Abstract
VP26 is the smallest capsid protein and decorates the outer surface of the capsid shell of herpes simplex virus. It is located on the hexons at equimolar amounts with VP5. Its small size (112 amino acids) and high copy number make it an attractive molecule to use as a probe to investigate the complex pattern of capsid protein interactions. An in vitro capsid binding assay and a green fluorescent protein (GFP) localization assay were used to identify VP26 residues important for its interaction with capsids. To test for regions of VP26 that may be essential for binding to capsids, three small in-frame deletion mutations were generated in VP26, Δ18-25, Δ54-60, and Δ93-100. Their designations refer to the amino acids deleted by the mutation. The mutation at the C terminus of the molecule, which encompasses a region of highly conserved residues, abolished binding to the capsid and the localization of GFP to the nucleus in characteristic large puncta. Additional mutations revealed that a region of VP26 spanning from residue 50 to 112 was sufficient for the localization of the fused protein (VP26-GFP) to the nucleus and for it to bind to capsids. Using site-directed mutagenesis of conserved residues in VP26, two key residues for protein-protein interaction, F79 and G93, were identified as judged by the localization of GFP to nuclear puncta. When these mutations were analyzed in the capsid binding assay, they were also found to eliminate binding of VP26 to the capsid structure. Surprisingly, additional mutations that affected the ability of VP26 to bind to capsids in vitro were uncovered. Mutations at residues A58 and L64 resulted in a reduced ability of VP26 to bind to capsids. Mutation of the hydrophobic residues M78 and A80, which are adjacent to the hydrophobic residue F79, abolished VP26 capsid binding. In addition, the block of conserved amino acids in the carboxy end of the molecule had the most profound effect on the ability of VP26 to interact with capsids. Mutation of amino acid G93, L94, R95, R96, or T97 resulted in a greatly diminished ability of VP26 to bind capsids. Yet, all of these residues other than G93 were able to efficiently translocate or concentrate GFP into the nucleus, giving rise to the punctate fluorescence. Thus, the interaction of VP26 with the capsid appears to occur through at least two separate mechanisms. The initial interaction of VP26 and VP5 may occur in the cytoplasm or when VP5 is localized in the nucleus. Residues F79 and G93 are important for this bi-molecular interaction, resulting in the accumulation of VP26 in the nucleus in concentrated foci. Subsequent to this association, additional amino acids of VP26, including those in the C-terminal conserved domain, are important for interaction of VP26 with the three-dimensional capsid structure.
The herpes simplex virus type 1 (HSV-1) genome is enclosed in an icosahedral proteinaceous capsid (36). This structure is assembled in the nucleus, resulting in the synthesis of three sedimentable particles: A, B, and C capsids (15). They differ in protein and DNA composition and in their eventual fate in the infected cell. B capsids contain internal scaffold proteins (proteins 22a and 21), the viral protease (VP24), and the capsid shell proteins (VP5, VP19C, VP23, and VP26). For C capsids, genomic DNA replaces the scaffold proteins. A capsids are devoid of any internal composition; that is, they are empty capsids (reviewed in references 23, 26 and 29). VP5 is the largest capsid protein and the major component of the capsid shell, forming the hexons and pentons of the capsid (19, 32, 35). VP26 is the smallest capsid protein (12 kDa) and decorates the capsid shell by virtue of its location on the hexons of the capsid structure (2, 33, 38, 39).
VP26 is encoded by the UL35 open reading frame (ORF) (9, 17). The UL35 gene product is expressed late in the infectious cycle after the onset of DNA replication and has been shown to be present in multiple phosphorylated forms (18). It localizes to the infected cell nucleus in a punctate pattern (18). Rixon and colleagues demonstrated that VP26 localizes to the nucleus of transfected cells only under conditions when VP5 was also nuclear, that is, in the presence of 22a or VP19C, both of which can transport VP5 to the nucleus (25). They concluded that since the distribution of VP26 was similar to that of VP5 in the nucleus, this indicated VP26 directly physically associates with VP5 (25). Because it is found only on hexons and not on pentons it has been calculated that there should be 900 copies of this protein per capsid (2, 33, 37-39). Although it interacts with VP5, it is not required for capsid formation in a baculovirus expression system (30, 31). VP26 is also dispensable for replication in cell culture. Capsid assembly and the composition of the virions synthesized appear to be unaffected by the absence of VP26, although infectious virus yield is decreased by twofold relative to the wild type (13). Therefore, VP26 is the only HSV capsid protein that is not required for replication in cell culture.
Capsid assembly precursors are formed in the cytoplasm of infected cells. Complexes composed of VP5, the scaffold protein, and VP26 assemble together and are translocated to the nucleus (20, 25). Similarly, complexes of the triplex proteins VP23 and VP19C form and then move to the nucleus. In the nucleus these two multiprotein complexes accumulate and participate in capsid assembly. The first complete structure that assembles is the procapsid, which unlike the mature capsid is more spherical and contains large spaces between the capsomeres (20, 24, 34). Capsid maturation occurs through a series of ill-defined steps, which result in the angularization of the structure and closure of the holes in the capsid shell. One observation, which relates to VP26 acquisition onto the capsid structure, is the absence of VP26 on the procapsid (21, 28). However, it is present in angular icosahedral capsids and in the open shells formed in the absence of the scaffold protein (11). The acquisition of VP26 onto the capsid structure appears to require energy because depletion of ATP eliminates VP26 localization to the nucleus (6).
The goal of the present study was to identify the residues of VP26 required for interaction with the capsid shell. Since VP26 is a small protein, it provides an excellent tool to identify specific residues involved in capsid protein interactions. Two assays were developed to carry out this goal. The first was an in vitro capsid binding protocol, and the second was a green fluorescent protein (GFP) localization assay. Using these assays and mutations that targeted the conserved residues of VP26 we identified a complex interactive interplay between this small protein and the capsid structure.
MATERIALS AND METHODS
Cells and viruses.
Vero and RPE-1 cells were grown in alpha minimum essential medium supplemented with 10% fetal calf serum (Gibco-Invitrogen) and passaged as described by Desai et al. (13). Virus stocks of KOS (HSV-1) and mutant viruses were prepared as previously described (13).
Antibodies.
Antisera (αVP26/C) to the UL35 ORF were generated in rabbits (Research Genetics). The peptide used for the antigen was a C-terminal peptide spanning from residue 95 to 112 of VP26 (13).
Plasmids.
