Abstract
SER virus is closely related to the paramyxovirus simian virus 5 (SV5) but is defective in syncytium formation. The SER virus F protein has a long cytoplasmic tail (CT) domain that has been shown to inhibit membrane fusion, and this inhibitory effect could be eliminated by truncation of the C-terminal sequence (S. Tong, M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, and H.-D. Klenk, Virology 301:322-333, 2002). To study the sequence requirements for regulation of fusion, codons for SER virus F protein residues spanning amino acids 535 to 542 and 548 were mutated singly to alanines, and the two leucine residues at positions 539 and 548 were mutated doubly to alanines. We found that leu-539 and leu-548 in the CT domain played a critical role in the inhibition of fusion, as mutation of the two leucines singly to alanines partially rescued fusion, and the double mutation L539, 548A completely rescued syncytium formation. Mutation of charged residues to alanines had little effect on the suppression of fusion activity, whereas the mutation of serine residues to alanines enhanced fusion activity significantly. The L539, 548A mutant also showed extensive syncytium formation when expressed without the SER virus HN protein. By constructing a chimeric SV5-SER virus F CT protein, we also found that the inhibitory effect of the long CT of the SER virus F protein could be partially transferred to the SV5 F protein. These results demonstrate that an elongated CT of a paramyxovirus F protein interferes with membrane fusion in a sequence-dependent manner.
The entry of enveloped viruses into host cells is accomplished by fusion of the viral envelope and the target cell membrane. For some enveloped viruses, exemplified by influenza virus, fusion occurs in the endosome after the viral fusion protein has been activated at low pH (18). Other viruses, such as paramyxoviruses, fuse with the cellular plasma membrane at neutral pH. Membrane fusion mediated by most paramyxoviruses requires the interaction of two virion-associated glycoproteins, the hemagglutinin-neuraminidase (HN) protein that mediates viral attachment and the F protein that mediates subsequent fusion. However, for some paramyxoviruses, including the W3A strain of simian virus 5 (SV5), the attachment proteins are not absolutely required for fusion activity (8, 21-23, 33).
Paramyxovirus F proteins are synthesized as an inactive precursor, F0, which must be proteolytically cleaved to form a disulfide-linked heterodimer of F1 and F2 to activate membrane fusion activity (20, 37). Earlier studies have implicated several domains, conserved in most F proteins, that direct membrane fusion. A hydrophobic sequence designated the fusion peptide, which is at the amino terminus of the F1 subunit, is thought to insert into the target membrane (18). Two heptad repeats—HR1, located just carboxyl terminal to the fusion peptide, and HR2, located adjacent to the transmembrane domain—have been shown to play essential roles in fusion (5). The importance of these domains for fusion was shown by mutational analysis (34, 41) and also by using peptide analogues which inhibit fusion, possibly because they interfere with the conformational changes in the molecule that are necessary for fusion (15). The cytoplasmic tail (CT) domain of some viral fusion proteins has also been shown to play a regulatory role in membrane fusion. In paramyxoviruses, F protein CT truncations in Newcastle disease virus show greatly reduced syncytium formation (39). Truncations in the CT of parainfluenza virus type 3 and SV5 F proteins abolished fusion activity (2, 44), whereas removal of the CT in human parainfluenza virus type 2 F protein did not affect its fusion activity (44). However, in measles virus, truncations of the CT caused increased syncytium formation (4). In SV5, deletion of the CT of the fusion protein inhibited fusion pore enlargement (9). Elongation of the CT of influenza virus hemagglutinin protein reduced fusion activity significantly (32). In simian immunodeficiency virus and human immunodeficiency virus types 1 and 2, truncation of the CT caused increased syncytium formation (7, 11, 14, 30, 35, 45). The C-terminal 16 amino acids of the envelope protein of murine leukemia virus (MuLV), called the R peptide, is also known to be inhibitory to membrane fusion (16, 25, 38).
SER virus is a recently identified virus belonging to the family Paramyxoviridae, genus Rubulavirus, that was isolated from aborted pigs with porcine reproductive and respiratory syndrome (17). It is very closely related to SV5 serologically and in its protein profile and nucleotide sequence. However, unlike SV5, SER virus is unable to induce syncytium formation in CV-1 or BHK-21 cells (42). Comparison of the deduced amino acid sequences revealed the SER virus F protein (551 amino acids) to be longer than the SV5 F protein (529 amino acids) by 22 residues in the CT domain, and truncation of the CT of SER virus F protein by 22 residues completely rescued the ability to cause syncytium formation (42).
In the present study, the sequence requirements in the CT domain for the regulation of fusion have been investigated by using site-directed mutagenesis. The expression and transport and the effect of mutations on fusion activity were investigated to determine the sequence requirements for the fusion-suppressive activity of the CT of the SER virus F protein.
MATERIALS AND METHODS
Cells, viruses, and vectors.
HeLa T4 cells were maintained in Dulbecco's modified minimal essential medium (DMEM) supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, Utah). The recombinant vaccinia virus vTF7-3 and the wild-type (wt) vaccinia virus strain IHD-J were kindly provided by Bernard Moss (National Institutes of Health, Bethesda, Md.). The vaccinia virus stocks were propagated and titers were determined on CV-1 cells. Plasmid pGINT7 β-gal was provided by Edward Berger (National Institutes of Health, Bethesda, Md.). The pGEM-3-SER F and pGEM-3-SER HN plasmids characterized previously (42) were expressed in HeLa T4 cells. The pGEM-3-SV5 F and pGEM-3-SV5 HN plasmids were described previously. Rabbit anti-SV5 antibody was a kind gift from R. A. Lamb (Northwestern University). The SER virus was obtained from H.-D. Klenk (University of Marburg, Marburg, Germany). SER virus was propagated in MDBK cells, and the virus titers were determined by hemagglutination assay using chicken erythrocytes (RBCs).
Generation of F mutant proteins. Mutations in the SER virus F gene were generated using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The pGEM-SER F plasmid was used as the template, and the two synthetic oligonucleotide primers (Sigma Genosys, Woodlands, Tex.) designed to carry the mutation at the desired site were complementary to each other. A schematic representation of the mutations in the CT region is shown in Fig. 1. A chimeric SV5-SER virus F CT mutant was also constructed by using overlap extension PCR as described earlier (19). The ectodomain and the transmembrane coding sequence from the SV5 F gene was PCR amplified, with the deletion of the sequence encoding the entire CT. A second PCR was done to amplify the complete CT region from the SER virus F gene with an overhang in the forward primer complementary to the 3′ end of the first PCR product. The two PCR-amplified fragments were annealed to each other and were further extended to generate the chimeric gene by five rounds of PCR. Finally, the chimeric fragment was used as a template to amplify the entire chimeric SV5-SER virus F gene CT mutant. A pGEM-3-EGFP plasmid was constructed by PCR amplifying the enhanced green fluorescent protein (EGFP) gene from pIRES-EGFP (Clontech, Palo Alto, Calif.) and cloning it into the pGEM-3 vector. This vector was subsequently used in the cytoplasmic-mixing assay using the vTF7-3 expression system as described below. All constructs were sequenced to confirm the presence of the desired mutations and the absence of additional mutations.
