Abstract
Budding of C-type retroviruses begins when the viral Gag polyprotein is directed to the plasma membrane by an N-terminal membrane-binding (M) domain. While dispersed basic amino acids within the M domain are critical for stable membrane association and consequent particle assembly, additional residues or motifs may be required for specific plasma membrane targeting and binding. We have identified an assembly-defective Rous sarcoma virus (RSV) Gag mutant that retains significant membrane affinity despite having a deletion of the fourth alpha-helix of the M domain. Examination of the mutant protein's subcellular distribution revealed that it was not localized to the plasma membrane but instead was mistargeted to intracytoplasmic membranes. Specific plasma membrane targeting was restored by the addition of myristate plus a single basic residue, by multiple basic residues, or by the heterologous hydrophobic membrane-binding domain from the cellular Fyn protein. These results suggest that the fourth alpha-helix of the RSV M domain promotes specific targeting of Gag to the plasma membrane, either through a direct interaction with plasma membrane phospholipids or a membrane-associated cellular factor or by maintaining the conformation of Gag to expose specific plasma membrane targeting sequences.
Retroviruses acquire their lipid envelopes from distinct locations along the cytoplasmic face of the plasma membrane. Specific targeting to the site of budding is directed by the retroviral Gag polyprotein, which is the only viral protein needed to drive the assembly process (reviewed in reference 49). Like many cellular proteins, the Gag polyprotein is initially synthesized on free ribosomes in the cytoplasm. Subsequently, Gag proteins are directed to the inner leaflet of the plasma membrane using targeting information located in the N-terminal membrane-binding domain (M domain) (7, 49, 51, 58). The mechanism whereby most Gag proteins are selectively directed to the plasma membrane rather than to intracytoplasmic membrane remains very poorly understood.
Retroviral morphogenesis follows one of several distinct pathways, characterized by where viral structures are initially visualized by electron microscopy (reviewed in reference 49). For viruses that follow the type C pathway, such as Rous sarcoma virus (RSV), human immunodeficiency virus (HIV), and murine leukemia virus (MLV), Gag proteins are first seen in dense aggregates just under the inner leaflet of the plasma membrane. Type B and type D retroviruses assemble viral cores within the cytoplasm, which are subsequently targeted to the plasma membrane for budding. A third pathway is followed by defective endogenous retroviruses, which form intracisternal A-type particles that are released into the endoplasmic reticulum (ER). The determinants for each morphogenic pathway are inherent to the Gag protein, as particle assembly can be redirected from one pathway to another by altering the amino acid sequence of the matrix (MA) domain (8, 10, 13, 40, 53).
While little is known about the mechanisms underlying how Gag proteins belonging to different morphogenetic families are directed to specific cellular membranes, the sequences involved in membrane binding have been identified for several viruses. For HIV type 1 (HIV-1), stable membrane association is accomplished using a bipartite membrane-binding domain consisting of the fatty acid myristate, added cotranslationally to the N terminus of Gag (2, 15, 45), in concert with a patch of basic residues between amino acids 15 and 31. Myristate provides a hydrophobic interaction with the lipid membrane, while the basic residues are believed to form electrostatic interactions with acidic phospholipids that are enriched at the cytoplasmic face of the plasma membrane (35, 58).
The biophysical basis underlying membrane binding for nonmyristoylated Gag proteins, such as those of RSV and equine infectious anemia virus (EIAV), is less obvious. Indeed, the M domain of RSV is much larger than that of HIV, comprising the first 87 amino acids of Gag (29, 51). Yet both the RSV M domain and the EIAV MA protein retain the conserved topology of retroviral matrix proteins, being composed of four overlapping alpha-helices packed around a central hydrophobic core (17, 25). The RSV M domain may exploit this three-dimensional conformation to bring basic residues together along one face of the molecule; this basic region might serve an analogous function in stabilizing interactions with the membrane phospholipids as the basic motif in the HIV MA sequence. Indeed, genetic evidence from RSV Gag and biochemical analysis of EIAV MA suggest that basic residues contribute to electrostatic interactions crucial to membrane association and viral assembly (3, 35, 37). However, it is possible that additional membrane-binding signals that do not rely on electrostatic interactions exist within nonmyristoylated Gag proteins. Evidence from studies of the EIAV MA protein support this idea, as EIAV MA binds equally well to electrically neutral and negatively charged lipid bilayers (37).
The microenvironment of the particular membrane targeted for particle assembly may be as important for budding as are the signals within Gag, since there appear to be specialized regions of the membrane that promote efficient particle release. This has been shown by the localization of Gag proteins at discrete, punctate regions of the plasma membrane by confocal microscopy (19) and by electron microscopic images of clusters of virions that appear to have been released from fixed positions on the membrane (36). Moreover, several investigators have implicated lipid “rafts,” microdomains of the plasma membrane enriched in cholesterol and sphingolipids, as sites of retrovirus particle assembly (23, 30, 31).