The VP26 ORF was derived using PCR assays and cloned into the EcoRI and BamHI sites of pGEM3Z and pEGFP-C2. Sequence analysis of the KOS UL35 gene revealed no differences at the nucleotide level with that of the published strain 17 sequence (16). The in-frame deletion mutations were generated by inverse-PCR protocols as described by Fisher and Pei (14). The template used for the inverse PCR was the pGEM3Z clone containing the UL35 gene. The forward and reverse primers used for Δ17-26 were 5′ GGGAGATCTGACGCTTATCGGTGGTAACGGT and 5′ CCCAGATCTGTCTTGGCCACCAATAACTCT, those for Δ53-61 were 5′ GGGAGATCTAAACTCCCCGCACGGCCCCTTG and 5′ CCCAGATCTCTGACGGACCTTGGTCTGGCC, and those for Δ92-101 were 5′ GGGAGATCTAAACGCGGGCCGCAACCAGGC and 5′ CCCAGATCTTTTGTCGTTCGAGAACCTTCG, respectively. Both of the primer pairs contained a BglII restriction site at the ends (underlined). PCR extension was in opposite directions so that the entire plasmid was synthesized. PCR assays were performed with a high-fidelity polymerase, Pfu turbo (Stratagene). The PCR product was subjected to digestion with DpnI to remove the original plasmid template and also with BglII to generate sticky ends. The DNA was then ligated at room temperature (RT) for 2 h and transformed into Escherichia coli (strain DH5α). DNA fragments were isolated from the plasmids that were shown by restriction digest to contain the desired mutation and cloned back into pGEM3Z plasmid cut with EcoRI and BamHI. Plasmid DNA was isolated, and the introduction of the mutation as well as the authentic synthesis of the rest of the gene by the polymerase was confirmed by DNA sequencing. N- and C-terminal truncation mutations were generated by PCR using Pfu turbo. The primers used to create VP26 mutant spanning residues 1 to 66 were 5′ GGAATTCATGGCCGTCCCGCAATTTCAC (forward) and 5′ CGGGATCCAGATCTCTACAGACCAAGGTCCGTCAGCGC (reverse), and the primers used to create the VP26 mutant spanning residues 50 to 112 were 5′ GGAATTCGTGCGGGAGTTTCTCCGCGGT (forward) and 5′ CGGGATCCAGATCTCTATCACGGGGTCCCGGGCGTCGA (reverse). The primers specified EcoRI (in italics) or BamHI (underlined) restriction enzyme sites. The template for the PCR assays was pEGFP-C2-UL35. The PCR fragments were purified, digested with EcoRI and BamHI, and cloned into the same sites of pEGFP-C2. The constructs were sequenced to confirm authentic amplification of the gene. The QuikChange (Stratagene) PCR mutagenesis protocol was used to create site-directed mutations. This method involves the use of complementary primers containing the mutation. Primers containing 15 nucleotides on either side of the mutation were made. The template for the QuikChange PCR was pEGFP-C2 containing the UL35 (VP26) ORF. Following PCRs according to the manufacturer's protocol, the DNA was digested with DpnI and transformed into E. coli. Positive clones were isolated, and the introduction of the mutation was confirmed by sequence analysis. All of the site-directed mutations resulted in the substitution of alanine for the wild-type residue except for amino acids at positions 58 and 80, which were alanine, and these were changed to valine. Mutations were transferred into pGEM3Z using the EcoRI and BamHI restriction enzyme sites.
Transient transfections.
RPE-1 cells (1.7 × 105 cells/well) in a 12-well tissue culture tray were transfected with 0.5 μg of the GFP plasmid DNA. The cells were transfected using Fugene-6 reagent according to the manufacturer's protocol (Roche). Twenty-four hours following transfection, the cells were superinfected with KΔ26Z at a multiplicity of infection (MOI) of 10 PFU per cell. Cells were visualized in an inverted fluorescence microscope 16 h after infection.
Sedimentation analysis of capsids.
Sedimentation analysis of capsids from infected cells was performed as described by Desai et al. (10) and Person and Desai (22). All gradients were made using a BioComp Gradient Mate. Generally lysates for sedimentation were prepared from cells (107) in 100-mm-diameter dishes. Infected cells were harvested by scraping into phosphate-buffered saline (PBS), pelleted, washed once in PBS, and repelleted. The cell pellet was resuspended in 2× CLB (capsid lysis buffer consisting of 2% Triton X-100, 2 M NaCl, 10 mM Tris [pH 7.5] and 2 mM EDTA), and the lysate was sonicated prior to sedimentation. This procedure was used to derive the total capsid population from infected cells. Capsids for use in binding assays were harvested using a fraction collector and pelleted by centrifugation in a Beckman SW41 rotor at 30,000 rpm for 60 min. The capsid pellets were resuspended in PBS at 4°C overnight.
In vitro protein translation and capsid binding.
Capsids that lack VP26 were derived from Vero cells infected with the VP26-null mutant, KΔ26Z. Typically, Vero cells in 100-mm-diameter dishes (4 × 107 cells) were infected with KΔ26Z at an MOI of 10 PFU/cell. One-fourth of the infected cells (107 cells in a 100-mm-diameter dish) were metabolically labeled with [35S]methionine from 8 to 24 h postinfection. The infected cells were harvested 24 h following infection and pooled together, and capsids were isolated following lysis of the cells and sedimentation in sucrose gradients (see below). All three capsid types (A, B, and C) derived from the infected cells were used for the capsid binding assay. Typically, four reactions were performed with capsids isolated in this manner. In vitro synthesis of [35S]methionine-labeled VP26 and mutants of VP26 were carried out in rabbit reticulocyte by using the coupled transcription and translation system (TNT T7 Quick; Promega) according to the manufacturer's protocol. The [35S]methionine-labeled capsids were incubated with the VP26 translation product for 90 min at RT with constant rotation. The capsids and bound translation product were then layered onto sucrose gradients (20 to 50%) and sedimented as described in Desai et al. (10). Fractions collected after sedimentation were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Radiolabeling, immunoprecipitation, and SDS-PAGE.
Radiolabeling of infected cells was carried out using preformed monolayers of Vero cells (107 cells) in 100-mm-diameter dishes. Following infection, the cells were overlaid with labeling media (70% methionine-free Dulbecco's modified Eagle's medium, 25% F-12 nutrient mixture [Ham], and 5% fetal calf serum [Gibco-Invitrogen]). Radioactivity (150 μCi of [35S]methionine [NEN-Dupont]) was added to this medium 8 h postinfection, and incubation continued for an additional 16 h. Immunoprecipitation of the in vitro-synthesized VP26 was performed by adding 5 μl of αVP26/C antibody to the TNT translation product diluted in 0.5× CLB and then incubated at 4°C overnight. The next day 50 μl of protein A-Sepharose beads (Sigma) swollen in 0.5× CLB buffer was added to the tube and incubated for 2 h at 4°C with continuous rotation. The beads were washed five times with 0.5× CLB buffer and resuspended in 25 μl of 2× Laemmli sample buffer. Samples (5 μl) were boiled prior to electrophoresis. SDS-PAGE was performed using the mini-protean gel system (Bio-Rad). Gels were cast according to the protocol supplied by the manufacturer, except N,N′-diallytartardiamide was the cross-linking agent. Gels were fixed in methanol (30%) and acetic acid (10%), treated with En3Hance (NEN-Dupont), and dried prior to autoradiography. The gel was exposed to X-ray film (Kodak Biomax) at −80°C using an intensifying screen.
Data preparation.
For figure preparation, autoradiographs were scanned at 600 dots per in. into Adobe Photoshop 6.0. The digital images for the light microscopy analysis were captured on a charge-coupled device camera as JPEG files and imported into Adobe Photoshop.
RESULTS
Development of assays to test for VP26 interaction with the capsid.
VP26 is located at the outer surface of hexons, but not pentons, at equimolar amounts with VP5; thus, there are 900 copies per capsid (2, 33, 37-39). Its small size (112 amino acids) and high copy number make VP26 an especially attractive molecule with which to identify residues that interact with VP5. Two assays were developed to identify key residues of VP26 that interact with the capsid, an in vitro capsid-binding procedure and a GFP localization assay.
Wingfield et al. (37) showed that purified VP26 derived from bacteria could bind to capsids that lack VP26, in vitro. They obtained capsids from insect cells infected with recombinant baculoviruses expressing all the essential capsid proteins except VP26. The assay we developed utilizes capsids derived from cells infected with a null mutant virus of VP26, KΔ26Z; these capsids consequently also lack VP26 (13). VP26 was synthesized using the in vitro transcription-translation system (TNT Promega). This allows one to synthesize radiolabeled VP26, which can then be added to the VP26-lacking capsids. Sedimentation of the capsids through sucrose gradients followed by SDS-PAGE analysis was used to detect the presence of in vitro translated VP26 in the same fractions as the capsid proteins indicative of VP26 capsid binding.