FIG. 1.
Schematic diagram of wt and mutant SER virus F proteins and the chimeric SV5 F protein. The residues mutated in each construct are underlined, and the designations assigned to each mutant are given on the left. TM, transmembrane domain; asterisk, stop codon; open box, SERF; hatched box, SVSF.
Transfection, radiolabeling, and immunoprecipitation.
The mutant and wt SER virus F proteins were expressed by using the vaccinia virus-bacteriophage T7 RNA polymerase transient-expression system (13). Briefly, 35-mm-diameter dishes of subconfluent cells were infected with the vTF7-3 virus at a multiplicity of infection (MOI) of 10 for 1 h and later transfected with 3 μg of plasmid DNA using Lipofectin (Invitrogen, Carlsbad, Calif.). Sixteen hours posttransfection, the transfected cells were starved in DMEM lacking methionine-cysteine for 45 min, pulse-labeled with 100 μCi of [35S]methionine-cysteine/ml for 30 min at 37°C, and then chased with DMEM containing 10% fetal calf serum for 2 h. The cells were washed three times and then lysed with cell dissociation buffer (10 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% Triton X-100, and 0.5% sodium deoxycholate). The cell lysate was incubated with SV5 hyperimmune serum for 2 h at 4°C, followed by precipitation using protein A-agarose (Immunopure; Pierce Chemical, Rockford, Ill.) for 2 h at 4°C. Proteins were characterized by sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE) and subsequently by autoradiography.
Cell surface biotinylation assay.
The expression of proteins at the cell surface was detected by a biotinylation assay as described earlier (28). Sixteen hours posttransfection, the transfected cells were starved in DMEM lacking methionine and cysteine for 45 min, pulse-labeled with 100 μCi of [35S]methionine-cysteine/ml for 30 min at 37°C, and then chased with DMEM containing 10% fetal calf serum for 2 h. The cells were washed three times with ice-cold phosphate-buffered saline (PBS) containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS-CM) and incubated with 1 ml of a 0.5-mg/ml solution of sulfosuccinimidyl-2-(biotin-amido) ethyl-1,3-dithiopropionate (Pierce) in PBS-CM at 4°C for 30 min. Free biotin was removed by a brief incubation with DMEM containing 10% fetal calf serum, and the cells were washed three times with PBS-CM. The biotinylated cell surface proteins were lysed and immunoprecipitated using anti-SV5 antibody (which also cross-reacts with SER virus-encoded proteins) and protein A-agarose beads (Pierce). The cell lysate beads were washed three times and divided into two aliquots. One aliquot was used for immunoprecipitation, and the other aliquot was boiled in 10 μl of 10% SDS and diluted with 1 ml of lysis buffer. The supernatant from the protein A-agarose beads was incubated with streptavidin-agarose beads for 2 h at 4°C. Proteins were characterized by SDS-8% PAGE and autoradiography. Quantitation of the cell surface expression of the mutant proteins was performed using a PhosphorImager (Molecular Dynamics, Amersham, Piscataway, N.J.) using the Storm860 software. The results are represented as percentages of the cell surface expression levels obtained for the corresponding F proteins.
Pulse-chase analysis.
Thirty-five-millimeter-diameter dishes of subconfluent HeLa T4 cells were infected for 1 h with vTF7-3 at an MOI of 10 and then transfected with wt and mutant SER virus F proteins. Sixteen hours posttransfection, the transfected cells were starved in DMEM lacking methionine and cysteine for 45 min, pulse-labeled with 100 μCi of [35S]methionine-cysteine/ml for 30 min at 37°C, and then chased with DMEM containing 10% fetal calf serum for 0-, 5-, 15-, 30-, 60-, 180-, or 300-min chase periods. The cells were washed three times with ice-cold PBS-CM at appropriate time points and incubated with 1 ml of a 0.5-mg/ml solution of sulfosuccinimidyl-2-(biotin-amido) ethyl-1,3-dithiopropionate in PBS-CM at 4°C for 30 min. The cells were washed and then lysed with cell dissociation buffer (10 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% Triton X-100, and 0.5% sodium deoxycholate) at the appropriate time points. The kinetics of the surface expression of the SER virus wt and mutant F proteins were followed by surface biotinylation studies as described above.
Cell fusion assays.
Thirty-five-millimeter-diameter dishes of subconfluent HeLa T4 cells were infected for 1 h with vTF7-3 at an MOI of 10 and then transfected with 3 μg of the wt or the mutant SER virus F construct with or without cotransfection of the wt SER virus HN plasmid using Lipofectin. Sixteen to 20 h posttransfection, the cells were monitored for syncytium formation under a light microscope and photographed at a magnification of 200×.
Quantitative cell fusion assays were performed according to the fusion-dependent reporter gene activation method described by Nussbaum et al. (31). In brief, one population of HeLa T4 cells was infected with the recombinant vaccinia virus vTF7-3 and transfected with plasmids encoding the wt SER virus HN and the wt or mutant SER virus F protein and then treated with neuraminidase before the two cell types were mixed. Cytosine arabinoside (40 μg/ml) was added to each plate. A second population of HeLa T4 cells was infected with wt vaccinia virus strain IHD-J and transfected with plasmid pGINT7 β-gal, which contains the β-galactosidase (β-Gal) gene under the control of the T7 promoter. Sixteen hours posttransfection, the two cell populations were collected and mixed by adding 100 μl of each cell population (106 cells/ml) to a flat-bottom 96-well tissue culture plate. The plate was incubated at 37°C for 5 h, and the cell fusion was measured by a colorimetric lysate assay as described previously (44). The quantity of β-Gal was calculated by comparing the hydrolysis rate for each sample with that of a standard commercial preparation of Escherichia coli β-Gal (Roche, Indianapolis, Ind.). Data were analyzed by using the Delta Soft II Microplate analysis program. The fusion activity of mutants was determined as a percentage of the β-Gal production observed in the cells coexpressing wt SV5 F and HN. A variation of the content-mixing assay was performed by infecting one set of HeLa T4 cells with vTF7-3 and cotransfecting them with plasmids as described above, and a second set of cells was infected with the wt vaccinia virus and transfected with the pGEM-3-EGFP construct. The cells were treated with neuraminidase before being mixed, and 106 cells from each set were mixed at 37°C and observed under an inverted fluorescence microscope after 24 h for green fluorescence.