Because the mechanism underlying the targeting of type C retroviruses to specific plasma membrane sites for budding remains elusive, we have undertaken the study of RSV Gag mutants to identify regions of the M domain that are involved in specific plasma membrane targeting. In the present study, we investigated whether mutants known to be impaired for particle assembly had defects in membrane association or membrane localization. We found that deletion of the fourth alpha-helix of the RSV M domain resulted in redirection of Gag proteins to intracellular membranes, leading to the release of particles through the endoplasmic reticulum via the secretory pathway. Specific plasma membrane targeting was restored by the addition of myristate and basic residues or by the substitution of the heterologous hydrophobic membrane-binding domain from the cellular Fyn protein, a member of the Src family of tyrosine kinases. Together these results suggest that membrane targeting and membrane binding are genetically separable and that the fourth alpha-helix of the M domain is needed for specific plasma membrane targeting of RSV Gag proteins.
MATERIALS AND METHODS
Plasmid construction.
Plasmids pMA.GFP (14), pGag.GFP (3), pT10C.GFP (Δ L domain) (36), pMyr0.BgBs.GFP (Δ I domain) (44), pSV.T14K.B1c (35), pSV.Myr0 (52), and pSV.SPG.D37S (21) were previously described. Plasmids pMyr2.B1c.MA.GFP, pMyr2.HB12.MA.GFP, and pMyr2.T14K.B1c.MA.GFP were created by digesting PCR products derived from PARE89 and USP19.263 (14) with Asp718, treating with Klenow, digesting with SstI, and exchanging similarly prepared fragments from pEGFP.N2 (Clontech).
The Fyn sequence was introduced into RSV Gag by using M13 oligonucleotide-directed mutagenesis (22) with primer PARE35 (5′-GGATCAAGCATGGGATGCGTCCAATGCAAGGATAAGGAGGGCCCTAAAACCTATTGCGGG), which introduces a diagnostic ApaI site. The Fyn sequence was transferred into the pSV.Myr0 plasmid between SstI and XhoI sites. The B1c deletion was incorporated into pSV.Fyn through exchange of an SstII fragment from pSV.Myr2.B1c (35). Fyn.myr− and Fyn.palm− derivatives were created by PCR using primers USP19.263 and PARE48 (5′-CTCCTTATCCTTGCATTGGACGCATGCCATGCTTGATCCA) or PARE49 (5′-CTCCTTATCCTTGGCTTGGACGGCTCCCATGCTTGATCCA), respectively, introducing SstI and SpeI sites for introduction into the pSV.Gag vector (20). All mutants of the RSV M domain were introduced into pMA.GFP and pGag.GFP by SstI-BspEI fragment exchange (35). The mutations were confirmed by automated dideoxy sequencing.
Cells, transfections, and confocal microscopy.
The chemically transformed quail fibroblast cell line (QT6) was maintained as previously described (6, 27). Cells were transfected with 1 μg of plasmid DNA by the calcium phosphate method, and 18 h posttransfection confocal microscopy was performed as previously described using a Zeiss LCM 10 (14). Photomicrographs of the cells shown are representative of the population of cells that were observed. Cells treated with leptomycin B (LMB) (Sigma) were incubated with 10 ng/ml (18 nM) for 2 h prior to observation with the confocal microscope. Indirect immunofluorescence was performed as described previously (44) using monoclonal antibodies against β-COP or the Golgi 58,000-molecular-weight (58K) protein (Sigma), or a polyclonal antibody against the C terminus of calnexin (Stressgen Biotechnologies, Victoria, British Columbia, Canada) and secondary antibodies conjugated to Cy3 (Sigma). Images of the same microscopic field were obtained using 488- and 643-nm lasers, and images were merged using Adobe Photoshop.
Radioimmunoprecipitation assay.
QT6 cells were transfected by the calcium phosphate method. Eighteen hours after transfection, cells were radiolabeled with [35S]methionine (>1,000 Ci/mmol; NEN Life Science Products) and lysed, and Gag proteins were immunoprecipitated from cell lysates (L) or culture media (M) using polyclonal anti-RSV serum as described previously (35, 52). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to analysis using a PhosphorImager (Molecular Dynamics). Budding efficiency [M/(L + M)] was calculated as previously described (52). Budding for wild type (Myr0.Gag) was assigned a value of 1.0, and the other mutants' budding efficiencies were normalized to that of the wild type. Statistical significance was determined by using the Student t test.
For brefeldin A (BFA) treatment, COS-1 cells were transfected by the DEAE-dextran-chloroquine method as previously described (55). Forty-eight hours after transfection, cells were treated either with BFA (0.5 μg/ml; Fluka) or with the solvent methanol for 1 h, after which cells were labeled for 2.5 h with l-[35S]methionine in the continued presence of BFA. Cell lysis and immunoprecipitation were performed as previously described (52) and analyzed by SDS-PAGE analysis and autoradiography. A budding ratio for both treated and untreated samples was calculated as the amount of p27CA present in the culture media following a 2.5-h labeling period divided by the amount of Pr76Gag in the cell lysates of a 15-min labeling. Budding efficiency in the presence of BFA was calculated as the fraction of the budding ratio in the treated versus the untreated cells.
Membrane fractionation.