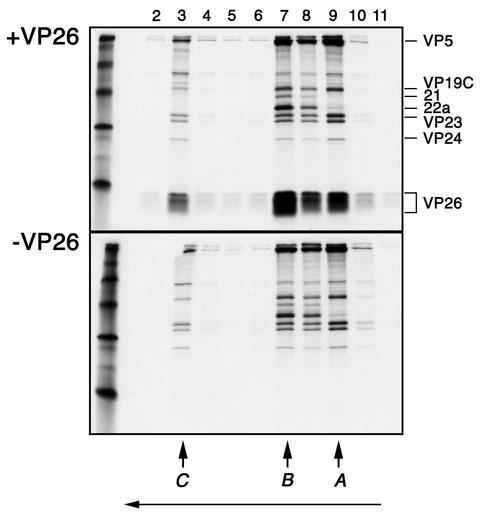
Radiolabeled ([35S]methionine) VP26-lacking capsids were obtained by sedimentation through sucrose gradients of lysates derived from KΔ26Z-infected Vero cells. We typically radiolabeled only 25% of the cells so that the radioactivity observed for the translated protein would be comparable to that for the other capsid proteins. A, B, and C capsids were harvested from the gradient using a fraction collector, pooled, pelleted, and resuspended in PBS. VP26 coding sequences were transferred into pGEM3Z (Promega) for coupled transcription-translation (TNT) reactions. The translation reaction utilized [35S]methionine. The VP26-lacking capsids present in a 500-μl volume were mixed with the in vitro reaction products in which the template was the UL35 (VP26) coding sequences (approximately 50 μl), incubated 90 min at RT (continuous rotation), layered on top of sucrose gradients, and sedimented to separate capsids from unbound VP26. Similar preparations of capsids were mixed with and were then translation reaction products that did not contain the UL35 (VP26) template assayed as described above. Following sedimentation the fractions were analyzed for protein composition by SDS-PAGE (Fig. 1). Data are shown for capsids to which VP26 reaction products were added (Fig. 1, top panel) and to which reticulolysate that lacked the VP26 template was added (lower panel). B capsid proteins were evident in fraction 7, A capsid proteins were evident in fraction 9, and C capsid proteins were evident in fraction 3. Radioactivity corresponding to VP26 was detected in the same fractions as the other capsid proteins, indicating cosedimentation as a consequence of VP26 binding to capsids. There are two distinct bands of radioactivity in the VP26 region of the gel. There is also some smearing of the radioactivity corresponding to VP26, and this was most likely due to gel resolution. The advantage of using this assay is that it measures interaction of VP26 with a three-dimensional template, the capsid that is a natural product of infection. This procedure assays directly for interaction between two protein entities. A similar procedure was also used to study the interaction of the cytomegalovirus (CMV) basic phosphoprotein with the capsid (1).
FIG. 1.
In vitro capsid binding assay to study interaction of VP26 with the capsid. Capsids that lack VP26 were isolated from sucrose gradients following sedimentation of [35S]methionine-radiolabeled lysates prepared from KΔ26Z-infected cells. The UL35 ORF in pGEM3Z was used as a template to synthesize [35S]methionine-labeled VP26. Capsids and in vitro-translated protein (+VP26) were incubated at RT for 90 min (continuous rotation) and then sedimented through 20 to 50% sucrose gradients (top panel). In parallel experiments, similarly prepared capsids were incubated with reticulolysate (−VP26) and analyzed as described above (lower panel). The fractions (2 to 11) collected following sedimentation were analyzed by SDS-PAGE (17% acrylamide). The gels were processed as described in Materials and Methods and dried prior to autoradiography, and the autoradiograph obtained is shown in the figure. The direction of sedimentation was from right to left (indicated by the arrow at the bottom of the figure). The positions at which C, B, and A capsids sediment are indicated at the bottom of the panel. The positions of the capsid proteins in the gel are indicated to the right of the top panel. Radioactivity corresponding to in vitro-synthesized VP26 was detected in the fractions containing capsid proteins, indicating VP26 binding to capsids. Protein standards were loaded in the extreme left lanes and correspond to 220, 97, 66, 45, 30, and 14.3 kDa.
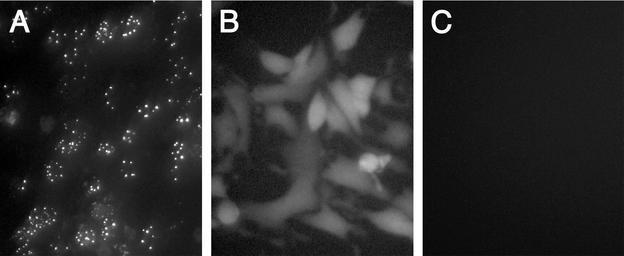
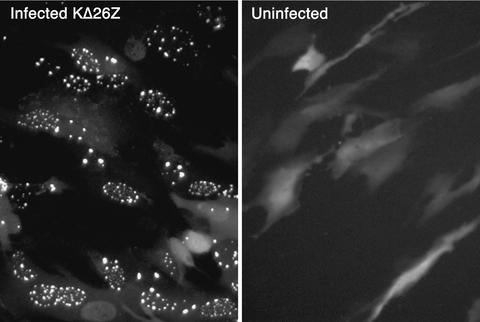
The second assay developed was a GFP localization procedure. We have previously isolated a virus, K26GFP, in which GFP (5) was fused to the N terminus of VP26 (12). The VP26-GFP fusion protein expressed by this virus participated in capsid assembly as judged by the localization of fluorescence in the infected cell nucleus in numerous puncta at early times after infection (12) (Fig. 2A). This interaction was mediated by the presence of VP5, because when the VP26-GFP marker was transferred into a VP5-null (10) background (recombined into KΔ5Z) the fluorescence observed was distributed throughout the cell (Fig. 2B). Thus, based on this genetic analysis of virus mutants a physical association between VP26 and VP5 must occur for VP26 to localize to the nucleus in these large puncta. The VP26-GFP fusion protein was also incorporated into the capsid shell (12). For the present experiments a transient-transfection infection protocol was used. The UL35 ORF was cloned into pEGFP-C2 (Clontech). In this plasmid, VP26 is fused downstream of and in frame with the GFP ORF, and expression of the fused protein was under the control of the human CMV immediate-early promoter. RPE-1 cell monolayers were transfected with this plasmid using Fugene-6 reagent (Roche). The next day when the cells were visualized in a fluorescence microscope, the fluorescence detected was diffusely distributed throughout the cell. The cells were then superinfected with the VP26-null mutant, KΔ26Z. This virus provides all the other capsid proteins necessary for assembly. The cells were visualized in an inverted fluorescence microscope 16 h postinfection. In the absence of any viral proteins, fluorescence was detected throughout the cell (Fig. 3, right panel). The distribution was similar to that of GFP when expressed by itself, that is, without fusion to VP26 (data not shown). Thus, VP26 on its own cannot direct localization of GFP to any specific cellular compartment. In infected cells, fluorescence was localized to the nucleus in large punctate spots (Fig. 3, left panel). The representative cells displayed in the figure were typical of the phenotype observed in the whole monolayer. Many of the cells photographed displayed this nuclear phenotype, whereas some exhibited a more cytoplasmic or diffuse nuclear pattern of fluorescence. We attribute this to the asynchronous nature of the infection process. This nuclear punctate phenotype was clearly indicative of sites of capsid assembly and the participation of VP26-GFP in this process. Thus, these two complementary assays, the capsid binding and GFP localization assays, should provide a basis to identify key residues of VP26 that are involved in its association with the capsid structure.