Hemifusion assay.
Guinea pig RBCs were labeled with the hydrophobic fluorescent dye R18 (Molecular Probes, Eugene, Oreg.) as described by Bagai and Lamb (1). In brief, 15 μl of a 1-mg/ml solution of R18 in ethanol was added to 10 ml of 1% fresh guinea pig RBC suspension in PBS with vortexing. After incubation at room temperature in the dark for 30 min, 30 ml of DMEM with 10% fetal calf serum was added, and the suspension was further incubated for 20 min to adsorb the unbound probe. After being labeled, the RBC suspension was washed five times with 50 ml of PBS to remove the unincorporated R18. The last washing step was in PBS containing an additional 1 mM CaCl2 and 1 mM MgCl2 (PBS++). To determine lipid mixing, HeLa T4 cell monolayers expressing SER virus F-HN or SER virus F mutants-SER virus HN at 16 h posttransfection at 32°C were washed twice with PBS and treated with 50 mU of neuraminidase per ml in DMEM for 1 h at 37°C. The neuraminidase-treated cells were washed twice with PBS, incubated with 1 ml of R18-labeled RBCs (0.2% hematocrit) in PBS++ at 4°C for 1 h in the dark with intermittent gentle agitation, washed, and then further incubated at 37°C for 30 min. Unbound RBCs were removed by washing the cells twice with PBS. Fusion was monitored and photographed at a magnification of 200× using an inverted fluorescence microscope. Vaccinia virus-infected or mock-transfected cells were used as negative controls.
Fusion of calcein-labeled RBCs with cells expressing SER virus F proteins.
Calcein-AM (Molecular Probes, Leiden, The Netherlands) dissolved in dimethyl sulfoxide was added to a concentration of 5 μM, immediately mixed, and then incubated at room temperature for 30 min in the dark. After the incubation, 30 ml of DMEM with 10% fetal calf serum was added, and the suspension was further incubated for 20 min in order to adsorb the unbound probe. The calcein-filled RBCs were washed five times and resuspended in PBS. Cell-RBC fusion was performed as described for the R18-labeled RBCs. The transfer of calcein from the RBCs to the cells expressing the wt or mutant fusion proteins was observed and photographed (200× magnification) using an inverted fluorescence microscope.
RESULTS
Construction and expression of mutant SER virus F proteins.
It was shown earlier that the elongated cytoplasmic domain of the SER virus F protein is inhibitory to the process of membrane fusion (42). Serial truncations in the CT of SER virus F protein indicated the importance of an 8-amino-acid stretch between amino acids 535 and 542 in suppressing membrane fusion activity. To determine the sequence requirements in this region for the regulation of fusion, SER virus F residues spanning amino acids 535 to 542 were mutated singly to alanines. Previous studies in our laboratory had shown that the C-terminal R peptide of the MuLV Env protein is a potent inhibitor of viral fusion activity and that a leucine residue at position 627 was particularly important for fusion inhibition (43). Hence, a mutation of the leucine residue at position 548 in the CT domain and a double mutation of leucines 539 and 548 to alanines were examined to study the role of these residues in the regulation of cell fusion.
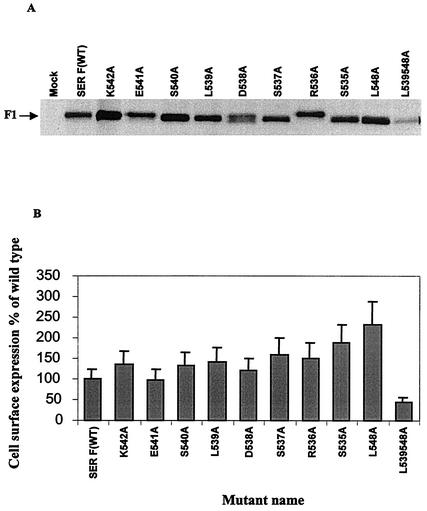
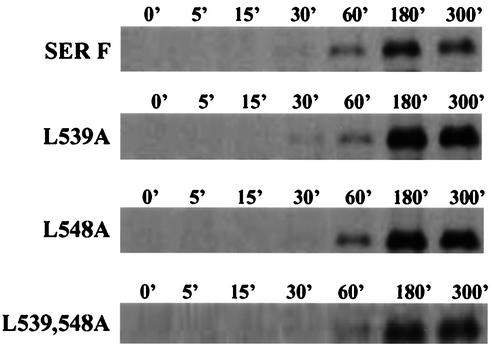
A transient vaccinia virus-T7 expression system was employed to express the wt and mutant SER virus F proteins in HeLa T4 cells. As shown in Fig. 2, most of the mutant SER virus F proteins were expressed efficiently and transported to the cell surface at levels comparable to those of the wt F protein, indicating that the mutations which we introduced did not affect the transport of the mutant proteins to the cell surface. The cell surface expression levels of the mutant proteins were estimated using a PhosphorImager as shown in Fig. 2B. Except for the double mutant L539, 548A which showed a surface expression level 45% of that of the wt SER virus F protein, all the other SER virus F protein CT substitution mutants had surface expression levels of 98 to 150%, except for the higher surface expression level (200%) shown by the leucine SER virus F protein mutant L548A. The differences in the sizes of the mutant proteins may be attributed to charge difference, which might lead to differential mobilities of the mutant proteins, as sequencing of the mutant genes ruled out the presence of additional mutations. The transport and stability of the mutant SER virus F proteins were examined by performing pulse-chase experiments. All the mutants were found to have stabilities on cell surfaces similar to or greater than that of the wt SER virus F protein. The L539A, L548A, and L539, 548A mutants showed greater stability after a 5-h chase period than the wt SER virus F protein (Fig. 3).
FIG. 2.
Expression of mutant SER virus F proteins. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with the mutant SER virus F constructs. Fourteen hours posttransfection, the cells were metabolically labeled with methionine and cysteine as described in Materials and Methods. Labeling was followed by biotinylation and immunoprecipitation using anti-SV5 antisera and protein A-agarose beads at 4°C for 2 h. The samples were prepared in reducing sample buffer and analyzed by SDS-8% PAGE and autoradiography. The surface quantitation was performed using a PhosphorImager, and the values are expressed as percentages of that of the wt F protein. The results are the averages of two independent experiments performed in triplicate. (A) Cell surface expression. (B) Cell surface quantitation of the wt SER virus F and mutant F proteins. The error bars indicate standard deviations from the mean.