Subcellular fractionation and membrane pelleting were performed according to published methods (42). Briefly, 16 to 18 h posttransfection 2.5 × 106 QT6 cells transfected with 15 μg of plasmid DNA were washed twice and rinsed from the culture plate in ice-cold NTE (100 mM NaCl, 10 mM Tris [pH 7.5], 1 mM EDTA). Cells were pelleted at 500 × g, resuspended in 1 ml of cold hypotonic lysis buffer (10 mM Tris [pH 7.5], 1 mM MgCl2, leupeptin [1 μg/ml], pepstatin [1 μg/ml], phenylmethylsulfonyl fluoride [100 μg/ml]) and allowed to swell on ice for 15 min. Cells were lysed with 15 to 20 strokes of a Dounce homogenizer, and cell disruption was monitored by trypan blue exclusion until approximately 90% of cells were disrupted. Lysates were adjusted to 150 mM NaCl, and nuclei were pelleted at 1,000 × g. Membranes were pelleted from postnuclear supernatants by centrifugation at 100,000 × g, and membrane pellets were vigorously vortexed in NTE with 0.5% Triton X-100. Clarified supernatants were adjusted to 0.5% Triton. Each fraction was analyzed by fluorometry in an Aminco-Bowman Series 2 spectrophotometer with an excitation maximum at 456 nm and an emission maximum at 508 nm. Membrane association was calculated as the fraction of total fluorescence intensity present in the pelleted fraction [P/(P + S), where P is the pellet fraction and S is the soluble fraction]. Membrane association for Myr0.Gag (wild type), which was 43%, was arbitrarily set at 1.0, and each of the mutants was normalized to this value. Statistical significance was analyzed by using the Student t test.
RESULTS
We previously characterized an assembly-defective mutant of the RSV Gag protein which has a deletion of the fourth alpha-helix of the M domain (Myr0.B1c, Fig. 1) (35, 55). Particle release was restored to the deletion mutant by the addition of myristic acid plus a cluster of basic residues inserted within the M domain (35) or by the well-characterized membrane-binding domains from the pp60 v-src [Src] oncoprotein (55) or the HIV-1 Gag M domain (59). To gain further insight into the mechanism of membrane binding for RSV Gag, we tested whether budding could also be rescued by addition of a membrane-binding domain that relies solely on hydrophobic interactions (e.g., that of the cellular Fyn protein) rather than one that consists of combined hydrophobic and electrostatic interactions (e.g., Src and HIV-1 Gag). As well, to determine whether RSV M domain mutants lacking the fourth alpha-helical region were defective in membrane binding or whether specific membrane targeting was disrupted, the ability of M domain mutants to associate with membranes in vivo was tested.
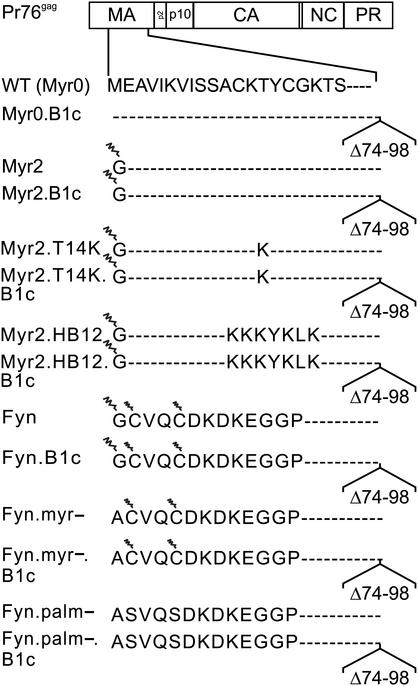
FIG. 1.
Mutants of the RSV M domain. The arrangement of the cleavage products produced from the wild-type [WT (Myr0)] Gag polyprotein is depicted at the top. The E2G substitution, termed Myr2, allows cotranslational addition of myristic acid (depicted as a zigzag line) to the N terminus of Gag. Mutations Myr2.T14K and Myr2.HB12 insert basic residues from the M domain of HIV either individually or within a cluster in the context of the myristoylated N terminus. The Fyn substitution replaces the first 10 amino acids of the RSV M domain with that of Fyn; a G2A change within this sequence, termed Fyn.myr−, prevents both the myristoylation of Fyn at position 2 and the reversible palmitoylation (indicated by the smaller zigzag line) at positions 3 and 6. The Fyn.palm− mutation substitutes alanines for cysteines that are the sites of Fyn palmitoylation. The deletion B1c removes the fourth alpha-helix of the M domain (amino acids 73 to 86) and extends through residue 98.
Budding efficiency and membrane binding of M domain mutants.
