Abstract
In the oral poliovirus vaccine, three attenuated virus strains generated by Albert Sabin are used. However, insufficient genetic stability of these strains causes major problems in poliovirus eradication. In infected cells, translation of the plus-strand poliovirus RNA genome is directed by the internal ribosome entry site (IRES), a cis-acting RNA element that facilitates the cap-independent binding of ribosomes to an internal site of the viral RNA. In each Sabin vaccine strain, a single point mutation in the IRES secondary-structure domain V is a major determinant of neurovirulence attenuation. Here we report how these decisive mutations in the IRES confer a reduction in poliovirus translation efficiency. These single-nucleotide exchanges impair the interaction of the standard translation initiation factor eIF4G with the IRES domain V. Moreover, binding of eIF4B and the polypyrimidine tract-binding protein and the association of ribosomes with the viral RNA are affected by these mutations. However, the negative effects of the IRES mutations are completely relieved by addition of purified eIF4F. This indicates that eIF4G is the crucial factor that initially binds to the poliovirus IRES and recruits the IRES to the other components of the translational apparatus, while impaired binding of eIF4G plays a key role in attenuation of poliovirus neurovirulence.
Three attenuated poliovirus live-vaccine strains that had been obtained after serial cell culture passages (40) are successfully used in the oral poliovirus vaccine (OPV) for fighting poliovirus. Nevertheless, the exclusive use of these OPV strains for globally eradicating the virus is hampered by its insufficient genetic stability (18), which causes major challenges for public health organizations (27). In many cases, the genetic instability of the OPV strains resides in the 5′ untranslated region (5′-UTR) of the poliovirus plus-strand genomic RNA (4, 43). Here we report how these decisive mutations in the viral 5′-UTR confer a reduction in poliovirus translation efficiency.
After infection of a susceptible cell, the plus-strand poliovirus RNA is directly used for translation of viral gene products. Initiation of translation is mediated by the internal ribosome entry site (IRES), a cis-acting element in the viral 5′-UTR. This highly structured RNA element (Fig. 1) (5, 37) is assumed to serve as a binding site for translation initiation factors which mediate the binding of ribosomes to an internal site of the viral RNA (15, 34). This strategy of cap-independent synthesis of the viral polyprotein allows poliovirus to shut down the cap-dependent cellular translation (for an overview, see reference 2).
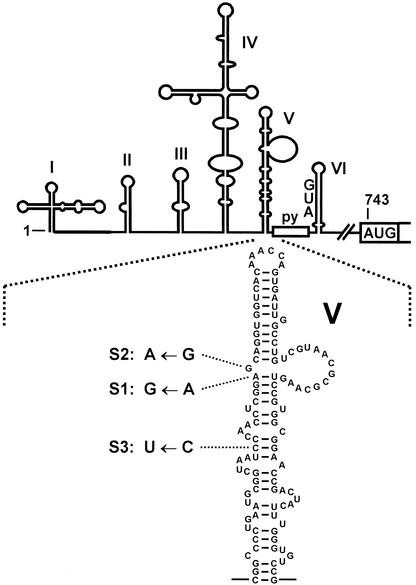
FIG. 1.
5′-UTR of poliovirus type 1. The IRES includes the predicted secondary-structure domains II to VI, the oligopyrimidine tract (py), and the silent AUG in domain VI. The authentic polyprotein start codon is at position 743. In this study, the single nucleotide exchanges that contribute to the attenuation of neurovirulence of the respective poliovirus Sabin vaccine strains were generated artificially in the domain V (blow-up) of the otherwise unchanged poliovirus type 1 IRES sequence. In these constructs (named S1, S2, and S3, respectively), the luciferase sequence is fused directly to the poliovirus initiator AUG at position 743.
According to their RNA secondary structures, the IRES elements of poliovirus and other members of the Picornaviridae family are classified in three groups, the type I elements of the enteroviruses and rhinoviruses including poliovirus, the type II elements of the cardioviruses and aphthoviruses including foot-and-mouth-disease virus (FMDV) and encephalomyocarditis virus (EMCV), and the type III element of hepatitis A virus. Picornavirus IRES elements interact with two different types of cellular RNA-binding proteins. On one hand, most of the standard eukaryotic translation initiation factors (eIFs) except the cap-binding protein eIF4E appear to be involved in picornavirus translation (35). Using the UV cross-link assay, direct physical interaction of initiation factors eIF4B and eIF4G has been shown for the IRES of FMDV (23, 25, 39, 41) and the closely related EMCV (35). In contrast, eIF4B and eIF4G are not essential for translation mediated by the IRES of the more distantly related hepatitis C virus (HCV) (36), a member of the Flaviviridae family.
On the other hand, several cellular RNA-binding proteins usually not involved in translation bind to picornavirus IRES elements and modulate their activity (2). The different groups of IRES elements appear to depend on these noncanonical factors to different extents, with the poliovirus IRES probably being the most demanding. In addition to the stimulation of the poliovirus IRES by the 57-kDa polypyrimidine tract-binding protein (PTB) (14), which also stimulates the EMCV (16) and FMDV (29) IRES, poliovirus IRES activity is modulated by the 52-kDa La protein (24) and the poly(rC)-binding protein PCBP2 (3). This obvious complexity of interactions with the poliovirus IRES may account for the fact that interaction of the standard initiation factors that basically guide the ribosome to the viral RNA is best understood for the IRES elements of EMCV and FMDV. While a possible eIF4G-binding site is not yet known for the poliovirus and rhinovirus IRES, it has been shown that eIF4G and eIF4B bind to a large Y-shaped RNA structure in the EMCV and FMDV IRES 3′ region (19, 23, 25, 35, 41).
