Abstract
Most spontaneously occurring mutations that cause extensive herpes simplex virus type 1 (HSV-1)-induced cell fusion are single amino acid changes within glycoprotein K (gK). Despite the strong genetic association of gK with virus-induced cell fusion, its direct involvement in cellular membrane fusion has been controversial, largely due to previously unsuccessful efforts to detect gK expression on virion and cellular surfaces. Recently, we showed that gK is expressed on HSV-1 virions and functioned in virus entry (T. P. Foster, G. V. Rybachuk, and K. G. Kousoulas, J. Virol. 75:12431-12438, 2001). To determine whether gK is expressed on cellular surfaces, as well as its membrane topology, we generated the recombinant viruses gKV5DI, gKV5DII, gKV5DIII, and gKV5DIVcontaining insertions of the V5 antigenic epitope within each of four domains of gK predicted to localize either in the cytoplasmic side or in the extracytoplasmic side of cellular membranes. Immunohistochemical and confocal microscopy analyses of infected cells showed that both wild-type and syncytial forms of gK were expressed on cell surfaces. Analysis of the topology of the V5-tagged gK revealed that gK domains I and IV were located extracellularly, whereas domains II and III were localized intracellularly. Transiently expressed gK failed to localize in cellular plasma membranes. In contrast, infection of gK-transfected cells with the gK-null virus ΔgK enabled expression of gK on cell surfaces, as well as gK-mediated membrane fusion. Transient-coexpression experiments revealed that the UL20 protein enabled cell surface expression of gK, but not gK-mediated cell-to-cell fusion, indicating that additional viral proteins are required for expression of the gK syncytial phenotype.
Viral glycoproteins are key determinants of membrane fusion events throughout the life cycle of herpesviruses. Herpes simplex viruses (HSVs) specify at least 11 glycoproteins: gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM, which are expressed in infected cells. These glycoproteins function in several important roles, including pH-independent virus entry via fusion of the virion envelope with cellular membranes, egress of infectious virion particles, cell-to-cell spread, and virus-induced cell fusion (31, 40, 43, 44).
Cell-to-cell transmission of HSV type 1 (HSV-1) occurs either by release of virions to extracellular spaces or through virus-induced cell-to-cell fusion. Wild-type (syn+) virions spread across cellular junctions of juxtaposed membranes and cause rounding and aggregation of cells, as well as limited virus-induced cell fusion. Certain spontaneous mutants of HSV-1 (syn) can rapidly spread into adjacent cells by inducing the formation of large multinucleated cells or syncytia. Mutations that cause extensive virus-induced cell fusion have been mapped to at least four regions of the viral genome: the UL20 gene (2, 29), the UL24 gene (21, 42), the UL27 gene encoding glycoprotein B (gB) (5, 34), and the UL53 gene coding for gK (3, 8, 35, 41). However, syncytial mutations (syn) in the UL53 gene are more frequently isolated than in any other gene (3, 4, 8, 10, 35, 36, 38, 41).
HSV-1 gK is a highly hydrophobic 338-amino-acid protein encoded by the UL53 open reading frame (8, 30). It has characteristics of a glycosylated membrane protein, including a cleavable 30-amino-acid NH2-terminal signal sequence, two potential sites for N glycosylation, and several hydrophobic domains (hpd) (8, 36). Initially, gK was predicted to have four transmembrane-spanning regions; however, experiments with in vitro-translated gK in the presence of microsomal membranes suggested that gK contained three instead of four membrane-spanning regions (8, 32, 37). Cleavage of the NH2-terminal signal peptide and the addition of carbohydrates at amino acid residues 48 and 58 suggested that the amino-terminal domain (domain I) is an ectodomain (10, 18, 37). The presence of multiple syncytial mutations within gK implied that gK is expressed on infected-cell surfaces where it could directly be involved in virus-induced cell fusion. However, anti-gK antibodies raised against gK peptides detected gK exclusively within the endoplasmic reticulum and nuclear membranes (20).
Mutant viruses that are deficient in gK expression have been isolated and characterized for HSV-1, pseudorabies virus (PRV) and varicella-zoster virus. These studies have indicated that gK is important in virion morphogenesis and egress of alphaherpesviruses. Deletion of HSV-1 gK resulted in a small plaque phenotype, reduced the production of infectious virions, and inhibited virion translocation from the cytoplasm to the extracellular space (12, 19, 22, 24, 33). Interestingly, PRV gK-null virions seemed to be able to reinfect cells immediately after their egress, implying that gK blocked fusion of virions with infected-cell membranes (24).
Recently, we significantly improved HSV-1 gK detection by generating recombinant viruses expressing gK containing a protein C (protC) epitope tag. Importantly, HSV-1 gK, like other alphaherpesviruses, was shown to be a structural component of purified virions and to function in virion entry (14). In the present study, we expanded upon these studies by constructing a panel of recombinant viruses expressing gK containing epitope tags within different domains of gK. We experimentally demonstrate that gK is expressed on cell surfaces in a topological orientation similar to that originally predicted by Debroy et al. (8). Furthermore, we found that the UL20 protein is necessary and sufficient for cell surface expression of gK but not for gK-mediated cell-to-cell fusion.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were propagated and maintained in Dulbecco modified Eagle medium (Sigma Chemical Co., St. Louis, Mo.) containing sodium bicarbonate and 15 mM HEPES and supplemented with 7% heat-inactivated fetal bovine serum. The parental wild-type strain used in the present study, HSV-1 (KOS), was originally obtained from P. A. Schaffer (Harvard Medical School). The gK-null virus (ΔgK) was propagated in Vero cells and was as described previously (22). Virus HSV-1 (KOS) d27-1, which has a 1.6-kb BamHI-StuI deletion of the ICP27 gene, was kindly provided by D. M. Knipe, Harvard Medical School, and was propagated in complementing V27 cells (39). The UL53-gK protC epitope-tagged virus (gKprotC) was propagated in Vero cells and was as described previously (14).
Plasmid construction.