FIG. 3.
Pulse-chase analysis of the wt and mutant SER virus F protein. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with plasmids expressing the designated proteins. Fourteen hours posttransfection, the cells were metabolically labeled with methionine or cysteine for 30 min and chased with cold methionine or cysteine for 5, 15, 30, 60, 180, or 300 min. At the end of the chase period, cell surface proteins were biotinylated and immunoprecipitated with anti-SV5 antibody. F proteins on the cell surface were detected by precipitating the cell immunoprecipitate with streptavidin-agarose beads as described in Materials and Methods.
Cell fusion activity of mutant proteins.
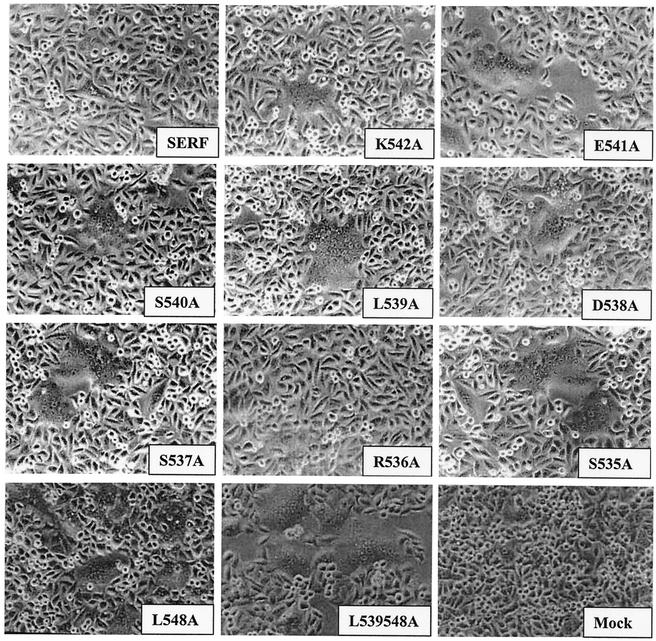
To investigate the effects of mutations on the induction of syncytium formation, we coexpressed the mutant F proteins with the SER virus HN protein using the recombinant vaccinia virus-T7 expression system. Sixteen hours posttransfection, cells were observed under an inverted Nikon microscope for syncytium formation. As shown in Fig. 4, all the mutant proteins were able to induce syncytium formation except the mutant R536A, which along with the wt SER virus F protein showed no fusion although they were expressed efficiently at the cell surface. The double mutant L539, 548A when coexpressed with SER virus HN protein showed extensive syncytium formation which was comparable to fusion levels observed in cells coexpressing SV5 F and HN proteins, indicating the important role played by these leucine residues in the suppression of SER virus F protein-induced fusion activity.
FIG. 4.
Cell fusion assay with mutant SER virus F proteins coexpressed with SER virus HN. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with the mutant SER virus F genes and cotransfected with SER virus HN plasmids using Lipofectin as described in Materials and Methods. Sixteen to 20 h posttransfection, the cells were observed using an inverted Nikon phase-contrast microscope and assayed for syncytium formation. The mutant F proteins cotransfected with the SER virus HN protein are shown in the insets.
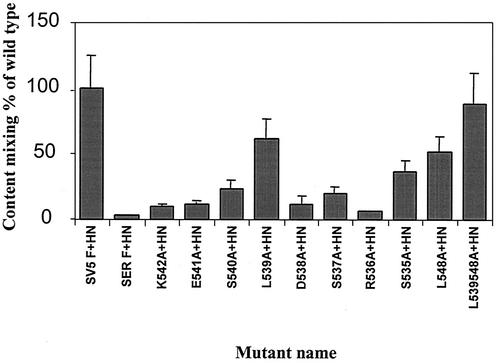
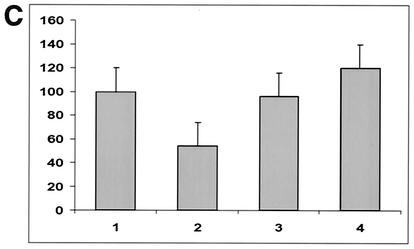
The extent of syncytium formation was quantitated by a reporter gene activation assay (31), and the results were expressed as the percentages of β-Gal activity of the mutant F proteins relative to the β-Gal activity of wt SVF HN. As shown in Fig. 5, mutants S540A, S537A, and S535A showed β-Gal activities ranging between 20 and 36% of that of the wt SV5 F protein, whereas other mutants, K542A, E541A, D538A, and R536A, showed much lower β-Gal activities of 6 to 15%. The lowest level (3%) of β-Gal activity over the mock-transfected background was detected when the wt SER virus F and HN proteins were coexpressed. The leucine mutants L548A and L539A showed fusion levels of >50%, and the double mutant L539, 548A showed β-Gal activity at 90% of that of the wt SV5 F protein, indicating that a critical role in fusion inhibition is played by the two leucine residues at these positions in the cytoplasmic domain.
FIG. 5.
Reporter gene activation assay with the SER virus F mutant proteins. One population of HeLa T4 cells was infected with recombinant vaccinia virus and transfected with the wt SER virus HN gene and a series of F mutants. The other cell population was infected with the wt vaccinia virus and transfected with the reporter gene construct. Sixteen hours posttransfection at 31°C, the two cell populations were mixed at a density of 106 cells/ml in a 96-well tissue culture plate and incubated for 5 h at 37°C, after which the cell fusion was quantitated by a colorimetric lysate assay. The data are represented as percentages of the β-Gal activity observed in the wt SV5 F- and HN-expressing cells. The results are the averages of three independent experiments. The error bars indicate standard deviations.
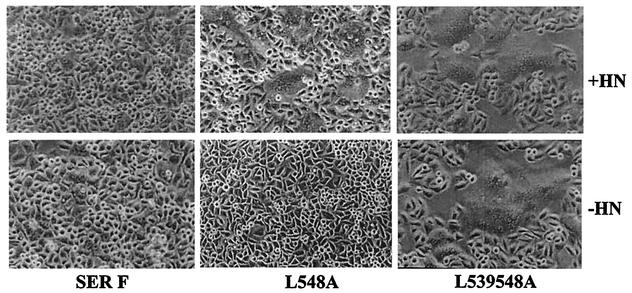
Fusion activity of mutant F proteins in the absence of HN.