Nevertheless, reasonable assumptions can be made about the region of the poliovirus IRES that is possibly involved in the binding of eIF4G. Domain I, the so-called cloverleaf, acts in viral RNA replication, whereas domains II to VI are involved mainly in translation (5, 28). Domain V appears to be the most important structure of the poliovirus IRES, since nearly any mutation affecting its structure is deleterious for IRES activity (10, 20, 33, 46). Moreover, attenuation of neurovirulence is conferred mainly by each a single point mutation in the IRES domain V sequence of the poliovirus Sabin vaccine strains (26) (Fig. 1). This is an A-to-G exchange in the Sabin 1 vaccine strain (nucleotide 480 according to the Sabin 1 nucleotide numbering) (17), a G-to-A exchange in the Sabin 2 strain (nucleotide 481 in the Sabin 2 strain) (38), and a C-to-U exchange in the Sabin 3 strain (nucleotide 472 in the Sabin 3 strain) (6). Consistent with a crucial role of domain V in poliovirus IRES function, we recently found that eIF4B binds strongly to domain V and is involved in ribosomal initiation complexes with the poliovirus IRES (32).
Here we identify the poliovirus IRES domain V to be required for binding of eIF4G and show that the single nucleotide exchanges involved in poliovirus attenuation reduce the binding of translation initiation factors eIF4B and eIF4G as well as the association of ribosomes with the poliovirus RNA.
MATERIALS AND METHODS
Plasmids.
pMPolio contains, downstream of an SP6 RNA polymerase promoter, the complete poliovirus type 1 (Mahoney) 5′-UTR sequence with the authentic initiator AUG (positions 743 to 745) fused to the firefly luciferase coding sequence (32). For construction of pMPolio-ΔI-III, pMPolio-ΔIV, pMPolio-ΔV, and pMPolio-ΔVI, the corresponding poliovirus 5′-UTR sequences were precisely removed from pMPolio by PCR mutagenesis. pMPolio-S1, pMPolio-S2, and pMPolio-S3 were derived from pMPolio by introducing single nucleotide exchanges (Fig. 1) in the domain V sequence of the otherwise unmodified poliovirus type 1 IRES.
Preparation of RNAs.
Plasmids were linearized with BsiWI in the luciferase sequence. Labeled RNAs were synthesized using SP6 RNA polymerase in the presence of 2.5 μM [α-32P]UTP (400 Ci/mmol; Amersham) plus 10 μM nonradioactive labeling nucleotide (30). The resulting RNAs contained 9 nucleotides of linker sequence at the 5′ end, the complete or mutated poliovirus 5′-UTR from positions 1 to 742, and 156 nucleotides of luciferase sequence. RNAs were separated from unincorporated nucleotides by gel filtration on Sephadex G-50 columns (Pharmacia). Unlabeled competitor RNAs were synthesized in the presence of a 500 μM concentration of each nucleotide. The RNA for translation of FMDV L protease was transcribed from pFMDV14 (41).
Translations, initiation complex formation, and sedimentation analysis.
Translation reactions were usually performed with 0.2 pmol of RNA and 4.4 μl of nuclease-treated rabbit reticulocyte lysate (RRL; Promega) in the presence of potassium at a final concentration of 80 mM (38 mM endogenous potassium acetate plus 42 mM added KCl) in a volume of 10 μl. Purified eIF4F (kindly provided by Adri Thomas) or RNA transcribed from pFMDV14 (41) linearized with BamHI were added if indicated. eIF4F was purified as described previously (42). Initiation complex formation and analysis were performed essentially as described before (31). Binding-reaction mixtures (75 μl) contained 25 μl RRL, 15 mM Tris-HCl (pH 7.5), 0.4 mM MgCl2, 10 mM dithiothreitol (DTT), 40 mM KCl, and 0.05 pmol of the corresponding radiolabeled IRES RNA. Reaction vials were kept strictly at 0°C all the time except for the incubation at 30°C. For the binding reaction, samples were incubated at 30°C for the times indicated. Each sample was loaded onto an ice-cold 10 to 35% sucrose gradient containing 50 mM Tris-HCl (pH 8.4), 6 mM MgCl2, 60 mM NaCl, and 10 mM DTT (1, 31) and centrifuged for 4.5 h at 200,000 × g at 4°C in a Beckman SW41 Ti rotor. Fractions were collected from the bottom and used for scintillation counting.
UV cross-linking assays and immunoblot analyses.
