced in bone marrow and splenic B cells from LMP2A transgenic mice compared to cells from wild-type littermates. In contrast to the RT-PCR results, Pax-5 levels were decreased in bone marrow B cells, but not in splenic B cells, from LMP2A transgenic mice. This is most likely because this B-cell-specific factor functions primarily during the B-cell commitment stage and is not highly expressed once B cells exit the bone marrow (51). It is possible that RT-PCR is much more sensitive than Western blotting and may detect much smaller differences in expression. Alternatively, Pax-5 expression may be regulated posttranscriptionally in splenic B cells, which would allow for detection of differences in mRNA by RT-PCR but not for detection of differences in protein by Western blotting. E2A is induced early during B-cell development and also in mature B cells upon BCR ligation and activation, which is consistent with our observations of decreased E2A expression mediated by LMP2A in both bone marrow and splenic B cells (59).
FIG. 3.
Decreased E2A and Pax-5 protein expression in B cells from LMP2A transgenic mice. Whole-cell lysates from CD19+ bone marrow (BM) and splenic (SP) B cells from wild-type (WT) and LMP2A transgenic mice were utilized for Western blot analysis. Each experiment was repeated three times, and a representative gel is shown. Antibodies were utilized to detect total E2A and Pax-5 protein levels. Antibodies against PI3-K were utilized as a control to demonstrate equal protein levels in each lane.
Increased expression of E2A inhibitors in B cells from LMP2A transgenic mice.
Expression of the E2A inhibitors Id2 and SCL was also verified by semiquantitative RT-PCR analysis (Fig. 4A). Experiments were repeated three times, and representative gels are shown. Again, the mRNA expression in wild-type B cells was set to a value of 100%, and the relative expression of each transcript in LMP2A-expressing B cells is shown graphically (Fig. 4B). In agreement with the microarray analysis, expression of Id2 and SCL was up-regulated in splenic B cells, but not in bone marrow B cells, from LMP2A transgenic mice. Id2 expression was more significantly up-regulated (four- to fivefold) than SCL expression (twofold). These results suggest that two mechanisms of E2A inhibition by LMP2A may exist, depending on the developmental stage of the cells. One mechanism may involve reducing E2A expression as demonstrated by decreased mRNA levels in bone marrow and splenic B cells. Another likely involves inhibition of E2A activity by up-regulation of Id2 and/or SCL expression as demonstrated in splenic B cells from LMP2A transgenic mice.
FIG. 4.
Increased expression of E2A inhibitors in splenic B cells from LMP2A transgenic mice. (A) Semiquantitative RT-PCR was performed with total RNA from CD19+ bone marrow (BM) (left) and splenic (SP) (right) B cells from wild-type (WT) and LMP2A transgenic mice. Primers specific for Id2 and SCL were utilized, along with GAPDH as a control for RNA levels. Each experiment was repeated three times, and a representative gel is shown. (B) The levels of expression of Id2 and SCL were normalized to the levels of GAPDH, and the amount of expression in wild-type B cells was set to 100%. Error bars indicate standard deviations.
Decreased E2A-specific DNA binding activity in B cells from LMP2A transgenic mice.
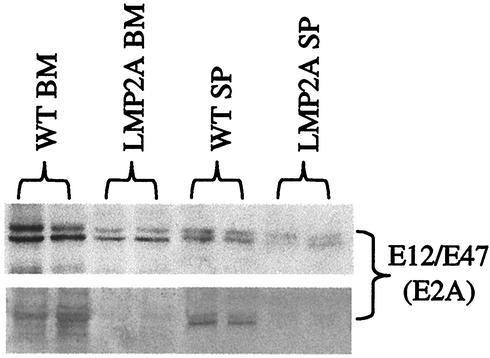
To determine whether reduced expression of E2A in B cells from LMP2A transgenic mice results in decreased E2A-specific DNA binding activity, we measured the amount of E2A bound to a κE2 Ig kappa-chain enhancer sequence (49). DNA-bound E2A was isolated by incubation of nuclear extracts with biotinylated target DNA, addition of streptavidin-coated magnetic beads, and collection on magnetic columns. The amount of E2A eluted from the columns was detected by Western blot analysis (Fig. 5, upper gel). Cytoplasmic extracts were also run as controls to demonstrate that there was no nonspecific DNA binding activity (data not shown). Correlating with the decreased E2A expression levels in bone marrow and splenic B cells from LMP2A transgenic mice, there was also a similar decrease in E2A activity in these cells. The decreased DNA binding activity of E2A in bone marrow and splenic B cells was further verified by silver staining of identical SDS-polyacrylamide gels (Fig. 5, lower gel). Taken together, these results indicate that LMP2A, when expressed during B-cell development, induces down-regulation of the transcription factors critical for proper B-cell development. Decreased expression and activity of E2A results in reduced expression of its downstream target genes, including those encoding EBF and Pax-5. This likely results in the inhibition of Ig heavy-chain gene rearrangement and BCR expression in developing B cells in LMP2A transgenic mice.