Plasmid pSJ1723, which contains gK-flanking sequences corresponding to the UL52 and UL54 (ICP27) genes to facilitate homologous recombination with viral DNA, was as described previously (22). The 14-amino-acid V5 epitope (GKPIPNPLLGLDST; Invitrogen/Life Technologies, Carlsbad, Calif.) was inserted in frame within gK domains I, II, III, and IV at sites predicted not to significantly affect the secondary structure of gK (Fig. 1). The recombinant gK genes coding for the epitope-tagged gKs were constructed by using PCR-based splice-overlap extension methodology as described previously except that either HSV-1 KOS gK or HSV-1 KOS gKsyn20 DNA was used as a template (14). The KOS gKsyn20 DNA specifies a single amino acid substitution at amino acid 40 of gK (Ala to Val) that results in extensive virus-induced cell-to-cell fusion. Wild-type UL53 gK genes specifying a V5 epitope tag within domain I, II, III, or IV were cloned into plasmid pSJ1723, generating plasmids pTFgKV5I, pTFgKV5II, pTFgKV5III, and pTFgKV5IV, respectively. Similarly, the syn20 UL53 gK genes specifying a V5 epitope tag within domain I, II, III, or IV were cloned into plasmid pSJ1723, generating plasmids pTFgKsyn20V5I, pTFgKsyn20V5II, pTFgKsyn20V5III, and pTFgKsyn20V5IV, respectively. Alternatively, gK genes containing tags were engineered to have a 5′ Kozak consensus sequence (A at position −3 of the ATG initiation codon) for optimal translation just prior to the start codon and cloned into pcDNA3.1 TOPO for eukaryotic expression (27). Plasmids pCMVgKV5DI, pCMVgKV5DIII, and pCMVgKV5DIV specify a V5 tag within domains I, III, or IV, respectively, whereas plasmid pCMVgKsyn20V5DI specifies both a V5 epitope tag within domain I and the syn20 mutation. Plasmid pCMV-UL20 contains the UL20 (KOS) open reading frame under the cytomegalovirus (CMV) immediate-early promoter in pcDNA3.1TA-TOPO (Invitrogen). Plasmid pCMV-gD was described previously (13). Plasmid pcDNA3-gB/gD was a gift from A. Minson (University of Cambridge, Cambridge, United Kingdom).
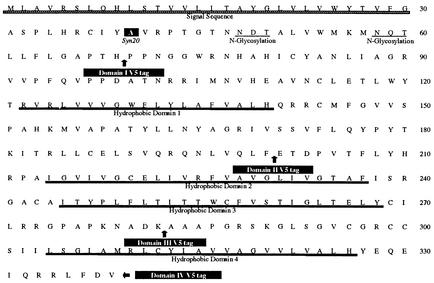
FIG. 1.
Primary structure of HSV-1 gK. The amino acid sequence of gK is shown with its signal sequence (▨), glycosylation sites, and predicted transmembrane domains (▪). The gK sites where the V5 epitopes were inserted, as well as the location of the engineered syncytial mutation syn20 (Ala40-to-Val40), are indicated.
Construction of recombinant viruses that specify V5 epitope tags within gK.
The plasmids that specified the V5 epitope tag and the gK-flanking sequences corresponding to the UL52 and UL54 (ICP27) genes were used to facilitate homologous recombination with viral DNA. Recombinant viruses gKV5DI, gKV5DII, gKV5DIII, and gKV5DIV specifying tags within domains I, II, III, and IV, respectively, were constructed by rescuing the mutant virus d27-1 (KOS) as shown previously (14, 22). Recombinant viruses gKsyn20V5DI, gKsyn20V5DII, gKsyn20V5DIII, and gKsyn20V5DIV were as described above except that they contained the syn20 gK mutation. Putative recombinant virus isolates were plaque purified and extensively tested by PCR and DNA sequencing for the presence of contaminating d27-1 virus and the engineered epitope-tagged gKs.
One-step growth kinetics.
Analysis of one-step growth kinetics was essentially as described previously (14). Briefly, each virus at a multiplicity of infection (MOI) of 5 was adsorbed to ca. 8 × 105 Vero cells at 4°C for 1 h. Thereafter, prewarmed media was added, and virus was allowed to penetrate for 2 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (0.1 M glycine, pH 3.0). Cells and supernatants were harvested immediately thereafter (0 h) or after 4-, 8-, 12-, or 24-h incubations. Virus titers were determined by endpoint titration of virus stocks on Vero cells.
Western analysis of gK expression.
Subconfluent Vero cell monolayers were infected with gKprotC-DIII at an MOI of 5. At 48 h postinfection (hpi) cells were collected by low-speed centrifugation, washed with Tris-buffered saline (TBS), and lysed at room temperature for 15 min in mammalian protein extraction reagent supplemented with a cocktail of protease inhibitors (Invitrogen/Life Technologies). Insoluble cell debris was pelleted, and samples were treated with endoglycosidase H (Endo-H) or peptide-N-glycosidase F (PNGase F) as described previously (14). Samples were electrophoretically separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with anti-protC monoclonal antibody HPC-4 at a 1:50 dilution (ATCC CRL HB-9892). Subsequently, blots were incubated for 1 h with a peroxidase-conjugated secondary antibody at a 1:50,000 dilution and then visualized on X-ray film by chemiluminescence (Pierce Chemicals, Rockford, Ill.) (11, 14). All antibody dilutions and buffer washes were performed in TBS supplemented with 0.135 M CaCl2 and 0.11 M MgCl2 (TBS-Ca/Mg).
Confocal microscopy.
Vero cell monolayers grown on coverslips in six-well plates were infected with the indicated virus at an MOI of 10. At 12 hpi, cells were washed with TBS and fixed with electron microscopy-grade 3% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, Pa.) for 15 min, washed twice with phosphate-buffered saline-50 mM glycine, and permeabilized with 0.1% Triton X-100. Monolayers were blocked for 1 h with 5% normal goat serum and 5% bovine serum albumin in TBS (TBS blocking buffer) before incubation for 5 h with either fluorescein isothiocyanate (FITC)-conjugated anti-V5 (Invitrogen) or anti-gB (Rumbaugh-Goodwin Institute, Plantation, Fla.) diluted 1:500 in TBS blocking buffer. Cells were then washed extensively and subsequently incubated for 1 h with a mixture of Alexafluor 488-conjugated anti-immunoglobulin G (IgG) and Alexafluor 488 anti-FITC (Vector Laboratories, Burlingame, Calif.) diluted 1:500 in TBS blocking buffer for FITC-conjugated anti-V5 or Alexafluor 488-conjugated anti-IgG diluted 1:500 for anti-gB. After incubation, excess antibody was removed by washing five times with TBS. In order to visualize the Golgi apparatus, cells were stained with a 1:1,500 dilution of AlexaFluor 594-conjugated lectin GS-II from Griffonia simplicifolia (Molecular Probes). Cell surface and Golgi-specific staining was visualized by using AlexaFluor 594-conjugated wheat germ agglutinin (WGA). For colocalization of gB with gK, an IgG1 isotype anti-gB antibody (Rumbaugh-Goodwin Institute) was labeled with AlexaFluor 568 by using zenon technology according to the manufacturer's directions (Molecular Probes). Sections were labeled with this antibody at a 1:500 dilution. The nucleus was counterstained with TO-PRO-3 iodide (1:5,000 dilution) and visualized in the blue channel. Cells were examined by using a Leica TCS SP2 laser-scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a ×100 Leica objective lens (Planachromatic; 1.4 numerical aperture). Individual optical sections in the z axis, averaged eight times, were collected simultaneously in the different channels at a 512-by-512 pixel resolution as described previously (28). Images were compiled and rendered in Adobe Photoshop.
Immunohistochemistry.
