Abstract
Alanine scanning mutagenesis was performed on monomeric gp120 of human immunodeficiency virus type 1 to systematically identify residues important for gp120 recognition by neutralizing and nonneutralizing monoclonal antibodies (MAbs) to the CD4 binding site (CD4bs). Substitutions that affected the binding of broadly neutralizing antibody b12 were compared to substitutions that affected the binding of CD4 and of two nonneutralizing anti-CD4bs antibodies (b3 and b6) with affinities for monomeric gp120 comparable to that of b12. Not surprisingly, the sensitivities to a number of amino acid changes were similar for the MAbs and for CD4. However, in contrast to what was seen for the MAbs, no enhancing mutations were observed for CD4, suggesting that the virus has evolved toward an optimal gp120-CD4 interaction. Although the epitope maps of the MAbs overlapped, a number of key differences between b12 and the other two antibodies were observed. These differences may explain why b12, in contrast to nonneutralizing antibodies, is able to interact not only with monomeric gp120 but also with functional oligomeric gp120 at the virion surface. Neutralization assays performed with pseudovirions bearing envelopes from a selection of alanine mutants mostly showed a reasonable correlation between the effects of the mutations on b12 binding to monomeric gp120 and neutralization efficacy. However, some mutations produced an effect on b12 neutralization counter to that predicted from gp120 binding data. It appears that these mutations have different effects on the b12 epitope on monomeric gp120 and functional oligomeric gp120. To determine whether monomeric gp120 can be engineered to preferentially bind MAb b12, recombinant gp120s were generated containing combinations of alanine substitutions shown to uniquely enhance b12 binding. Whereas b12 binding was maintained or increased, binding by five nonneutralizing anti-CD4bs MAbs (b3, b6, F105, 15e, and F91) was reduced or completely abolished. These reengineered gp120s are prospective immunogens that may prove capable of eliciting broadly neutralizing antibodies.
Broadly neutralizing antibodies can protect against intravenous and mucosal challenges with immunodeficiency viruses in animal models (3, 16, 21, 32, 34, 43, 47, 49, 64). It has, therefore, become increasingly clear that eliciting such antibodies should be a major goal of efforts to develop a human immunodeficiency virus type 1 (HIV-1) vaccine (7, 9, 33, 42, 61, 76, 78). Animal model studies have provided a number of guidelines regarding the types of antibodies that should be elicited. First, protection is generally provided by antibodies that effectively neutralize virus in vitro (43, 46). Second, serum neutralizing antibody levels at the time of virus challenge need to be relatively high (about 1:100) to achieve sterile protection, although lower levels can provide benefit in terms of delayed and/or decreased viremia (43, 49, 64). Third, protection by broadly neutralizing human monoclonal antibodies (MAbs) against a number of viruses suggests that protection against many different strains of HIV-1 may be achievable (3, 48, 49).
The major problem to date, from a vaccine standpoint, is that no immunogen has been generated that can elicit reasonable levels of such broadly neutralizing antibodies. These antibodies should be targeted to relatively conserved and exposed regions of the HIV-1 envelope, but the paucity of broadly neutralizing antibodies in natural infection suggests that the virus presents these regions to the immune system in such a way as to minimize an effective antibody response (9, 51, 76, 78). A molecular understanding of regions on the HIV-1 envelope that are exposed and conserved and how they can be recognized by antibodies would be invaluable in the design of immunogens that can elicit broadly neutralizing antibodies.
The CD4 binding site (CD4bs) on HIV-1 surface glycoprotein gp120 is a highly conserved region that is known to be exposed for ligand binding (12, 23). In theory, this would seem to form an excellent target for neutralizing antibodies. Many MAbs that bind with a high affinity to the CD4bs of monomeric gp120 from various primary and T-cell-line-adapted (TCLA) HIV-1 isolates have been isolated (http://resdb.lanl.gov/ABDB/antibody_id.htm). These MAbs are characterized by their ability to compete with soluble CD4 and with one another (41). Anti-CD4bs MAbs typically neutralize TCLA viruses with moderate efficacy but neutralize primary isolates of HIV-1 very weakly if at all (52). However, one MAb, b12, which interacts with the CD4bs does neutralize many primary and TCLA viruses very efficiently (10, 13, 22, 35). MAb b12 and nonneutralizing anti-CD4bs MAbs typically have very similar binding affinities for monomeric gp120 from a number of isolates (40, 41). The differences between b12 and the other MAbs in neutralizing activity against TCLA viruses, therefore, have been associated with different affinities for the mature envelope trimer expressed on virions (50, 57, 60, 63). Typically, MAb b12 is able to bind with comparable affinities to monomeric gp120 and the mature trimer on the surface of infected cells (50), which is believed to be identical to the functional envelope molecule on the surface of virions (60). Nonneutralizing anti-CD4bs MAbs, on the other hand, bind with a lower affinity to the mature trimer. The implication, therefore, is that b12 is able to bind similarly to monomeric gp120 and to the native TCLA trimer and neutralize the virus effectively, whereas the other anti-CD4bs MAbs suffer some impediment in their access to the CD4bs on the mature TCLA trimer and, therefore, neutralize virus less effectively (53). Lower levels of envelope expression have made the investigation of the correlation between binding to the mature trimer and neutralization more troublesome for primary HIV-1. One study reports such a correlation (14), and we have considered that the explanation for the efficacy of b12 against primary viruses is likely to be similar to that for TCLA viruses. However, other studies have suggested that other, undefined mechanisms may be important for primary viruses (15, 65, 66, 80).
Irrespective of the underlying mechanisms responsible for neutralization differences, b12 and nonneutralizing anti-CD4bs MAbs constitute probes to distinguish presentations of gp120 that are desirable for vaccine purposes from those that are less desirable. We reasoned that although monomeric gp120 binds b12 and nonneutralizing anti-CD4bs antibodies equivalently, it might be possible to mutate monomeric gp120 so that it would bind b12 well but nonneutralizing MAbs less well. Such a molecule would be an interesting immunogen. As a first step, we decided to identify by alanine scanning mutagenesis amino acid residues on gp120 which modulate binding by b12 and to compare these to residues which affect the binding of two representative nonneutralizing anti-CD4bs MAbs that we previously characterized in detail, namely, b3 and b6. Amino acid changes which had an effect on antibody binding were compared to those that affected CD4 binding. The results show that the MAbs and CD4 bind to gp120 with similar footprints. The footprint for CD4 is in fact very close to that which would be expected from the crystal structure of a complex of CD4 and the core of gp120 (30). The footprint for antibody b12 also corresponds well with that expected from a docking model of the b12 structure and the gp120 core structure (44). However, a number of differences observed in the epitope maps between b12 and the nonneutralizing anti-CD4bs MAbs suggest that it may indeed be possible to engineer a gp120 molecule which is more disposed to eliciting b12-like antibodies.
MATERIALS AND METHODS
Antibodies.
MAbs b3, b6, and b12 were isolated as Fab fragments from a phage display library derived from a single donor and were characterized in previous studies (4, 5, 8, 10, 57). All three antibodies recognize discontinuous epitopes overlapping the CD4bs on gp120. CD4-immunoglobulin G2 (IgG2), a recombinant antibody-like fusion protein in which the heavy- and light-chain variable domains of human IgG2 have been replaced with the D1D2 domains of human CD4 (1), was a kind gift from William Olson. This molecule was used in the present study as a surrogate for CD4. MAbs 15e and F91, which react with epitopes overlapping the CD4bs on gp120 (20, 39, 40, 58, 71), and MAb 17b, which recognizes an epitope overlapping the coreceptor binding site on gp120 (68, 70, 77), were kindly provided by James Robinson. Human IgG purified from pooled plasma obtained from healthy asymptomatic seropositive individuals and MAb F105, another anti-CD4bs antibody (55, 56, 71), were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (NIH ARRRP). MAb 2G12, which recognizes an epitope involving α1→2 mannose residues on the carbohydrate-rich “silent” face (30, 76) of gp120 (59, 62, 73), was a generous gift from Hermann Katinger.
Plasmid constructs and mutagenesis.