FIG. 5.
E2A activity is inhibited in B cells from LMP2A transgenic mice. DNA binding assays were performed with nuclear extracts prepared from bone marrow (BM) and splenic (SP) B cells from wild-type (WT) and LMP2A transgenic mice. Nuclear extracts were examined for binding to a biotinylated κE2 DNA probe in order to detect E2A-specific DNA binding activity. Bound protein was selected by incubating with streptavidin-coated magnetic beads followed by purification on magnetic columns as described in Materials and Methods. Western blotting was then performed with antibodies specific for E2A (upper gel). Duplicate SDS-polyacrylamide gels were run, and DNA-bound E2A was identified by silver staining (lower gel). Each experiment was repeated two times, and a representative gel is shown.
Upstream regulation of E2A function.
Upstream regulation of E2A activity has been best demonstrated in studies involving T cells. Notch signaling in T cells has been shown to block a pathway involving Ras-mediated activation of E2A (54). Although in most systems activation of effectors in the Ras pathway occurs posttranslationally, we have observed changes in gene expression which suggest that this pathway is altered in LMP2A-expressing B cells. As shown in Table 2, several Ras-associated genes were affected by LMP2A expression. In bone marrow B cells cultured in IL-7, several Ras-associated genes were down-regulated over twofold, including PAC-1 ERK/MAPK phosphatase, Rho and Raf effectors, GBP-2 GTPase, Vav and DAB adaptor molecules, SOS-2 guanine nucleotide exchange factor, and MEK and ERK kinases (for a review, see reference 1). This suggests that Ras-mediated activation of E2A may be repressed by LMP2A in developing bone marrow B cells. In these same cells, we observed increased expression of two Notch-associated molecules, Delta and enhancer of split (ESG) (for a review, see reference 56). Additionally, expression of the SEL1L inhibitor of Notch was down-regulated threefold in bone marrow B cells from LMP2A transgenic mice, suggesting that Ras-mediated activation of E2A may be inhibited by Notch signaling.
TABLE 2.
Expression of genes in the Ras and Notch pathways
| Gene | Fold changea
|
|
|---|---|---|
| BM | Spleen | |
| Ras-associated genes | ||
| Ki-Ras | −1.5 ± 0.1 | 2.7 ± 0.2 |
| PAC-1 ERK/MAPK phosphataseb | −2.3 ± 0.1 | −2.2 ± 0.2 |
| Kinase suppressor of Ras (KSR1)b | 2.5 ± 0.3 | −2.5 ± 0.2 |
| Rho A GTPase | −2.8 ± 0.3 | −1.1 ± 0.2 |
| Rho B GTPase | −2.4 ± 0.3 | 2.4 ± 0.2 |
| Raf | −5.2 ± 0.5 | −1.0 ± 0.3 |
| GBP-2 GTPase | −3.3 ± 0.3 | 3.4 ± 0.2 |
| SOS-2 | −2.7 ± 0.3 | 1.3 ± 0.2 |
| Vav | −2.8 ± 0.3 | 1.7 ± 0.2 |
| DAB-2 | −3.2 ± 0.3 | 1.0 ± 0.2 |
| ERK-1 | −2.2 ± 0.2 | −1.2 ± 0.2 |
| ERK-5 | −5.6 ± 0.3 | 1.7 ± 0.5 |
| MEK-5 | −3.3 ± 0.2 | −1.3 ± 0.1 |
| Notch-associated genes | ||
| Delta (Dlk1-like) | 2.7 ± 0.2 | −2.3 ± 0.2 |
| Enhancer of Split (mESG) | 2.2 ± 1.0 | −1.2 ± 0.2 |
| Suppressor of Hairless (RBP-L) | 1.4 ± 0.2 | 2.3 ± 0.3 |
| Sel 11 inhibitor of Notchb | −3.0 ± 0.2 | 4.2 ± 0.1 |
Identical microarray experiments were performed in duplicate with CD19+ bone marrow (BM) and splenic B cells from wild-type and LMP2A transgenic mice. The average fold change in gene expression in cells from LMP2A transgenic mice versus wild-type mice from the two experiments is shown, along with the standard deviation. Negative values indicate decreased expression.