Vero cell monolayers in six-well plates were infected with the indicated virus at an MOI of 5 and then incubated at 37°C for 12 h. Infected monolayers were washed with TBS-Ca/Mg and either fixed with electron microscopy-grade 3% paraformaldehyde or left unfixed (live). Immunohistochemistry was performed by utilizing the Vector Laboratories Vectastain Elite ABC kit essentially as described in the manufacturer's directions. Briefly, cells were washed with TBS-Ca/Mg and incubated in TBS blocking buffer supplemented with normal horse serum at room temperature for 1 h. After a blocking step, cells were reacted with anti-V5 antibody (1:500) in TBS blocking buffer for 2 h, washed four times with TBS blocking buffer, and incubated with biotinylated horse anti-mouse antibody. Excess antibody was removed by four washes with TBS-Ca/Mg and subsequently incubated with Vectastain Elite ABC reagent for 30 min. Finally, cells were washed three times with TBS-Ca/Mg, and reactions were developed with NovaRed substrate (Vector Laboratories) according to the manufacturer's directions.
Transfection and transfection-complementation assay.
Subconfluent Vero cells in six-well plates were transfected with the indicated plasmids utilizing the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's directions. At 16 h posttransfection (hpt) cells were either infected with ΔgK virus at an MOI of 3 or left uninfected. The infections were incubated for 12 h at 37°C, and the cells were then processed as indicated for immunohistochemistry.
For transient-coexpression experiments, gKDIV5 was cotransfected at 1:1 ratio with either pCMVUL20 or pcDNA3-gB/gD. The cell surface expression of gK was assessed at 36 hpt by immunohistochemistry of live cells.
RESULTS
Construction and characterization of HSV-1 recombinant viruses containing an in-frame epitope tag within each of the four putative gK domains.
At least 11 HSV-1 syncytial mutants have mutations within the UL53 gene encoding gK (3, 10). Despite the strong genetic association of gK with virus-induced cell fusion, previous reports indicated that HSV-1 wild-type and syncytial gKs were detected exclusively within the endoplasmic reticulum and nuclear membranes and not on the plasma membranes of infected cells, eliminating a direct role of gK in membrane fusion (20). Recently, we detected HSV-1 gK as a structural component of purified virions by using a protC-tagged gK and demonstrated that gK functioned in virus entry (14). This finding suggested that gK might also be expressed on cell surfaces, where it may function in virus-induced cell fusion. Unfortunately, the protC epitope tag was not efficiently detected in immunofluorescence or immunohistochemical assays. In order to facilitate the detection of gK, we generated a series of viruses that specified the V5 epitope tag within each of the four previously defined domains of gK (Fig. 1) (14). The recombinant gK genes coding for the epitope-tagged gKs were constructed by using PCR-based splice-overlap extension methodology as described previously (14) and cloned into plasmid pSJ1723, generating plasmids pTFgKV5I, pTFgKV5II, pTFgKV5III, and pTFgKV5IV. These plasmids contained gK-flanking sequences corresponding to the UL52 and UL54 (ICP27) genes to facilitate homologous recombination with viral DNA (22). Recombinant viruses gKV5DI, gKV5DII, gKV5DIII, and gKV5DIV were constructed by rescuing the mutant virus d27-1 (KOS) (39), which has a lethal deletion within the UL54 gene specifying the immediate-early protein ICP27 as detailed previously (12, 14, 22). Putative recombinant virus isolates were plaque purified and tested by PCR and DNA sequencing for the presence of contaminating d27-1 virus and the engineered epitope-tagged gK (not shown).
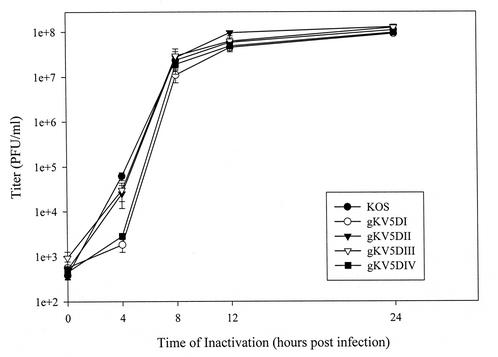
To evaluate whether insertion of the epitope tags within gK affected the replication of recombinant HSV-1 viruses in vitro, one-step replication kinetics for each of the recombinant viruses were analyzed relative to the wild-type KOS virus. Vero cells were inoculated with each virus and at the designated time points, infected monolayers were harvested, and the virus titers were determined by endpoint dilution of virus stocks on Vero cells (Fig. 2). All recombinant viruses exhibited replication kinetics similar to that of wild-type KOS virus, as evidenced by the final slopes of their virus production kinetics. Furthermore, the plaque morphology of each virus was indistinguishable from that of the parental KOS virus (data not shown). Similarly, insertion of the V5 tags within gK carrying the syn20 mutation (Ala40-to-Val40) did not affect either the replication kinetics of these recombinant viruses or the syncytial plaque morphology in comparison to their parental virus KOS (gKsyn20). These results indicated that insertion of the V5 epitope tag within the tagged gK regions did not significantly affect the structure and function of gK with regard to virus replication, cell-to-cell spread, and virus-induced cell fusion.
FIG. 2.
Comparison of replication characteristics of V5-tagged recombinant viruses to the wild-type strain KOS: one-step kinetics of infectious virus production after infection of Vero cells at an MOI of 5 and incubation at 37°C.
Golgi-dependent glycosylation and cell surface expression of gK.
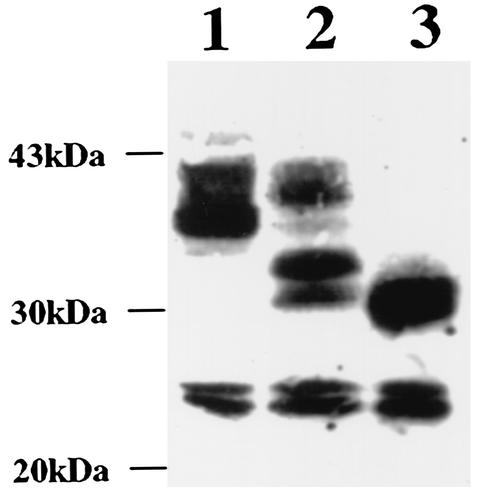
Previously, it was shown that HSV-1 gK exclusively localized to the rough endoplasmic reticulum (RER) and did not contain peripheral carbohydrates, which are normally added in the Golgi apparatus (20). To assess the level of Golgi-dependent glycosylation and distribution of gK in infected cells, we characterized gK tagged with the protC tag or the V5 tag inserted within domain III in Western analyses or immunofluorescence assays, respectively. The level of Golgi-dependent glycosylation of gK in infected cells was assessed in Western analyses optimized for the detection of highly glycosylated gK species (Fig. 3). Vero cells were infected with HSV-1 gKprotC virus, and cellular extracts were prepared at 24 hpi and were treated with either PNGase F or Endo-H or mock treated. Mock-treated samples contained gK species migrating with apparent molecular masses of 40 to 43, 36 to 38, 23, and 25 kDa (Fig. 3, lane 1). Treatment with Endo-H, which is specific for high-mannose carbohydrate containing precursor glycoproteins, substantially reduced the 36- to 38-kDa gK species, whereas it did not appreciably alter the relative amounts of the 40- to 43-, 23-, and 25-kDa gK species. In addition, Endo-H treatment resulted in the appearance of new gK species migrating with apparent masses of 31 and 33 kDa (Fig. 3, lane 2). Treatment of cellular extracts with PNGase F, which cleaves most carbohydrates from both underglycosylated and fully glycosylated glycoproteins, resulted in the appearance of a new gK species of 29 to 31 kDa, whereas the 23- and 25-kDa proteins remained unaffected (Fig. 3, lane 3). These results clearly demonstrated that gK was processed by Golgi enzymes to fully glycosylated forms resistant to Endo-H treatment.