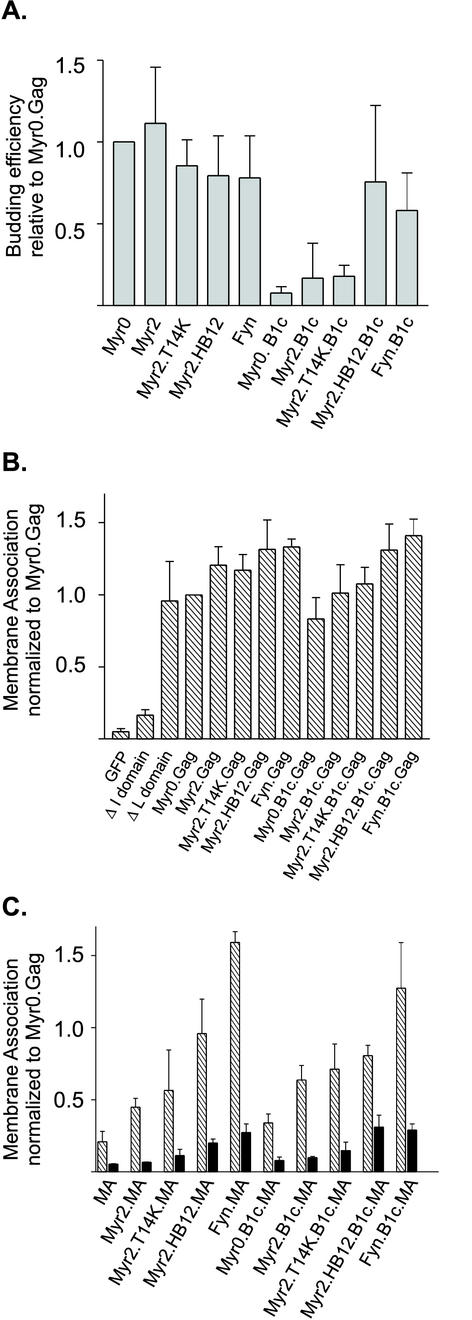
In an attempt to separate nonspecific membrane association from specific plasma membrane localization, wild-type and mutant Gag proteins were fused in frame to the reporter green fluorescent protein (GFP) and tested for particle assembly and in vivo membrane binding (Fig. 2). To assess the efficiency of budding, Gag proteins were expressed in QT6 cells and the amount of Gag protein released into the medium during a 2.5-h labeling period was compared to the level of intracellular Gag proteins, as detected by immunoprecipitation (Fig. 2A). Budding efficiency for wild type (Myr0.Gag) was 36%. Addition of myristate alone (Myr2.Gag), myristate plus a single basic residue substitution (Myr2.T14K.Gag), or myristate plus a cluster of basic residues (Myr2.HB12.Gag) did not significantly alter particle assembly for the full-length Gag protein, as expected. In addition, a mutant containing a substitution of the RSV M domain with 10 amino acids from the Fyn protein was capable of producing virus-like particles (Fyn.Gag). The Fyn membrane-binding domain is myristoylated at the penultimate glycine residue and reversibly palmitoylated on cysteines at positions 3 and 6. For the Fyn protein, specific association with caveolae, microdomains of the plasma membrane involved in signal transduction and regulation of lipid metabolism, depends on both myristate and palmitate modifications (24, 38, 56). Thus, hydrophobic membrane interactions provided by the Fyn sequence can substitute for the RSV M domain to mediate plasma membrane targeting and particle release. Presumably, the particles formed by the Fyn.Gag.GFP fusion protein bud from raft-containing caveolae in the plasma membrane.
FIG. 2.
Particle assembly and membrane association of the wild-type and mutant Gag.GFP and MA.GFP fusion proteins. (A) Budding efficiency. To determine the ability of each Gag derivative to assemble virus-like particles, QT6 cells transfected with indicated Gag.GFP derivatives were labeled for 2.5 h with l-[35S]methionine, lysed, and immunoprecipitated with polyclonal serum against RSV. Budding efficiency was calculated by dividing the amount of Gag detected in the extracellular medium by the sum of the Gag protein present in the cell lysate plus that in the medium. Budding for each mutant was normalized to Gag.GFP, which was assigned the value of 1.0. (B) Membrane association of wild-type and mutant Gag.GFP proteins. Transfected QT6 cells were fractionated by hypotonic lysis, and membranes were pelleted by differential centrifugation. Membrane association was determined by quantification of the amount of fluorescence present in the membrane (P100) fraction divided by the amount present in both the membrane and soluble fractions (P100 + S100). (C) Membrane association of wild-type and mutant MA.GFP proteins. Membrane association was determined as explained in the legend to panel B. Black bars represent samples that were pretreated with 0.5% Triton X-100 prior to membrane pelleting. Error bars (A to C) represent the standard deviation of three or more independent experiments.
Disruption of the RSV M domain by deleting the fourth alpha-helical region, known as mutant B1c, severely impaired particle production for the Myr0.B1c.Gag.GFP fusion protein; budding efficiency was approximately 7% of the amount of budding for full-length Myr0.Gag (Fig. 2A). Addition of myristate or myristate plus a single lysine substitution did not restore efficient budding (Myr2.B1c and Myr2.T14K.B1c, 15 and 21%, respectively) in QT6 cells, although there was a slight improvement in particle production. In contrast, we previously found that Myr2.T14K.B1c was capable of more efficient particle release in COS-1 cells, reflecting either a cell-type-specific factor affecting budding or a difference in intracellular protein levels, which are markedly increased in the COS-1 overexpression system (35). Similar to our results in COS-1 cells, adding myristate plus a cluster of basic residues derived from the HIV-1 M domain restored budding in QT6 cells as well (Myr2.HB12. B1c, 95% budding efficiency compared to Myr2.HB12). The hydrophobic membrane-binding domain of Fyn (Fyn.B1c) also suppressed the effects of the B1c deletion, returning particle assembly to 75% of full-length Fyn.Gag (Fig. 2A).