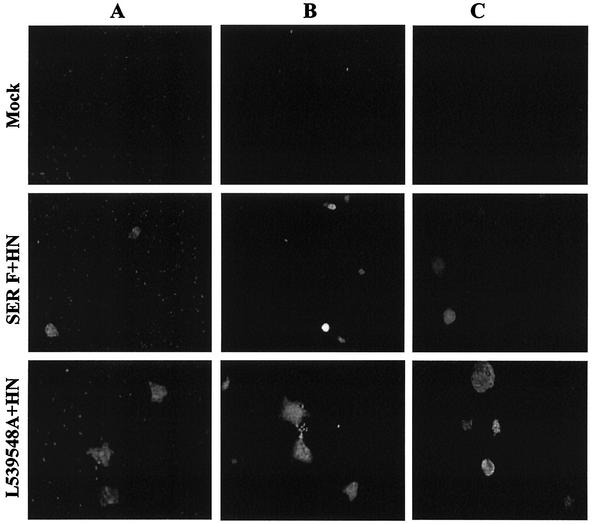
All the mutant SER virus F proteins generated were also assayed for syncytium formation in the absence of the SER virus HN protein. Most of the mutant proteins showed no fusion activity in the absence of HN. However, we observed that the double leucine mutant, L539, 548A, could induce cell fusion in the absence of the SER virus HN protein (Fig. 6). Upon quantification of the fusion activity by the cytoplasmic-mixing assay described previously, the L539, 548A mutant by itself showed 25% fusion activity compared to the wt SV5 F-HN protein-induced fusion (data not shown). These results indicate that changes in the F cytoplasmic domain can eliminate the requirement for HN in membrane fusion.
FIG. 6.
HN-independent fusion by the SER virus L539, 548A mutant protein. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with wt SER virus F, L548A, and L539, 548A constructs with (+) or without (−) cotransfection with the SER virus HN gene using Lipofectin as described in Materials and Methods. Sixteen to 20 h posttransfection, the cells were observed using an inverted Nikon phase-contrast microscope and assayed for syncytium formation.
Effects of other amino acid substitutions in inhibition of membrane fusion.
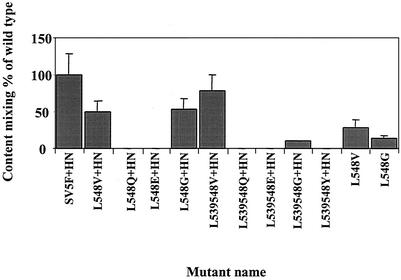
To further study whether the hydrophobicity of the leucine residues is involved in the inhibitory role in membrane fusion, a number of substitution mutants were generated in which the leucine residues at position 548 or at 539 and 548 were mutated to valine, glutamine, glutamic acid, serine, glycine, tryptophan, or isoleucine. The mutant proteins were expressed using the vTF7-3 transient-expression system in HeLa T4 cells. All the mutant proteins were expressed at the cell surface with wt efficiencies as shown by surface biotinylation analysis, but mutants L548S; L548Y; L548I; L539, 548S; and L539, 548I were found to be defective in proteolytic cleavage of F0 into F1 and F2 (Fig. 7). The mutant proteins were also analyzed for transport and stability by pulse-chase analysis as described earlier. All the mutant proteins were transported efficiently to the cell surface and were as stable as the wt SER virus F protein but less stable than the L539, 548A double mutant (Fig. 8). All the single and double mutants generated, except for the cleavage-defective mutants, were assayed for syncytium formation and were quantified by the cytoplasmic-mixing assay when coexpressed with or without the coexpressed SER virus HN protein. Of particular interest were mutants L548V and L548G, as well as L539, 548V and L539, 548G, which showed fusion efficiencies of 51, 52, 77, and 9%, respectively, when coexpressed with SER virus HN, whereas other substitutions had no effect on syncytium formation or cytoplasmic-content mixing (Fig. 9).
FIG. 7.
Cell surface expression of the mutant SER virus F proteins. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with the mutant SER virus F genes (as listed above the lanes). Fourteen hours posttransfection, the cells were metabolically labeled with methionine-cysteine as described in Materials and Methods. Labeling was followed by biotinylation and immunoprecipitation using anti-SV5 antisera and protein A-agarose. The samples were analyzed by SDS-8% PAGE and autoradiography. The surface quantitation was performed using a PhosphorImager, and the values are expressed as percentages of the wt F protein expression. (A) Single leucine mutations. (B) Double leucine mutations.
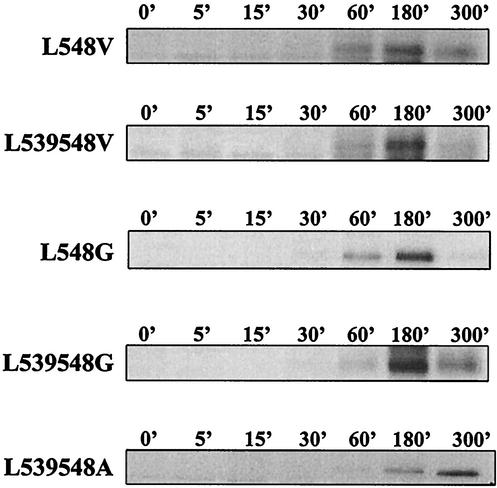
FIG. 8.
Pulse-chase analysis of mutant SER virus F proteins. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with plasmids expressing L548V; L539, 548V; L548G; L539, 548G; and L539, 548A. Fourteen hours posttransfection, the cells were metabolically labeled with methionine or cysteine for 30 min and chased with cold methionine or cysteine for 5, 15, 30, 60, 180, or 300 min. At the end of the chase period, the cell surface proteins were biotinylated and immunoprecipitated with anti-SV5 antibody. F proteins on the cell surface were detected by precipitation with streptavidin-agarose beads as described in Materials and Methods.
FIG. 9.
Reporter gene activation assay with mutant proteins. One population of HeLa T4 cells was infected with recombinant vaccinia virus and transfected with the wt SER virus HN gene and a series of plasmids expressing F protein mutants, and the other cell population was infected with wt vaccinia virus and transfected with the reporter gene construct. Sixteen hours posttransfection at 31°C, the two cell populations were mixed at a density of 106 cells/ml in a 96-well tissue culture plate and incubated for 5 h at 37°C, after which the cell fusion was quantitated by a colorimetric lysate assay. The data are represented as percentages of the β-Gal activity observed in the wt SV5 F- and HN-expressing cells. The results are the averages of three independent experiments. The error bars indicate standard deviations.