Shown to be a positive or negative regulator, depending on the model system.
T-cell receptor (TCR) signaling through the Ras-ERK-MAPK pathway has been shown to induce up-regulation of Id proteins, which inhibit the activity of E2A (6). As shown in Table 2, alterations in the expression of Ras-associated signaling molecules were observed in splenic B cells from LMP2A transgenic mice. Particularly, expression of the potentially oncogenic Ki-Ras was increased 2.7-fold, while the kinase suppressor of Ras (KSR1) was down-regulated 2.5-fold, in splenic B cells. These results suggest that in peripheral B cells, LMP2A may utilize the Ras pathway to induce Id expression, which results in decreased activity of E2A.
DISCUSSION
By utilizing DNA microarray technology and a transgenic mouse model, we have demonstrated that LMP2A, when expressed in developing B cells, alters the expression of critical transcription factors involved in normal B-cell development. In particular, the transcription factors E2A, EBF, and Pax-5 are each down-regulated two- to threefold in bone marrow and splenic B cells from LMP2A transgenic mice. Additionally, the DNA binding activity of E2A is significantly inhibited in bone marrow and splenic B cells from LMP2A transgenic mice, and expression of two E2A inhibitors, Id2 and SCL, is up-regulated in splenic B cells. Other transcription factors known to play a role in B-cell development by cooperating with these three factors to modulate proper gene expression, such as PU.1 and LEF-1, are also down-regulated in B cells expressing LMP2A. Interestingly, similar changes affecting global gene transcription have been noted in Reed-Sternberg cells from Hodgkin's lymphoma (19, 33, 34, 39). Approximately 40 to 50% of Hodgkin's lymphoma cells are EBV positive and express LMP2A (37, 39). Our research suggests that LMP2A may be responsible for these transcriptional changes and may provide information regarding the importance of LMP2A in Hodgkin's disease. In addition, the ability of LMP2A to repress cellular gene transcription has important implications regarding its role in establishment of viral latency in EBV-infected B lymphocytes.
Inhibition of cellular signaling by LMP2A.
It has been demonstrated that LMP2A blocks the activation of downstream signaling molecules following BCR ligation and may provide constitutive signals to mimic an activated BCR (42, 46, 47). Thus, LMP2A maintains a constant state of viral latency by preventing BCR-mediated induction of lytic EBV replication and subsequent immune recognition (42, 46). When expressed early during B-cell development, before expression of a functional BCR, LMP2A may have a similar inhibitory effect on signaling from the pre-BCR. In our transgenic mouse model, LMP2A expression is driven by the Ig heavy-chain promoter and enhancer and is thus turned on at the pro-B stage of development. The results of this study suggest that LMP2A may then inhibit or bypass necessary signaling through the pre-BCR during development. LMP2A was shown to down-regulate the expression of proteins involved in recombination as well as components of the pre-BCR (Table 1). These genes are normally up-regulated prior to Ig rearrangement and pre-BCR formation and then down-regulated by receptor feedback to prevent further Ig heavy-chain gene rearrangements (for a review, see reference 27). Down-regulated expression of components of the pre-BCR complex by LMP2A may simply inhibit the formation of a functional pre-BCR. Alternatively, LMP2A may somehow allow a bypass of the required pre-BCR signaling in the absence of Ig heavy-chain gene rearrangement and cause a premature reduction in the levels of these transcripts, allowing for progression through the pre-B stage and induction of Ig light-chain rearrangement. Interestingly, induction of light-chain gene rearrangement, which has been shown not to require signaling from the pre-BCR, appears to be normal in LMP2A transgenic mice (13, 27).