FIG. 3.
Differentiation of fully and underglycosylated gK species specified by gKprotC-DIII virus. Immunoblots of gKprotC-DIII (lanes 1 to 3)-infected-cell extracts reacted with anti-protC monoclonal antibody HPC-4. Cellular extracts were either treated with Endo-H (lane 2) or PNGase F (lane 3) or were mock treated (lane 1).
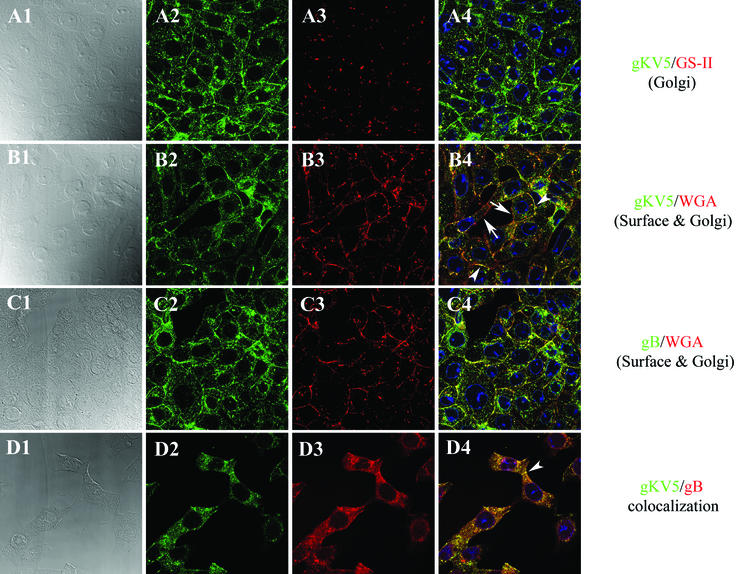
To visualize and compare the cellular distribution of gK and gB within infected cells, Vero cells were infected with the gKV5DIII virus, and these glycoproteins were detected by confocal immunofluorescence microscopy with anti-V5 and anti-gB antibodies, as well as specific Golgi apparatus and plasma membrane markers, including lectin GS-II, which specifically stains the Golgi apparatus (45) and WGA, which stains both cell surfaces and the Golgi apparatus (7) (Fig. 4). gK was detected throughout the cytoplasm, as well as on the surface of infected cells (Fig. 4A2, B2, and D2). The GS-II Golgi marker stained the Golgi apparatus as punctate-appearing structures in the cytoplasm of infected cells (Fig. 4A3). In agreement with the Endo-H and PNGase results cited above, gK colocalized within the Golgi apparatus with lectin GS-II (Fig. 4A4). Lectin WGA efficiently stained both Golgi and plasma membranes of infected cells (Fig. 4B3 and C3). Superimposition of the gK-specific fluorescence onto the WGA-specific fluorescence clearly showed that gK and WGA colocalized both in Golgi membranes and on cell surfaces (Fig. 4B4). The cell surface colocalization was detected on the juxtaposed (demarcated by arrowheads) but not on noncontacting cellular surfaces (demarcated by arrows). A similar cellular distribution was observed for gB (Fig. 4C1 to C4), which localized to both Golgi and plasma membranes (Fig. 4C4) and was used in these experiments as an additional reference standard. Both gB and gK are intimately involved in virus-induced cell fusion, inasmuch as single amino acid replacements of both proteins are known to cause extensive cell fusion. Costaining experiments showed that gB and gK colocalized throughout the infected cells and, particularly, at contacting cell surfaces (Fig. 4D1 to D4), indicating that gK localizes to similar cellular membranes and cellular organelles as HSV-1 gB.
FIG. 4.
Subcellular distribution of gK and gB in gKV5DIII virus-infected cells. Vero cells were infected with gKV5DIII at an MOI of 5. Cells were fixed and processed for confocal microscopy; panels show staining with anti-V5 (green: A2, A4, B2, B4, D2, and D4), anti-gB (green: C2 and C4), or anti-gB (red: D3 and D4) antibodies at 12 hpi. Cellular organelles were counterstained with TO-PRO3 to specifically stain the nucleus (blue: A4, B4, C4, and D4) and with either lectin GS-II, which specifically stains the Golgi apparatus (red: A3 and A4), or lectin WGA to label Golgi and plasma membranes (red: B3, B4, C3, and C4). Corresponding DIC images of cells are as shown (A1, B1, C1, and D1). Superimpositions of red and green images for each group of images are shown (A4, B4, C4, and D4).
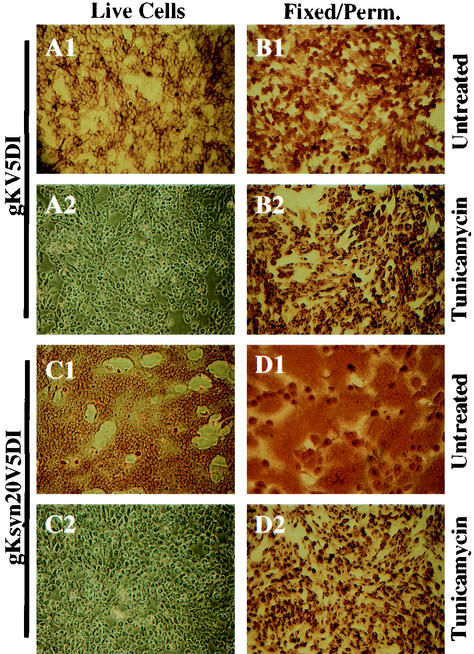
Conclusive detection of gK on cell surfaces requires the restriction of anti-V5 antibody reactivity to gK portions exposed on the extracellular side of cellular membranes. Previously, we used immunohistochemical assays to isolate monoclonal antibody-resistant mutant viruses (mar) by selecting nonreacting viruses under live cellular conditions (26, 34). We employed a similar, but improved immunohistochemical methodology to detect gK on plasma membranes of live cells, as well as within fixed and permeabilized cells. Cleavage of the amino-terminal 30 amino acids and the addition of carbohydrate residues at amino acids 48 and 58 of gK predict that the amino-terminal domain I of gK is an ectodomain that should localize to extracellular spaces, if gK is expressed in plasma membranes of infected cells. Furthermore, the gK syn20 mutation (Ala40-to-Val40), which is located within domain I, causes extensive cell fusion also implying that gK domain I is exposed to extracellular spaces, where it could be involved in gK-mediated cell fusion. Therefore, we investigated whether gK and gKsyn20 tagged with the V5 epitope within domain I could be detected on cell surfaces. Vero cells were infected with either gKV5DI (Fig. 5A1, A2, B1, and B2) or gKsyn20V5DI (Fig. 5C1, C2, D1, and D2) viruses and incubated in the presence (Fig. 5A2, B2, C2, and D2) or absence (Fig. 5A1, B1, C1, and D1) of 5 μg of tunicamycin (TM)/ml at 37°C. TM inhibits the transfer of carbohydrate residues to the backbone polypeptide onto Asn-X-Ser/Thr sites of proteins in the RER and, thus, prevents protein transport out of the endoplasmic reticulum to Golgi and plasma membranes (6, 46, 47). Immunohistochemical assays were performed on either live infected cells (Fig. 5A1, A2, C1, and C2) or paraformaldehyde-fixed and Triton X-100-permeabilized cells (Fig. 5B1, B2, D1, and D2) prepared at 12 hpi. HSV-1 gK localized to cell surfaces in both the wild type (Fig. 5A1)- and syn20 (Fig. 5C1)-infected live cells, whereas there was no gK detected within plasma membranes of infected live cells treated with TM (Fig. 5A2 and C2). Furthermore, treatment of syn20-infected cells with TM inhibited cell fusion (Fig. 5C2) and caused blocking of gK to perinuclear regions of infected cells (Fig. 5B2 and D2).