Mutants L548V and L548G showed fusion activities of 30 and 15% when expressed in the absence of HN protein, which clearly supports the idea that the nature of residues present at positions 539 and 548 in the CT of SER virus F protein is important in the regulation of fusion activity.
Effects of CT mutations on stages of membrane fusion.
The process of membrane fusion involves several steps before the complete fusion of two membranes takes place. First, the outer leaflets of the membranes fuse with each other, bringing about the mixing of the lipid molecules between the two membranes, which has been described as lipid mixing or hemifusion (27). This is followed by the formation of fusion pores, leading to the exchange of cytoplasmic content between cells.
To further define the nature of the block in syncytium formation observed for SER virus, we performed dye redistribution assays using the fluorescent label R18 for lipid mixing and calcein for the complete content mixing. We labeled guinea pig RBCs with either R18 or calcein as described by Kemble et al. (27), and 16 h posttransfection, cells were incubated with the labeled RBCs at 4°C for 1 h and then transferred to 37°C for different lengths of time. As shown in Fig. 10A, the cells expressing wt SER virus F and HN proteins showed minimal R18 transfer, and labeled SER virus F- and HN-expressing HeLa T4 cells were seen as single cells. In contrast, we observed syncytia showing dye transfer in the cells coexpressing L548A or L539, 548A and SER virus HN. These results indicate that even though the wt SER virus F protein does not induce any syncytium formation, there is a residual level of mixing of the lipid membranes between labeled RBCs and individual cells expressing the F and HN proteins. Similarly, as shown in Fig. 10B, cells expressing the wt SER virus F and HN proteins were assayed for calcein dye transfer. Some dye transfer from the calcein-filled RBCs to the F-expressing cells was observed, but it was found to be restricted to single cells, whereas an extensive calcein transfer was seen in the syncytia formed in L548A- (not shown); L539, 548A-; and SER virus HN-expressing cells. Dye transfer to singly infected cells has also been observed in SER virus-infected cells (36).
FIG. 10.
Fusion activities of SER virus F proteins monitored by fluorescent-dye transfer. Sixteen hours posttransfection, SER virus F- and HN-expressing HeLa T4 cells were incubated with guinea pig RBCs labeled with either R18 or Calcein-AM at 4°C for 1 h and then washed to remove unbound RBCs and incubated at 37°C for 30 min. The fluorescent-dye transfer was observed under an inverted fluorescence microscope. (A) R18 transfer (hemifusion). (B) Calcein transfer (complete fusion). (C) Reporter gene activation assay using the EGFP expression system. One population of HeLa T4 cells infected with vTF7-3 and cotransfected with plasmids expressing wt or mutant SER virus F and SER virus HN proteins 16 h posttransfection was mixed with the cells infected with wt vaccinia virus and transfected with T7-driven EGFP. Twenty-four hours postmixing, the cells were observed for EGFP fluorescence in mock-transfected cells and cells transfected with plasmids encoding SER virus F plus HN or L539, 548A plus HN.
These results were confirmed by performing an EGFP reporter assay. The EGFP gene was PCR amplified and cloned in the pGEM-3 vector and was used as a reporter gene plasmid. Cell populations infected with vTF7-3 and coexpressing wt or mutant SER virus F and SER virus HN proteins were mixed with cells infected with wt vaccinia virus and expressing EGFP. Twenty-four hours postmixing, the cells were observed for the ability to show fluorescence, and as shown in Fig. 10C, the wt SER virus F-HN cells showed low levels of EGFP expression compared to the EGFP expression observed in L539, 548A or other mutants, which showed significant green fluorescence in multinucleate syncytia. The restricted dye transfer in wt SER virus F-expressing cells suggests that the block in fusion might be at the level of fusion pore enlargement.
Effect of an elongated CT in SV5-SER virus F CT chimeric proteins.
To investigate whether the elongation of the CT would also suppress fusion induced by the SV5 F protein, a chimeric SV5-SER virus gene was constructed which encoded the SV5 F protein carrying an elongated CT derived from the SER virus F protein. Genes encoding a chimeric SV5-SER virus F L548A protein and a chimeric SV5-SER virus F L539, 548A protein were also constructed. A transient vaccinia virus-T7 expression system was employed to express the wt and the mutant SV5-SER virus F CT chimeric proteins in HeLa T4 cells. The expression levels of the chimeric SV5-SER virus F CT mutants ranged between 35 and 45% of that of the wt SV5 F protein (Fig. 11A, lanes 1 to 3). The syncytium formation assay with the chimeric SV5-SER virus F protein CT showed reduced syncytium formation compared to the wt SV5 F protein when coexpressed with the SER virus HN protein (Fig. 11B, a). On the other hand, the chimeric mutant proteins L548A and L539, 548A showed no suppressive effect, as the mutants induced extensive multinucleate cell formation (Fig. 11B, b and c). The cytoplasmic-mixing assay with the chimeric SV5-SER virus F mutant protein showed a reduction in β-Gal activity of 40% compared to the wt SV5 F protein (Fig. 11C, lane 2); this was compensated for by the leucine mutations at positions 539 and 548, which showed extensive fusion comparable to that of the wt SV5 F protein (Fig. 11C, lanes 3 and 4). The chimeric SV5-SER virus F and SV5-SER virus F L539, 548A mutant proteins also showed lipid mixing and cytoplasmic mixing, though the dye transfer was reduced in chimeric SV5-SER virus F-SV5 HN-expressing cells (Fig. 11D, lane 1). These results indicate that the suppressive effect of the elongated CT of SER virus F protein could be partially transferred to the SV5 F protein, in which the CT of the SV5 F protein was replaced by the CT of the SER virus F protein. However, the chimeric L548A and the L539, 548A mutants showed fusion as efficient as that of the wt SV5 F protein.
FIG. 11.