We presumed that the budding defect associated with the B1c deletion resulted from a disruption of membrane binding for Gag. If so, then we predicted that addition of myristate plus basic residues would promote hydrophobic and electrostatic interactions, thereby recreating a functional membrane-binding domain. To test this idea directly, we analyzed membrane association for wild-type Gag.GFP and its derivatives in QT6 cells using subcellular fractionation followed by membrane pelleting. Membrane-bound and soluble GFP proteins were detected by fluorometry, and membrane association was expressed as a ratio of the pelletable material to the total fluorescence in the pelleted and soluble fractions (Fig. 2B). Each of the mutants was normalized to the level of membrane association of wild-type Gag.GFP (Myr0.Gag), which was assigned a value of 1.0. As controls, we utilized GFP alone (soluble), a C-terminal deletion of Gag that deletes interaction (I) domains important for protein-protein interactions (soluble), and a late (L) domain mutant of Gag that associates efficiently with membranes (pelletable). Addition of myristate alone (Myr2.Gag) modestly increased membrane association when compared to Myr0.Gag, as did myristate plus basic residues (Myr2.T14K and Myr2.HB12.Gag), as well as the hydrophobic membrane-binding domain of Fyn (Fyn.Gag).
Because we had attributed the assembly defect of Myr0.B1c to a general decrease in membrane affinity, we expected that membrane association of the B1c deletion proteins would parallel the budding efficiency profiles illustrated in Fig. 2A. To our surprise, the Myr0.B1c.Gag, Myr2.B1c.Gag, and Myr2.T14K.B1c.Gag proteins (Fig. 2B) retained significant membrane affinity despite severe defects in particle production. Addition of myristate and a cluster of basic residues (Myr2.HB12.B1c.Gag) or the membrane-binding domain of Fyn (Fyn.B1c.Gag) suppressed the mild decrease in membrane affinity associated with the B1c deletion.
To separate membrane association mediated by the M domain from potential membrane-stabilizing effects contributed by downstream regions of Gag, we examined the ability of each mutant to promote membrane association in the context of the mature MA protein (Fig. 2C). Membrane association was normalized to that of wild-type Gag (Myr0.Gag, assigned a value of 1.0) to illustrate the membrane-binding capability of MA compared to full-length Gag (Fig. 2C). The MA.GFP fusion protein was highly soluble compared to full-length Gag, likely due to its lack of I domains that are believed to strengthen membrane binding through cooperative interactions. In support of this interpretation, the degree of membrane association for MA.GFP is similar to that of the I domain mutant shown in Fig. 2B (52).
In the context of MA, the addition of myristate (Myr2.MA) or myristate plus basic residues, either singly (Myr2.T14K.MA) or in a cluster (Myr2.HB12.MA), substantially increased membrane binding (P = 0.01, P = 0.05, and P < 0.0001, respectively). Strikingly, substitution of the membrane-binding domain of Fyn for the RSV M domain in MA (Fyn.MA) enhanced membrane affinity more significantly than the myristate plus basic residue substitutions (P < 0.0001), achieving the level obtained for Fyn.Gag. Thus, the strong hydrophobic interactions provided by the Fyn domain appeared to surpass the contribution of the I domains of Gag to promote stable membrane association of the MA protein.
To ensure that the amount of viral protein in the pelleted fraction was the result of membrane association and not simply the formation of large pelletable protein complexes, cell lysates were treated with Triton X-100 prior to membrane pelleting, as indicated by the solid black bars in Fig. 2C. In each case, there was a dramatic reduction in the amount of pelleted viral protein after detergent treatment, suggesting that Gag protein detected in the pelleted fraction was stably bound to membranes.
Subcellular localization of M domain mutants.
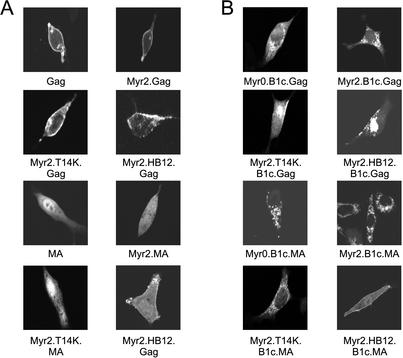
The degree of membrane association retained by Gag proteins in the presence of the B1c deletion was unexpected, given that this deletion severely impairs particle production (compare Fig. 2A and B for Myr0.B1c, Myr2.B1c, and Myr2.T14K.B1c). This discrepancy suggested that the block to budding was not accounted for by a simple defect in global membrane affinity, but instead might reflect targeting to a membrane location that could not properly support particle assembly. The membrane-pelleting assay does not provide information about the site on the membrane at which Gag is accumulating, nor does it verify that Gag is associating with the plasma membrane rather than with intracellular membranes. We therefore employed the GFP marker on our fusion proteins to examine the intracellular distribution of wild-type and mutant Gag and MA proteins by confocal microscopy (Fig. 3A). As reported previously, the wild-type RSV Gag.GFP protein localizes to the cytoplasm and to punctate stretches along the plasma membrane (3, 44). Introduction of myristate (Myr2.Gag), plus a single basic residue (Myr2.T14K.Gag) or multiple basic residues (Myr2.HB12.Gag), did not alter the plasma membrane localization of Gag.GFP. The wild-type MA protein, on the other hand, was present throughout the cell with accumulation within the nucleus of transfected cells (Fig. 3A) (14, 44). Introduction of myristate alone (Myr2.MA) slightly reduced the amount of MA in the nucleus, while myristate in combination with a single basic residue (Myr2.T14K.MA) slightly enhanced nuclear localization. More dramatic was the recruitment of MA to extended patches along the plasma membrane, mediated by the addition of myristate and a charged patch of basic residues (Myr2.HB12.MA).