FIG. 2.
Localization of VP26-GFP to the nucleus requires the presence of VP5. RPE-1 cell monolayers (106 cells) in 35-mm-diameter dishes were infected with K26GFP (A) or KΔ5Z-26GFP (B) at an MOI of 10 PFU/ml or were mock infected (C). The infected cells were visualized under a fluorescence microscope 12 h postinfection (40× objective).
FIG. 3.
GFP localization assay. The UL35 ORF was fused to the C terminus of GFP (pEGFP-C2), and this plasmid (0.5 μg) was transfected into RPE-1 cells. The following day the cells were either infected with KΔ26Z at an MOI of 10 PFU/cell or were mock infected. Sixteen hours following infection, the infected cells and uninfected cells were visualized in a fluorescence microscope (40× objective).
Conserved regions and residues of VP26.
Sequence alignment of the UL35 ORFs from representative members of the alphaherpesviruses was performed to determine regions and residues of VP26 that can be targeted for mutagenesis. The sequences were aligned using the Pustell program in MacVector (Oxford Molecular Ltd.). This analysis revealed a number of conserved amino acids and in addition a large block of conserved amino acids at the C terminus of the protein, which spanned from residues 87 to 100 (Fig. 4).
FIG. 4.
Amino-acid sequence alignment of the UL35 ORFs of alphaherpesviruses. The UL35 ORFs of representative members of the alphaherpesvirus family were aligned to each other, using the Pustell program in MacVector. The residue numbering at the top of the sequence corresponds to HSV-1 numbering. Conserved residues are shaded in gray. The residues boxed in rectangles were deleted in mutants Δ18-25, Δ54-60, and Δ93-100. The virus genomes used for this analysis were equine herpesvirus type 1 (EHV-1), EHV-4, varicella-zoster virus (VZV), HSV-1, HSV-2, pseudorabies virus (PRV), and bovine herpesvirus type 1 (BHV-1).
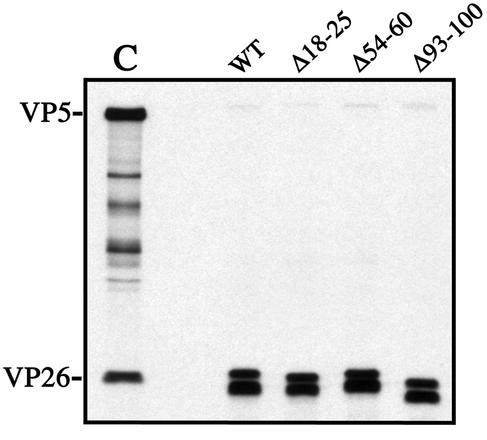
Using this sequence alignment, three in-frame deletion mutants were made using inverse-PCR protocols. They were constructed to delete blocks of conserved amino acids, as in the case of the C-terminus deletion, and to crudely define the important regions of VP26 by dispersing the deletions throughout the sequence (Fig. 4). The mutants constructed were Δ18-25, Δ54-60, and Δ93-100. The numbers refer to the amino acids deleted by each mutation. A BglII restriction site sequence was inserted at the site of deletion in order to screen for the mutation. The insertion of this restriction site sequence resulted in the insertion of arginine and serine residues in the protein sequence. These three deletion mutations were analyzed using the capsid binding assay. The in vitro-synthesized proteins were first immunoprecipitated with antisera to VP26 to confirm the expression of the mutant polypeptide (Fig. 5). All three mutants synthesized the VP26 polypeptide and at amounts comparable to that of the wild-type construct. The mobility differences between the different immunoprecipitated polypeptides were probably due to the deletion of the amino acids in each construct. When VP26 was synthesized by coupled TNT, multiple forms of the polypeptide were detected. There were two distinct bands observed in the gels. The polypeptide with the lowest mobility was the form detected in C capsids of HSV (Fig. 5). The polypeptide with the higher mobility may be comprised of two bands; this was more evident in some of the other gels (see Fig. 10). These multiple forms of VP26 most likely correspond to the phosphorylated forms of this protein. Three different phosphorylated polypeptides corresponding to VP26 were detected in infected cells (18). The deletions do not affect the synthesis of the multiple forms of VP26. We could not analyze directly the synthesis of VP26 in the reticulolysate, because it comigrates with an abundant protein in the lysate, which creates a distortion in the band corresponding to VP26 in SDS-PAGE.
FIG. 5.
Three in-frame deletion mutants of the VP26 polypeptide. Three in-frame deletion mutants, Δ18-25, Δ54-60, and Δ93-100, were constructed (Fig. 4). The three deletion mutants and the wild-type construct were expressed in vitro using coupled transcription-translation (TNT) reactions in the presence of [35S]methionine. The TNT products were reacted with rabbit polyclonal antisera to the C terminus of VP26 (αVP26/C). The immunoprecipitates were analyzed by SDS-PAGE (17% acrylamide). The gel was dried, and the autoradiograph obtained from exposure of this gel to X-ray film is shown. Shown in lane C are C capsid proteins; the positions of VP5 and VP26 in these samples are indicated on the left.
FIG. 10.
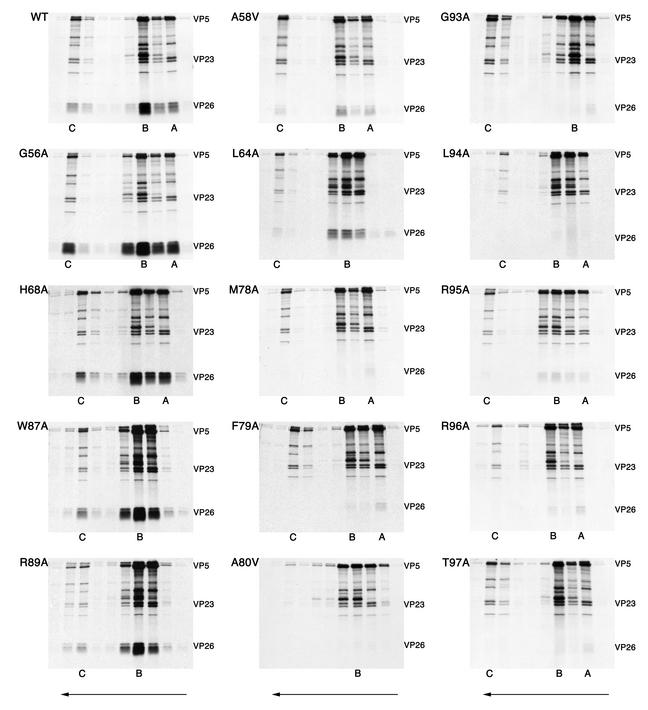
Capsid binding of the VP26 amino acid substitution mutants. The in vitro translation protein products of the VP26 mutants were mixed with radiolabeled capsids as described in the legend to Fig. 1. Following sedimentation of the mixture, fractions were analyzed by SDS-PAGE (17% acrylamide). The gels were dried and processed for autoradiography, the results of which are shown in the figure. The direction of sedimentation was from right to left (indicated by the arrows at the bottom of the figure). The positions in the gradient where capsids sedimented are indicated at the bottom of each panel. For some mutants there was not a clear demarcation between B and A capsids, so only the B capsid position is indicated. The positions of VP5, VP23, and in vitro-translated VP26 are indicated to the right of each panel.