Alanine mutations (Table 1 and Fig. 1A) were generated by using a Quikchange mutagenesis kit (Stratagene). For antibody mapping experiments, plasmid pSVIIIexE7pA−JR-CSF was used as a template. This plasmid was derived from plasmid pSVIIIexE7pA−HxB2 (19), which was modified as described previously (82) in order to subclone the env gene of HIV-1JR-CSF, a molecularly cloned primary HIV-1 strain (28). Two variable-loop deletion mutants were also generated by using pSVIIIexE7pA−JR-CSF as a PCR template. A V1 deletion (ΔV1) mutant (deletion of residues 134 to 154) was constructed by PCR with primers csf120-f (5′-GTCTGAGTCGGAGCTAGCGTAGAAAAGTTGTGGGTCA-3′) and csfdV1-r (5′-GTCTGAGTCGGAACCGGACCCATCTTTGCAATTTAAAGTA-3′) and primers csfdV1-f (5′-GTCTGAGTCGGATCCGGTTCTGGGAAAAACTGCTCTTT-3′) and csf120-r (5′-GTCTGAGTCGGACTCGAGTTTTCTCTTTGCACCACTCTTC-3′). Primers csfdV1-f and csfdV1-r both contain a BsaWI restriction site (underlining). The PCR products were cloned into pSVIIIexE7pA−JR-CSF by using KpnI, BsaWI, and MfeI in a two-step ligation reaction. A ΔV3 mutant (deletion of residues 303 to 324) was generated in a similar manner by using primers csf120-f and csfdV3-r (5′-GTCTGAGTCGGAACCGGACCCATTGTTGCTGGGCCTTGT-3′) and primers csfdV3-f (5′-GTCTGAGTCGGATCCGGTTCTGGGGATATAAGACAAGCCC-3′) and csf120-r. Primers csfdV3-f and csfdV3-r contain unique BsaWI restriction sites (underlining). To generate a ΔV1/V2 mutant (V2 loop deleted from residues 160 to 193), the pSVIIIexE7pA−JR-CSF-ΔV1 mutant was used as a template. First, the BsaWI site introduced into the ΔV1 mutant was changed by site-directed mutagenesis so that the amino acid sequence was retained. Deletion of the V2 sequence was performed in a manner analogous to that used for the ΔV1 and ΔV3 mutants by using primers csf120-f and csfdV2-r2 (5′-GTCTGAGTCGGAACCGGACCCGAAAGAGCAGTTTTT-3′) and primers csfdV2-f (5′-GTCTGAGTCGGATCCGGTTCTGGGATAAGTTGTAACACC-3′) and csf120-r. (The unique BsaWI restriction sites in primers csfdV2-r2 and csfdV2-f are underlined.) The PCR fragments were cloned into pSVIIIexE7pA−JR-CSF by using KpnI, BsaWI, and MfeI. In all variable-loop deletion mutants, the deleted sequences were replaced by a Gly-Ser-Gly-Ser-Gly linker.
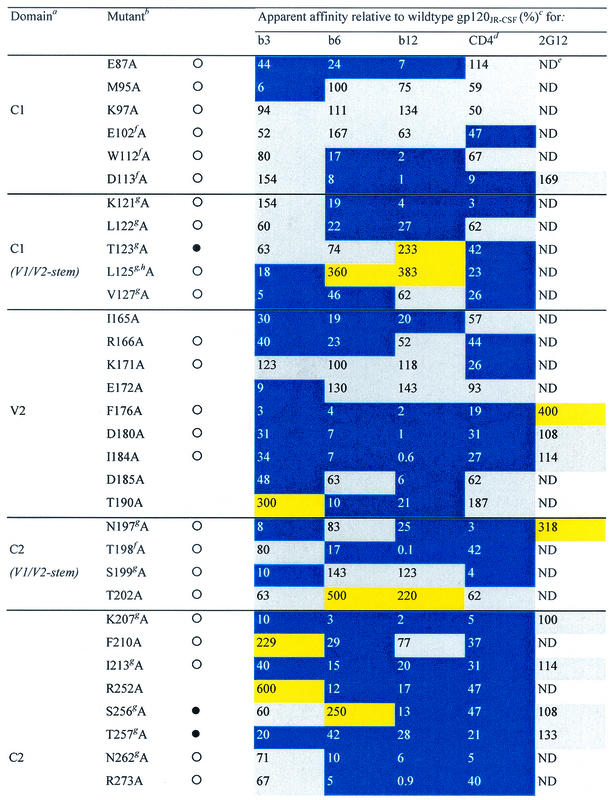
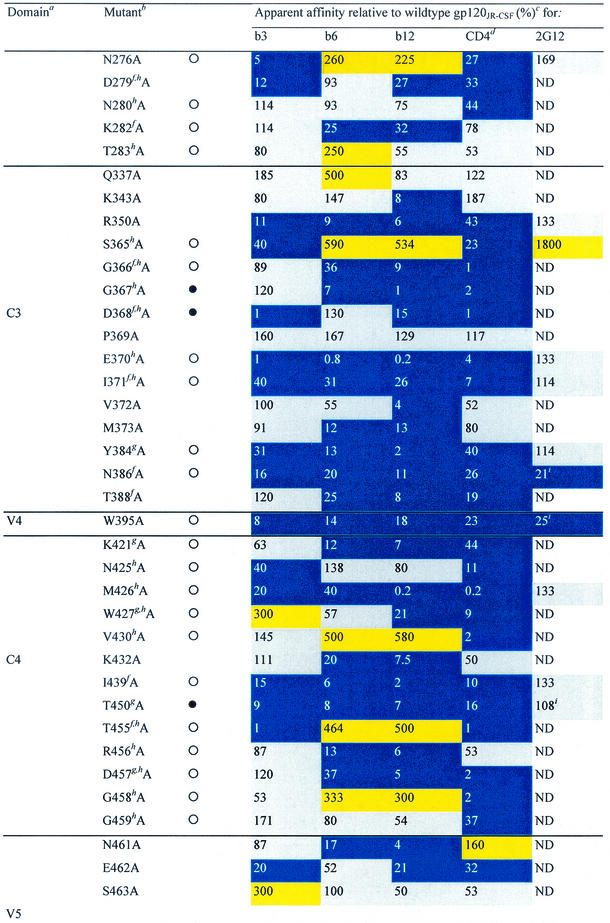
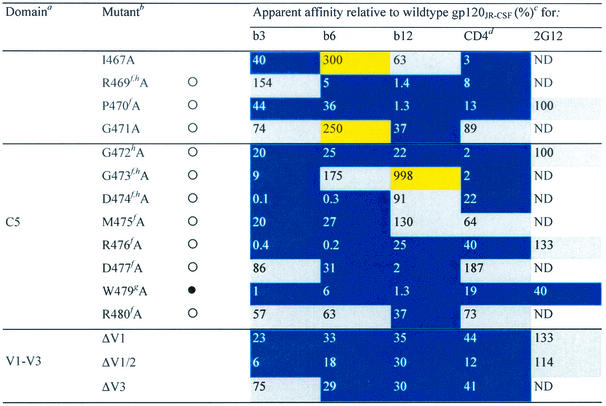
TABLE 1.
Alanine and variable loop deletion mutants generated in this study and their binding to MAbs and CD4
gp120 domain. C, constant domain; V, variable loop. Amino acid numbering is relative to that of HIV-1HxB2, where 1 is the initial methionine (26). Δ, amino acid deletions: ΔV1, amino acids 134 to 154; ΔV1/V2, amino acids 134 to 154 and 160 to 193; and ΔV3, amino acids 303 to 324. White circles indicate that an amino acid is identical among 51 to 98% of all HIV-1 isolates. Black circles indicate that an amino acid is identical among 99 to 100% of all HIV-1 isolates. Amino acid identity was determined from a sequence alignment of HIV-1 isolates listed in the HIV sequence database at http://hiv-web.lanl.gov/content/hiv-db/mainpage.html. Apparent affinities were calculated as the antibody concentration at half-maximal binding. Apparent affinities relative to those for wild-type gp120 were calculated with the formula (apparent affinity for the wild type/apparent affinity for the mutant) × 100. The color scheme is the same as that in Fig. 3; substitutions which resulted in an apparent affinity of <50% relative to that for the wild type are colored blue, those which resulted in an apparent affinity of 50 to 200% relative to that for the wild type are colored grey, and those that resulted in an apparent affinity of >200% relative to that for the wild type are colored yellow. CD4-IgG2 was used as a surrogate for CD4 in this study. ND, not determined. Amino acid residues conserved among all HIV-1 isolates. Amino acid conservation is defined as in reference 30: single amino acid changes are allowed, as are larger substitutions, as long as the character of the side chain is maintained (e.g., Lys to Arg or Phe to Leu). Amino acid residues conserved among all primate immunodeficiency viruses. CD4 contact residues (as determined from the crystal structure of the gp120-CD4-17b complex by Kwong et al. [30]). Data were derived from reference 62.
FIG. 1.
Locations of amino acid substitutions on gp120, spatial relationships of epitopes on gp120, and variability of gp120 residues among primate immunodeficiency viruses and HIV-1. (A) Ribbon amino acid sequence diagram of gp120JR-CSF indicating the locations of the alanine substitutions in this study. Arrows indicate amino acids that were mutated to alanine. Variable-loop deletions are indicated by white letters encircled by blue spheres. Light blue dots indicate the positions of amino acid mutations in b12 neutralization escape mutants (38, 54). Red and yellow dots indicate primary and secondary b12 contact residues, respectively, based on a computational docking model of b12 and the gp120HxB2 core (44). Primary b12 contacts are gp120 residues that, based on our docking model, contact MAb b12; secondary contacts are gp120 residues within 3 to 5 Å of MAb b12. (B) Locations of mutations (shown in blue) mapped onto the structure of the gp120 core of HIV-1HxB2. The view is from the perspective of CD4. (C) Approximate locations of the faces of the gp120 core, defined by the interactions of gp120 and antibodies (30, 76, 78). The region in the CD4bs that is accessible to neutralizing ligands on primary HIV-1 isolates (i.e., MAbs CD4-IgG2 and b12), termed the neutralizing face, is shown in yellow. The region that is believed to be poorly accessible on oligomeric gp120 and that elicits nonneutralizing antibodies is shown in cyan. The location of the immunologically silent face, which encompasses the epitope recognized by broadly neutralizing MAb 2G12 (59, 62, 73), is shown in magenta. The coreceptor binding site is shown in light grey. Modeled carbohydrate chains are shown in dark grey and black. The approximate areas that are believed to be covered by the V2 and V3 loops (primarily the coreceptor binding site) are indicated. The locations of the Phe43 cavity (30), involved in CD4 binding, and the stem of the V1/V2 loop are also indicated. (D) Molecular surface of gp120 depicting the sequence variability of the amino acid residues among primate immunodeficiency viruses and HIV-1: green, residues that are conserved among all primate immunodeficiency viruses; yellow, residues that are conserved among all HIV-1 isolates but not among all primate immunodeficiency viruses; grey, residues that are variable among HIV-1 isolates. Amino acid conservation is defined as in reference 30.