Both the BCR and pre-BCR utilize similar signaling pathways to mediate downstream transcriptional activation. Similar to signaling through the BCR, the pre-BCR utilizes signals from molecules such as Syk, Lyn, Btk, and BLNK for progression to the pre-B stage of development and establishment of allelic exclusion (14, 20, 29, 40). We have previously demonstrated the importance of Syk, Btk, and BLNK in mediating the effects of LMP2A on B-cell development and survival in these transgenic mice (23, 43, 44). It is therefore likely that LMP2A utilizes these same signaling molecules not only to interfere with transcription factor regulation during B-cell development but also to inhibit activation in mature B cells.
Although reduced expression of the E2A, EBF, and Pax-5 transcription factors in LMP2A-expressing B cells was modest (two- to threefold), studies have shown that this is a strong enough signal to inhibit B-cell development. B cells from mice heterozygous for both E2A and EBF display a phenotype very similar to that of the mice in our study in that the B cells fail to express IgM (55). Upon B-cell activation, E2A activity has been shown to be necessary for class switch recombination; therefore, it is interesting to speculate that E2A activity may also be affected by LMP2A expression in latently infected mature B cells (59). Id3, when overexpressed, can inhibit E2A-mediated isotype switching (22). Additionally, research indicates that Id2 negatively controls mature B-cell differentiation, which is essential for B cells to become responsive to antigen, by inhibiting E2A activity (9). Inhibition of E2A activity during latency may allow LMP2A to inhibit B-cell activation and subsequent induction of EBV lytic gene expression. Interestingly, E2A heterozygosity has been shown to result in reduced expression of the cell cycle inhibitor p21 and increased pro- and pre-B-cell proliferation (30). This may explain our previous observation of a two- to threefold increase in the colony size of bone marrow B cells from LMP2A transgenic mice compared to those of wild-type mice (12, 13).
LMP2A regulation of E2A activity.
Regulation of E2A activity by upstream signaling molecules during B-cell development is largely unknown. The most well defined pathway has been described for T cells and involves TCR signaling. In this pathway, TCR signaling through the Ras-ERK-MAPK pathway can induce up-regulation of Id proteins, specifically Id3, which inhibit the activity of E2A (6). Notch signaling has also been shown to block a pathway involving Ras-JNK-mediated activation of E2A (54). Both pathways are involved in mediating T-cell activation and/or development and may be employed for E2A regulation in B cells as well.
It appears that LMP2A may regulate E2A activity in at least two distinct ways. E2A was shown to be down-regulated transcriptionally in bone marrow and splenic B cells. Microarray analysis of bone marrow B cells cultured in IL-7 suggests that the Notch signaling pathway is activated in bone marrow B cells, while components of Ras signaling pathways are down-regulated (Table 2). It is possible that these events result in E2A down-regulation in B cells as well as T cells. Recently, Notch activation was shown to contribute to tumor proliferation of Reed-Sternberg cells in Hodgkin's lymphoma; however, that study did not include a correlation between Notch activation and the presence of EBV in these cells (34).
An additional mechanism of E2A inhibition by LMP2A appears to involve increased expression of two inhibitors of E2A, Id2 and SCL, in splenic B cells. Id proteins are normally expressed only in pro-B cells and are absent in more differentiated precursors. Similarly, SCL expression decreases as B cells mature (9, 10, 22). In agreement with this and as shown in Fig. 4, there was virtually no expression of Id2 in wild-type splenic B cells; however, there was abundant expression in splenic B cells from LMP2A transgenic mice. Based upon the microarray results presented in Table 2, it is likely that Id2 is up-regulated in peripheral B cells as a result of Ras activation by LMP2A.
Many studies have demonstrated the importance of Ras signaling during B-cell development. Expression of a dominant-inhibitory mutant of Ha-ras in mouse B-cell precursors results in an almost complete loss of pro-B and pre-B cells in the bone marrow (50). Interestingly, expression of a constitutively activated form of Ras in the RAG-1−/− background results in the production of B cells with a phenotype nearly identical to that of our LMP2A transgenic mice. Activated Ras expression induces the progression of B cells beyond normal developmental checkpoints, allowing B220+ IgM− cells to accumulate in the periphery (65). By utilizing embryonic cells unable to synthesize heavy-chain variable-region genes, this same group has demonstrated that B-cell populations expressing constitutively activated Ras display extensive Ig light-chain rearrangement in the absence of heavy-chain rearrangement, similar to what we have observed in LMP2A transgenic mice (64).