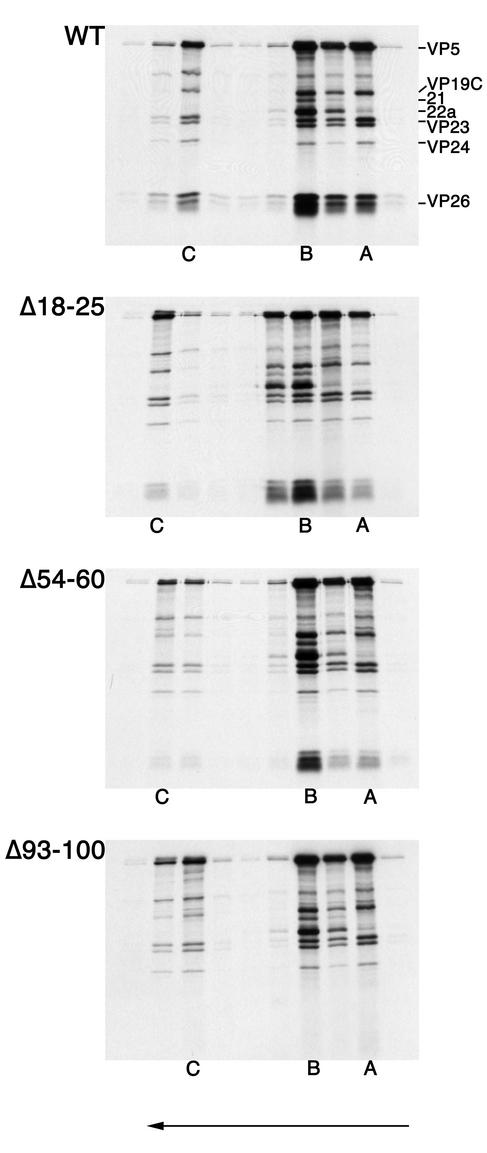
The in vitro-translated products of the wild-type construct and the three mutant constructs were added to [35S]methionine-labeled capsids and analyzed as described above. The data from these experiments are shown in Fig. 6. The results of the capsid binding assay showed that deletion of residues 18 to 25 did not appear to affect binding. Deletion of VP26 residues 93 to 100 abolished binding. Deletion of residues 54 to 60 enabled VP26 to bind capsids, albeit with reduced efficiency. This result demonstrates the importance of the C-terminal conserved domain for capsid association of VP26. Because this procedure directly assays a physical interaction between two protein complexes, the C-terminal conserved domain is important for this VP26-VP5 interaction.
FIG. 6.
Deletion of the conserved residues in the C terminus of VP26 eliminates capsid binding. All three nuclear capsids were isolated and purified from lysates of KΔ26Z-infected cells. The infected cells were metabolically labeled with [35S]methionine. The capsids were mixed with radiolabeled translation products of wild-type VP26 or the three in-frame deletion mutants. Following incubation of capsids and in vitro-translated protein, the mixture was sedimented through 20 to 50% sucrose gradients. The fractions collected were analyzed by SDS-PAGE (17% acrylamide). The gels were processed as described in Materials and Methods and dried prior to autoradiography, and the autoradiograph obtained is shown in the figure. The direction of sedimentation was from right to left (indicated by the arrow at the bottom of the figure). The positions at which A, B, and Ccapsids sedimented are indicated at the bottom of the panels. The positions of VP5, VP19C, 21, 22a, VP23, VP24, and VP26 are indicated on the left of the top panel.
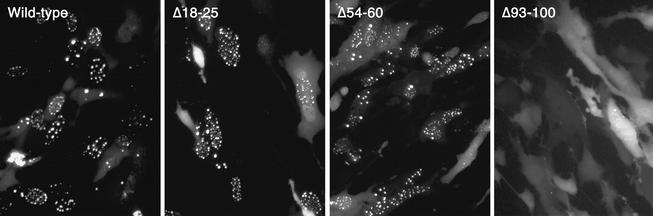
The mutations were also tested for their ability to translocate GFP to the nucleus and display the characteristic punctate fluorescence (Fig. 7). In this assay the phenotype of the mutants was similar to the results obtained from the capsid binding assay. Thus, the Δ93-100 mutation in VP26 abolished the punctate nuclear localization of GFP; the distribution of this fusion protein in the cell was similar to that observed in uninfected cells transfected with the wild-type construct (Fig. 3), that is, in the absence of VP5. The Δ18-25 mutation has no effect on the pattern of nuclear fluorescence displayed by VP26-GFP. The VP26 polypeptide containing the Δ54-60 deletion appears to localize GFP to the nucleus in the characteristic punctate pattern in this experiment. If there was decreased or impaired activity of this mutant in this GFP assay it was not evident, unlike the capsid binding assay which revealed a discernible reduction in VP26 binding to capsids. Thus, the capsid binding assay may be more sensitive at detecting and discriminating differences in association of VP26 with the capsid. From the results of both the capsid binding assay and the GFP localization assay, it was evident that the C terminus of VP26 was important for capsid interaction, but other regions of the protein were also important for this interaction.
FIG. 7.
Deletion of the C-terminal conserved region abolishes nuclear punctate localization of GFP. The three deletion mutants of VP26 were fused to GFP, and these constructs were transfected into RPE-1 cells. The wild-type control was also transfected into cells. The next day the cells were infected with KΔ26Z, which provides the remaining capsid proteins. Following infection for 16 h, the live infected cell cultures were visualized in a fluorescence microscope (40× objective).
We next designed oligonucleotides that would represent amino acid sequences from the C terminus of the protein. The goal was to test if this region was sufficient for capsid interaction. The oligomers made represented amino acid domains 93 to 100, 91 to 102, and 89 to 105 and were inserted into pEGFP-C2. When the domains fused to GFP were tested in the nuclear localization assay, none of the domains were able to transport GFP into the nucleus (data not shown). Thus, protein domains representing the conserved C terminus were not sufficient for interaction with VP5.
The C-terminal half of VP26 is sufficient for GFP nuclear localization and capsid binding.
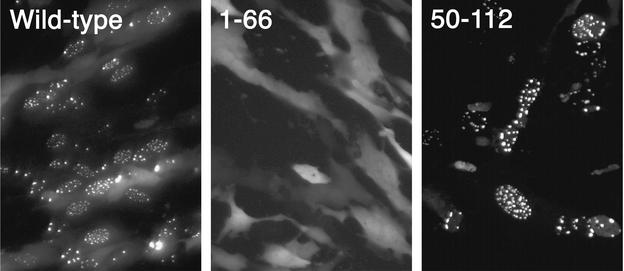
Since the VP26 C-terminus domains were insufficient for localization of GFP to the nucleus, other regions of the molecule must play a role in this interaction. In order to discover this, N- and C-terminal truncation mutants of VP26 were generated by PCRs. The mutants were cloned into pEGFP-C2, that is, fused downstream of and in-frame with GFP, and the GFP localization of these mutants was monitored by fluorescence microscopy (Fig. 8). VP26 sequences spanning from residues 1 to 66 could not localize GFP to the nucleus; the fluorescence observed was distributed throughout the cell. VP26 residues 50 to 112 could localize GFP to punctate spots in the nucleus. The behavior of this mutant protein was similar to that of the wild-type polypeptide as judged by this assay. Further shortening of the construct containing residues 50 to 112 from the N terminus resulted in decreased nuclear localization and increased cytoplasmic fluorescence (data not shown). Thus, the ability to localize GFP to the nucleus resides in the C-terminal half of VP26. The construct containing residues 50 to 112 fused to GFP was also expressed in the TNT system and tested for capsid binding. This fusion protein was also capable of binding to capsids (data not shown).