Mutants containing multiple alanine substitutions were also generated by using Quickchange, except that plasmid pCMV-Tag-tpaJR-FLgp120 was used as a template. This plasmid was derived from plasmid pCMV-Tag4A (Stratagene). First, a DNA fragment encoding the tissue plasminogen activator protein (Tpa) was generated by PCR with primer tpa1 (5′-CGTTGAATTCGCCGCCACCATGGATGCAATGAAGAGAGGGCTCTGCTGTGTGCTGCTGCTGTGTGGAGCAGTCTTCGTTTCG-3′), which contains an EcoRI restriction site (underlining), and primer tpa2 (5′-GCACCTCGAGGCGCGCTCCTCTTCTGAATCGGGCATGGATTTCCTGGCTGGGCGAAACGAAGACTGCTCCACACAG-3′), which contains XhoI and BssHII restriction sites (single and double underlining, respectively). This fragment was cloned into pCMV-Tag4A by using EcoRI and XhoI to generate plasmid pCMV-Tag-tpa. The env gene from HIV-1JR-FL (28) was amplified by PCR with plasmid pSyngp140JR-FL (obtained from the NIH ARRRP and contributed by Eun-Chung Park and Brian Seed) (2, 17) as a template; in this plasmid, most wild-type gp140 codons have been replaced with codons from highly expressed human genes. For PCR, primer jr-fl5′ (5′-CGTTGCGCGCGTGGAGAAGCTGTGGGTG-3′), which contains a BssHII restriction site (underlining), and primer flXho-r (5′-GCAGAGGGAGAAGCGCCTCGAGGCTGTGGGCATTGG-3′), which contains an XhoI restriction site (underlining), were used. The PCR fragment was cloned into pCMV-Tag-tpa by using BssHII and XhoI to generate pCMV-Tag-tpaJR-FLgp120. All plasmids and mutations generated in this study were verified by DNA sequencing.
Generation of recombinant HIV-1 virions.
To produce recombinant virions, 293T cells grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with penicillin, streptomycin, l-glutamine, and fetal bovine serum (10%) were transiently transfected with wild-type or mutant pSVIIIexE7pA−JR-CSF plasmids (2 μg) along with the luciferase reporter plasmid pNL4.3.Luc.R−E− (4 μg) (obtained from the NIH ARRRP and contributed by Nathaniel Landau) (11, 18) by using FuGENE6 transfection reagent (Roche). At 24 h posttransfection, the culture supernatant was replaced with serum-free medium, and incubation was continued for another 24 h. Cell culture supernatants containing pseudovirions were subsequently harvested and stored at −80°C for neutralization assays (see below). Alternatively, recombinant virions were lysed by the addition of detergent to the harvested culture supernatants, which were then stored at −20°C until further use.
Expression of recombinant gp120.
pCMV-Tag-tpaJR-FLgp120 plasmids expressing wild-type or mutant gp120 were used to transiently transfect subconfluent 293T cells grown in serum-containing medium as described above, except that no pNL4.3Luc was used. At 2 days posttransfection, culture supernatants containing recombinant gp120 were collected and stored at −20°C.
ELISAs.
For enzyme-linked immunosorbent assays (ELISAs), microtiter plate wells (flat bottom; Costar type 3690; Corning Inc.) were coated overnight at 4°C with anti-gp120 antibody D7324 (International Enzymes, Inc.) at a concentration of 5 μg/ml (250 ng/well; diluted in phosphate-buffered saline [PBS]). Subsequent incubation steps were performed at room temperature. Coated plates were washed twice with PBS supplemented with 0.05% Tween, blocked for 1 h with PBS supplemented with 3% bovine serum albumin, and then incubated for 2 to 4 h with cell culture supernatants that had been diluted 1:3 in PBS containing 1% bovine serum albumin and 0.02% Tween (PBS-B-T). Plates were washed with PBS supplemented with 0.05% Tween (10 times) and then incubated with MAbs serially diluted in PBS-B-T (starting at a concentration of 10 μg/ml). Human IgG purified from pooled plasma obtained from healthy asymptomatic seropositive individuals (1 μg/ml; diluted in PBS-B-T) was used as a control to ensure that similar amounts of envelope protein were captured. After plates were washed as described above, peroxidase- or alkaline phosphatase-conjugated goat anti-human IgG [F(ab′)2-specific; Pierce] was added (diluted 1:1,000 in PBS-B-T), and incubation was continued for another hour. Plates were washed again and then incubated with tetramethylbenzidine substrate (Pierce) when peroxidase-conjugated secondary antibody was used or p-nitrophenyl phosphate (Sigma) when alkaline phosphatase-conjugated secondary antibody was used. The color reaction was stopped by adding 2 M sulfuric acid (when tetramethylbenzidine was used), and absorbances were measured at 450 nm. Absorbances in assays developed with p-nitrophenyl phosphate were measured at 405 nm without stopping the reaction. Apparent binding affinities were calculated as the antibody concentration at half-maximal binding; percent changes in affinity relative to that of the wild type were expressed as (apparent affinity of the wild type/apparent affinity of the mutant) × 100.
HIV-1 neutralization assays.
Recombinant virions competent for a single round of infection were generated as described above. Neutralization assays were performed essentially as described previously (82) by using an initial seeding density of between 1 × 104 and 3 × 104 target cells (U87.CD4.CCR5; obtained from the NIH ARRRP). The degree of virus neutralization by antibody was determined by measuring luciferase activity. The percent neutralization at a given antibody concentration was expressed as [(luciferase activity in the absence of antibody − luciferase activity in the presence of a given antibody concentration)/luciferase activity in the absence of antibody] × 100. To determine the degree of correlation between neutralization efficiency and the change in antibody binding affinity for each mutant relative to the wild type, a neutralization index was defined. This index was expressed as [(antibody concentration required to achieve 90% neutralization of the wild type × apparent antibody affinity for wild-type gp120)/(antibody concentration required to achieve 90% neutralization of the mutant × apparent antibody affinity for mutant gp120)]. Neutralization indices of between 0.2 and 5 were considered indicative of a reasonable correlation between the change in antibody affinity for monomeric gp120 and neutralization efficiency.
RESULTS
Effect of alanine substitutions and variable-loop deletions on CD4 and anti-CD4bs antibody binding to monomeric gp120.
A number of previous mutagenesis studies reported on the effects of amino acid substitutions on binding by anti-CD4bs antibodies (20, 36, 67, 69, 71). However, only the study by Roben et al. (57) described such an analysis applied to MAb b12. In that study, wild-type and mutant gp120s were tested for their ability to bind a saturating concentration of each antibody, the results being expressed as the ratio of antibody bound to mutant gp120 to that bound to wild-type gp120. This approach is most satisfactory for substitutions that produce large effects in binding but may be less reliable for detecting smaller effects, since a change in saturation does not necessarily indicate a change in antibody affinity.
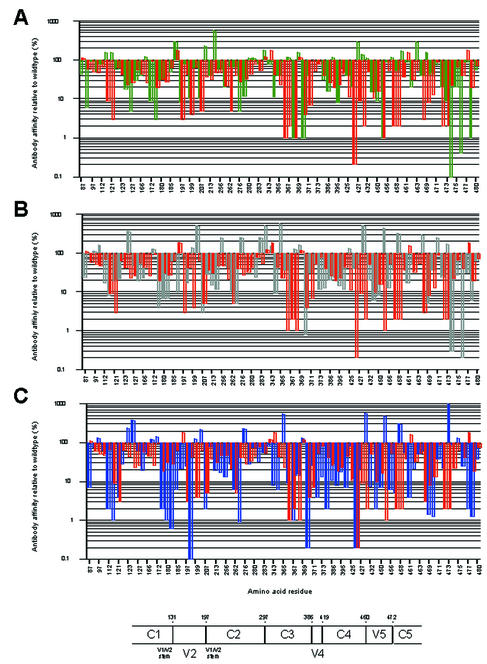
In the present study, we therefore opted for a more rigorous approach by determining the change in apparent antibody affinity for gp120 of each mutant relative to that for wild-type gp120. Alanine scanning mutagenesis was performed on monomeric gp120 to define in more detail which residues on gp120 influence or modulate b12 reactivity. In parallel, CD4-IgG2 (used as a surrogate for CD4) and two nonneutralizing anti-CD4bs MAbs (b3 and b6) (4) were assayed to compare the effects of alanine mutations on binding by neutralizing versus nonneutralizing MAbs, as well as to distinguish between amino acid substitutions that uniquely affect MAb binding and mutations that affect the binding of anti-CD4bs ligands in general. Mutagenesis was performed by using gp120 from HIV-1JR-CSF (gp120JR-CSF) as the parent. A total of 81 mutants containing single alanine substitutions and 3 variable-loop deletion mutants were generated (Fig. 1A). The residues that were mutated to alanine were selected primarily from a list of likely contact residues based on the docking model of the crystal structure of MAb b12 and the CD4-complexed gp120 core structure of HxB2 (44). As might be expected, most mutations were in or adjacent to the CD4bs (Fig. 1B), encompassing the neutralizing face as well as the nonneutralizing face of gp120 (Fig. 1C). However, a number of amino acids that were selected for mutagenesis were also located on the silent face. As shown in Fig. 1D, amino acids that were selected for mutagenesis ranged from those that are highly conserved among primate immunodeficiency viruses to residues that vary significantly among HIV-1 isolates.