UV cross-linking assays were performed with 4.4 μl of RRL in the presence of 15 mM Tris-HCl (pH 7.5), 10 mM DTT, 0.5 mM MgCl2, 2% glycerol, 0.01% Tween 20, 0.1 μg of tRNA per μl, KCl to a final potassium concentration of 80 mM, and 0.2 pmol of labeled IRES RNA in a volume of 10 μl. Unlabeled competitor RNA or purified eIF4F was added if indicated. The reaction mixtures were incubated for 10 min at 30°C and irradiated with UV light for 20 min. Excess RNA was digested with RNase A at 0.1 mg/ml at 37°C for 60 min. Proteins were separated on sodium dodecylsulfate-8% polyacrylamide gels and analyzed by autoradiography. Immunoblot detection of eIF4G and its C-terminal cleavage product was performed as described previously (41).
RESULTS
Translation of poliovirus IRES domain V mutants.
Attenuation of the poliovirus Sabin vaccine strains is mainly conferred by a single point mutation in the IRES domain V sequence (6, 17, 26, 38). We have introduced these single nucleotide exchanges in the otherwise unmodified context of the poliovirus type 1 (Mahoney) IRES sequence which was fused at the authentic poliovirus initiator AUG at nucleotide 743 to the firefly luciferase reporter gene sequence. The mutations change an A to a G (referred to as the S1 construct), mimicking the decisive mutation in the Sabin 1 vaccine strain IRES sequence, a G to an A in the S2 construct, and a C to a U in the S3 construct (Fig. 1).
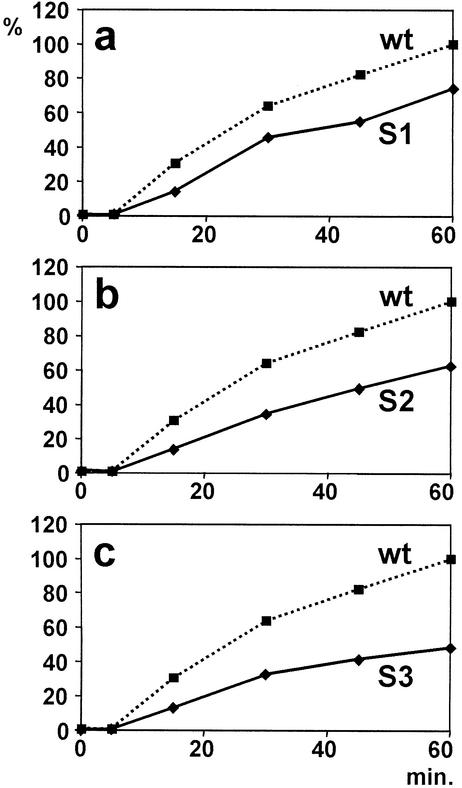
The translation activity of these artificial Sabin-like IRES elements was analyzed in an RRL which is competent for poliovirus translation and for the formation of ribosomal initiation complexes with the poliovirus IRES (32). In the reaction mixtures containing 44% RRL, the RNA concentration was about 20 nM, which was revealed to be not limiting in test reactions (data not shown). Luciferase reporter protein activity was measured after 15, 30, 45, and 60 min. As expected, the translational activity of all three Sabin-like IRES mutants was decreased compared with that of the unmodified poliovirus type 1 wild-type IRES during the entire incubation period (Fig. 2), confirming that these single point mutations indeed account for the described reduction in translation efficiency of the Sabin vaccine strains (44, 45).
FIG. 2.
Translation efficiency of the artificial Sabin-like poliovirus mutants. Translation reactions with the respective IRES followed by the firefly luciferase sequence were performed at 30°C in RRL. Luciferase activity readings were standardized by setting the 60-min reaction with wild-type (wt) IRES as 100% and plotted against incubation time.
Domain V mutants affect ribosome association.
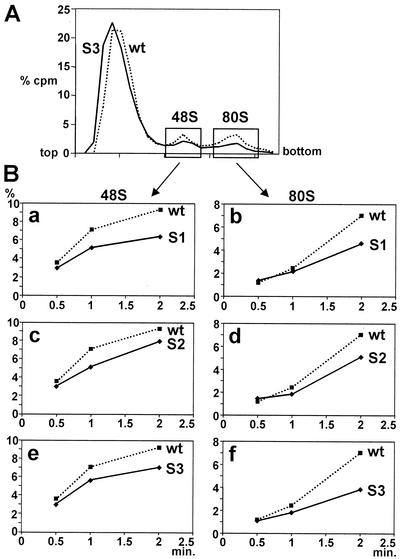
To test if the decreased translation efficiency of the Sabin-like poliovirus IRES constructs is due to decreased efficiency of ribosome association with the viral RNA, we analyzed the efficiency of formation of ribosomal initiation complexes with the modified IRES RNAs. Association of ribosomal subunits with the poliovirus IRES as well as with the distantly related FMDV and EMCV IRES elements is an energy-dependent process (31, 32, 35). Accordingly, radioactive mutant Sabin-like and wild-type poliovirus IRES RNAs were incubated in reticulocyte lysate for different times at 30°C whereas all other steps were performed strictly at 0°C. The total magnesium concentration in these binding reactions was in the physiological range (0.5 mM) and therefore allows dissociation of ribosomes (7) to generate empty ribosomal subunits, which are, in turn, required for the association of ribosomal subunits driven by the RNA to be translated and the initiation factors involved. Once formed, ribosomal initiation complexes were separated on sucrose gradients under ionic conditions stabilizing ribosomal subunits associated with the RNA (6 mM magnesium) (1), thereby preserving the initiation complexes which had been formed with the viral RNA during the previous incubation at 30°C. The efficiency of formation of ribosomal initiation complexes was measured by the amount of radioactive IRES RNA incorporated in the complexes. The radioactivity profiles of typical gradient runs are shown as examples for the poliovirus wild-type IRES and the S3 IRES in Fig. 3A. 48S complexes represent IRES RNA associated with the small ribosomal 40S subunit, and 80S complexes represent RNA associated with completely assembled ribosomes also containing the large ribosomal 60S subunit.
