Abstract
The human immunodeficiency virus type 1 (HIV-1) gp120 exterior glycoprotein is conformationally flexible. Upon binding the host cell receptor, CD4, gp120 assumes a conformation that is able to bind the chemokine receptors CCR5 or CXCR4, which act as coreceptors for the virus. CD4-binding-site (CD4BS) antibodies are neutralizing antibodies elicited during natural infection that are directed against gp120 epitopes that overlap the binding site for CD4. Recent studies (S. H. Xiang et al., J. Virol. 76:9888-9899, 2002) suggest that CD4BS antibodies recognize conformations of gp120 distinct from the CD4-bound conformation. This predicts that the binding of CD4BS antibodies will inhibit chemokine receptor binding. Here, we show that Fab fragments and complete immunoglobulin molecules of CD4BS antibodies inhibit CD4-independent gp120 binding to CCR5 and cell-cell fusion mediated by CD4-independent HIV-1 envelope glycoproteins. These results are consistent with a model in which the binding of CD4BS antibodies limits the ability of gp120 to assume a conformation required for coreceptor binding.
Human immunodeficiency virus type 1 (HIV-1) is the etiological agent of AIDS in humans (1, 9). There are 40 million people in the world currently infected by HIV-1. Curtailing the continued spread of HIV-1 infection will probably require an effective vaccine, which should ideally elicit both virus-neutralizing antibodies and cellular immune responses.
The HIV-1 envelope glycoproteins serve as the only viral target for neutralizing antibodies (32). The trimeric envelope glycoprotein complex contains three gp120 exterior envelope glycoproteins and three gp41 transmembrane glycoproteins. Virus binding to target cells is mediated by the interaction between the gp120 glycoprotein and cellular receptors, CD4 and coreceptor molecules (CCR5 or CXCR4) that are members of the chemokine receptor family (29). The gp120 glycoprotein consists of a core and surface-exposed variable loops (V1 to V5). Structural studies of gp120-CD4 complexes have shown that the gp120 core molecule consists of an inner domain, an outer domain, and a bridging sheet (14). All three gp120 elements contact the most amino terminal of the four immunoglobulin domains of CD4.
Binding of the HIV-1 envelope glycoproteins to CD4 triggers conformational changes that allow the binding of gp120 to the chemokine coreceptor, ultimately leading to membrane fusion and virus entry (20, 27). One of the important conformational changes induced by CD4 is the movement of the gp120 V1/V2 variable loops, which are thought to mask the chemokine receptor-binding site on gp120 (31). It has been shown that gp120 proteins with deletions or alterations in the V1 and V2 loops often exhibit the ability to bind coreceptor in the absence of the receptor CD4 (12). In some cases, viruses with such altered envelopes can infect CD4-negative cells that express the appropriate coreceptor (12, 13).
CD4 binding also induces conformational changes in the gp120 core, as revealed by isothermal titration microcalorimetry (17, 21). These studies suggest that both full-length and core gp120 glycoproteins exhibit a high entropy in the free state, apparently sampling multiple conformations. CD4 binding entails an unusually large reduction in entropy, presumably locking the gp120 core into a specific conformation.
The HIV-1 envelope glycoproteins elicit an antibody response during natural infection (4-6); however, the elicitation of broadly reactive neutralizing antibodies is inefficient. Many of the naturally elicited antibodies do not recognize the functional oligomeric envelope protein and fail to neutralize the virus (19). Neutralizing antibodies are raised against both the variable and conserved regions of the envelope glycoproteins (16). The conserved gp120 neutralization epitopes consist of regions near the CD4-binding site (CD4BS epitopes) (22, 24), regions induced by CD4 binding (CD4-induced [CD4i] epitopes) (23), and the carbohydrate-dependent 2G12 epitope (25). The location of these epitopes on the crystallized gp120 core has been mapped by mutagenesis (30). The CD4BS epitopes are located at the interface of the gp120 inner and outer domains, and antibodies raised against this site can compete with CD4 for binding gp120 (22). The CD4i antibodies recognize conserved bridging sheet structures on gp120 that are induced by CD4 binding and are near a conserved gp120 region that has been shown to be involved in coreceptor binding (23). The 2G12 antibody binds the carbohydrate-rich gp120 outer domain region (25).