FIG. 8.
The C-terminal half of VP26 is sufficient for localization of GFP to punctate spots in the nucleus. PCR-generated sequences representing the N-terminal (residues 1 to 66) and the C-terminal (residues 50 to 112) regions of VP26 were fused to the GFP ORF, and the constructs were transfected into RPE-1 cells. Following transfection, cells were infected with KΔ26Z and visualized under a fluorescence microscope 16 h after infection (40× objective). Also shown is the result obtained with the full-length wild-type polypeptide fused to GFP.
Single amino acid substitutions of conserved residues of VP26.
The analysis of the deletion and truncation mutants of VP26 indicated the C-terminal half of the protein as being required for nuclear punctate localization of GFP and capsid binding. Data from both assays were generally in agreement with this result. To define residues of VP26 that were important for this interaction and to avoid the possible global effects deletion and truncation mutations have on polypeptide structure, site-directed mutagenesis was carried out. We again used the sequence alignment of VP26 to identify which residues to target (Fig. 4). All the conserved residues were mutated as well as the two conserved residues P9 and N30 in the N-terminal half of VP26. The only mutant we could not derive was P90A. Other residues that were highly conserved in the different members of the family, such as A58 and L64, were also subjected to mutation. The residue R94 was also mutated even though HSV-1 is the only member to specify an arginine at this position; however, there is conservation of the positive charge at this position in all the other members. The amino acids were mutated to alanine except for A58 and A80, which were mutated to valine. The mutation of each amino acid to alanine allows one to determine the contribution of the side chain functional group for activity (7).
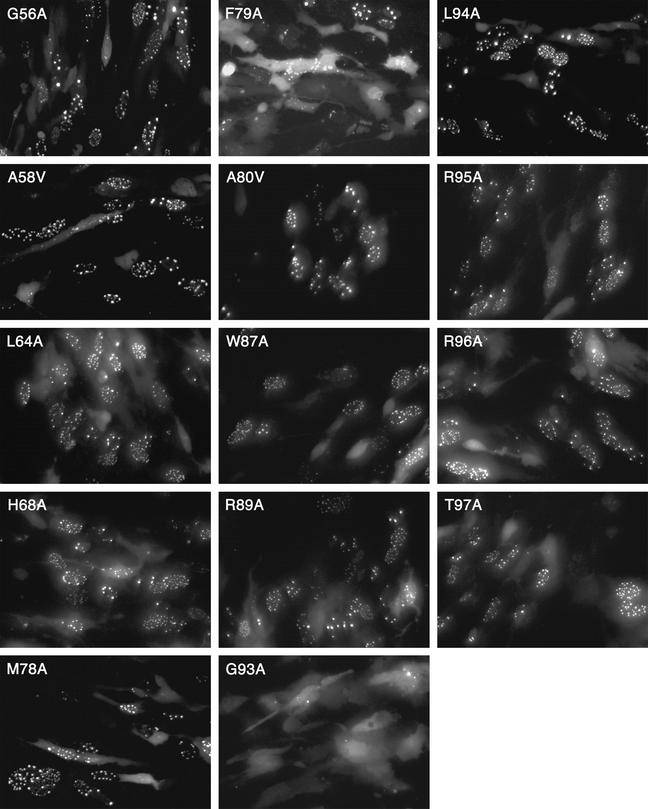
The mutant constructs were initially tested for their ability to localize GFP to the nucleus since this assay would allow one to rapidly screen the many mutations generated. The data are shown in Fig. 9. The results were obtained from transfected RPE-1 cells. The majority of the mutations were capable of translocating GFP into the nucleus, and the cells displayed the characteristic nuclear punctate fluorescence. Because of the transient nature of the assay and the asynchronous feature of the infection process, some cells displayed diffuse cytoplasmic fluorescence or diffuse nuclear fluorescence. The photographed cells in Fig. 9 were representative of the overall phenotype observed in the culture. Mutations at amino acids P9, N30, P100, and P106 were also analyzed in this assay, and all were capable of efficiently localizing GFP to the nucleus as judged by the observation of punctate fluorescence (data not shown). Two mutations, G93A and F79A, had a more dramatic effect on the distribution of fluorescence in the transfected cell. Most of the cells transfected with the G93A mutant construct exhibited a diffuse cytoplasmic and nuclear pattern of fluorescence, indicating disruption of interaction with VP5. Of the cells transfected with the F79A construct, most displayed fluorescence distributed throughout the cell; however, a few puncta of fluorescence were detected, but this was observed in only a small percentage (less than 5%) of the cells. The GFP localization assay indicated that G93A and F79A for the most part eliminate translocation of VP26-GFP to puncta in the nucleus. Thus, residues F79 and G93 are important for interaction with VP5, which acts as the mediator to relocate or concentrate VP26 in the nucleus.
FIG. 9.
Residues F79 and G93 are important for the accumulation of VP26-GFP in the nucleus. The site-directed mutations generated in VP26 were fused to GFP and assayed in the GFP localization assay for activity. The plasmids were transfected into RPE-1 cells, and the following day the cells were infected with KΔ26Z. Infected cells were visualized under the fluorescence microscope 16 h after infection (40× objective).
All the mutants were also analyzed in the same manner in Vero cells by using the calcium phosphate transfection reagent CellPhect (Pharmacia). Although the transfection efficiency was lower in these cells, the phenotypes observed were consistent with those observed in the RPE-1 cells.
Analysis of the ability of VP26 mutants to bind to the capsid.
Since the previous mutational analysis revealed a similar correlation between the two assays, it seemed that the capsid binding assay could be used to confirm the data obtained from the light microscopy analysis. In addition, since this assay is more quantitative than the GFP assay we could derive some numbers with respect to the binding ability of the mutants. All the amino acid substitution mutations in the C-terminal half of the protein were analyzed for capsid binding. Prior to examining the individual mutants for capsid binding, the mutant VP26 constructs were translated in vitro and radiolabeled protein was immunoprecipitated with antisera to VP26 to confirm the synthesis of each mutant. The results of the immunoprecipitation assay (data not shown) revealed that all of the mutant VP26 proteins are synthesized in vitro at levels comparable to that of the wild-type translation product.
Next, the mutants were assayed for their ability to bind to capsids in the in vitro assay. The procedure was performed as described above, and in most cases each mutant was analyzed two to three times. In addition, the wild type was always included when a set of mutants was assayed. The data are shown in Fig. 10. Mutations G56A, H68A, W87A, and R89A have no effect on the ability of the VP26 polypeptide to bind to the capsid as judged by the appearance of abundant radiolabeled VP26 in the same fractions as the other capsid proteins. Mutations at positions P100A and P106A were also analyzed in the capsid binding assay. The mutations at these positions did not have any effect on the ability of VP26 to bind capsids (data not shown). Mutations at position A58 and L64 result in a reduced ability of VP26 to bind to capsids. This result was consistent with that observed with the in-frame deletion mutant Δ53-60, which displayed a reduction in capsid binding. The three mutations at residues M78, F79, and A80 all eliminated the capsid binding ability of VP26. These mutations specify a hydrophobic domain, which is centered around F79, the residue that was important for GFP localization to nuclear puncta. The mutation at G93, which also abolished the nuclear punctate localization of fluorescence, also eliminated binding of VP26 to the capsid. Interestingly, mutations in the conserved region of the C terminus, which appeared not to affect capsid interaction of VP26 as judged by the light microscopy assays, did affect capsid binding. These mutants—L94A, R95A, R96A, and T97A—all had a greatly reduced ability to bind to capsids as judged by the greatly reduced levels of radioactivity that correspond to the translated VP26 present in the same fractions as the capsid proteins.