FIG. 3.
Subcellular localization of Gag and MA mutant proteins. QT6 cells transfected with full-length (A) and B1c deletion (B) GFP fusion proteins were examined by confocal microscopy 16 to 20 h posttransfection as indicated. Representative images are shown for each construct.
Examination of the localization of B1c-deleted GFP fusion proteins confirmed the results of the membrane-pelleting assay; these proteins remained associated with cellular membranes but were mistargeted within the cell, accumulating at intracellular membranes (Fig. 3B). The Myr0.B1c.Gag protein adopted a reticulated pattern within the cell that excluded both the nucleus and the plasma membrane. Addition of myristate (Myr2.B1c.Gag) conferred a more regular pattern of association with intracellular membranes. While both the Myr2.T14K.B1c.Gag and the Myr2.HB12.B1c.Gag proteins remained localized to intracellular membranes, the addition of basic residues was able to rescue a portion of the protein to the plasma membrane at extended patches. The targeting of a subpopulation of the Myr2.HB12.B1c.Gag protein to the plasma membrane is consistent with the ability of this construct to efficiently form virus-like particles (Fig. 2A). However, the discrepancy between the partial restoration of the Myr2.T14K.B1c.Gag protein to plasma membrane localization and the failure of this construct to efficiently release extracellular particles in QT6 cells (Fig. 2A) suggests that addition of a single basic residue does not promote sufficient plasma membrane association to drive budding.
The B1c deletion not only mislocalized the Gag protein to intracellular membranes but also resulted in redistribution of the previously soluble MA protein to intracellular membranes (Fig. 3B). The relocalization of MA.B1c was preserved following the addition of myristate alone (Myr2.B1c.MA) or in combination with a single basic residue (Myr2.T14K.B1c.MA). Addition of myristate plus multiple basic residues (Myr2.HB12.B1c.MA) restored the association of the mutant protein with the plasma membrane. The localization of B1c-deleted proteins with intracellular membranes suggests either that the deletion creates a new membrane-binding domain specific for intracellular membranes or that the deletion impairs the ability of Gag to properly select the plasma membrane. The contributions of basic residues to membrane binding were subtle, but their effects on subcellular distribution were dramatic, suggesting that reestablishment of plasma membrane targeting by Myr2.T14K.B1c.Gag, Myr2.HB12.B1c.Gag, and Myr2.HB12.B1c.MA is the result of restoring proper membrane selectivity to the deleted Gag protein.
Intracellular distribution of Fyn substitution mutants.
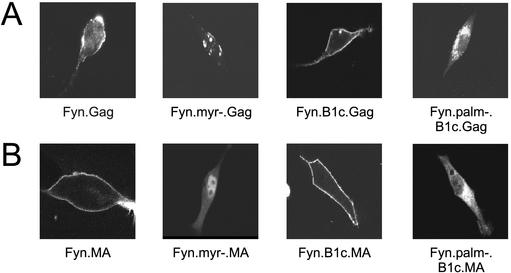
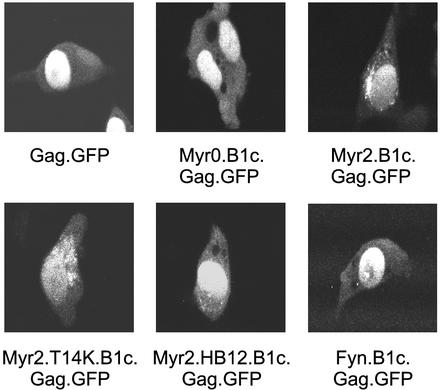
Because the extent of plasma membrane rescue varies with the addition of single or multiple basic residues to the Myr2.B1c.Gag and Myr2.B1c.MA proteins, we examined the effect of hydrophobic membrane-binding properties of the Fyn domain on subcellular localization of Gag. The Fyn membrane-binding domain has been shown to confer plasma membrane localization to another heterologous protein (50), and we found that the Fyn-substituted Gag and MA proteins were strongly associated with membranes (Fig. 2). When examined by confocal microscopy, Fyn.Gag.GFP accumulated almost exclusively at the plasma membrane (Fig. 4). The Fyn.MA.GFP protein also localized to the plasma membrane, likely due to a reduced dependence on I domains for membrane binding (Fig. 4B). The Fyn substitution also restored plasma membrane targeting of the Myr0.B1c.Gag and MA.B1c proteins, as demonstrated by a pattern of continuous plasma membrane accumulation (Fyn.B1c.Gag and Fyn.B1c.MA).