Features of the gp120 structure are thought to contribute to its inefficiency in eliciting neutralizing antibodies. These include the high degree of glycosylation, exposed variable loops, and the lability of the trimeric envelope glycoprotein complex (4-6, 30, 32). More recently, it has been appreciated that the high level of conformational flexibility of the gp120 core (17) creates entropic barriers to the binding of antibodies (CD4BS and CD4i antibodies) directed against the receptor-binding regions of the molecule (15). Moreover, mutagenesis studies have revealed that CD4 and CD4i antibodies induce a closely related conformation in gp120, whereas CD4BS antibodies preferentially bind other gp120 conformations (33). Thus, a model emerges in which free gp120 samples many conformations. CD4 binding locks the gp120 core into a conformation that is competent for chemokine receptor binding and is recognized by CD4i antibodies. This conformational transition occurs on the pathway to virus entry. CD4BS antibodies bind free gp120 and restrict its ability to assume the CD4-bound conformation, thus disrupting the entry process. This model predicts that, even when CD4 is not involved in the process, CD4BS antibodies should exert inhibitory activity on coreceptor binding and envelope glycoprotein function. Herein, we test this prediction.
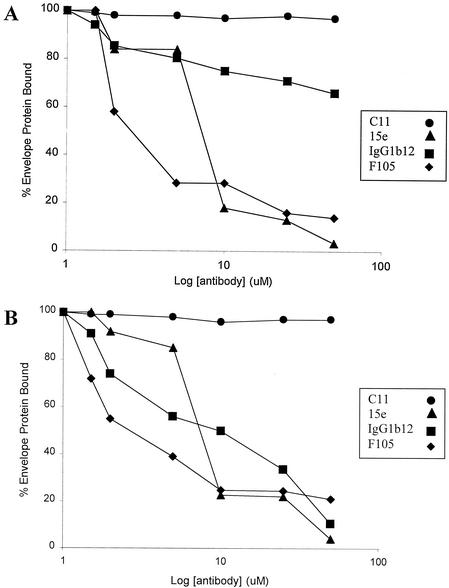
We wished to study the effects of CD4BS antibodies on the HIV-1 envelope glycoprotein-chemokine receptor interaction in a situation where the presence of CD4 is not required. Removal of the gp120 V1/V2 variable loops from the envelope glycoproteins of the ADA primary HIV-1 isolate allows CD4-independent binding to CCR5 and infection of CCR5-expressing cells lacking CD4 (12). More recently, CD4-independent binding of V1/V2 loop-deleted gp120 glycoproteins from other HIV-1 isolates to either CCR5 or CXCR4 has been documented (2; A. Raja and J. Sodroski, unpublished observations). To examine the effect of CD4BS antibodies on CD4-independent CCR5 binding, radiolabeled V1/V2 loop-deleted gp120 glycoproteins from three R5 HIV-1 isolates, ADA, JR-FL, and YU2, were produced transiently in 293T cells. The amount of the ΔV1/V2 gp120 present in the supernatants was estimated by precipitation, using a pool of sera from HIV-1-infected individuals. For an assessment of the CD4-independent binding of the ΔV1/V2 gp120 glycoproteins to CCR5, the supernatants were incubated at 37°C with Cf2Th cells expressing CCR5. After washing, bound envelope glycoproteins were detected by lysis of the cells and precipitation with pooled sera from HIV-1-infected individuals and protein A-Sepharose beads. The precipitated proteins were analyzed on sodium dodecyl sulfate-polyacrylamide gels, which were autoradiographed as described previously (12, 13). The effect of CD4BS antibodies on CD4-independent CCR5 binding was assessed by incubation of the