FIG. 3.
Poliovirus IRES domain V mutations affect ribosome association. (A) Principle of the assay. Radiolabeled wild-type or mutated IRES RNA, respectively, was incubated with RRL for 0.5, 1, or 2 min at 30°C. Initiation complexes were separated on 10 to 35% sucrose gradients, and fractions collected from the bottom were used for scintillation counting. The relative amount of radioactivity in each fraction (% cpm) is plotted against the fraction number. The plot exemplifies the radioactivity profiles obtained after 2 min with the wild-type (wt) poliovirus 1 IRES (dotted line) and with the artificial Sabin 3-like IRES (S3) (solid line). (B) Efficiency of formation of 48S (a, c, and e) and 80S complexes (b, d, and f) with the Sabin-like S1, S2, and S3 construct, respectively, compared with the wild-type IRES.
The efficiency of association of ribosomes with the mutant IRES RNAs is seriously impaired compared with the wild-type poliovirus IRES. This effect is already evident at the stage of 48S complex formation. After 2 min of incubation at 30°C, markedly reduced amounts of 48S complexes were formed with the mutated S1-, S2- and S3-IRES RNAs (Fig. 3B, panels a, c, and e). The reduction in ribosome association also extends to the 80S complexes (Fig. 3B, panels b, d, and f). These results correlate with the reduced translation efficiencies of these mutated IRES RNAs (Fig. 2), indicating that the serious reduction in translation efficiency observed with the Sabin vaccine strains is caused at the stage of association of the small ribosomal subunit with the viral RNA.
Initiation factor eIF4G binds to the poliovirus IRES.
From the above results, we suspected that the Sabin-like mutations would impair the binding of cellular protein factors to the viral RNA that mediate the association of the ribosomal 40S subunit with the IRES. With the distantly related FMDV and EMCV IRES elements, it had been shown that eIF4B, eIF4G, and eIF4A bind synergistically to the IRES 3′ region (19, 23, 25, 35, 41). Moreover, eIF4B is involved in ribosomal initiation complexes with the poliovirus IRES (32), suggesting that eIF4G may also bind to the poliovirus IRES.
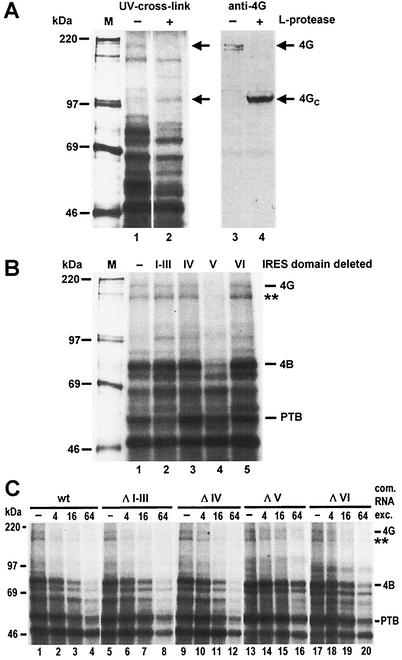
Therefore, we used the UV cross-link assay in combination with immunoblotting to identify eIF4G binding to the poliovirus IRES. After optimizing UV cross-link assay conditions, we detected a band migrating at an apparent molecular mass of about 200 kDa, which is supposed to be identical to eIF4G (Fig 4A, lane 1). On preincubation of the lysate with an RNA expressing the leader (L) protease of FMDV (41), which specifically cleaves eIF4G (22), this band disappeared (lane 2), while an additional band of about 110 kDa, most probably representing the C-terminal cleavage product of eIF4G, appeared. This was confirmed by an anti-eIF4G immunoblot analysis using an antiserum directed against a C-terminal sequence of eIF4G (41). In the untreated lysate, eIF4G was detected (lane 3) comigrating with the band detected in the UV cross-link (lane 1). Following cleavage of eIF4G with L-protease (lane 4), this 200-kDa band disappeared while a new band of about 110 kDa appeared (lane 4) that exactly comigrates with the band appearing in the UV cross-link assay (lane 2). These results indicate that the band of about 200 kDa which binds to the poliovirus IRES is identical to eIF4G.