To determine apparent antibody affinities, mutant monomeric gp120 from pseudovirions was captured on ELISA plate wells and probed with various concentrations of antibodies to generate a binding curve for each mutant. Apparent binding affinities were determined from the antibody concentration at half-maximal binding. The apparent antibody affinity for each mutant gp120 was then related to that for wild-type gp120 (Table 1). Changes in the relative affinity of greater than 200% were designated increases, whereas those of less than 50% were designated decreases. Intermediate values were recorded as having no effect or a limited effect on antibody binding.
Three variable-loop deletion mutants of gp120 (ΔV1, ΔV1/V2, and ΔV3) were investigated. Deletion of the V1 loop alone or together with the V2 loop had an adverse effect on the binding of CD4 and all three MAbs to gp120, whereas deletion of the V3 loop decreased the binding affinity for CD4 and MAbs b6 and b12 but not b3 (Table 1). Nineteen alanine substitutions in gp120 reduced the affinity for CD4 and all three MAbs. Three substitutions (at D180, I184, and F176) are located in the V2 loop; one (at K207) is located at the base of the stem of the V1/V2 loop; one (at I213) is located on the nonneutralizing face at the putative gp120-gp41 interface (30, 76); six (at T257, E370, I371, Y384, M426, and G472) line the so-called Phe43 cavity (30) of gp120; four (at N386, P470, R476, and W479) are in close proximity to the CD4 binding pocket (30); three (at R350, W395, and T450) are on the carbohydrate-rich silent face of gp120; and one (at I439) is close to the junction of the silent and neutralizing faces (30, 76), adjacent to the base of the stem of the V1/V2 loop. Except for R350 and W395, these residues are all conserved among primate immunodeficiency viruses or HIV-1 isolates in terms of identity or similarity of the amino acid side chain (Table 1). Four of the nineteen residues listed above (E370, I371, M426, and G472) are CD4 contact residues (Table 1) (30), and their conservation is probably required for the optimal interaction of gp120 with CD4. For the remaining residues, conservation may be associated with maintenance of the structural integrity of gp120 (30, 76). To determine whether monomeric gp120 from these mutants was globally perturbed, we also investigated the binding of MAb 2G12, which recognizes a carbohydrate-dependent conformational epitope on the silent face of gp120 (59, 62, 73). For most mutant glycoproteins, 2G12 binding was unchanged (Table 1). It would thus appear that these proteins are not globally misfolded. Interestingly, mutating residue F176 (V2 loop) to alanine caused a fourfold increase in 2G12 binding relative to that seen with wild-type gp120, indicating that mutations in the V2 loop of gp120 can have some effect on 2G12 binding to its carbohydrate epitope. Alanine substitutions at three residues (N386, W395, and W479) caused moderate to significant decreases in 2G12 relative affinity (Table 1). None of these residues is believed to be a contact residue for MAb 2G12 (59, 62), suggesting that these alanine replacements may cause significant perturbations of gp120 structure.
The decrease in CD4 binding affinity observed with the W395A mutant is somewhat striking, considering that the residue at position 395 shows significant variability among HIV-1 isolates (30). However, alignment of the sequences from HIV-1 clade B isolates shows that this amino acid is identical in 106 of the 107 isolates listed in the HIV sequence database at http://hiv-web.lanl.gov/content/hiv-db/mainpage.html. In the single clade B isolate in which this amino acid is not Trp, it is replaced by a His residue. Position 395 may thus play a role in preserving a structural conformation that is required for optimal CD4 binding in clade B isolates and that is achieved by the incorporation of aromatic amino acids. The same may hold true for R350, which is 60% conserved as arginine in clade B isolates but 80% conserved as a positively charged side chain.
The remaining 62 alanine substitutions had various effects on CD4 and antibody reactivity (Fig. 2). A noticeable difference in the effects of alanine substitutions on ligand binding was that, whereas some substitutions enhanced MAb binding, CD4 binding was always either reduced or unchanged. Many substitutions produced similar effects on MAb and CD4 binding (Table 1 and Fig. 2). For 31 mutations, the effects on b3 binding were similar to the effects on CD4 binding. In comparison, the effects on b6 and b12 binding were similar to the effect on CD4 binding for 24 and 26 mutations, respectively. However, when we focused on substitutions that decreased ligand binding, we found the greatest correspondence between MAb b12 and CD4, closely followed by MAb b6 and CD4, and the greatest discrepancy between MAb b3 and CD4. Thus, when we excluded the 19 alanine substitutions that uniformly reduced binding by CD4 and the three MAbs, we found that 18 substitutions decreased the binding of both b12 and CD4, 17 decreased the binding of both b6 and CD4, and only 11 decreased the binding of both b3 and CD4.
FIG. 2.
Apparent affinity of MAbs for alanine mutants of gp120JR-CSF relative to the wild type. Numbering is based on the sequence of HIV-1HxB2 (26). (A) b3 binding (green bars). (B) b6 binding (grey bars). (C) b12 binding (blue bars). On the x axis, only every second amino acid residue listed in Table 1 is numbered. Orange bars represent CD4 binding. A schematic of the conserved and variable regions of HIV-1 gp120 is also shown. Numbers indicate amino acid residues (HxB2 numbering). C, conserved domain. V, variable region.
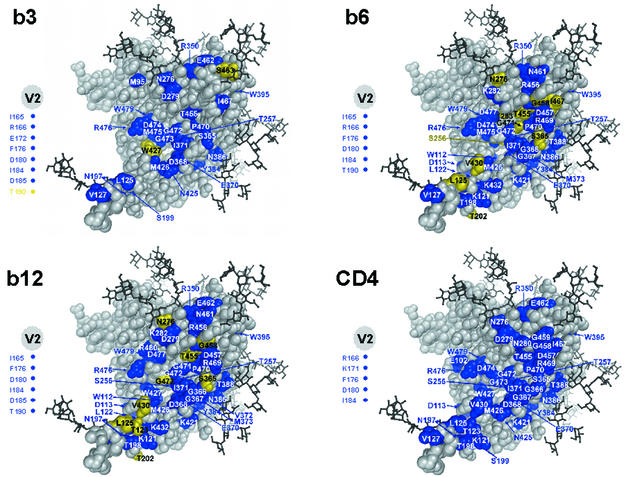
Amino acid substitutions that affected MAb binding were mapped onto the crystal structure of the gp120 core of HIV-1HxB2 to obtain a better understanding of the spatial arrangement of the amino acid residues that were mutated (Fig. 3). Although the mutagenesis studies were done with gp120JR-CSF, this approach was thought to be valid since the structure of the core seems to be highly conserved among HIV-1 isolates (29). One caveat to note is that the structure of gp120 is that of a core molecule complexed to CD4 and Fab 17b; some differences in the conformations of this molecule and the corresponding noncomplexed molecule have been proposed (30, 76, 78), but no structural data to address this issue yet exist. The epitope maps for the antibodies, particularly MAbs b6 and b12, were highly similar (Fig. 3). This result is not surprising, given that anti-CD4bs antibodies compete with each other for binding to gp120 (41). The map observed for CD4 was in good agreement with the CD4 footprint derived from the crystal structure of the CD4-gp120 complex (30, 76). Notably, the map for b12 was also in agreement with the footprint obtained from a docking model of the b12 structure and the gp120 core structure (44).
FIG. 3.
Effects of alanine substitutions on antibody binding mapped onto the structure of the gp120 core of HIV-1HxB2. The view is from the perspective of CD4. Only substitutions that affected antibody binding are colored and labeled. Alanine substitutions of residues colored yellow significantly enhanced MAb binding (>200% affinity relative to the wild type), whereas those colored blue significantly reduced MAb or CD4 binding (<50% affinity relative to the wild type). Amino acid substitutions in the V2 loop that affected antibody or CD4 binding are indicated by colored circles to the left of each structure.