radiolabeled supernatants containing the ΔV1/V2 gp120 glycoproteins with CD4BS antibodies at various concentrations at 37°C for 1 h prior to addition to the target cells. Figure 1A shows that all three CD4BS antibodies tested, F105, 15e, and immunoglobulin G1b12 (IgG1b12), inhibited the binding of the ADA ΔV1/V2 gp120 glycoprotein to Cf2Th cells expressing CCR5. The IgG1b12 antibody was less potent than either the F105 or 15e antibodies. An enzyme-linked immunosorbent assay of IgG1b12 antibody recognition of the ADA ΔV1/V2 gp120 glycoprotein revealed that the IgG1b12 antibody did not recognize this protein as efficiently as the other CD4BS antibodies (data not shown). By contrast, the IgG1b12 antibody recognized the JR-FL and YU2 ΔV1/V2 gp120 glycoproteins efficiently (data not shown). Deletion of the V1 and V2 variable loops from the gp120 glycoproteins of some HIV-1 strains decreases recognition by the IgG1b12 antibody (3, 16); apparently, this effect depends upon the strain of origin of the gp120 molecule. Figure 1B shows that all three CD4BS antibodies equivalently inhibited the CD4-independent binding of the JR-FL ΔV1/V2 gp120 glycoprotein to CCR5-expressing cells. Similar results were seen for the ΔV1/V2 gp120 glycoprotein derived from the YU2 strain of HIV-1 (data not shown). A control antibody, C11, did not block the binding of the ΔV1/V2 gp120 glycoproteins to CCR5 (Fig. 1 and data not shown). The C11 antibody recognizes a discontinuous epitope composed of elements derived from the conserved N and C termini of gp120 (16), efficiently binds variable loop-deleted gp120 proteins (3), and has previously been shown not to interfere with gp120-coreceptor interactions (29). We conclude that CD4BS antibodies can interfere with the gp120-CCR5 interaction, even when CD4 is not involved in promoting this interaction.
FIG. 1.
Inhibition of variable loop-deleted gp120 glycoprotein binding to CCR5 by CD4BS antibodies. Radiolabeled ΔV1/V2 gp120 glycoproteins from the ADA strain (A) or JR-FL strain (B) of HIV-1 were incubated at 37°C with Cf2Th cells expressing CCR5 in the presence of the indicated amounts of CD4BS antibodies (F105, IgG1b12, and 15e) or a control antibody, C11. The amount of the ΔV1/V2 gp120 glycoproteins bound to the Cf2Th-CCR5 cells is presented as the percentage of the amount bound in the absence of antibody. The experiment was performed twice with comparable results.
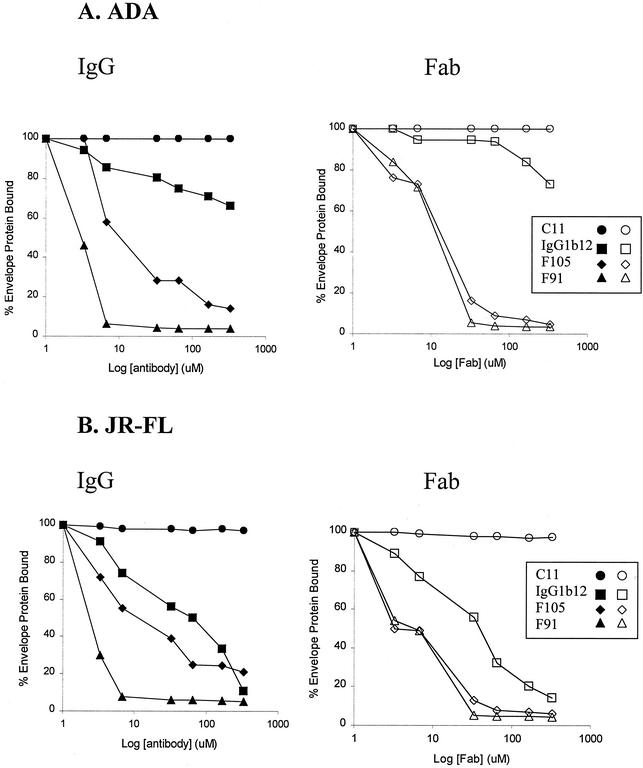
To determine if smaller, monovalent moieties directed against the CD4BS epitopes could also interfere with CD4-independent CCR5 binding, the above experiments were repeated using complete antibodies or Fab fragments of the F105, F91, and IgG1b12 CD4BS antibodies. Figures 2A and B show that Fab fragments of the CD4BS antibodies were able to inhibit the binding of ΔV1/V2 gp120 glycoproteins from the ADA and JR-FL HIV-1 strains, respectively, to CCR5-expressing Cf2Th cells.