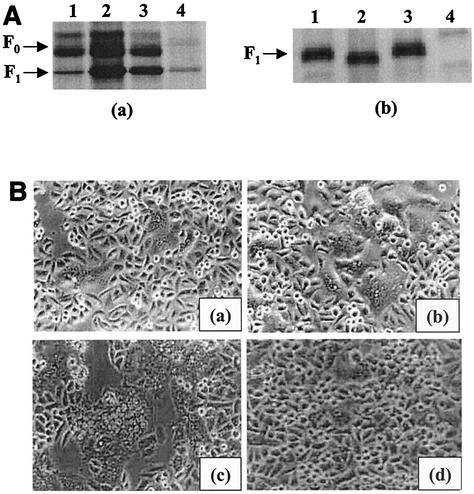
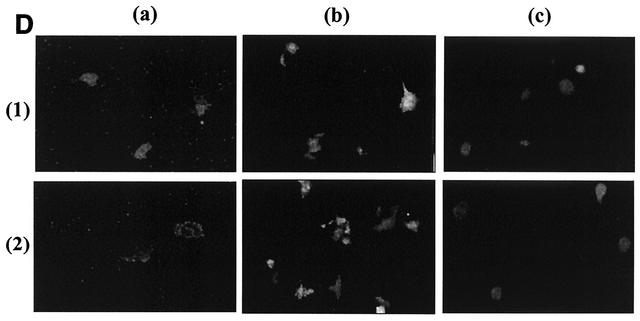
Analysis of chimeric SV5-SER virus F CT proteins. (A) Expression of chimeric SV5-SER virus F CT proteins. HeLa T4 cells were infected with vTF7-3 at an MOI of 10 for 1 h at 37°C and then transfected with the mutant SER virus F or chimeric SV5-SER virus constructs. Fourteen hours posttransfection, the cells were metabolically labeled with methionine and cysteine as described in Materials and Methods. Labeling was followed by biotinylation and immunoprecipitation using anti-SV5 antisera and protein A-agarose beads at 4°C for 2 h each. The samples were prepared in reducing sample buffer and analyzed on SDS-8% PAGE. (a) Cell lysate; (b) cell surface. Lanes: 1, SV5-SER virus F protein CT chimera; 2, SV5-SER virus F CT chimera L548A; 3, SV5-SER virus F CT chimera L539, 548A; 4, pGEM-3 vector control. (B) Cell fusion assay with chimeric SV5-SER virus F CT mutant proteins coexpressed with SV5 HN. Sixteen to 20 h posttransfection, the cells were observed with an inverted Nikon phase-contrast microscope and assayed for syncytium formation. (a) Chimeric SV5-SER virus F-CT plus HN; (b) chimeric SV5-SER virus F CT L548A plus HN; (c) chimeric SV5-SER virus F CT L539548A plus HN; (d) mock-transfected control. (C) Reporter gene activation assay with chimeric mutant proteins as described in Materials and Methods. 1, SV5 F plus HN; 2, chimeric SV5-SER virus F CT plus HN; 3, chimeric SV5-SER virus F CT L548A plus HN; 4, chimeric SV5-SER virus F CT L539548A plus HN. The error bars indicate standard deviations. (D) Fusion activities of chimeric SV5-SER virus F proteins monitored by fluorescent-dye transfer. Sixteen hours posttransfection, chimeric SV5 F-SV5 HN-expressing HeLa T4 cells were incubated with guinea pig RBCs labeled with either R18 or Calcein-AM at 4°C for 1 h and then washed to remove unbound RBCs and incubated at 37°C for 30 min. The fluorescent-dye transfer was observed under an inverted fluorescence microscope. (a) R18 transfer (hemifusion); (b) calcein transfer (complete fusion); (c) reporter gene activation assay using the EGFP expression system as described in Materials and Methods. (1) Chimeric SV5-SER virus F CT plus HN; (2) chimeric SV5-SER virus F CT L539, 548A plus HN.
DISCUSSION
SER virus, a recently characterized paramyxovirus, exhibits minimal cell fusion activity in several cell types, and this lack of fusion activity was shown to be the result of an extended F protein CT domain (42). In this study, we have defined the roles of specific amino acids in the SER virus cytoplasmic domain that modulate fusion activity. Previous studies with CT truncation mutants of SER virus F showed that truncation of 22 or 17 residues from the C terminus restores the fusion to 100 and 80%, respectively, whereas the truncation of 9 residues resulted in recovery of fusion to only 20% of the level of wt SV5 F and HN (42). In the present study, we therefore constructed mutant SER virus F genes encoding proteins with substitutions between amino acids 542 and 535 to define the molecular features that are important for the inhibitory role of this portion of the cytoplasmic domain in membrane fusion. Additional leucine mutations in the CT domains of SER virus F, L548A, and L539, 548A were also studied for their effects on membrane fusion. We found that Leu-539 and Leu-548 played critical roles in the inhibition of fusion by the CT domain, as mutation of the two leucines singly to alanines partially rescued fusion and the double mutant L539, 548A completely rescued syncytium formation. Mutation of most of the charged amino acids had little effect on fusion inhibition, whereas mutation of serine residues had a slightly greater effect on suppression of fusion activity, signifying that other residues besides leucines in the CT domain play a less significant role in fusion inhibition.
The present work was further extended by investigating the requirement for HN in membrane fusion by the mutant SER virus F proteins. Most of the mutants were unable to show fusion in the absence of the HN protein; however, the mutant L539, 548A eliminated the requirement for HN protein expression for membrane fusion. This is the first evidence that a change in the cytoplasmic domain of F can confer HN-independent fusion activity; other studies have associated HN-independent fusion with residues located in the ectodomain of paramyxovirus F proteins. It has been shown that a single-amino-acid difference in the F2 polypeptide of SV5 (L22P) can account for the different requirements for the HN protein for fusion by different strains of SV5 (23). An amino acid in the heptad repeat 1 domain (Glu-132) was also shown to be important for the HN-independent fusion activity of the SV5 F protein (24). In the Newcastle disease virus F ectodomain, a single-amino-acid change, L289A, results in fusion in the absence of the HN protein (40).
The nature of residues introduced at positions 539 and 548 was shown to affect syncytium formation, as the single mutations L548V and L548G and the double mutation L539, 548V were able to rescue fusion quite efficiently in the presence of the HN protein. The mutations L548V and L548G also showed fusion in the absence of the SER virus HN protein, indicating the critical role of smaller uncharged amino acids at positions 539 and 548 in recovering membrane fusion. Other mutations, such as L548Q/E/Y, had no enhancing effect on the fusion activity. These mutations might alter the conformation of SER virus F, affecting its ability to undergo transition to a fusogenic form. The finding of a cleavage defect in mutants such as L548S/Y/I and L539, 548S/I strongly indicates that these mutations in the CT affect the conformation of the external domain.
The block in syncytium formation that is observed with SER virus led us to perform hemifusion and calcein binding assays. Our results indicated that there was a low level of dye transfer (R18 and calcein) in the cells expressing wt SER virus F and HN proteins, but it was restricted to single cells (signifying a block in cell-to-cell fusion) compared to dye transfer to giant cells expressing mutant SER virus F and HN proteins. Dye transfer to single cells was also observed in SER virus-infected cells (36), indicating that there is some level of fusion induced by the wt SER virus F and HN proteins that does not result in multinucleate cell formation. This minimal fusion activity may play a role in viral entry.