FIG. 5.
Cell surface immunohistochemical detection of gK in gKV5DI- or gKsyn20V5DI-infected cells. Vero cells were infected with gKV5DI (A1, B1, A2, and B2) or gKsyn20V5DI (C1, D1, C2, and D2) at an MOI of 5 and were either treated with TM (A2, B2, C2, and D2) or mock treated (A1, B1, C1, and D1). At 12 hpi, infected cells were immunohistochemically processed under either live (A1, A2, C1, and C2) or fixed and permeabilized (B1, B2, D1, and D2) conditions with anti-V5 antibody.
Determination of the cell surface membrane topology of gK.
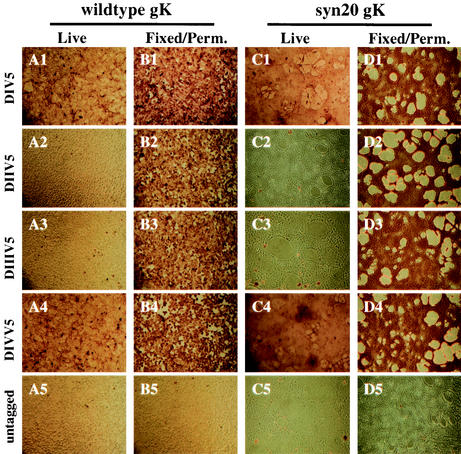
HSV-1 gK was initially predicted to contain four hpd capable of transversing cellular membranes (8). This predicted orientation placed both the gK amino and carboxyl termini on the extracellular or luminal sides of cellular membranes. However, more recently, Mo and Holland (32) concluded on the basis of in vitro-transcribed and -translated deletion, insertion, and truncation mutants of gK that gK contained three active transmembrane regions. This gK arrangement placed domains I and III of gK on the extracellular or luminal side of cellular membranes, whereas domains II and IV were located on the cytoplasmic side of cellular membranes. To ascertain which of the two models accurately depicted the gK arrangement in infected-cell membranes, we determined the molecular topography of individual gK domains on cell surfaces. Confluent Vero cells in six-well plates were infected with the indicated epitope-tagged viruses and incubated at 37°C. At 12 hpi, cells were washed and then either incubated under live conditions (Fig. 6A1 to A5 and C1 to C5) or fixed and permeabilized with 0.1% Triton X-100 (Fig. 6B1 to B5 and D1 to D5). Immunohistochemical analysis indicated that wild-type HSV-1 gK domains I and IV were exposed on extracellular surfaces (Fig. 6A1 and A4, respectively), whereas domains II and III were not accessible to the antibody, suggesting that they localized intracellularly (Fig. 6A2 and A3, respectively). Similarly, gKsyn20 domains I and IV were detected on the extracellular surfaces (Fig. 6C1 and C4, respectively), whereas domains II and III were not detected on cell surfaces (Fig. 6C2 and C3, respectively). HSV-1 gK could be clearly detected in permeabilized cells infected with all epitope-tagged recombinant viruses (Fig. 6B1 to B4 and D1 to D4). As expected, there was no immunohistochemical staining produced by wild-type HSV-1 KOS- or MP (syn20)-infected cells, which served as negative controls (Fig. 6A5, B5, C5, and D5).
FIG. 6.
Topological arrangement of wild-type and syn20 gK domains on virus-infected-cell surfaces. Vero cells were infected with gKV5DI (A1 and B1), gKsyn20V5DI (C1 and D1), gKV5DII (A2 and B2), gKsyn20 V5DII (C2 and D2), gKV5DIII (A3 and B3), gKsyn20V5DIII (C3 and D3), gKV5DIV (A4 and B4), gKsyn20V5DIV (C4 and D4), KOS (A5 and B5), or MP (C5 and D5) at an MOI of 5. At 12 hpi, infected cells were immunohistochemically processed under either live (A1 to A5 and C1 to C5) or fixed and permeabilized (B1 to B5 and D1 to D5) conditions with anti-V5 antibody.
Cell surface expression of gK requires viral functions.
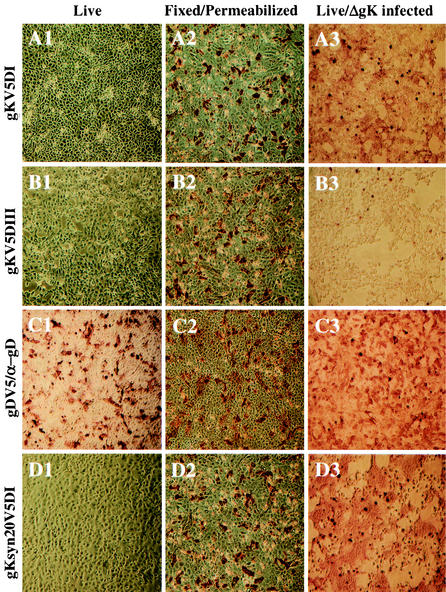
Previous attempts to incorporate gKsyn20 into a virus-free cell fusion system were not successful, indicating that perhaps gK required additional viral proteins or other viral functions for transport to cellular membranes and expression of its syncytial properties (13, 25). Therefore, we investigated whether plasma membrane expression of gK could be induced after infection of transfected cell surfaces by immunohistochemistry. Subconfluent Vero cells were transfected with the indicated plasmids and incubated at 37°C for 16 h. Thereafter, the cells were left uninfected (Fig. 7A1, A2, B1, B2, C1, C2, D1, and D2) or were infected with the gK-null virus ΔgK (Fig. 7A3, B3, C3, and D3) and incubated at 37°C for an additional 12 h. Live cells (Fig. 7A1, B1, C1, D1, A3, B3, C3, and D3) or fixed and permeabilized cells (Fig. 7A2, B2, C2, and D2) were immunohistochemically stained with anti-V5 (Fig. 7A, B, and D) or anti-gD (Fig. 7C) antibodies to determine cell surface localization of gK or gD, respectively. All transfections produced similar transfection efficiencies and levels of glycoprotein expression (Fig. 7A2, B2, C2, and D2). In contrast to gK epitope-tagged HSV-1-infected cells, neither gK domain I (Fig. 7A1) nor gK domain IV (data not shown) could be detected on cell surfaces when expressed in the absence of viral infection. Similarly, the gKsyn20 tagged with V5 in domain I was not detected on the plasma membrane (Fig. 7D1). As expected, both the cytoplasmically localized V5-tagged gD and gK domain III were not detected on surfaces of transfected cells (data not shown and Fig. 7B1, respectively). In contrast, anti-gD antibody readily detected gD on transfected cell surfaces (Fig. 7C1), indicating that gD did not require any viral functions for cell surface expression.