FIG. 2.
Fab fragment and antibody inhibition of variable loop-deleted gp120 binding to CCR5. Radiolabeled ΔV1/V2 gp120 glycoproteins from the ADA strain (A) or JR-FL strain (B) were incubated with Cf2Th cells expressing CCR5 (Cf2Th-CCR5) in the presence of the indicated amounts of antibodies (left panels) or Fab fragments (right panels). The amount of the ΔV1/V2 gp120 glycoprotein bound to the Cf2Th-CCR5 cells is presented as the percentage of the amount bound in the absence of added antibody or Fab. The experiment was performed twice with comparable results.
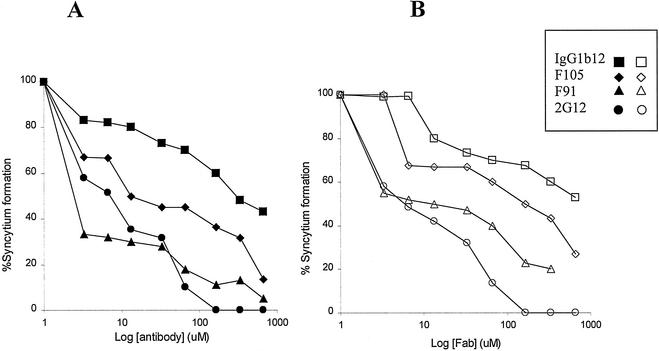
The V1/V2 loop-deleted ADA envelope glycoproteins can mediate HIV-1 entry into target cells lacking CD4 but expressing CCR5 (12). Similarly, the V1/V2-deleted ADA envelope glycoproteins can fuse envelope-expressing cells and CD4-negative cells that express CCR5. To examine whether CD4BS antibodies could inhibit this CD4-independent cell-cell fusion, CD4BS antibodies were added to mixtures of 293T cells expressing the ΔV1/V2 ADA envelope glycoproteins and Cf2Th cells expressing CCR5 but not CD4. Figure 3A shows that the F105 and F91 antibodies efficiently reduced the number of syncytia formed by the ΔV1/V2 ADA envelope glycoproteins in a CD4-independent setting. The IgG1b12 antibody was less efficient at inhibiting syncytium formation in this assay, consistent with its inefficient recognition of the V1/V2 loop-deleted ADA envelope glycoproteins (see above). Fab fragments of the CD4BS antibodies inhibited syncytium formation with efficiencies comparable to those seen with the complete antibodies (Fig. 3B). The formation of syncytia in this assay was also inhibited by the 2G12 antibody and Fab fragment (25). We conclude that CD4BS antibodies can inhibit HIV-1 envelope glycoprotein function, even when CD4 is not required for such function.
FIG. 3.
Fab fragment and antibody inhibition of CD4-independent syncytium formation. 293T cells expressing the ΔV1/V2 envelope glycoproteins from the ADA HIV-1 strain were cocultivated with Cf2Th cells expressing CCR5 in the presence of antibodies (A) or Fab fragments (B). IgG1b12, F105, and F91 are CD4BS antibodies, whereas 2G12 recognizes a distinct, carbohydrate-dependent gp120 epitope (25). The percentage of syncytia observed, compared with the number seen in the absence of added antibody or Fab, is reported. The experiment was performed twice with comparable results.