In order to quantify the binding efficiencies of the different VP26 mutants to capsids, we exposed the gels to a phosphorimaging plate and derived the photostimulated luminescence (PSL) units for the radioactivity corresponding to VP26 and normalized it to the radioactivity corresponding to VP23. Since VP23 is a triplex protein, its copy number per capsid would be invariant. The results of this analysis are shown in Table 1. Results for two amino acids, G56 and H68, that showed no visible reduction in capsid binding were quantified. Taking the wild-type level as 100%, values for G56A and H68A mutants were 104 and 114%, respectively. Based on these data, G56A and H68A mutants bind to capsids as well as or slightly better than the wild-type protein. For mutations at amino acids W87 and R89 that also had no visual effect on capsid binding, data were not obtained. The polypeptides encoding mutations A58V and L64A resulted in a reduction in VP26 capsid binding to 9.2 and 15% of wild-type levels, respectively. The mutations in the hydrophobic triplet (amino acids M78, F79, and A80) reduced capsid binding of VP26 to less than 2% of the wild-type value. Similarly, mutation of residues between 93 and 97 gave values that were 2% or less of the wild-type level, except for mutation at R95, which gave a value of 5.7%. The quantitative data clearly show the importance of these two domains for the interaction of VP26 with the capsid.
TABLE 1.
Relative capsid binding efficiencies of VP26 mutants
| Mutant | Relative binding of VP26 (% of wild type)a |
|---|---|
| Wild type | 100 |
| G56A | 104 |
| A58V | 9.2 |
| L64A | 15 |
| H68A | 114 |
| M78A | 1.9 |
| F79A | 1.6 |
| A80V | 1.6 |
| W87A | ND |
| R89A | ND |
| G93A | 2.0 |
| L94A | 1.9 |
| R95A | 5.7 |
| R96A | 1.1 |
| T97A | 1.2 |
Values were calculated as PSL units for the radioactivity corresponding to VP26 divided by that corresponding to VP23. Data for C capsids and B/A capsids were combined, since the values obtained for C or B/A capsids relative to wild-type values were similar, and then used to derive the mean ratio of VP26 PSL units to VP23 PSL units. Data are expressed as a percentage of the wild-type units. The numbers derived for the wild type were averaged from three separate experiments. Data from three independent experiments were used to derive the values for F79A, and data from two independent experiments were used to derive the values for G56A, L64A, M78A A80V, and G93A. The remaining values were derived from single experiments. ND, not determined.
In summary there are two key amino acids of VP26, F79 and G93, that are important for localization of GFP to nuclear foci (Table 2). The same residues are also important for capsid binding; however, additional residues were revealed that are also required for capsid binding (Table 2). Some of these amino acids (L94, R95, R96, and T97) are clustered in the C-terminal conserved domain, which also includes G93. The residues on either side of F79 (M78 and A80) comprise a second important domain for capsid association.
TABLE 2.
Summary of key residues of VP26 for capsid binding and GFP nuclear localization
| Assay | VP26 residues |
|---|---|
| GFP nuclear puncta | F79, G93 |
| Capsid bindinga | A58, L64 |
| Capsid bindingb | M78, F79, A80, G93, L94, R95, R96, T97 |
Mutation of residue that results in reduced binding.
Mutation of residue that eliminates binding.
DISCUSSION
One of the goals of our research is to identify the complex network of capsid protein interactions and uncover the specific residues of the capsid proteins that mediate these interactions. In the case of VP26, its relatively small size made it ideal as a target for the study of its interaction with the capsid and also as a template for mutagenesis. There were two other properties of VP26 that made it suitable for this analysis. The first was that, in vitro, VP26 could readily bind capsids that are devoid of this protein (37) and the second was that the autofluorescent protein GFP could be fused to VP26 without affecting its ability to interact with the capsid, thus affording a visual tracking of the protein in animal cells (12). These two properties were the basis of the two assays we developed to identify VP26 interactive residues. An in vitro capsid binding assay and a GFP localization assay were used for this analysis. The nuclear localization of VP26-GFP to punctate regions within this structure required the presence of VP5, because in the absence of VP5 alone the fluorescence detected was diffusely distributed throughout the cell. Thus, this assay is based on an interaction between VP5 and VP26. This assay was used to quickly screen the many mutations that were generated. This advantage and the fact that live cultures are analyzed by light microscopy make it a powerful screen for VP26 mutants. The capsid-binding procedure is more laborious and time-consuming, but it has the advantage that the interaction is between VP26 and a three-dimensional assembled shell and thus allows direct assay of protein-protein binding. As expected, capsid binding proved to be more sensitive at uncovering key VP26 interactive residues and consequently was more informative due to the quantitative nature of the assay.
The alignment of the UL35 ORFs from different members of the alphaherpesvirus family was important and insightful for revealing regions and residues to mutate. The first mutations made were three small in-frame deletions, Δ18-25, Δ54-60, and Δ93-100. When these mutations were analyzed in the two assays, it was apparent that the conserved domain of the C terminus was important for the ability of VP26 to interact with the capsid. However, it also became apparent that this region, although it was necessary for interaction, was not sufficient for this interaction. Analysis of N- and C-terminal truncation mutants established that the C-terminal half of VP26 (amino acids 50 to 112) was necessary and sufficient for capsid interaction as judged by both GFP localization and capsid binding. In the end we generated amino acid substitution mutations in the conserved residues of VP26, and this analysis was key to uncovering a complex interactive profile between this small protein and the capsid. When the single-amino-acid-substitution mutants of VP26 were assayed in the GFP localization assay, two mutants, F79A and G93A, had an effect on the distribution of fluorescence in the transfected cell. These two mutations essentially eliminated the interaction with VP5 that normally localizes VP26 to the nucleus (Table 2). When the same mutations were analyzed in the capsid binding assay, they resulted in a 50-fold decrease in the binding capability of VP26.
Eight additional amino acids were identified that affect VP26 interaction with the capsid as judged by the in vitro capsid binding assay (Table 2). They were A58, L64, M78, V80, L94, R95, R96, and T97. Amino acids A58 and L64 contribute significantly to the binding ability of VP26 since mutations at these positions result in a 10-fold reduction in the amounts of in vitro-synthesized VP26 bound to capsids. This finding would explain why the in-frame deletion mutant Δ54-60 displayed a reduction in VP26 binding relative to wild-type VP26. Mutations at amino acids M78 and V80 that surround F79 result in an elimination of VP26 binding. Thus, these three hydrophobic residues comprise an important interactive domain that is required for both localization of VP26 to nuclear puncta and capsid binding. The most interesting finding was that in the conserved domain of the C terminus, the string of amino acids G93, L94, R95, R96, and T97 eliminates binding of VP26 to the capsid when changed to alanine, even though all of these residues with the exception of G93 could readily interact with VP5 and localize GFP to the nucleus in punctate spots. This result demonstrates that the interaction of VP26 with the capsid is more complex than that which allows VP26 to accumulate in the nucleus. There is an interaction between VP26 and VP5 in the cytoplasm that translocates or concentrates VP26 in the punctate spots of the nucleus. Amino acids F79 and G93 are important for this interaction, because mutation results in a reduction or absence of the puncta in the nucleus. Subsequent to this interaction, the C-terminal conserved domain and amino acids around F79 are important for interaction of VP26 with the three-dimensional template of the icosahedral capsid.