FIG. 4.
eIF4G binds to the poliovirus IRES domain V. (A) Identification of eIF4G. (Left) UV cross-linking assay using normal RRL (−) or RRL treated with FMDV leader protease (+ L-protease). L-protease RNA transcribed from plasmid pFMDV14 was preincubated with RRL at 30°C for 15 min. Samples were then used for UV cross-linking reactions with [α-32P]UTP-labeled wild-type poliovirus IRES. Reaction mixtures were incubated at 30°C for 10 min and UV irradiated for 20 min, and excess RNA was digested with RNase A. Then 50% of each sample was applied to a sodium dodecyl sulfate-8% polyacrylamide gel and analyzed by autoradiography. (Right) The other 50% was separated on a gel, proteins were transferred to nitrocellulose, and eIF4G was immunostained using anti-eIF4G antiserum. Molecular masses of marker proteins (M) are given in kilodaltons. 4G, eIF4G; 4GC, C-terminal fragment of eIF4G. (B) Mapping of the eIF4G-binding site. Either domains I to III or single domains IV, V, or VI, were deleted from the poliovirus IRES. [α-32P]UTP-labeled domain deletion mutants were transcribed from templates linearized with BsiWI 156 nucleotides downstream of the poliovirus AUG in the luciferase sequence and used in UV cross-linking assays with RRL. 4G, eIF4G; 4B, eIF4B. The putative p170 subunit of eIF3 is indicated by a double asterisk. (C) Competitions. The binding of labeled wild-type (wt) poliovirus IRES to RRL proteins was competed by unlabeled wild-type or deletion mutant IRES RNAs in the molar excesses (exc.) indicated.
eIF4G binds to domain V of the poliovirus IRES.
To identify the binding site for eIF4G in the poliovirus IRES, we precisely removed the sequences of the predicted RNA secondary-structure domains (5, 37) from the poliovirus 5′-UTR (Fig. 1). In the first mutant, domains I to III were removed, whereas in each of the other deletion mutants, only one of the domains IV, V, or VI, respectively, was removed. In UV cross-link assays using the wild-type poliovirus IRES and these domain deletion mutants, we found that domain V is essential for the interaction of eIF4G with the poliovirus IRES (Fig. 4B, lane 4). Although label transfer from the radioactive RNA to the eIF4G protein is not very efficient, binding of eIF4G to domain V is specific, since no other deletion affected eIF4G binding. Confirming our previous results (32), binding of eIF4B was also seriously affected by the deletion of domain V. In conclusion, the poliovirus IRES domain V is the major determinant for the binding of both standard initiation factors eIF4G and eIF4B.
Also, the binding of other proteins to the poliovirus IRES is affected by the domain deletion mutants. At least two separate regions appear to contribute to the binding of PTB, since deletion of domains I to III as well as deletion of domain V slightly reduced the efficiency of PTB binding (Fig. 4B, lanes 2 and 4, respectively). Moreover, a band of about 170 kDa that migrates close to the eIF4G band completely disappeared when domain V was deleted. This band most probably represents the p170 subunit of eIF3 (L. Saleh and M. Niepmann, unpublished observations), which appears also to be involved in specific recognition of domain V.
To confirm that the loss of detection of eIF4G in the UV cross-link-assay was due to the loss of eIF4G binding to the IRES and not to the loss of label transfer from the radioactive RNA to the protein, we performed competition assays (Fig. 4C). Unlabeled wild-type, ΔI-III, ΔIV and ΔVI competitor RNAs competed efficiently with the binding of eIF4G and eIF4B to radiolabeled wild-type RNA. In contrast, even a 16-fold excess of unlabeled competitor RNA ΔV competed the binding of labeled wild-type RNA to both factors only slightly (Fig. 4C, lanes 13 to 15). Moreover, eIF4G and eIF4B were still detected with a 64-fold excess of competitor (lane 16). Thus, the loss of detection of eIF4G with the domain V deletion observed in Fig. 4B is actually due to the loss of eIF4G binding. Taken together, the above results indicate that the major determinant for binding of eIF4G resides in the domain V of the poliovirus IRES.
The attenuating mutations in domain V directly affect binding of eIF4G and eIF4B.
The observation that the Sabin-like point mutations in domain V of the poliovirus IRES seriously affect ribosome association with the IRES and the fact that domain V is the major determinant for binding of eIF4G and eIF4B led us to the conclusion that these single nucleotide exchanges may directly affect the binding of these translation initiation factors and hence impair the association of the ribosome with the viral RNA. When UV cross-link assays were performed with the Sabin-like IRES RNAs, we found that the binding of eIF4B (Fig. 5 A, left panel) and eIF4G (right panel) was indeed seriously impaired by these mutations compared with the effect of the wild-type IRES, while the binding of another protein of about 50 kDa was not affected (left panel). This effect of reduced binding of eIF4G and eIF4B was most dramatic with the S3 construct (lanes 4 and 4′) and thus correlates with the reduced translation efficiencies of these constructs (Fig. 2). Also in this experiment, the intensities of bands representing PTB and the band supposed to be identical to the p170 subunit of eIF3 were affected by the mutations. Competition experiments (Fig. 5B) confirmed that the observed reductions in the intensities of the eIF4G, eIF4B, PTB, and p170 bands were due to reduced binding of these factors and not to reduced label transfer only, while other bands were less strongly affected by the competitions.
FIG. 5.