FIG. 7.
Transport of gK to cell surfaces and gK-mediated cell fusion require additional viral functions. Vero cells were transfected with plasmids expressing gKV5DI (A1 to A3), gKV5DIII (B1 to B3), gKsyn20V5DI (D1 to D3), and gDV5 (C1 to C3). At 16 hpt, cells were either mock infected (A1, B1, C1, D1, A2, B2, C2, and D2) or infected with ΔgK (A3, B3, C3, and D3). At 12 hpi, infected cells were immunohistochemically processed under either live (A1, B1, C1, D1, A3, B3, C3, and D3) or fixed and permeabilized (A2, B2, C2, and D2) conditions with anti-V5 antibody (A1 to A3, B1 to B3, and D1 to D3) or anti-gD (C1 to C3).
Next, we investigated whether plasma membrane expression of gK could be induced after infection of transfected cells with the gK-null virus, ΔgK. HSV-1 gK tagged within domains I was readily detected on cell surfaces after infection with ΔgK (Fig. 7A3), whereas gK and gD tagged intracellularly with V5 were not detected on cell surfaces (Fig. 7B3 and data not shown, respectively). Similarly, infection of gKsyn20-transfected cells with ΔgK virus resulted in detection of gKsyn20 on cell surfaces. Importantly, cell surface expression of gKsyn20 was associated with extensive cell fusion (Fig. 7D3). These results indicate that plasma membrane expression of gK and the gK-associated syncytial phenotype requires the presence of virally encoded functions.
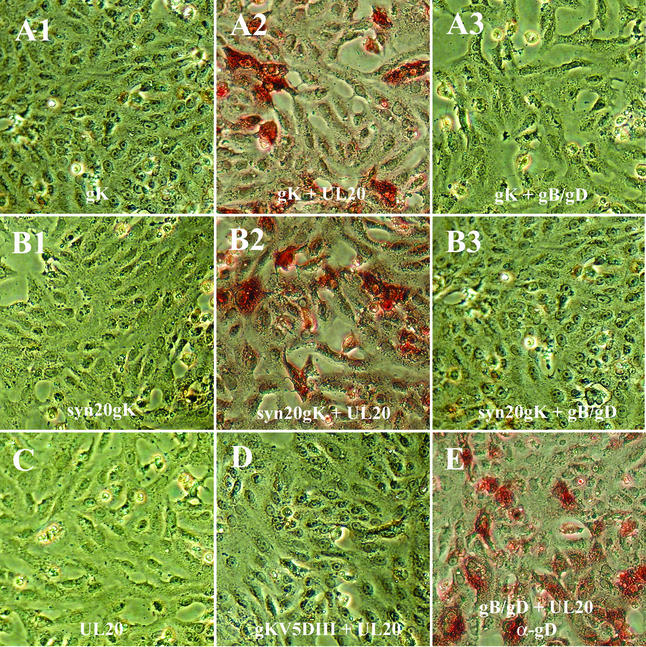
Expression of the PRV UL20 protein enhanced PRV gK intracellular transport and Golgi-dependent glycosylation (9). To investigate whether HSV-1 UL20 enabled cell surface expression of gK, Vero cells were cotransfected with either individual plasmids for control purposes or with combinations of two different plasmids. Coexpression of UL20 with either gKV5DI or gKsyn20V5DI enabled efficient detection of gK on transfected cell surfaces (Fig. 8A2 and B2). However, cell surface expression of the gKsyn20V5DI did not cause any cell-to-cell fusion (Fig. 8B2). As expected, gKV5DIII was not detected on cell surfaces when cotransfected with UL20 (Fig. 8D), since the V5 epitope inserted in gK domain III is localized intracellularly. Coexpression of gB/gD with gK failed to transport gK to cell surfaces (Fig. 8A3 and B3). As a positive control, gD was detected on cell surfaces by using anti-gD antibody (Fig. 8E) and was not dependent on UL20 for cell surface localization as shown in Fig. 7C1. Control transfections with the UL20 plasmid alone did not produce any cell surface signal (Fig. 8C).
FIG. 8.
Coexpression of the UL20 protein enables transport of gK to cell surfaces but not gK-mediated cell-to-cell fusion. Vero cells were transfected with either individual plasmids or combinations of plasmids. Transfections with individual plasmids were as follows: pCMV-gKV5DI (A1), pCMVgKsyn20V5DI (B1), pCDNA3-gB/gD (E), and pCMVUL20 (C). Transfections with combinations of plasmids were as follows: pCMV-gKV5DI + pCMVUL20 (A2), pCMVgKsyn20V5DI + pCMVUL20 (B2), pCMV-gKV5DIII + pCMVUL20 (D), pCDNA3-gB/gD + pCMV-gKV5DI (A3), and pCDNA3-gB/gD + pCMVgKsyn20V5DI (B3). Cell surfaces of transfected cells were stained under live conditions with anti-V5 antibody for detection of gK, except for the cells shown in panel E, which were stained with anti-gD antibody for the detection of gD.
DISCUSSION
Most spontaneous mutations that cause virus-induced cell fusion are single amino acid changes in gK. Furthermore, gK syncytial mutants cause fusion of a broader range of cell types in comparison to syncytial mutations found in gB or elsewhere in the viral genome. Despite the well-defined genetic association of gK with cellular membrane fusion phenomena, the function of gK in these events has remained elusive since its discovery in 1985 (8), largely due to lack of appropriate immunological reagents necessary for its characterization. Early reports that gK could not be detected in virions and on infected-cell surfaces further complicated the role of gK in virus-associated membrane fusion (20). Recently, we constructed a recombinant virus specifying an antigenic epitope within gK and demonstrated that gK was a structural component of the HSV-1 virion functioning in virion entry into susceptible cells (14). In the present study, we focused on determining whether wild-type and syncytial forms of gK are expressed on infected-cell surfaces, thereby enabling their direct involvement in membrane fusion. In addition, we compared the topological arrangement of wild-type and syncytial gK in plasma membranes to assess whether syncytial mutations significantly altered the plasma membrane topology of gK. We demonstrate here that (i) insertion of 13-amino-acid antigenic epitopes within each of the four gK domains does not significantly affect the structure and function of gK, (ii) gK is expressed on infected-cell surfaces, (iii) gK expression on cell surfaces is required for gK-mediated cell fusion, (iv) gK assumes a membrane topology that places the amino and carboxyl termini in the extracellular spaces while domains II and III are located in the cytoplasm, (v) coexpression of the UL20 protein is necessary and sufficient for gK cell-surface expression, and (vi) additional viral functions are required for expression of the syncytial phenotype of gK, since coexpression of UL20 enabled cell surface expression of gK but not gK-mediated cell-to-cell fusion.