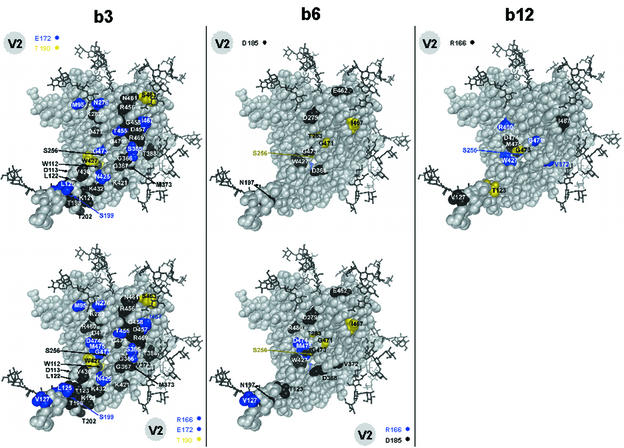
To gain a better understanding of the differences between the MAbs, only those alanine substitutions for which the effect (or lack of effect) on antibody reactivity was unique compared to the effects on the other two antibodies were mapped onto the structure of the gp120 core of HIV-1HxB2 (Fig. 4, top panels). The differential map for MAb b3 shows clusters of several alanine mutations on the neutralizing face (30, 76) (G366A, G367A, T388A, K421A, D457A, G458A, and R469A) and close to the stem of the V1/V2 loop (30, 76) (K121A and V430A) of gp120 that uniquely do not affect b3 binding. This result implies that the epitope recognized by b3 may not involve these particular regions. The differential map for MAb b6 shows that, in contrast to b3 and b12, b6 is not affected by certain substitutions (D279A and E462A) in the upper region of the neutralizing face of gp120 or by some alanine mutations which cluster around the Phe43 cavity (D368A, W427A, and G473A). The differential map for MAb b12, in contrast to the maps for the other two antibodies, shows little clustering of amino acid mutations that uniquely do not affect b12 binding. Rather, alanine substitutions to which b12 is uniquely insensitive are located at the tip of the stem of the V1/V2 loop (V127A), at the junction of the neutralizing and nonneutralizing faces (D474A and M475A), and in the upper half of the neutralizing face (I467A). Interestingly, most mutations that uniquely reduce b12 binding were located in close proximity to the Phe43 cavity (Fig. 4, top panels).
FIG. 4.
Epitope maps of the unique effects of alanine substitutions on MAb binding affinity. Color scheme and labeling are as described in the legend to Fig. 3, except that amino acid mutations that did not significantly affect antibody binding are also indicated (colored black). (Top panels) Differential maps of alanine point mutations for which the effect on MAb binding was unique compared to the effects on the binding of the other two MAbs. (Bottom panels) Differential maps of the unique effects of alanine substitutions on binding by MAbs b3 and b6 in comparison to MAb b12.
In addition, we also mapped those alanine substitutions that had a unique effect on b3 or b6 (nonneutralizing antibody) binding compared to b12 (neutralizing antibody) binding (Fig. 4, bottom panels). The differential map for b3 shows that it is not affected by many alanine substitutions, particularly those on the perimeter of the CD4bs, compared to b12. The strikingly high number of colored residues in the map for b3 suggests that b3 and b12 differ significantly in gp120 contact residues. Indeed, many of the residues that, when replaced by alanine, uniquely cause a decrease in the binding of b3 compared to b12 are located on the nonneutralizing face and the inside face of the bridging sheet which faces the nonneutralizing face. The differential map for b6 shows fewer colored residues than the map for b3, suggesting that there are few differences between b6 and b12 in their interactions with gp120.
Effect of alanine substitutions on susceptibility of pseudovirions to neutralization.
Mutants which are able to escape neutralization by MAb b12 have been generated in vitro and in vivo (38, 54) and are characterized by amino acid mutations in the V2 loop (D167N and D185N), as well as in the C3 region (P369L and P369Q) adjacent to the Phe43 cavity. To determine the extent to which alanine substitutions in the present study resulted in neutralization escape, 19 alanine mutants with various effects on b12 binding affinity for monomeric gp120 were selected to encompass the entire gp120 envelope and were used in an assay in which an env-defective HIV-1 provirus encoding the firefly luciferase gene (pNL4.3Luc) was complemented for a single round of infection by a plasmid encoding wild-type or mutant envelope glycoproteins (Table 2). A neutralization index was defined to determine the degree of correlation between antibody affinity for monomeric gp120 and neutralization efficiency.
TABLE 2.
Neutralization of wild-type and mutant pseudovirions of HIV-1JR-CSF by MAb b12
| Pseudovirus | IC90 (μg/ml)a | Apparent affinity relative to that for wild-type gp120JR-CSF (%) | Neutralization indexb |
|---|---|---|---|
| Wild type | 14 | 100 | 1 |
| K97A | 12 | 134 | 0.87 |
| D113A | 6 | 1 | 233 |
| T123A | 4 | 233 | 1.50 |
| L125A | 5 | 383 | 0.73 |
| V127A | 4 | 62 | 5.65 |
| R166A | 8 | 52 | 3.37 |
| D180A | 10 | 1 | 140 |
| N197A | 1 | 25 | 56 |
| F210A | 4 | 77 | 4.55 |
| R252A | 43 | 17 | 1.92 |
| S256A | 16 | 13 | 6.73 |
| N276A | 46 | 225 | 0.14 |
| T283A | 13 | 55 | 1.96 |
| Q337A | 9 | 83 | 1.87 |
| S365A | 64 | 534 | 0.04 |
| P369A | 8 | 129 | 1.36 |
| G459A | 9 | 54 | 2.88 |
| G471A | 11 | 37 | 3.44 |
| D474A | 11 | 91 | 1.40 |
IC90, antibody concentration yielding 90% neutralization efficiency.
See the text for details.
For 12 mutants (K97A, T123A, L125A, R166A, F210A, R252A, T283A, Q337A, P369A, G459A, G471A, and D474A), there was a reasonable correlation between the neutralization sensitivity of pseudovirions expressing mutant gp120s and the antibody affinity for monomeric gp120 of the corresponding mutants. For the remaining seven alanine mutants, discrepancies were observed between the change in b12 binding affinity for monomeric gp120 and neutralization efficiency (Table 2 and Fig. 5). Strikingly, for five mutants (D113A, V127A, D180A, N197A, and S256A), neutralization efficiency was maintained or increased despite a decrease in antibody affinity for monomeric gp120. For instance, whereas the binding affinity of b12 for monomeric gp120 from the D113A virus was reduced 100-fold compared to the binding affinity for wild-type gp120, the D113A virus was neutralized by b12 as efficiently as the wild-type virus. In contrast, the neutralization indices for the remaining two mutants, N276A and S365A, were low (0.14 and 0.04, respectively), because of decreased neutralization efficiency despite increased binding affinity for monomeric gp120. These results suggest that antibody affinity changes observed with amino acid substitutions for monomeric gp120 are not always maintained in the context of functional oligomeric gp120 present on the surface of the virus.
FIG. 5.
Alanine mutants that were selected for neutralization assays. Labeled amino acids colored grey indicate mutant pseudovirions for which there was a reasonable correlation between b12 binding to monomeric gp120 and neutralization efficiency. Residues labeled and colored yellow indicate pseudovirions which were neutralized equally as well as or better than wild-type virus despite a decrease in the b12 affinity for monomeric gp120 of the respective mutant. Residues labeled and colored red indicate pseudovirions which were not neutralized as well as the wild-type virus despite an increase in the b12 affinity for monomeric gp120 of the respective mutant.
To determine whether the observed discrepancies between binding to monomeric gp120 and neutralization were applicable only to b12, we also tested MAbs CD4-IgG2, 2G12, 15e, and 17b with six of the seven mutants described above in neutralization assays. Mutant V127A was excluded because the observed neutralization index was only slightly higher than the cutoff value and thus was considered only marginally discrepant. MAb 15e recognizes an epitope overlapping the CD4bs on gp120 (58), whereas MAb 17b recognizes an epitope which overlaps the coreceptor binding site on gp120 and is better exposed upon CD4 binding (70). Both MAbs, which neutralize HIV-1 primary isolates poorly (14, 37, 72), were not able to achieve 90% neutralization of any of the mutant viruses or wild-type virus at concentrations of up to 100 μg/ml, although both MAbs bound monomeric gp120 from all mutants with affinities comparable to that for wild-type gp120 (data not shown). These results suggest that the introduced mutations do not dramatically increase the sensitivity of JR-CSF to neutralization by these MAbs. Further, all six mutants showed sensitivity to neutralization by MAb 2G12 similar to that of the wild-type virus (Table 3). Therefore, it seems likely that the changes in neutralization efficiency observed with mutant pseudovirions are restricted to the b12 epitope. Interestingly, for mutant S365A, a neutralization index of <0.2 with MAb 2G12 was observed; 2G12 had a higher affinity for monomeric gp120 from this mutant, but the corresponding pseudovirions were not neutralized better than the wild-type virus. In view of the location of the 2G12 epitope on gp120 (59, 62, 73) and considering that residue S365 is located in the CD4bs of gp120, it would appear that mutating this residue to alanine causes a conformational change in monomeric gp120 which leads to a better presentation of one or more glycans on the silent face of gp120, to which 2G12 binds. However, this conformational change does not appear to take effect on the viral spike, since mutant viruses are not neutralized more efficiently than wild-type viruses.
TABLE 3.
Neutralization of wild-type and selected mutant pseudovirions of HIV-1JR-CSF by MAbs CD4-IgG2 and 2G12
| Pseudovirus | IC90 (μg/ml) for MAba:
|
Apparent affinity relative to that for wild-type gp120JR-CSF (%) for MAb:
|
Neutralization index for MAb:
|
|||
|---|---|---|---|---|---|---|
| CD4-IgG2 | 2G12 | CD4-IgG2 | 2G12 | CD4-IgG2 | 2G12 | |
| Wild type | 13 | 6 | 100 | 100 | 1 | 1 |
| D113A | 5 | 3 | 9 | 169 | 28.9 | 1.18 |
| D180A | 1 | 14 | 31 | 108 | 41.9 | 0.40 |
| N197A | 1 | 6 | 3 | 318 | 433 | 0.31 |
| S256A | 20 | 6 | 47 | 108 | 1.38 | 0.93 |
| S365A | 6 | 9 | 23 | 1,800 | 9.42 | 0.04 |
| N276A | 30 | 3 | 27 | 169 | 1.60 | 1.18 |
IC90, antibody concentration yielding 90% neutralization efficiency.