Ras has been shown to stimulate many downstream signaling events following BCR activation, including the activation of PI3-K and Raf/MAPK (for reviews, see references 20 and 40). In splenic B cells, it is possible that LMP2A activates the Ras-ERK-MAPK pathway in the absence of BCR or pre-BCR signaling and this results in increased Id2 expression and subsequent inhibition of E2A activity. Two serine residues (S15 and S102) in the LMP2A amino terminus have been shown to be phosphorylated by the ERK1 MAPK in vitro, although the significance of this has not been elucidated (57). Studies involving LMP2A-expressing HaCaT keratinocytes have demonstrated that LMP2A does not activate the MAPK pathway; however, a recent report indicates that LMP2A may activate the ERK/JNK MAPK pathways in 293 cells (17, 63). A potential cell-type-specific induction of these pathways by LMP2A may account for these differences, as our data indicate that LMP2A may repress Ras activation in developing bone marrow B cells and activate the Ras pathway in peripheral B cells. Experiments involving the identification and characterization of the effects of LMP2A on Ras and Notch signaling pathways in transgenic mice are under way.
Additional B-cell regulatory transcription factors.
Two Ets transcription factors, PU.1 and Spi-B, were down-regulated in LMP2A-expressing B cells. PU.1 was more significantly down-regulated in bone marrow B cells than in splenic B cells, which correlates with its critical role early in B-cell development (for reviews, see references 8 and 53). Spi-B, a factor implicated in the proliferation of mature B cells in response to BCR stimulation, was shown to be down-regulated in splenic B cells from LMP2A transgenic mice (27). Spi-B-deficient mice are defective in BCR-stimulated proliferation, and this effect is further exacerbated in PU.1 heterozygous, Spi-B deficient mice, suggesting that these transcription factors may function synergistically in mature B-cell activation (8, 27). Since peripheral B cells in this transgenic mouse model do not express a functional BCR, reduced Spi-B expression may result simply from LMP2A-mediated alterations in another upstream pathway normally targeted by this viral protein. Alternatively, an LMP2A-mediated reduction in Spi-B expression may result in decreased cellular proliferation in response to immune stimulation through molecules other than the BCR. LEF-1, a Wnt component involved in B-cell development, was down-regulated in bone marrow B cells from LMP2A transgenic mice. In contrast, the related TCF transcription factor, normally involved in thymocyte development, was up-regulated in bone marrow B cells from LMP2A transgenic mice (for reviews, see references 72 and 73). Although the significance of these findings is not clear, it is intriguing that the Wnt pathway appears to be altered by LMP2A expression, since inappropriate activation of TCF target genes has been shown to be a primary event for cellular transformation in colon carcinomas (for reviews, see references 72 and 73).
In summary, we have utilized DNA microarray technology to study changes in gene transcription induced upon LMP2A expression in murine B lymphocytes. The results of this study demonstrate that LMP2A interferes with global transcription factor regulation for proper B-cell development when expressed during B lymphopoiesis. These results indicate that LMP2A may be responsible for the recent observation of global changes in gene expression demonstrated in Hodgkin's Lymphoma (19, 33, 34, 39). In particular, this may allow LMP2A to promote the persistence of cells that lack a functional BCR, which is a common feature of Reed-Sternberg cells (37, 39). Future studies that address the mechanisms by which LMP2A-induced changes in transcription factor gene expression promote altered development and survival of B cells may provide insight into how LMP2A maintains latency in EBV-infected B lymphocytes.
Acknowledgments
R.L. is supported by Public Health Service grants CA62234, CA73507, and CA93444 from the National Cancer Institute and DE13127 from the National Institute of Dental and Craniofacial Research and is a Stohlman Scholar of the Leukemia and Lymphoma Society of America. T.P. is a Special Fellow of the Leukemia and Lymphoma Society of America.
We thank members of the Longnecker laboratory for help with these studies. In addition, we thank Michelle Swanson-Mungerson, Patrick Dennis, and Rebecca Katzman for helpful discussions and for kindly reading the manuscript prior to submission.