In most natural settings, in which HIV-1 is dependent upon CD4 for entry into target cells, CD4BS antibodies block gp120-CD4 interaction and thus inhibit virus replication. Here we show that CD4BS antibodies and Fab fragments can block the CD4-independent interaction of gp120 and CCR5 and can block envelope glycoprotein-induced fusion of CD4-negative cells. The comparable potency of inhibition seen for Fab fragments and the complete CD4BS antibodies in most instances suggests that steric factors contribute only modestly to the observed inhibitory effects. The inhibitory effects of Fab fragments in these assays contrast with the stimulatory effects of two- or four-domain soluble CD4 molecules (29), which are similar in size to Fab fragments. The results obtained are consistent with recently proposed models of conformational flexibility within the HIV-1 gp120 exterior envelope glycoprotein (15, 17, 33) (Fig. 4). Free gp120, certainly as a monomer and probably to some extent in the assembled envelope glycoprotein trimer, exhibits a high entropy and is thought to sample many conformations (17). CD4 and the chemokine receptors, as well as the CD4i antibodies, apparently bind closely related or identical conformations of gp120, the CD4-bound conformation (33). The acquisition of the CD4-bound conformation promotes the virus entry process by allowing coreceptor binding and perhaps by triggering additional conformational changes in the gp120 and/or gp41 glycoproteins important for the membrane fusion process. Our results suggest that the binding of CD4BS antibodies to the HIV-1 envelope glycoproteins inhibits coreceptor binding and envelope glycoprotein-mediated syncytium formation, even when CD4 is not present. In conjunction with recent data suggesting that CD4BS antibodies recognize gp120 conformations distinct from the CD4-bound conformation (33), the observed inhibition may result from an inability of gp120, once bound to a CD4BS antibody, to negotiate the transition required to assume the CD4-bound conformation. This model raises the possibility of identifying other inhibitors of HIV-1 receptor binding by searching for molecules that restrict the ability of gp120 to achieve a CD4-bound conformation. This mechanism of inhibition might be compatible with a greater range of chemical structures and potential gp120 binding sites than steric interference with key gp120-CD4 contacts. Thus, compared with steric hindrance, conformational restriction may be easier to achieve with low-molecular-weight compounds. It is encouraging that the IgG1b12 antibody, which in binding gp120 induces only a small decrease in entropy (15), was able to block CD4-independent CCR5 binding. This suggests that gp120 ligands that prevent the transition to a CD4-bound conformation need not encounter a large entropic barrier to binding; i.e., they need not lock gp120 into a fixed conformational state.
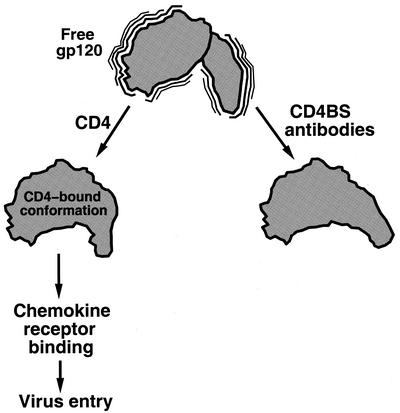
FIG. 4.
Model of the mechanism of neutralization by CD4BS antibodies. Free gp120 may sample multiple conformations but is locked into the CD4-bound conformation by CD4 (17). This gp120 conformation is competent for chemokine receptor binding and virus entry. Because CD4BS antibodies prefer to bind other conformations of gp120 (33), they prevent gp120 from assuming the CD4-bound conformation and from binding the chemokine receptor.
The proximity of the HIV-1 receptor-binding structures (14) and their location in a conformationally flexible region of gp120 (17) provide opportunities for complex mechanisms of neutralization. Simple steric effects of CD4BS antibodies on CD4 binding and CD4i antibodies on chemokine receptor binding appear to be accompanied by inhibitory conformational effects on the binding of the heterologous receptor. In addition to the example provided by these studies, the binding of CD4i antibodies has been shown to inhibit gp120-CD4 binding, but only if the V1/V2 variable loops are present (3, 16). This inhibition could result from a restriction on V1/V2 loop movement that occurs upon CD4 binding (31).