For MAb CD4-IgG2, neutralization efficiency for two (S256A and N276A) of the six mutant pseudovirions that were tested was decreased with respect to that for the wild-type virus (Table 3). This decrease correlates well with the observed decrease in CD4-IgG2 affinity for monomeric gp120 (Table 3). For the remaining four mutants, there was a discrepancy between neutralization efficiency and antibody affinity for gp120. For mutants D113A, D180A, and N197A, we observed the same discrepancy as that seen with b12: an increase in neutralization efficiency despite a decrease in binding affinity for the monomer. Interestingly, for mutant S365A, the effects on CD4-IgG2 and b12 binding and neutralization diverged. Monomeric gp120 of mutant S365A showed enhanced affinity for b12 and reduced affinity for CD4-IgG2. However, whereas the corresponding virus was neutralized by CD4-IgG2 equally as well as the wild-type virus, a decrease in neutralization efficacy was observed for b12 with pseudovirions from mutant S365A. Thus, although mutations in gp120 can affect the binding of b12 and CD4-IgG2 to monomeric gp120 differently, these differences are apparently not necessarily maintained when gp120 is oligomerized.
Generation and expression of recombinant gp120 containing multiple alanine substitutions.
Although MAb b12 and nonneutralizing anti-CD4bs antibodies bind monomeric gp120 with similar affinities, we hypothesized that it might be possible to mutate monomeric gp120 so that it preferentially binds b12 but not nonneutralizing MAbs. Based on our structural analysis (Fig. 4), it was apparent that a region encompassing amino acids G473 to R476, which partially line the Phe43 cavity, uniquely affected MAb b12 binding compared to MAb b3 and MAb b6 binding (Fig. 4, right panel). We therefore decided to generate a small panel of recombinant gp120s with multiple alanine substitutions at these amino acid positions to determine whether the unique differences in the effects observed with the single alanine mutations could be retained. For this experiment, we constructed a plasmid encoding the gp120 segment of a codon-optimized env gene of primary isolate JR-FL, which is 94% identical in amino acid sequence to JR-CSF. A tissue plasminogen activator leader sequence was placed upstream of the env gene to ensure the secretion of gp120 into the culture medium. Four mutants (GDMR, DMR, DR, and GM) were generated and tested with a panel of anti-CD4bs MAbs (Table 4). The results showed that b12 binding affinity for the double (DR and GM) and triple (DMR) mutants, was similar to that for wild-type gp120, whereas b12 binding affinity was increased for the quadruple (GDMR) mutant. In contrast, the binding affinities of MAbs b3, b6, and F105 were severely reduced for all four mutants. CD4 binding was also severely diminished for the GDMR, DMR, and GM mutants, but not for the DR mutant (twofold reduction in affinity relative to that for the wild type). Two other anti-CD4bs antibodies, MAbs F91 and 15e, were not as susceptible to alanine substitutions in this region as the other MAbs; none of the mutations produced a decrease of more than 50% with either of the two antibodies. The binding affinity of MAb F91 was affected most by mutant DMR, whereas the binding affinity of MAb 15e was affected most by mutants GDMR and GM. These results suggest that these MAbs may bind to gp120 in a manner different from that of other anti-CD4bs antibodies. Indeed, the binding of MAb F91 has been shown to be uniquely enhanced in the presence of anti-V2 and anti-V3 loop antibodies (41), whereas both MAbs have been shown to enhance the binding of several anti-V2 and anti-V3 loop MAbs (41). The other anti-CD4bs antibodies tested here do not generally display similar effects. The binding of MAb b12, for example, is decreased in the presence of a number of anti-V2 loop antibodies (41).
TABLE 4.
Binding affinities of a panel of anti-CD4bs MAbs for recombinant gp120JR-FL containing multiple alanine substitutions
| Mutanta | Apparent affinity relative to that for wild-type gp120JR-FL (%) for:
|
||||||
|---|---|---|---|---|---|---|---|
| b12 | b6 | b3 | CD4 | F91 | 15e | F105 | |
| GDMR | 250 | 0.1 | 0.1 | 0.1 | 82 | 63 | 0.1 |
| DMR | 167 | 0.1 | 0.1 | 0.1 | 64 | 112 | 0.1 |
| DR | 167 | 0.1 | 0.1 | 43 | 90 | 112 | 0.1 |
| GM | 120 | 9 | 0.1 | 0.1 | 75 | 68 | 0.1 |
Mutants are denoted by the amino acids that were changed to alanine: G, G473; D, D474; M, M475; R, R476.
DISCUSSION
The CD4bs on gp120 is a particularly attractive target for HIV-1 vaccine design because CD4 is the primary receptor on target cells for virtually all naturally occurring viruses studied to date (12, 23). This fact suggests a common structural framework for the binding sites of all HIV-1 isolates. This notion is also supported by the limited sequence variability among amino acids which make up the CD4 binding pocket (27, 30, 45) as well as by the high degree of similarity between the crystallized gp120 core structures of a TCLA strain and a primary isolate (29). Of the three broadly neutralizing anti-gp120 MAbs that have been described to date, namely, b12, 2G12, and Fab X5, only the first binds to an epitope directly overlapping the CD4bs (4, 8, 10, 57, 59, 62, 73; A. F. Labrijn et al., unpublished results).
The aim of this study was to systematically define which residues on gp120 affect binding by broadly neutralizing MAb b12, CD4, and two nonneutralizing anti-CD4bs antibodies. Selected residues were changed to alanine because alanine generally does not significantly alter the main-chain conformation or impose extreme electrostatic or steric effects and so permits the identification of amino acid side chains which may be important for ligand binding. To determine antibody affinity changes, antibody binding curves were generated from ELISA data, and the apparent antibody affinity for each gp120 mutant was determined and related to that for wild-type gp120.
The effects of alanine substitutions on CD4 binding were determined by using CD4-IgG2, because it could be used with the same detection system as that used for the other antibodies. Interestingly, no increase in affinity for CD4 was observed with any of the alanine mutants tested in the present study (Table 1 and Fig. 2 and 3), consistent with a drive toward the selection during viral evolution of a particular ensemble of residues for optimal CD4 binding. In fact, of the 28 amino acid substitutions that diminished CD4 binding to less than 20% of wild-type levels (Table 1), all but one (I467) are conserved (as defined by amino acid identity or conservation of the amino acid type) among primate immunodeficiency viruses or HIV-1 isolates and/or are CD4 contact residues. The decrease in CD4 reactivity may therefore be a reflection of the loss of certain functional or structural features required to maintain the integrity of the CD4bs. Twenty-nine mutations caused a moderate decrease (20 to 50%) in CD4 binding affinity. Of the 29 affected residues, 21 are conserved among primate immunodeficiency viruses or HIV-1 isolates and/or are CD4 contact residues. The other eight residues (K171, F210, R252, R273, N276, R350, W395, and E462) show moderate to significant variability among HIV-1 isolates; the affinity changes observed with CD4 when these amino acids were mutated to alanine may therefore relate specifically to the HIV-1 isolate, i.e. JR-CSF, used in this study.
The results obtained here are largely in agreement with those obtained previously by Olshevsky et al. (45), who tested a panel of gp120 mutants for binding to CD4+ target cells. However, direct comparison between that study and the study reported here is difficult due to the different assay formats used; in the study of Olshevsky et al., densitometric quantitation of autoradiograms of immunoprecipitated gp120 mutants after incubation of the mutants with CD4+ target cells was performed. Minor discrepancies between this study and the previous study may also result from differences in amino acid substitutions; in the study of Olshevsky et al., selected residues were replaced by many different amino acids, whereas in this study, all residues were replaced by alanine.
Many amino acid changes were found to affect b3, b6, and b12 binding similarly (Fig. 2 and 3), indicating a high degree of overlap in gp120 determinants recognized by these anti-CD4bs antibodies. This finding is not surprising, considering that anti-CD4bs antibodies compete with each other for binding (41). Noticeably, increases in gp120 binding affinity were observed with a number of alanine substitutions, in contrast to what was seen for CD4. An increase in affinity may indicate that the amino acid in the wild type is sterically hindering antibody binding. Typically, alanine substitutions result in a smaller side chain at a given position, a change which may decrease steric interference and increase antibody affinity. However, most mutations had an adverse effect on binding ability (Fig. 2 and 3). Deletion of the V1 and/or V2 loops diminished binding by all three MAbs, as well as CD4. Deletion of the V3 loop also diminished binding by CD4 and by MAbs b6 and b12. These findings support the notion that the variable loops are able to affect the binding of anti-CD4bs antibodies (79). Alanine substitutions in the V2 loop and the C-terminal strand of the stem of the V1/V2 loop generally decreased the binding affinity of b12 more severely than those of b3 and b6. Previous studies showed that b12 is sensitive to changes in the V1/V2 stem-loop structure (6, 40), to V1/V2 deletion (38), and to mutations in the V2 loop (38). The results obtained here are thus consistent with previous observations, although it was noted that MAb b12 does not appear to be uniquely sensitive to V1/V2 deletion. These results are further supportive of the assumption (76) that the V2 loop in particular is in close proximity to the Phe43 cavity on the gp120 core. The mutagenesis results are also consistent with those from previous studies, since many of the substitutions that adversely affected antibody binding (e.g., those at residues D113, S256, T257, N262, E370, D368, K421, and D477) diminish binding by other anti-CD4bs MAbs (20, 36, 67, 69, 71).