FIG. 4.
Subcellular localization of Fyn.MA and Fyn.Gag derivatives. The intracellular distributions of Fyn.Gag.GFP (row A) and Fyn.MA.GFP (row B) chimeric proteins were determined by confocal microscopy of transfected QT6 cells. The localizations of the Fyn.Gag and Fyn.MA proteins were also examined in the absence of palmitoylation (Fyn.palm−.B1c) and the absence of both myristoylation and palmitoylation of Fyn (Fyn.myr−). The individual cells shown are representative of the population of cells that were observed.
The ability of the Fyn sequence to promote plasma membrane association was dependent entirely upon fatty acid modifications; a G2A mutation that abolished myristoylation also prevented subsequent palmitoylation (Fyn.myr−.Gag) and resulted in the formation of protein aggregates within the cell (Fig. 4A). However, the nuclear transport of MA was not affected by the loss of acylation (Fyn.myr−.MA) (Fig. 4B). Disruption of Fyn palmitoylation independently of myristoylation (Fyn.palm−) prevented the ability of Fyn to restore plasma membrane targeting specificity; the Fyn.palm−.B1c.Gag and Fyn.palm−.B1c.MA proteins revealed the intracellular membrane localization characteristic of proteins bearing the B1c deletion. Failure of both the Fyn.palm−.B1c.Gag (Fig. 4A) and the Myr2.B1c.Gag (Fig. 3B) proteins to localize to the plasma membrane suggests that myristoylation alone is not a sufficient signal for plasma membrane localization of Gag.
Colocalization of M domain mutants with intracellular membranes.
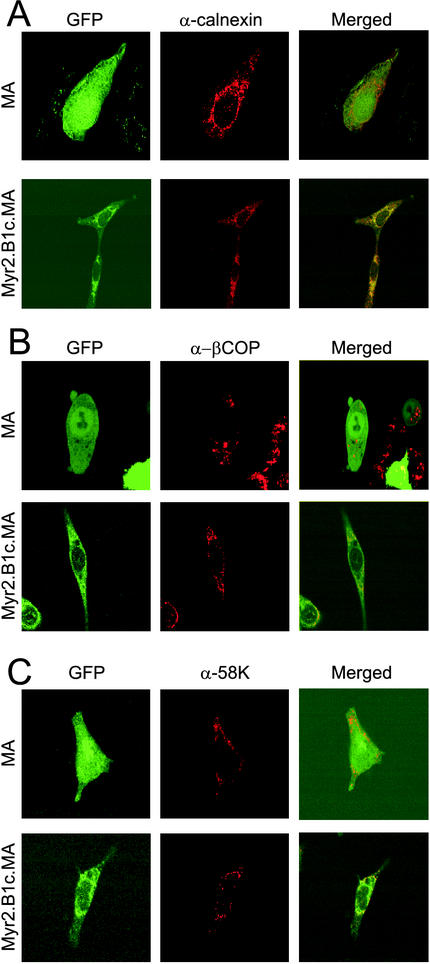
If the B1c deletion prevents the ability of Gag to select the plasma membrane, then we predicted that the association of B1c deletion mutants with intracellular membranes would reflect a nonspecific membrane association rather than targeting to a specific membrane. To determine whether Gag proteins bearing the B1c deletion accumulated at specific intracellular membrane locations, we employed indirect immunofluorescence with markers of the ER and Golgi complex to look for colocalization of Gag protein derivatives with these subcellular organelles. The wild-type MA.GFP protein localized to the cytoplasm and nucleus following fixation, and there was no colocalization of staining with markers of the ER membrane (calnexin) (Fig. 5A), ER COP-1 vesicle component (β-COP) (Fig. 5B), or Golgi complex (58K) (Fig. 5C). In contrast, there was partial overlap of the pattern of epifluorescence seen with the Myr2.B1c.MA.GFP protein and the fluorescence seen by staining with all three antibodies, with the greatest colocalization seen when using the calnexin antibody (see merged view, Fig. 5A). Indeed, the fluorescence pattern of Myr2.B1c.MA.GFP encompassed a greater area of the intracellular space than any of the individual organelle markers, further suggesting not that the B1c deletion has introduced a targeting signal but rather that the protein simply accumulated at the most accessible intracellular membrane sites.
FIG. 5.
Colocalization of the Myr2.B1c.MA protein with markers of the ER and Golgi. Cells expressing the MA.GFP or Myr2.B1c.MA.GFP fusion protein were fixed in 3:1 acetone-methanol and stained with antibodies against calnexin (α-calnexin) (A), β-COP (α-βCOP) (B), or the 58K Golgi protein (α-58K) (C) and a secondary antibody conjugated to Cy3 and viewed by confocal microscopy. Localization of the MA and Myr2.B1c.MA proteins is displayed as GFP epifluorescence in the left-hand panels, while the subcellular distribution of endogenous marker proteins is displayed in corresponding fields in the center panels. The right-hand panels show merged views of the GFP and Cy3 images.