Localization of gK within Golgi and plasma membranes.
We clearly demonstrate here that gK is expressed within Golgi and plasma membranes of infected cells. However, it was previously shown with rabbit anti-peptide antibodies that gK was exclusively localized in the perinuclear spaces of infected cells and contained immature carbohydrates added in RER, indicating that gK was not transported to the Golgi apparatus (20). It is possible that anti-peptide antibodies used in these studies detected only immature forms of gK (high-mannose precursors) that were localized in the perinuclear spaces of cells. Furthermore, because gK is a γ2 viral protein (8), relatively low levels of gK will be expressed at early times after infection (9 to 12 hpi), further limiting its detection. Finally, anti-peptide antibodies may not react with their targeted epitopes because these epitopes are masked by fully processed gK interactions with one or more viral or cellular proteins.
Immunohistochemical and confocal analysis confirmed that gK was expressed within Golgi and plasma membranes of virus-infected cells with a distribution similar to that of gB, which is also a principal player in virus-induced cell fusion. Both wild-type and syncytial gK (gKsyn20) were efficiently expressed on infected-cell surfaces, indicating that the syn20 mutation did not appreciably alter cell surface expression of gK. Therefore, it is likely that both gB and gK function in the fusion of plasma membranes via direct interaction with apposed membranes. The preferential localization of gK in juxtaposed cellular membranes further supports a direct role of gK in cell-to-cell fusion.
The finding that gK is expressed in the Golgi apparatus, as well as on cell surfaces, suggests that gK may function in post-Golgi virion transport and egress. This is in agreement with our previous findings that truncated or mutated gK caused accumulation of virions within cytoplasmic vesicles and drastic inhibition of virus spread (12). It remains to be investigated whether the cytoplasmic vesicles, which contained gK-null virions, are Golgi apparatus derived.
Cell surface-expressed gK may recirculate to the trans-Golgi network in a manner analogous to other herpes glycoproteins. Indeed, gK domain II, predicted to localize within the cytoplasm, contains a YXXΦ motif, which is known to function in internalization and Golgi-targeted retrieval of surface proteins. In this regard, it is notable that mutagenesis of the tyrosine residue of this motif caused a gK-null phenotype (12). It remains to be tested whether modification of the YXXΦ motif causes substantial inhibition of gK internalization and/or significant alteration of cell surface distribution, especially in polarized cells. In this context, it has been suggested that expression of gK in PRV-infected cells may prevent reinfection of extracellularly found virions by inhibiting fusion between viral envelopes and the plasma membrane. This conclusion was based on the use of a gK-null PRV mutant, which seemed to allow the reentry of virions found in the extracellular spaces (24). HSV-1 gK, by virtue of the fact that it is expressed on cell surfaces, may act in a similar manner to inhibit virus reentry into infected cells.
Topological arrangement of gK domains in plasma membranes.
Insertions of the 13-amino-acid V5 epitope tag within the four different sites of gK did not appreciably affect virus replication or phenotypic properties. Furthermore, insertion of the epitope tags in gK carrying the syn20 gK mutation did not alter the ability of gK to cause extensive virus-induced cell fusion. These results suggest that gK possesses multiple functional domains that are topologically and functionally distinct. For example, the extracellular domains I and IV may function in virus-induced cell fusion and virion entry, whereas domains II and III may function in glycoprotein transport and virion egress, as suggested by site-directed mutagenesis and deletion analysis of gK (12, 14).
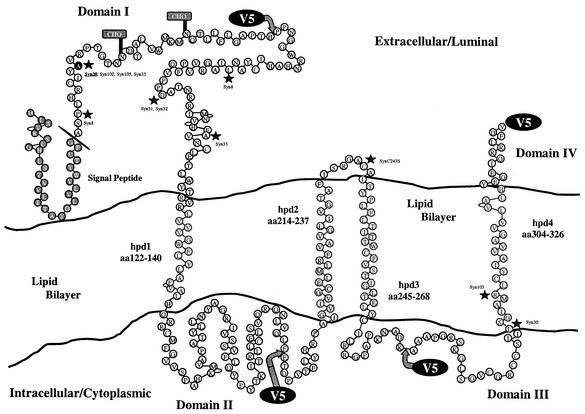
The initial prediction of the secondary structure of gK suggested the presence of four membrane-spanning domains (8). Subsequent studies utilizing in vitro-translated gK, which contained truncations, as well as in-frame deletions of putative transmembrane regions, showed that gK domain III localized to luminal or extracellular sides of membranes, whereas domain IV was a cytoplasmic domain. This gK membrane topology was particularly appealing because it placed all syncytial mutations in gK in the extracellular spaces. These results were based on membrane protection experiments of gK carrying in-frame deletions of either hpd1 and hpd2 or hpd1, hpd2, and hpd4 (32). It is likely that deletion of these membrane-spanning domains caused substantial alteration of gK's structure, leading to entrapment of the truncated gK within the lumen of the microsomal membranes. It is also possible that hpd3 can only act as a membrane-spanning region in the presence of hpd2. These conclusions are corroborated by additional evidence obtained in our laboratory that gK carrying a deletion of hpd4 did not localize to cell surfaces (data not shown), implying that this truncated gK may be substantially misfolded and entrapped in the RER (T. P. Foster and K. G. Kousoulas, unpublished results). Immunohistochemical detection of V5-tagged gK expressed during infection conclusively demonstrates that domains I and IV assumed an extracellular orientation, whereas domains II and III were oriented toward the cytoplasm (Fig. 9). This gK orientation places the syncytial mutations within gK domain III in the cytoplasm. Furthermore, the syn20 mutation did not alter gK membrane topology, since both the syn20 and wild-type gKs assumed the same topological orientation. Recalculation of the hydrophobic and membrane-spanning potential of all hpd's by using the SPLIT, TMPred, and SOSUI computer algorithms (15, 16, 23) resulted in slight adjustments of the membrane-spanning domains (Fig. 1 and 9), but otherwise the gK topography was largely in agreement with the original prediction that gK had four hpd's (8). Interestingly, the syncytial mutations syn30 and syn103 (3, 4, 32) are contained within the new hpd4 membrane-spanning region. Therefore, these mutations must function indirectly by perturbing fusogenic domains of gK presumably located extracellulary. It is likely that domain I directly participates in membrane fusion phenomena because it contains the majority of the known syncytial mutations. Preliminary evidence in our laboratory indicates that domain IV may also regulate membrane fusion between the viral envelope and cellular membranes during entry. This conclusion stems from the observation that mutant virus ΔgKhpd4, which carries a deletion of the 13-amino-acid carboxyl tail of gK, is defective in virus entry, although it exhibits replication and cell-to-cell transmission characteristics similar to those of its parental wild-type virus (G. V. Rybachuk, T. P. Foster, and K. G. Kousoulas, unpublished data).