Unique differences between antibodies were mapped onto the gp120 core structure to obtain better insight into how neutralizing antibody b12 differs from the other, nonneutralizing anti-CD4bs antibodies (Fig. 4). The large cluster of residues on the neutralizing face of gp120, which uniquely do not affect b3 binding, suggests that the nonneutralizing face and the inner domain (76) of gp120 and, to a lesser extent, the neutralizing face form a major contact region for this antibody. This notion is also supported by the insensitivity of MAb b3 to removal of the V3 loop. For MAb b6, some substitutions that uniquely affected antibody binding were located close to or facing the nonneutralizing face. This finding implies that b6 may interact with an epitope extending across the nonneutralizing and neutralizing faces, but at an angle inclined toward the nonneutralizing face. Based on its unique differences from MAbs b3 and b12, MAb b6 appears to contact very few residues which line the Phe43 cavity, suggesting that this region is less involved in the b6 epitope. For MAb b12, no spatial clustering was observed for residues which uniquely do not affect binding (Fig. 4). Most mutations that uniquely affect b12 binding are located on the neutralizing face. This observation and a consideration of the epitope map obtained for b12 suggest that the epitope recognized by b12 is located primarily in this region. This notion supports our computational docking model of the gp120 core structure and the MAb b12 structure (44), in which b12 binds to an epitope extending from the stem of the V1/V2 loop across the neutralizing face of gp120 and has little contact with the nonneutralizing face. Figure 4 (top panels) also shows that a number of mutations around the Phe43 cavity on gp120 uniquely diminished b12 binding, supporting recent results that residues comprising the antigen binding region, particularly those in the extended finger-like loop of the third complementarity-determining region (CDR) of the heavy chain of Fab b12, make crucial contacts with the residues close to the CD4 binding pocket on the gp120 surface (44; M. B. Zwick, P. W. H. I. Parren, E. Ollmann Saphire, M. Wang, J. K. Scott, P. E. Dawson, I. A. Wilson, and D. R. Burton, submitted for publication).
We sought to derive a model to explain the differences between b12 and nonneutralizing antibodies, based on the data and the epitope maps obtained here. Figure 6 depicts how two hypothetical nonneutralizing antibodies may be excluded from interacting with trimeric gp120, in contrast to b12, which binds effectively. As shown in Fig. 6C and D, nonneutralizing antibodies binding to gp120 molecules in a functional trimer may be hindered by the close proximity of a neighboring gp120 molecule. In contrast, b12, in this view, is not hindered by adjacent gp120 protomers because the angle of interaction permits binding to both monomeric gp120 and oligomeric gp120 (Fig. 6B).
FIG. 6.
Antibody binding in the context of the functional envelope trimer. (A) Trimeric gp120 model depicted as proposed by Kwong et al. (31). Here, gp120 is depicted as viewed from the virus. (B) Model of docking of MAb b12 (yellow) to gp120. (C and D) Models of how two hypothetical nonneutralizing anti-CD4bs MAbs (pink and green) may interact with gp120. Note that b12 and the nonneutralizing anti-CD4bs MAbs are able to interact with monomeric gp120 but that only b12 binds at an orientation that also allows an interaction with gp120 in the context of the functional envelope trimer. For clarity, only antibody Fab fragments are shown.
The neutralization sensitivity of pseudovirions from a selection of alanine mutants was determined to investigate how well changes in b12 binding affinity for monomeric gp120 correlated with neutralizing ability. For this experiment, a neutralization index was defined: mutants with high indices are neutralized by MAb b12 better than or equally as well as wild-type virus, despite a decrease in b12 binding affinity for monomeric gp120, whereas mutants with very low indices are neutralized worse than wild-type virus, despite an increase in b12 binding affinity for the monomer. Although there was a reasonable correlation for most mutants, e.g., an increase in b12 binding affinity and an increase in neutralization efficacy, discrepancies were observed for 7 of the 19 mutants that were selected (Table 2). This result implies that the effects of certain substitutions on antibody binding to monomeric gp120 are nullified or reversed in the context of a functional oligomeric gp120 complex on the viral surface, because the ability of an antibody to bind to functional oligomeric gp120 is generally believed to correlate with neutralization efficacy (14, 46, 51, 60). Interestingly, mutants for which there was a discrepancy between binding to the monomer and neutralization were located on opposite sides of the core (Fig. 5). Three of the five mutants (D113A, N197A, and V127A) for which b12 binding affinity for monomeric gp120 was reduced, but which were still neutralized efficiently by the antibody (neutralization indices of 233, 56, and 5.65, respectively; Table 2), were located in close proximity to or on the stem of the V1/V2 loop on the inside face of the bridging sheet which faces the nonneutralizing face. Considering that the location of the V1/V2 loop may affect b12 binding, we speculate that the substitutions described for the stem region lead to a repositioning of the V1/V2 loop, the effect of which is markedly different for monomeric gp120 and functional oligomeric gp120; i.e., the substitutions lead to increased obstruction of the CD4bs for monomeric gp120 but have little effect for oligomeric gp120. The observations are reminiscent of results obtained recently by Kolchinsky et al. (24, 25). In one of their studies, mutant pseudovirions of primary isolate ADA with an N→K or Q substitution at position 197 became highly sensitive to neutralization by various anti-gp120 MAbs (25). These mutations, which eliminate an N-linked glycan at position 197, are believed to cause movement of the V2 loop, as inferred from the ability of the viruses to infect target cells independent of CD4 (24, 25). Surprisingly, the N197A mutant generated here was not sensitive to neutralization by MAbs 15e and 17b (data not shown), which neutralize ADA viruses containing the N197K mutation well (25). The exceptional sensitivity of the ADA mutant pseudovirions to antibody neutralization may therefore relate specifically to ADA and may not apply to other HIV isolates to the same extent. In fact, preliminary results indicate that the N197A mutant of JR-CSF is not able to infect target cells which do not express CD4.
The other two mutants, D180A and S256A, for which a similar discrepancy was observed between binding to the monomer and virus neutralization were located in the V2 loop and lined the Phe43 cavity, respectively. It is likely that changing residue D180 to alanine also influences the position of the V2 loop. The reason for the observed discrepancy with mutant S256A is not readily apparent. Residue S256 lies recessed in the Phe43 cavity of the CD4-complexed conformation of gp120, at the interface between the inner and the outer domains (30, 76), and, therefore, is not likely to contact b12 directly. Replacing this residue with alanine may affect the spatial orientation of the inner and outer domains on monomeric gp120, whereas on functional envelope spikes this effect may be counteracted by oligomerization, although this notion is speculative.
The effect on binding to monomeric gp120 and neutralization observed for mutants N276A and S365A was opposite that observed for the previous five mutants; i.e., although the affinity of binding to monomeric gp120 was increased, neutralization efficiency was lost (neutralization indices were 0.44 and 0.04, respectively). Residue N276 is an N-linked glycosylation site that is part of the D loop on gp120 (30), which is believed to provide important residues for b12 binding (81). Removal of the glycan may facilitate the interaction of b12 with monomeric gp120, whereas on the native trimer, the absence of the glycan may adversely affect the conformation of the D loop (74, 75) and lead to the observed decrease in neutralization efficiency. Residue S365 is part of a ridge formed by residues 364 to 368, which make direct contact with CD4 (30) and, based on our b12-gp120 docking model, may fit into a cleft formed by heavy-chain CDR2 and CDR3 of b12 (44). The S365A mutation, like the N276A mutation, may cause a conformational change on monomeric gp120 that favors b12 binding to the monomer but has a negative effect on b12 binding to functional oligomeric gp120.
A feature of b12 neutralization escape mutants selected in vivo is a mutation at position 369 that changes Pro to Gln or Leu (38, 54). When this residue was mutated to alanine (P369A), we observed no change in the binding affinity for monomeric gp120 and no significant change in neutralization sensitivity (Table 2). These results imply that this residue does not normally form part of the b12 epitope. Rather, it is more likely that mutating this amino acid to a larger residue causes steric impairment of the interaction between b12 and gp120, either directly or by altering peptide backbone conformation.
One purpose of this study was to determine by alanine scanning mutagenesis which amino acids on gp120 modulate b12 binding. We expected to derive a clearer picture of how b12 may differ from other anti-CD4bs antibodies in its ability to bind to oligomeric spikes on virion surfaces and thus efficiently neutralize primary HIV-1 isolates. At the same time, we reasoned that, although monomeric gp120 binds b12 and nonneutralizing anti-CD4bs antibodies equivalently, it might be possible to mutate monomeric gp120 so that b12 binding would be maintained at the expense of nonneutralizing MAb binding. Therefore, based on the mutagenesis results, four recombinant gp120 molecules containing multiple alanine mutations between amino acid residues G473 and R476 were generated and tested with a panel of five nonneutralizing anti-CD4bs MAbs, b12, and CD4 (Table 4). The quadruple mutant GDMR was found to increase b12 binding but to completely abolish binding by three (b3, b6, and F105) of the five nonneutralizing anti-CD4bs MAbs tested. Of the two remaining MAbs, the binding affinity of one MAb, 15e, was decreased for two of the four mutants, whereas for the other MAb, F91, a slight decrease in binding affinity was observed.