Weaker binding of initiation factors to mutated poliovirus IRES elements. (A) UV cross-linking assay using RRL and either the [α-32P]UTP-labeled wild-type (wt) poliovirus IRES or the artificial Sabin-like mutants. (Left) Short exposure of X-ray film to the gel (right) long exposure. 4G, eIF4G; 4B, eIF4B. (B) Competitions. The binding of labeled wild-type poliovirus IRES to RRL proteins was competed by unlabeled complete (wt) or mutated IRES RNAs in the molar excesses (exc.) indicated. A double asterisk indicates the putative p170 subunit of eIF3; single asterisks indicate bands that are not differentially affected by the competitions.
Purified eIF4F relieves the negative effects of the IRES mutations.
Since the Sabin mutations affected the binding of more than one protein to the poliovirus IRES, we used purified eIF4F to confirm that reduced binding of eIF4G is the primary cause of impaired translation of the mutated IRES elements. The eIF4F preparation (Fig. 6A) contains eIF4E, eIF4A, and eIF4G, including two additional high-molecular-weight bands also recognized by the antibody directed to the C-terminus of eIF4G (Fig. 6A, lane 2), which represent proteolytic degradation products of eIF4G that often appear in eIF4F preparations (42).
FIG. 6.
Effects of purified eIF4F on poliovirus IRES mutants. (A) Coomassie blue stain (lane 1) and immunoblot (lane 2) of the eIF4F preparation. In the immunoblot, an antiserum directed against the C terminus of eIF4G was used. (B) Translation directed by 0.2 pmol of the wild-type and mutant poliovirus IRES elements in RRL, either with no additional eIF4F or in the presence of increasing amounts of purified eIF4F (shown in panel A) as indicated. Values are means of data from two experiments for wt, S1, and S2 and from four experiments for S3. (C) UV cross-linking assay with the wild-type and mutant poliovirus IRES elements in RRL, either with no additional eIF4F or in the presence of increasing amounts of purified eIF4F as indicated. A double asterisk indicates the putative p170 subunit of eIF3. (D) Translation of the S3 IRES in RRL, either with no additional eIF4F or in the presence of increasing amounts of purified eIF4F as indicated. Before translation, the lysate was preincubated for 30 min with RNA from pFMDV14 that expresses the L-protease of FMDV. (E) UV-cross-linking assay with the S3 IRES in RRL treated with L-protease as in panel D, either with no additional eIF4F or in the presence of increasing amounts of purified eIF4F as indicated. 4GC, C-terminal fragment of eIF4G.
When increasing concentrations of eIF4F were used in translation reactions with the poliovirus IRES RNAs (Fig. 6B), translation of the wild-type IRES was slightly improved, reflecting a slight increase in product concentration according to the equilibrium of the reaction driven by increased eIF4F concentrations. In contrast, the moderately impaired translation of the S1 and S2 constructs, and particularly the seriously impaired translation of the S3 construct, were improved significantly, in each case reaching translation efficiency levels close to that of the wild-type IRES. Similar results were obtained in these assays using either 0.02 or 0.2 pmol of RNA per translation reaction, indicating that even at the higher RNA concentration only the RNA or eIF4F, respectively, but not other factors, was limiting. When the effect of added eIF4F was monitored in the UV cross-linking assay (Fig. 6C), addition of eIF4F resulted in significant improvement of eIF4G and eIF4B binding to the S3 IRES (Fig. 6C, lane 16), approaching the levels observed with the wild-type IRES. With the wild-type or the S1 and S2 IRES elements, no increase in eIF4G band intensity was observed (lanes 1 to 12). However, the binding of eIF4B, as well as the binding of a band migrating close to eIF4G, which corresponds to the p170 subunit of eIF3 (Saleh and Niepmann, unpublished), and another band of about 110 kDa, which may represent the p116 or the p110 subunit of eIF3, was improved. This effect is particularly evident with the S3 construct (lane 16). Similar results were obtained when the N-terminal portion of eIF4G, which is required for binding of the cap-binding protein eIF4E, was cleaved off by the L-protease of FMDV (Fig. 6D and E). Also in this experiment, the translation efficiency of the S3 construct was completely relieved (Fig. 6D) and binding of eIF3 p170 was improved, while a faint band representing the 110-kDa C-terminal cleavage product of eIF4G (4GC in Fig. 6E) appeared at high eIF4F concentrations (Fig. 6E, lane 4).
These results indicate that increased concentrations of eIF4F drive the recruitment of the poliovirus IRES to the ribosome. eIF4G is most probably mediating the interaction of the IRES with the ribosome-bound eIF3 and perhaps other components of the translational apparatus, irrespective of whether eIF4G is in its unmodified or its cleaved form. Accordingly, increased eIF4F concentrations can relieve the disadvantage of the IRES Sabin mutations that cause impaired binding of eIF4G.
DISCUSSION
Here we demonstrate that the reduction in poliovirus translation efficiency caused by the single nucleotide exchanges in the IRES of the three poliovirus Sabin vaccine strains is mediated by impaired binding of the standard translation initiation factor eIF4G to the poliovirus IRES domain V. This in turn causes impaired association of ribosomes with the viral RNA. The disadvantage of the Sabin constructs, particularly that observed with the S3 construct, is relieved by addition of purified eIF4F, which results in improved binding not only of eIF4G but also of eIF4B and the p170 subunit of eIF3 to the mutated IRES. This indicates that eIF4G is the crucial factor that initially binds to the poliovirus IRES and recruits the IRES to the other components of the translational apparatus, particularly to the p170 subunit of the ribosome-bound eIF3, thereby mediating association of the viral RNA with the small ribosomal subunit. Our findings may have major implications for understanding the attenuation of poliovirus neurovirulence and the pathogenicity of other members of the enteroviruses and rhinoviruses.