We also thank Eric Bremer and his staff at CMIER for use of their Affymetrix scanner and fluidics station and for performing microarray hybridizations and data analysis for us.
REFERENCES
- 1.Adjei, A. A. 2001. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 93:1062-1074. [DOI] [PubMed] [Google Scholar]
- 2.Akao, I., Y. Sato, K. Mukai, H. Uhara, S. Furuya, T. Hoshikawa, Y. Shimosato, and I. Takeyama. 1991. Detection of Epstein-Barr virus DNA in formalin-fixed paraffin-embedded tissue of nasopharyngeal carcinoma using polymerase chain reaction and in situ hybridization. Laryngoscope 101:279-283. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, F. E., C. P. Daniel, A. A. Armstrong, D. A. Clark, D. E. Onions, R. A. Cartwright, and R. F. Jarrett. 1995. Case clustering, Epstein-Barr virus Reed-Sternberg cell status and herpes virus serology in Hodgkin's disease: results of a case-control study. Eur. J. Cancer 9:1479-1486. [DOI] [PubMed] [Google Scholar]
- 4.Ambinder, R. F. 2001. Epstein-Barr virus associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1209-1216. [DOI] [PubMed] [Google Scholar]
- 5.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 6.Bain, G., C. B. Cravatt, C. Loomans, J. Alberola-Ila, S. M. Hedrick, and C. Murre. 2001. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2:165-171. [DOI] [PubMed] [Google Scholar]
- 7.Bain, G., E. C. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, and M. van Roon. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 8.Bartel, F. O., T. Higuchi, and D. D. Spyropoulos. 2000. Mouse models in the study of the Ets family of transcription factors. Oncogene 19:6443-6454. [DOI] [PubMed] [Google Scholar]
- 9.Becker-Herman, S., F. Lantner, and I. Shachar. 2002. Id2 negatively regulates B cell differentiation in the spleen. J. Immunol. 168:5507-5513. [DOI] [PubMed] [Google Scholar]
- 10.Begley, C. G., and A. R. Green. 1999. The SCL gene: from case report to critical hematopoietic regulator. Blood 93:2760-2770. [PubMed] [Google Scholar]
- 11.Burkhardt, A. L., J. B. Bolen, E. Kieff, and R. Longnecker. 1992. An Epstein-Barr virus transformation-associated membrane protein interacts with Src family tyrosine kinases. J. Virol. 66:5161-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldwell, R. G., R. C. Brown, and R. Longnecker. 2000. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EμLMP2A transgenic mice. J. Virol. 74:1101-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, M. A., and B. M. Sefton. 1992. Association between B-lymphocyte membrane immunoglobulin and multiple members of the Src family of protein tyrosine kinases. Mol. Cell. Biol. 12:2315-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, J. K., W. Y. Tsang, C. S. Ng, C. S. Wong, and E. S. Lo. 1995. A study of the association of Epstein-Barr virus with Burkitt's lymphoma occurring in a Chinese population. Histopathology 26:239-245. [DOI] [PubMed] [Google Scholar]
- 16.Chen, F., J. Z. Zou, and L. di Renzo. 1995. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J. Virol. 69:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, S.-Y., J. Lu, Y.-C. Shih, and C.-H. Tsai. 2002. Epstein-Barr virus latent membrane protein 2A regulates c-Jun protein through extracellular signal-regulated kinase. J. Virol. 76:9556-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, J. I. 2000. Epstein-Barr virus infection. N. Engl. J. Med. 343:481-492. [DOI] [PubMed] [Google Scholar]
- 19.Cossman, J., C. M. Annunziata, S. Barash, L. Staudt, P. Dillon, W.-W. He, P. Ricciardi-Castagnoli, C. A. Rosen, and K. C. Carter. 1999. Reed-Sternberg cell genome expression supports a B-cell lineage. Blood 94:411-416. [PubMed] [Google Scholar]