These results thus substantiate the aforementioned postulate that gp120 can be engineered in such a way as to make it less prone to recognition by nonneutralizing antibodies. For vaccine design, it will now be interesting to test the performance of these and other modified gp120 molecules in the induction of neutralizing antibodies with b12-like properties.
Acknowledgments
We thank J. Sodroski (Dana-Farber Cancer Institute, Harvard Medical School) for providing plasmid pSVIIIexE7pA−HxB2; J. Robinson (Tulane University) for generously providing MAbs 17b, 15e, and F91; W. Olson and P. Maddon (Progenics Pharmaceuticals Inc.) for providing CD4-IgG2; H. Katinger (Institute of Applied Microbiology, University of Agriculture, Vienna, Austria) for providing MAb 2G12; and A. Hessell, E. Schultz, and D. Slifka for technical assistance.
This study was supported by NIH grants GM46192 (to I.A.W.), AI40377 (to P.W.H.I.P.), and AI33292 (to D.R.B.) and through the Neutralizing Antibody Consortium of the International AIDS Vaccine Initiative and by The Universitywide AIDS Research Program (support given to E.O.S.).
REFERENCES
- 1.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M.-C. Gauduin, R. A. Koup, J. S. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 11:533-539. [DOI] [PubMed] [Google Scholar]
- 2.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 4.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, E. Norrby, and D. R. Burton. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. Wyatt, E. Desjardins, P. D. Kwong, W. Hendrickson, J. P. Moore, and J. Sodroski. 1998. Analysis of the interaction of antibodies with a conserved, enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R., C. F. Barbas III, M. A. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 10.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Dalgleish, A. G., P. C. L. Beverly, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, M. P., D. Livnat, J. A. Bradac, S. Bridges, The AIDS Clinical Trials Group Antibody Selection Working Group, and Collaborating Investigators. 1997. Evaluation of monoclonal antibodies to HIV-1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 14.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouts, T. R., A. Trkola, M. S. Fung, and J. P. Moore. 1998. Interactions of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate: relationship to neutralization. AIDS Res. Hum. Retrovir. 14:591-597. [DOI] [PubMed] [Google Scholar]
- 16.Gauduin, M. C., P. W. H. I. Parren, R. Weir, C. F. Barbas III, D. R. Burton, and R. A. Koup. 1997. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 3:1389-1393. [DOI] [PubMed] [Google Scholar]
- 17.Haas, J., E.-C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 18.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, B. A. Smith, T. W. Baba, V. Liska, F. Ferrantelli, D. C. Montefiori, H. M. McClure, D. C. Anderson, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, T. C. Chou, J. Andersen, and R. M. Ruprecht. 2001. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J. Virol. 75:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kessler, J. A., P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1 b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-581. [DOI] [PubMed] [Google Scholar]
- 23.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Glukman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 24.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single n-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolchinsky, P., E. Kiprilov, and J. Sodroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber, B., B. T. Foley, C. Kuiken, S. K. Pillai, and J. G. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG, p. III-102-III-111. In B. Korber, C. L. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N. Mex.
- 27.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 28.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 29.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 30.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic SHIV-89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascola, J. R., S. Schlesinger Frankel, and K. Broliden. 2000. HIV-1 entry at the mucosal surface: role of antibodies in protection. AIDS 14(Suppl. 3):S167-S174. [PubMed] [Google Scholar]
- 34.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 35.McInerney, T. L., L. McLain, S. J. Armstrong, and N. J. Dimmock. 1997. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a postfusion event. Virology 233:313-326. [DOI] [PubMed] [Google Scholar]
- 36.McKeating, J. A., M. Thali, C. Furman, S. Karwowska, M. K. Gorny, J. Cordell, S. Zolla-Pazner, J. Sodroski, and R. A. Weiss. 1992. Amino acid residues of the human immunodeficiency virus type 1 gp120 critical for the binding of rat and human neutralizing antibodies that block the gp120-sCD4 interaction. Virology 190:134-142. [DOI] [PubMed] [Google Scholar]
- 37.McKeating, J. A., Y. J. Zhang, C. Arnold, R. Frederiksson, E. M. Fenyö, and P. Balfe. 1996. Chimeric viruses expressing primary envelope glycoproteins of human immunodeficiency virus type 1 show increased sensitivity to neutralization by human sera. Virology 220:450-460. [DOI] [PubMed] [Google Scholar]
- 38.Mo, H., L. Stamatatos, J. E. Ip, C. F. Barbas III, P. W. H. I. Parren, D. R. Burton, J. P. Moore, and D. D. Ho. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody immunoglobulin G1 b12. J. Virol. 71:6869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nabel, G. J., and N. J. Sullivan. 2000. Antibodies and resistance to natural HIV infection. N. Engl. J. Med. 343:1263-1265. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura, Y., T. Igarashi, N. Haigwood, R. Sadjadpour, R. J. Plishka, A. Buckler-White, R. Shibata, and M. A. Martin. 2002. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titer neutralizing antibodies. J. Virol. 76:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ollmann Saphire, E., P. W. H. I. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 45.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parren, P. W. H. I., and D. R. Burton. 2001. The anti-viral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W. H. I., H. J. Ditzel, R. J. Gulizia, J. M. Binley, C. F. Barbas, I. I. I., D. R. Burton, and D. E. Mosier. 1995. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 9:F1-F6. [DOI] [PubMed] [Google Scholar]
- 48.Parren, P. W. H. I., M. C. Gauduin, R. A. Koup, P. Poignard, Q. J. Sattentau, P. Fisicaro, and D. R. Burton. 1997. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol. Lett. 58:125-132. [DOI] [PubMed] [Google Scholar]
- 49.Parren, P. W. H. I., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parren, P. W. H. I., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 52.Poignard, P., P. J. Klasse, and Q. J. Sattentau. 1996. Antibody neutralization of HIV-1. Immunol. Today 17:239-246. [DOI] [PubMed] [Google Scholar]
- 53.Poignard, P., E. Ollmann Saphire, P. W. H. I. Parren, and D. R. Burton. 2001. GP120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 54.Poignard, P., S. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. H. I. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10:431-438. [DOI] [PubMed] [Google Scholar]
- 55.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 6:7-14. [PubMed] [Google Scholar]
- 56.Posner, M. R., H. S. Elboim, T. Cannon, L. Cavacini, and T. Hideshima. 1992. Functional activity of an HIV-1 neutralizing IgG human monoclonal antibody: ADCC and complement-mediated lysis. AIDS Res. Hum. Retrovir. 8:553-558. [DOI] [PubMed] [Google Scholar]
- 57.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson, J. E., D. Holton, S. Pacheco-Morell, J. Liu, and H. McMurdo. 1990. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS Res. Hum. Retrovir. 6:567-579. [DOI] [PubMed] [Google Scholar]
- 59.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattentau, Q. J., M. Moulard, B. Brivet, F. Botto, J. C. Cuillemot, I. Mondor, P. Poignard, and S. Ugolini. 1999. Antibody neutralization of HIV-1 and the potential for vaccine design. Immunol. Lett. 66:143-149. [DOI] [PubMed] [Google Scholar]
- 62.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schønning, K., O. Lund, O. S. Lund, and J. E. S. Hanssen. 1999. Stoichiometry of monoclonal antibody neutralization of T-cell-line-adapted human immunodeficiency virus type 1. J. Virol. 73:8364-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 65.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatatos, L., S. Zolla-Pazner, M. K. Gorny, and C. Cheng-Mayer. 1997. Binding of antibodies to virion-associated gp120 molecules of primary-like human immunodeficiency virus type 1 (HIV-1) isolates: effect on HIV-1 infection of macrophages and peripheral blood mononuclear cells. Virology 229:360-369. [DOI] [PubMed] [Google Scholar]
- 67.Sugiura, W., C. C. Broder, B. Moss, and P. L. Earl. 1999. Characterization of conformation-dependent anti-gp120 murine monoclonal antibodies produced by immunization with monomeric and oligomeric human immunodeficiency virus type 1 envelope proteins. Virology 254:257-267. [DOI] [PubMed] [Google Scholar]
- 68.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4 binding region of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thali, M., U. Olshevshy, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 73.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watkins, B. A., S. Buge, K. Aldrich, A. E. Davis, J. Robinson, M. S. Reitz, Jr., and M. Robert-Guroff. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 70:8431-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watkins, B. A., M. S. Reitz, Jr., C. A. Wilson, K. Aldrich, A. E. Davis, and M. Robert-Guroff. 1993. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: evidence for multiple pathways. J. Virol. 67:7493-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 77.Wyatt, R., J. Moore, M. Accola, E. Desjardins, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 79.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell-line-adapted isolates of human immunodefieciency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zwick, M. B., L. L. C. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas, P. W. H. I. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]