Studies of the mechanism of viral membrane fusion have led to a common theme involving conformational transitions that have been observed in several viral fusion proteins (6, 12, 18). In some viral fusion proteins, as influenza virus hemagglutinin, vesicular stomatitis virus G protein, and Semliki Forest virus fusion protein, conformational changes take place upon treatment at low pH (18, 36). The trigger for membrane fusion promoted by paramyxovirus fusion proteins is not known, but it is thought to involve interaction with receptors. The SV5 F protein was shown to undergo a conformational change after proteolytic cleavage into F1 and F2 using a panel of anti-peptide antibodies (9). These conformational changes include the extracellular domain and the transmembrane anchor, in which the heptad repeats are known to form a thermostable trimer of heterodimers (3, 10, 26). However, besides the ectodomain, the CT of some viruses is known to modulate fusion activity. Truncations in the CT of parainfluenza virus type 3 F protein and SV5 F protein abolished fusion activity (2, 44), whereas the removal of the CT in human parainfluenza virus type 2 F protein did not affect its fusion activity (44). In MuLV, the C-terminal 16 amino acids, called the R peptide, were found to be inhibitory to membrane fusion (16, 25, 38). It was shown recently that mutations in the CT of the MuLV envelope protein can suppress fusion inhibition by the C-terminal R peptide and that Leu-627 plays an important role in fusion inhibition by the R peptide (29, 43). Our results also suggest that the CT appears to affect the conformation of the extracellular domain of SER virus F protein, as mutations in the elongated CT rescue the ability of SER virus F protein to induce fusion. In addition, the finding that mutations in the CT can affect cleavage of the external domain without blocking transport to the cell surface also demonstrates that these changes affect the conformation of the external domain.
Modeling studies in the Emory Molecular Modeling Core Facility comparing the CT regions of the SER virus F and MuLV Env proteins suggest that the proteins might share some common structural features, including a helical membrane-proximal region and an extended distal loop containing highly charged residues. Thus, they may share a common regulatory mechanism for membrane fusion. One plausible explanation for the inhibitory effect of the cytoplasmic domain on membrane fusion is that the presence of the long CT in the SER virus F protein may serve to stabilize the metastable conformation of the protein whereas the leucine-to-alanine mutations in the elongated CT region lower the activation energy for the conversion from a fusion-inactive to a fusion-active conformation upon receptor binding.
In conclusion, we found that mutations in the long CT of the SER virus F protein can rescue its lack of fusion activity depending on the positions and the nature of the residues in the CT domain. Two leucine residues were found to play particularly important roles in the suppression of SER virus fusion activity and to eliminate the requirement for HN in membrane fusion. These results, as well as the finding that mutations in the CT domain inhibit cleavage of the F protein, support the conclusion that the CT regulates fusion activity through an effect on the conformation of the extracellular domain of the F protein via the transmembrane domain.
Acknowledgments
This study was supported by grant CA 18611 from the National Institutes of Health.
We thank Kim Gernert, Molecular Modeling Core Facility (Emory University School of Medicine, Atlanta, Ga.), for her advice on molecular modeling. We thank Tanya Cassingham for assistance in preparing the manuscript.
REFERENCES
- 1.Bagai, S., and R. A. Lamb. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J. Virol. 69:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1996. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J. Cell Biol. 135:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 4.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 71:3075-3080. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutch, R. E., and R. A. Lamb. 2001. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. J. Virol. 75:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutch, R. E., G. P. Leser, and R. A. Lamb. 1999. Paramyxovirus fusion protein: characterization of the core trimer, a rod-shaped complex with helices in anti-parallel orientation. Virology 254:147-159. [DOI] [PubMed] [Google Scholar]
- 11.Earl, P. L., S. Koenig, and B. Moss. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 65:31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, J. K., S. G. Peisajovich, M. Ovadia, and Y. Shai. 1998. Structure-function study of a heptad repeat positioned near the transmembrane domain of Sendai virus fusion protein which blocks virus-cell fusion. J. Biol. Chem. 273:27182-27190. [DOI] [PubMed] [Google Scholar]
- 16.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinen, E., W. Herbst, and N. Schmeer. 1998. Isolation of a cytopathogenic virus from a case of porcine reproductive and respiratory syndrome (PRRS) and its characterization as parainfluenza virus type 2. Arch. Virol. 143:2233-2239. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Homma, M., and M. Ouchi. 1973. Trypsin action on the growth of Sendai virus in tissue culture cells. 3. Structural difference of Sendai viruses grown in eggs and tissue culture cells. J. Virol. 12:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath, C. M., and R. A. Lamb. 1992. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 66:2443-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, M., M. Nishio, M. Kawano, S. Kusagawa, H. Komada, Y. Ito, and M. Tsurudome. 1997. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J. Virol. 71:9855-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, M., M. Nishio, H. Komada, Y. Ito, and M. Tsurudome. 2000. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81:719-727. [DOI] [PubMed] [Google Scholar]
- 25.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 27.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 28.Le Bivic, A., A. Quaroni, B. Nichols, and E. Rodriguez-Boulan. 1990. Biogenetic pathways of plasma membrane proteins in Caco-2, a human intestinal epithelial cell line. J. Cell Biol. 111:1351-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, M., C. Yang, and R. W. Compans. 2001. Mutations in the cytoplasmic tail of murine leukemia virus envelope protein suppress fusion inhibition by R peptide. J. Virol. 75:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohuchi, M., C. Fischer, R. Ohuchi, A. Herwig, and H. D. Klenk. 1998. Elongation of the cytoplasmic tail interferes with the fusion activity of influenza virus hemagglutinin. J. Virol. 72:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reitter, J. N., T. Sergel, and T. G. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69:5995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 36.Ruigrok, R. W., N. G. Wrigley, L. J. Calder, S. Cusack, S. A. Wharton, E. B. Brown, and J. J. Skehel. 1986. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 5:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, A., and A. Rein. 1985. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology 145:335-339. [DOI] [PubMed] [Google Scholar]
- 39.Sergel, T., and T. G. Morrison. 1995. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology 210:264-272. [DOI] [PubMed] [Google Scholar]
- 40.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong, S., M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi and H.-D. Klenk. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301:322-333. [DOI] [PubMed] [Google Scholar]
- 43.Yang, C., and R. W. Compans. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J. Virol. 71:8490-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, Q., and R. W. Compans. 1995. Differences in the role of the cytoplasmic domain of human parainfluenza virus fusion proteins. J. Virol. 69:7045-7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]