- 20.DeFranco, A. L. 1997. The complexity of signaling pathways activated by the BCR. Curr. Opin. Immunol. 9:296-308. [DOI] [PubMed] [Google Scholar]
- 21.Dykstra, M. L., R. Longnecker, and S. K. Pierce. 2001. Epstein-Barr virus coopts lipid rafts to block the signaling and antigen transport functions of the BCR. Immunity 14:57-67. [DOI] [PubMed] [Google Scholar]
- 22.Engel, I., and C. Murre. 2001. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 23.Engels, N., M. Merchant, R. Pappu, A. C. Chan, R. Longnecker, and J. Wienands. 2001. Epstein-Barr virus latent membrane protein 2a (LMP2A) employs the SLP-65 signaling module. J. Exp. Med. 194:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Era, T., M. Ogawa, S. Nishikawa, M. Okamoto, T. Honjo, K. Akagi, J. Miyazaki, and K. Yamamura. 1991. Differentiation of growth signal requirement of B lymphocyte precursor is directed by expression of immunoglobulin. EMBO J. 10:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fruehling, S., and R. Longnecker. 1997. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 235:241-251. [DOI] [PubMed] [Google Scholar]
- 26.Fruehling, S., R. Swart, K. M. Dolwick, E. Kremmer, and R. Longnecker. 1998. Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J. Virol. 72:7796-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glimcher, L. H., and H. Singh. 1999. Transcription factors in lymphocyte development—T and B cells get together. Cell 96:13-23. [DOI] [PubMed] [Google Scholar]
- 28.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, A., and K. Calame. 1998. Transcriptional regulation during B cell development. Annu. Rev. Immunol. 16:163-200. [DOI] [PubMed] [Google Scholar]
- 30.Herblot, S., P. D. Aplan, and T. Hoang. 2002. Gradient of E2A activity in B-cell development. Mol. Cell. Biol. 22:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horcher, M., A. Souabni, and M. Busslinger. 2001. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity 14:779-790. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, H. L., I. Wadman, J. T. Tsan, and R. Baer. 1994. Positive and negative transcriptional control by the TAL1 helix-loop-helix protein. Proc. Natl. Acad. Sci. USA 91:5947-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jundt, F., K. Kley, I. Anagnostopoulos, K. S. Probsting, A. Greiner, S. Mathas, C. Scheidereit, T. Wirth, H. Stein, and B. Dorken. 2002. Loss of PU.1 expression is associated with defective immunoglobulin transcription in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease. Blood 99:3060-3062. [DOI] [PubMed] [Google Scholar]
- 34.Jundt, F., I. Anagnostopoulos, R. Forster, S. Mathas, H. Stein, and B. Dorken. 2002. Activated Notch 1 signaling promotes tumor cell proliferation and survival in Hodgkin's and anaplastic large cell lymphoma. Blood 99:3398-3403. [DOI] [PubMed] [Google Scholar]
- 35.Kee, B. L., and C. Murre. 1998. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 188:699-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kee, B. L., and C. Murre. 2001. Transcription factor regulation of B lineage commitment. Curr. Opin. Immunol. 13:180-185. [DOI] [PubMed] [Google Scholar]
- 37.Khan, G., and P. J. Coates. 1994. The role of Epstein-Barr virus in the pathogenesis of Hodgkin's disease. J. Pathol. 174:141-149. [DOI] [PubMed] [Google Scholar]
- 38.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 1109-1162. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, Lippincott-Raven Publishers, Philadelphia, Pa.