Other examples exist of neutralizing antibodies that prevent conformational changes in viruses that are important for infection. It has been suggested that the 2F5 antibody inhibits postattachment, conformational changes in the HIV-1 gp41 envelope glycoprotein that are important for virus-cell membrane fusion (8, 26). Neutralizing antibodies directed against influenza virus, poliovirus, rabies virus, and adenoviruses have been suggested to block conformational changes that are required for virus entry into the target cell (7, 10, 11, 18, 28).
Understanding the conformational flexibility available to the functional HIV-1 envelope glycoproteins should provide insights into strategies for limiting mechanistically important envelope glycoprotein conformational transitions. This knowledge should assist attempts to intervene in the virus entry process and to elicit more effective immune responses.
Acknowledgments
We thank Yvette McLaughlin and Sheri Farnum for manuscript preparation.
This study was supported by grants from the National Institutes of Health (AI24755, AI31783, AI41851, and AI24030), by a Center for AIDS Research grant (AI42848), and by gifts from the G. Harold and Leila Y. Mathers Charitable Foundation, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 2.Basmaciogullari, S., G. Babcock, D. Van Ryk, W. Wojtowicz, and J. Sodroski. 2002. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein important for CXCR4 binding. J. Virol. 76:10791-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J., R. Wyatt, E. Desjardins, P. D. Kwong, W. A. Hendrickson, J. Moore, and J. Sodroski. 1997. Analysis of the interaction of antibodies with a conserved enzymatically deglycosylated core of the HIV-1 gp120 envelope glycoprotein. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 4.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11:S87-S98. [PubMed] [Google Scholar]
- 6.Burton, D. R., and J. P. Moore. 1998. Why do we not have an HIV vaccine and how can we make one? Nat. Med. 4:495-498. [DOI] [PubMed] [Google Scholar]
- 7.Emini, E. A., P. Ostapchuk, and E. Wimmer. 1983. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J. Virol. 48:547-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Follis, K., S. Larson, M. Lu, and J. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 10.Imai, M., K. Sugimoto, K. Okazaki, and H. Kida. 1998. Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Res. 53:129-139. [DOI] [PubMed] [Google Scholar]
- 11.Kida, H., S. Yoder, M. Kuwabara, and R. Yanagawa. 1985. Interference with a conformational change in the haemagglutinin molecule of influenza virus by antibodies as a possible neutralization mechanism. Vaccine 3:219-222. [DOI] [PubMed] [Google Scholar]
- 12.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubenstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. Leavitt, S. Majeed, T. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. Parren, J. Robinson, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. HIV-1 evades neutralization through conformational masking of receptor-binding sites. Nature, in press. [DOI] [PubMed]
- 16.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raux, H., P. Coulon, F. Lafay, and A. Flamand. 1995. Monoclonal antibodies which recognize the acidic configuration of the rabies glycoprotein at the surface of the virion can be neutralizing. Virology 210:400-408. [DOI] [PubMed] [Google Scholar]
- 19.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein, B. S., S. D. Gowda, J. D. Lifson, R. C. Penhallow, K. G. Bensch, and E. G. Engleman. 1987. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell 49:659-668. [DOI] [PubMed] [Google Scholar]
- 21.Stites, W. E. 1997. Protein-protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 97:1233-1250. [DOI] [PubMed] [Google Scholar]
- 22.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ugolini, S., I. Mondor, P. W. Parren, D. R. Burton, S. A. Tilley, P. J. Klasse, and Q. Sattentau. 1997. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 28.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 31.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt, R., and J. Sodroski. 1998. The human immunodeficiency virus (HIV-1) envelope glycoproteins: fusogens, antigens and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 33.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]