FIG. 9.
Experimentally validated model of HSV-1 gK membrane topology. The originally predicted four hpd's of gK (8) were adjusted according to computer-based predictions and are shown as embedded within the lipid bilayer. The locations of the sites where the four V5 tags were inserted and the experimentally derived topology of each tagged domain are as shown. The schematic includes demarcations for the signal peptide and its cleavage site, the two N-linked glycosylation sites within domain I, and the location of syncytial mutations.
Requirements for cell surface expression of gK.
Transient expression of gK after transfection of cells indicated that gK was unable to be expressed on cell surfaces in the absence of viral infection, despite high levels of gK expression relative to those achieved during viral infection. Infection of transfected cells with a gK-null virus resulted in efficient translocation of gK to the plasma membrane, as well as substantial gK-mediated cell-to-cell fusion. Similarly, transient coexpression of the UL20 protein enabled efficient cell surface localization of gK but not gK-mediated cell fusion. Therefore, the UL20 protein was necessary and sufficient for gK transport to plasma membranes of transfected cells but not for gK-mediated membrane fusion, indicating that additional viral functions are required for expression of the gK syncytial phenotype. These results are in agreement with recent findings that the PRV UL20 protein enhanced Golgi-dependent processing of PRV gK (9). There is substantial scientific precedence for the coordinate expression and localization of glycoproteins in herpesvirus-infected cells. For example, glycoprotein H requires heterodimer formation with gL for proper transport and cell surface expression (17). Glycoproteins gE and gI form multimers that affect the membrane targeting of gI (1). Therefore, the UL20 protein may physically interact with gK and facilitate gK intracellular transport and cell surface expression, suggesting that the UL20 protein is also localized to plasma membranes.
Fundamental questions about the role of gK in multiple virus-associated membrane fusion events occurring during virus entry, virus-induced cell fusion, and intracellular virion transport remain unanswered. Is gK a membrane fuser or a modulator of membrane fusion caused by other glycoproteins such as gB and the gH/gL heterodimer? Development of a virus-free membrane fusion system, which incorporates the syncytial phenotypes of both gB and gK, would help define fundamental mechanisms that govern alphaherpesvirus glycoprotein-mediated membrane fusion phenomena. In this regard, the inability of syncytial gK to cause cell-to-cell fusion in the absence of viral infection explains previous findings that syncytial gK did not enhance cell fusion in the virus-free cell fusion assay when coexpressed with gB, gH/gL, and gD (13, 25, 48) or with the UL20 protein (Foster and Kousoulas, unpublished). Elucidation of additional molecular partners of gK necessary for expression of its syncytial phenotype may ultimately enable the generation of both gB- and gK-specific virus-free cell fusion systems.
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (AI43000) to K.G.K. and by a base grant to the Tulane National Primate Research Center (RR00164).
REFERENCES
- 1.Alconada, A., U. Bauer, L. Baudoux, J. Piette, and B. Hoflack. 1998. Intracellular transport of the glycoproteins gE and gI of the varicella-zoster virus: gE accelerates the maturation of gI and determines its accumulation in the trans-Golgi network. J. Biol. Chem. 273:13430-13436. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 4.Bond, V. C., S. Person, and S. C. Warner. 1982. The isolation and characterization of mutants of herpes simplex virus type 1 that induce cell fusion. J. Gen. Virol. 61:245-254. [DOI] [PubMed] [Google Scholar]
- 5.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Bzik, D. J., S. Person, and G. S. Read. 1982. The active inhibition of herpes simplex virus type 1-induced cell fusion. Virology 117:504-509. [DOI] [PubMed] [Google Scholar]
- 7.Cottin, V., A. Van Linden, and D. W. Riches. 1999. Phosphorylation of tumor necrosis factor receptor CD120a (p55) by p42mapk/erk2 induces changes in its subcellular localization. J. Biol. Chem. 274:32975-32987. [DOI] [PubMed] [Google Scholar]
- 8.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 9.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolter, K. E., R. Ramaswamy, and T. C. Holland. 1994. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J. Virol. 68:8277-8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, T. P., V. N. Chouljenko, and K. G. Kousoulas. 1999. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology 258:365-374. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An α-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 14.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMbase—database of membrane spanning segments. Biol. Chem. 347:166. [Google Scholar]
- 17.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchinson, L., C. Roop-Beauchamp, and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J. Virol. 69:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson, J. G., S. L. Martin, and D. M. Coen. 1989. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63:1839-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juretic, D., D. Zucic, B. Lucic, and N. Trinajstic. 1998. Preference functions for prediction of membrane-buried helices in integral membrane proteins. Comput. Chem. 22:279-294. [DOI] [PubMed] [Google Scholar]
- 24.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kousoulas, K. G., P. E. Pellett, L. Pereira, and B. Roizman. 1984. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology 135:379-394. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, B. S., X. Alvarez, S. Ishido, A. A. Lackner, and J. U. Jung. 2000. Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi's sarcoma-associated herpesvirus K1. J. Exp. Med. 192:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897-906. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 32.Mo, C., and T. C. Holland. 1997. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K. J. Biol. Chem. 272:33305-33311. [DOI] [PubMed] [Google Scholar]
- 33.Mo, C., J. Suen, M. Sommer, and A. Arvin. 1999. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J. Virol. 73:4197-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 36.Pogue-Geile, K. L., and P. G. Spear. 1987. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67-74. [DOI] [PubMed] [Google Scholar]
- 37.Ramaswamy, R., and T. C. Holland. 1992. In vitro characterization of the HSV-1 UL53 gene product. Virology 186:579-587. [DOI] [PubMed] [Google Scholar]
- 38.Read, G. S., S. Person, and P. M. Keller. 1980. Genetic studies of cell fusion induced by herpes simplex virus type 1. J. Virol. 35:105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Ruyechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 43.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 44.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press, Boca Raton, Fla.
- 45.Suzaki, E., and K. Kataoka. 1999. Three-dimensional visualization of the Golgi apparatus: observation of Brunner's gland cells by a confocal laser scanning microscope. J. Struct. Biol. 128:131-138. [DOI] [PubMed] [Google Scholar]
- 46.Takatsuki, A., K. Arima, and G. Tamura. 1971. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. 24:215-223. [DOI] [PubMed] [Google Scholar]
- 47.Tkacz, J. S., and O. Lampen. 1975. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem. Biophys. Res. Commun. 65:248-257. [DOI] [PubMed] [Google Scholar]
- 48.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]