- 39.Kuppers, R., I. Schwering, A. Brauninger, K. Rajewsky, and M.-L. Hansmann. 2002. Biology of Hodgkin's lymphoma. Ann. Oncol. 13:11-18. [DOI] [PubMed] [Google Scholar]
- 40.Kurosaki, T. 1999. Genetic analysis of B cell antigen receptor signaling. Annu. Rev. Immunol. 17:555-592. [DOI] [PubMed] [Google Scholar]
- 41.Kurth, J., T. Spieker, J. Wustrow, G. J. Strickler, L. M. Hansmann, K. Rajewsky, and R. Kuppers. 2000. EBV-infected B cells in infectious mononucleosis: viral strategies for spreading in the B cell compartment and establishing latency. Immunity 13:485-495. [DOI] [PubMed] [Google Scholar]
- 42.Longnecker, R. 2000. Epstein-Barr virus latency: LMP2, a regulator or means for Epstein-Barr virus persistence? Adv. Cancer Res. 79:175-200. [DOI] [PubMed] [Google Scholar]
- 43.Merchant, M., R. G. Caldwell, and R. Longnecker. 2000. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 74:9115-9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merchant, M., and R. Longnecker. 2001. LMP2A survival and developmental signals are transmitted through Btk-dependent and Btk-independent pathways. Virology 291:46-54. [DOI] [PubMed] [Google Scholar]
- 45.Miller, C. L., A. L. Burkhardt, J. H. Lee, B. Stealey, R. Longnecker, J. B. Bolen, and E. Kieff. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155-166. [DOI] [PubMed] [Google Scholar]
- 46.Miller, C. L., J. H. Lee, E. Kieff, and R. Longnecker. 1994. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. USA 91:772-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, C. L., R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J. Virol. 67:3087-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 50.Nagaoka, H., Y. Takahashi, R. Hayashi, T. Nakamura, K. Ishii, J. Matsuda, A. Ogura, Y. Shirakata, H. Karasuyama, T. Sudo, S.-I. Nishikawa, T. Tsubata, T. Mizuochi, T. Asano, H. Sakano, and T. Takemori. 2000. Ras mediates effector pathways responsible for pre-B cell survival, which is essential for the developmental progression to the late pre-B cell stage. J. Exp. Med. 192:171-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nutt, S. L., B. Heavey, A. G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556-562. [DOI] [PubMed] [Google Scholar]
- 52.Nutt, S. L., P. Urbanek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476-491. [DOI] [PubMed] [Google Scholar]
- 53.Oikawa, T., T. Yamada, F. Kihara-Negishi, H. Yamamoto, N. Kondoh, Y. Hitomi, and Y. Hashimoto. 1999. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Diff. 6:599-608. [DOI] [PubMed] [Google Scholar]
- 54.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 56.Osborne, B., and L. Miele. 1999. Notch and the immune system. Immunity 11:653-663. [DOI] [PubMed] [Google Scholar]
- 57.Panousis, C. G., and D. T. Rowe. 1997. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J. Virol. 71:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu, L., and D. Rowe. 1992. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J. Virol. 66:3715-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quong, M. W., D. P. Harris, S. L. Swain, and C. Murre. 1999. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 18:6307-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 61.Rothenberg, E. V., J. C. Telfer, and M. K. Anderson. 1999. Transcriptional regulation of lymphocyte lineage commitment. BioEssays 21:726-742. [DOI] [PubMed] [Google Scholar]
- 62.Schaniel, C., M. Gottar, E. Roosnek, F. Melchers, and A. G. Rolink. 2002. Extensive in vivo self-renewal, long-term reconstitution capacity, and hematopoietic multipotency of Pax5-deficient precursor B-cell clones. Blood 99:2760-2766. [DOI] [PubMed] [Google Scholar]
- 63.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaw, A. C., W. Swat, L. Davidson, and F. W. Alt. 1999. Induction of Ig light chain gene rearrangement in heavy chain-deficient B cells by activated Ras. Proc. Natl. Acad. Sci. USA 96:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaw, A. C., W. Swat, R. Ferrini, L. Davidson, and F. W. Alt. 1999. Activated Ras signals developmental progression of recombinase-activating gene (RAG)-deficient pro-B lymphocytes. J. Exp. Med. 189:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen, C.-P., and T. Kadesch. 1995. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 15:4518-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 68.Sun, X.-H. 1994. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell 79:893-900. [DOI] [PubMed] [Google Scholar]
- 69.Sun, X. H., N. G. Copeland, N. A. Jenkins, and D. Baltimore. 1991. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 11:6185-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nature Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 71.Tierney, R. J., N. Steven, L. S. Young, and A. B. Rickinson. 1994. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection in the carrier state. J. Virol. 68:7374-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van de Wetering, M., W. de Lau, and H. Clevers. 2002. WNT signaling and lymphocyte development. Cell 109:S13-S19. [DOI] [PubMed] [Google Scholar]
- 73.van Noort, M., and H. Clevers. 2002. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev. Biol. 244:1-8. [DOI] [PubMed] [Google Scholar]
- 74.Young, L. S., and M. Rowe. 1992. Epstein-Barr virus, lymphomas and Hodgkin's disease. Semin. Cancer Biol. 3:273-284. [PubMed] [Google Scholar]
- 75.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]