The observation that a glycine residue has a significant contribution to capsid protein-protein interactions is surprising. The importance of the hydrophobic amino acids M78, F79, and A80 for this interaction is consistent with such protein-protein interactions. Glycine residues do provide a flexible “hinge” or “pivot” between protein modules, and this may be the role G93 plays; that is, it positions the C terminus domain in close apposition to VP5 both in the cytoplasm and VP5 in the capsid shell. The importance of such glycine hinges has been demonstrated for fusion events between virus and cell membranes (8). However, there are other instances in which glycine-rich regions together with interspersed aromatic residues constitute an important interaction domain (4).
The initial interaction between VP5 and VP26 probably occurs in the cytoplasm, and this complex together with the scaffold protein translocates to the nucleus, where there is accumulation of VP26 at the sites of capsid assembly. It is also possible that VP26 associates with VP5 when VP5 becomes concentrated in the nucleus. Both the VP26 (12-kDa) and the VP26-GFP (39-kDa) proteins are small enough to traverse the nuclear pore. When VP5 accumulates in the nucleus, the interaction between these proteins results in the concentration of VP26 in the nuclear puncta. The light microscopy results of cells infected with the virus expressing VP26-GFP (K26GFP) (Fig. 2) demonstrate an almost-exclusive nuclear distribution of this protein, and this was seen at early times postinfection (data not shown). This could suggest an active translocation of VP26-GFP into the nucleus rather than a diffusion or concentration process for nuclear localization. This question could be resolved by excluding VP26-GFP from the nucleus and then determining its distribution when VP5 is present. This initial interaction of VP26 may occur with VP5 that is conformationally different from that found in capsids. Or VP26 may interact with VP5 that is in a monomer/dimer configuration rather than the hexamer complex present in capsomeres. Reports in the literature have shown that VP26 may not interact with or is not incorporated onto the procapsid structure (6, 21, 28). This structure has a more spherical shape on the surface of the shell, unlike the more angular B capsid (34). When the capsid proteins assemble in the nucleus and produce the procapsid, the interaction of VP26 with VP5 would have to be reversed to explain the above observations. It would subsequently have to “re-interact” with VP5 and the capsid shell once the structure has angularized. VP26 is also found in the open shells formed in the absence of the scaffold protein (11). Thus, an icosahedral structure is not necessary for this binding. The VP5 capsomeres in these shells are probably in the conformation present in the mature capsid, thus facilitating VP26 association. This second interaction between VP26 and VP5 is mediated by a different and expanded set of amino acids. However, these additional residues are adjacent and in close proximity to amino acids F79 and G93, which are important for nuclear localization. We have observed that when in vitro-translated VP26 is added to wild-type capsids, that is, capsids that contain VP26, radioactivity corresponding to the in vitro-translated protein binds to these capsids (data not shown). This observation suggests that VP26 can be displaced from the capsid by the addition of an exogenous source of VP26. If this is the case, it shows that the association of VP26 is highly fluid and may explain the complex interactive properties of this small protein.
Since VP26 binds only to hexons, it has been suggested that binding of this protein to the capsid occurs only when VP26 is in a hexameric multimer (39). Studies by Wingfield et al. (37) demonstrated that bacterially expressed VP26 could be detected in both a monomer and dimer configuration. This oligomerization of VP26 could occur after the accumulation of this protein in the nucleus when there are sufficient concentrations of this protein and may occur independently of procapsid formation. Once the capsid structure has attained an angular conformation, the VP26 oligomer could then bind to the hexameric capsomeres. Thus, angularization of the VP5 hexamer may be the trigger that regains VP26 binding. Analysis of the VP26 polypeptide sequence using computer-assisted programs (PHD Predict [27]) indicates very little structural conformation in the C terminus of the protein. There is a region, however, that spans from residues 46 to 66 and that displays a high propensity to conform to an alpha-helical configuration. This region encompasses the residues A58 and L64, which appear to play some role in VP26 interaction with the capsid. This region could also be required for the self-association of VP26 through helix-helix interactions.
The role of VP26 in the virus life cycle is poorly understood. It is not required for capsid assembly (13, 30, 31). It is necessary for productive replication of HSV-1 in the mouse central nervous system; its absence results in a 100-fold reduction in infectious virus yield in trigeminal ganglia (13). In CMV it appears to be essential for replication of the virus in cell culture (3). Borst et al. (3) made an attempt to generate a virus recombinant that expresses a fusion of GFP to the VP26 counterpart; however, they failed to obtain replication-competent virus. The fusion protein did localize to the nucleus during productive infection, but it seemed that a function specified by this small protein subsequent to capsid assembly was eliminated (3). Thus, the role of this protein may be very complex in the different herpesvirus families and in vivo infections.
Acknowledgments
This research was supported by grant AI33077 from the NIH.
We thank Mercy Okoye and Tamika Redfern for technical assistance. Also, we acknowledge Wade Gibson for support and discussions of the data.
REFERENCES
- 1.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booy, F. P., B. L. Trus, W. W. Newcomb, J. C. Brown, F. F. Conway, and A. C. Steven. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. USA 91:5652-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 6.Chee, J. H. I., and D. W. Wilson. 2000. ATP-dependent localization of the herpes simplex virus capsid protein VP26 to sites of procapsid maturation. J. Virol. 74:1468-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clackson, T., and J. A. Wells. 1995. A hot spot of binding energy in a hormone-receptor interface. Science 267:383-386. [DOI] [PubMed] [Google Scholar]
- 8.Cleverly, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J. Gen. Virol. 73:2709-2713. [DOI] [PubMed] [Google Scholar]
- 10.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai, P., S. C. Watkins, and S. Person. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, C. L., and G. K. Pei. 1997. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23:570-571, 574. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 17.McNabb, D. S., and R. J. Courtney. 1992. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J. Virol. 66:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNabb, D. S., and R. J. Courtney. 1992. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J. Virol. 66:4839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., F. L. Homa, F. P. Booy, D. R. Thomsen, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Person, S., and P. Desai. 1998. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frame of HSV-1 but are not formed in a null mutant of UL38 (VP19C). Virology 242:193-203. [DOI] [PubMed] [Google Scholar]
- 23.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 24.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rixon, F. J., C. Addison, A. McGregor, S. J. McNab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 26.Roizman, B., and A. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 28.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steven, A. C., and P. G. Spear. 1996. Herpesvirus capsid assembly and envelopment, p. 312-351. In R. Burnett, W. Chiu, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 30.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliot, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 31.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Stevens. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP 26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Stevens. 1996. The herpes simplex virus procapsid structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 35.Vernon, S. K., M. Ponce de Leon, G. H. Cohen, R. J. Eisenberg, and B. A. Rubin. 1981. Morphological components of herpesvirus. III. Localization of herpes simplex virus type 1 nucleocapsid polypeptides by immune electron microscopy. J. Gen. Virol. 54:39-46. [DOI] [PubMed] [Google Scholar]
- 36.Wildly, P., W. C. Russell, and R. W. Horne. 1960. The morphology of herpes virus. Virology 12:204-222. [DOI] [PubMed] [Google Scholar]
- 37.Wingfield, P. T., S. J. Stahl, D. R. Thomsen, F. L. Homa, F. P. Booy, B. L. Trus, and A. C. Steven. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J. Virol. 71:8955-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, Z. H., J. He, J. Jakana, J. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, Z. H., B. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]