In recombination experiments, the single point mutations in domain V were found to contribute to the attenuation of neurovirulence of the respective Sabin vaccine strains (6, 17, 38), and experiments using chimeras of the poliovirus and rhinovirus IRES elements revealed that the IRES domains V and VI may contain determinants of neuropathogenicity (8). Nevertheless, studies that clearly delimit the effects of these mutations to translation deficiencies are rare. Translation directed by the authentic Sabin 1 IRES was reduced in extracts from reticulocytes (45) and from cells of neuronal origin but not from HeLa cells (12). Translation of Sabin 3 RNA is reduced in reticulocyte lysate (45), and this effect is attributed to the C472U mutation (44). Moreover, poliovirus type 3 with a U residue at position 472 is unable to replicate in mouse brain, whereas poliovirus with a C residue at this position is neurovirulent in mice (21).
Probably due to the possible complexity of protein-RNA interactions involved in the activity of the poliovirus IRES, the initiation factor requirements have been investigated so far mainly with the distantly related picornavirus type II IRES elements. EMCV IRES activity depends on eIF2, eIF3, eIF4G, eIF4B, and eIF4A (35), with eIF4G, eIF4B, and eIF4A binding synergistically (19). Extending these results to the type I IRES of poliovirus, we showed recently that eIF4B binds strongly to the poliovirus IRES domain V (32). Consequently, it can be expected that the basic apparatus of initiation factors acting on the poliovirus IRES is the same as with the EMCV IRES and that eIF4G and eIF4B are directly and functionally involved in the process of translation initiation at the poliovirus RNA.
In this work we mapped the binding site for eIF4G to reside in domain V, suggesting that all initiation factors that are involved in formation of initiation complexes with the EMCV IRES also act on the poliovirus IRES. We have generated the relevant single nucleotide exchanges in the context of the otherwise unmodified sequence context of the poliovirus type 1 IRES sequence. In the absence of an in vitro system using purified ribosomal subunits, initiation factors, and possibly other cellular proteins required for the formation of ribosomal initiation complexes with the poliovirus IRES RNA, we developed an assay that allowed us to measure the efficiency of ribosome association with the mutated IRES elements. This system is suitable for the formation of functional ribosomal initiation complexes that are sensitive to translation inhibitors (32). In this system, the efficiency of ribosome association with the mutated IRES elements was considerably reduced compared to that observed with the wild-type poliovirus type IRES. This effect was most dramatic with the Sabin 3-like mutation, correlating with the most severe decrease of translation efficiency with this construct. During preparation of the manuscript, we became aware of a complementary study by Kean and coworkers, who showed that introduction of the Sabin 3 point mutation into the poliovirus IRES domain V resulted in a significantly defective IRES, correlating with poor growth in HeLa and neuroblastoma cells (K. Kean, personal communication).
While it is known that additional cellular proteins, such as PTB, that bind to the IRES are indeed involved in stimulating the translation of some picornaviruses, their actual mode of action remains unclear. It can only be speculated if PTB exerts a so-called RNA-chaperone function by stabilizing the IRES secondary or tertiary structure. PTB appears to bind mainly to two separate regions of the poliovirus IRES, including determinants both in the 5′ region and in domain V of the 5′-UTR, whereas sequences further downstream bind PTB more weakly (13, 32). A mild stimulation of poliovirus translation by PTB was shown only recently (14). Interestingly, PTB binding to the authentic Sabin 3 IRES sequence is reduced in extracts from neuroblastoma cells but not from HeLa cells (9). Our finding that the single point mutations reduce the binding of PTB to the poliovirus IRES further support the idea that reduced binding of PTB may also contribute to the attenuation of neurovirulence. Moreover, domains V and VI have been reported to cooperatively determine poliovirus neuropathogenicity (8), and binding of three cellular proteins to these domains was reported (11). A 39-kDa protein present in HeLa and neuronal cells, as well as a 60-kDa protein present only in neuronal cells, was shown to bind to domain VI, whereas binding of a 36-kDa protein from HeLa cells and PTB was shown to require the presence of both domains V and VI. However, it can only be speculated if these proteins additionally modulate poliovirus IRES activity in neuronal tissue, if their binding to Sabin-like mutants is reduced, and if their possibly reduced binding contributes to the attenuated phenotype of the Sabin strains.
Our results presented here implicate that the main effect of the mutations in the viral 5′-UTR that contribute to the attenuation of poliovirus neurovirulence is impaired binding of the standard translation initiation factors. This may cause slower translation of the viral RNA in some tissues, e.g., in neuronal cells and thus may contribute to the attenuated phenotype of the poliovirus vaccine strains.
Acknowledgments
We thank Adri Thomas for the generous gift of eIF4F, Tatyana Pestova and Christopher Hellen for plasmids, Denise Egger and Kurt Bienz for critically reading the manuscript, and Ewald Beck for helpful discussions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 535 and GK 370).
REFERENCES
- 1.Anthony, D. D., and W. C. Merrick. 1992. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 267:1554-1562. [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divizia, M., L. Palombi, E. Buonomo, D. Donia, V. Ruscio, M. Equestre, L. Leno, A. Pana, and A. M. Degener. 1999. Genomic characterization of human and environmental polioviruses isolated in Albania. Appl. Environ. Microbiol. 65:3534-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrenfeld, E., and B. L. Semler. 1995. Anatomy of the poliovirus internal ribosome entry site. Curr. Top. Microbiol. Immunol. 203:65-83. [DOI] [PubMed] [Google Scholar]
- 6.Evans, D. M. A., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 7.Goss, D. J., and T. Harrigan. 1986. Magnesium ion dependent equilibria, kinetics, and thermodynamic parameters of Artemia ribosome dissociation and subunit association. Biochemistry 25:3690-3695. [DOI] [PubMed] [Google Scholar]
- 8.Gromeier, M., B. Bossert, M. Arita, A. Nomoto, and E. Wimmer. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez, A. L., M. Denova-Ocampo, V. R. Racaniello, and R. M. del Angel. 1997. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J. Virol. 71:3826-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller, A. A., and B. L. Semler. 1995. Stem-loop structure synergy in binding cellular proteins to the 5′ noncoding region of poliovirus RNA. Virology 206:923-934. [DOI] [PubMed] [Google Scholar]
- 12.Haller, A. A., S. R. Stewart, and B. L. Semler. 1996. Attenuation stem-loop lesions in the 5′ noncoding region of poliovirus RNA: neuronal cell-specific translation defects. J. Virol. 70:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen, C. U., T. V. Pestova, M. Litterst, and E. Wimmer. 1994. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J. Virol. 68:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt, S. L., and R. J. Jackson. 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5:344-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang, S. K., H. G. Kräusslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. Jackson. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van Der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine- derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 19.Kolupaeva, V. G., T. V. Pestova, C. U. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 20.Kuge, S., and A. Nomoto. 1987. Construction of viable deletion and insertion mutants of the Sabin strain type I poliovirus: function of the 5′ noncoding sequence in viral replication. J. Virol. 61:1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Monica, N., J. W. Almond, and V. R. Racaniello. 1987. A mouse model for poliovirus neurovirulence identifies mutations that attenuate the virus for humans. J. Virol. 61:2917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 23.Lopez de Quinto, S., and E. Martinez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, K., A. Petersen, M. Niepmann, and E. Beck. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minor, P. D. 1993. Attenuation and reversion of the Sabin vaccine strains of poliovirus. Dev. Biol. Stand. 78:17-26. [PubMed] [Google Scholar]
- 27.Nathanson, N., and P. Fine. 2002. Virology. Poliomyelitis eradication—a dangerous endgame. Science 296:269-270. [DOI] [PubMed] [Google Scholar]
- 28.Niepmann, M. 1999. Internal initiation of translation of picornaviruses, hepatitis C virus and pestiviruses. Recent Res. Dev. Virol. 1:229-250. [Google Scholar]
- 29.Niepmann, M. 1996. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 388:39-42. [DOI] [PubMed] [Google Scholar]
- 30.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochs, K., R. C. Rust, and M. Niepmann. 1999. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 73:7505-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochs, K., L. Saleh, G. Bassili, V. H. Sonntag, A. Zeller, and M. Niepmann. 2002. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 76:2113-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 35.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 38.Ren, R. B., E. G. Moss, and V. R. Racaniello. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 65:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rust, R. C., K. Ochs, K. Meyer, E. Beck, and M. Niepmann. 1999. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J. Virol. 73:6111-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabin, A. B. 1985. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J. Infect. Dis. 151:420-436. [DOI] [PubMed] [Google Scholar]
- 41.Saleh, L., R. C. Rust, R. Füllkrug, E. Beck, G. Bassili, K. Ochs, and M. Niepmann. 2001. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth-disease virus internal ribosome entry site. J. Gen. Virol. 82:757-763. [DOI] [PubMed] [Google Scholar]
- 42.Scheper, G. C., H. O. Voorma, and A. A. Thomas. 1992. Eukaryotic initiation factors 4E and 4F stimulate 5′ cap-dependent as well as internal initiation of protein synthesis. J. Biol. Chem. 267:7269-7274. [PubMed] [Google Scholar]
- 43.Shulman, L. M., Y. Manor, R. Handsher, F. Delpeyroux, M. J. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. M. Kew, R. Crainic, and E. Mendelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitkin, Y. V., N. Cammack, P. D. Minor, and J. W. Almond. 1990. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472—U. Virology 175:103-109. [DOI] [PubMed] [Google Scholar]
- 45.Svitkin, Y. V., T. V. Pestova, S. V. Maslova, and V. I. Agol. 1988. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology 166:394-404. [DOI] [PubMed] [Google Scholar]
- 46.Trono, D., R. Andino, and D. Baltimore. 1988. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 62:2291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]