Budding pathway of B1c.Gag proteins.
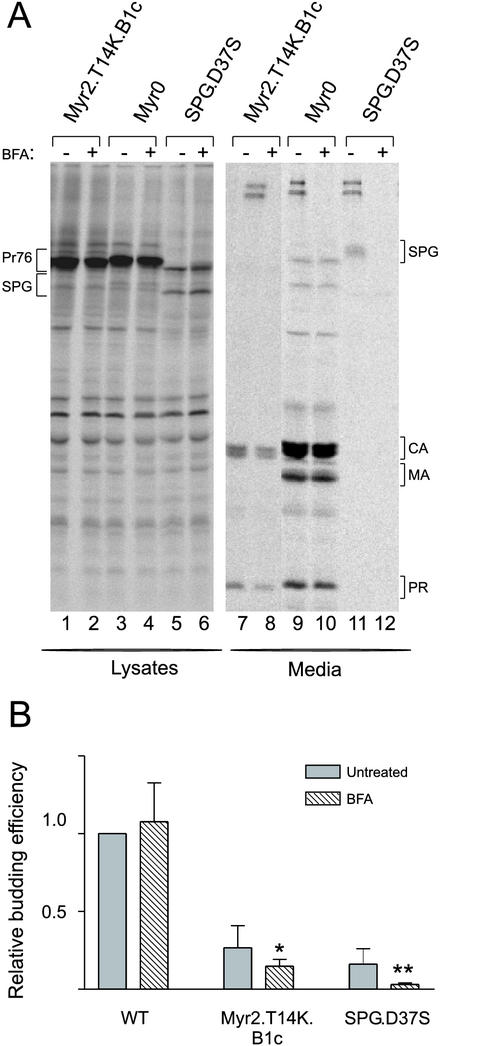
Although Myr2.T14K.B1c.Gag accumulated primarily at intracellular membranes, this mutant Gag protein could be immunoprecipitated from transfected cell culture supernatants to a limited extent. Because no visible budding structures were visible by electron microscopy at the plasma membrane or along cytoplasmic membranes (data not shown), we wondered whether the particles released into the culture medium were released from the plasma membrane or through bulk secretory flow resulting from an association with ER and Golgi membranes. To address this question, we expressed Myr2.T14K.B1c.Gag in COS-1 cells, since budding is more readily detectable for this mutant in these cells than in QT6 cells (Fig. 6) (35). We examined the effect of BFA, a drug which disrupts the structure of the Golgi complex (12), on the release of wild-type and Myr2.T14K.B1c Gag proteins. Transfected COS-1 cells were pretreated with BFA (0.5 μg/ml) for 1 h prior to radiolabeling for 2.5 h, and Gag proteins were immunoprecipitated from cell lysates and culture media (Fig. 6A). As a control for BFA activity, release of an Env-Gag fusion protein (SPG.D37S), which is known to follow the secretory pathway, was severely inhibited by the drug (21), with protein levels in the media reduced to 23% of those in the untreated control (P = 0.0047) (Fig. 6). Importantly, particle release for the wild-type Gag protein was insensitive to BFA, indicating that drug treatment had not disrupted all cellular membranes and subcellular targeting processes. The detection of Myr2.T14K.B1c protein in the media was significantly reduced upon treatment with BFA (Fig. 6B [65% of its untreated control; P = 0.027]). This finding confirmed that the Myr2.T14K.B1c protein does associate with ER and Golgi membranes. Thus, there are two trafficking pathways for this mutant: (i) the BFA-resistant pathway, which may reflect the inefficient targeting to and budding from the plasma membrane, and (ii) the BFA-sensitive pathway, which is associated with intracellular membranes that serve as the precursors for secretory vesicles.
FIG. 6.
Particle assembly in the presence of BFA. COS-1 cells transiently transfected with simian virus 40 promoter-based expression vectors were pretreated with either methanol alone (−) or with BFA (+) for 1 h. (A) SDS-PAGE analysis of immunoprecipitated wild-type and mutant Gag.GFP constructs treated with BFA. The Env-Gag fusion (SPG) contains a point mutation in PR (D37S) and appears as two protein bands within the cell and a highly glycosylated form in the medium (21). Bands corresponding to the Gag precursor Pr76gag, the cleavage proteins CA, MA, and PR, and the SPG.D37S chimera are indicated. (B) Efficiency of Gag protein release into the medium was calculated as described in Materials and Methods following either a pulse-label or 2.5-h labeling of transfected COS-1 cells. Error bars indicate the standard deviation of three or more independent experiments. Student's t test was performed to determine statistical significance (*, P = 0.027; **, P = 0.0047). WT, wild type.
Nuclear trafficking pathway of RSV M domain mutants.
It was possible that the B1c deletion, which removes the fourth alpha-helix of the M domain, could affect additional trafficking pathways other than the plasma membrane targeting of Gag. Indeed, we previously reported that the RSV Gag protein enters and exits the nucleus, and that the fourth alpha-helix of the M domain is necessary for the nuclear localization of the RSV MA protein (44). While the role of nuclear trafficking is not yet known, it does appear that transient nuclear localization of Gag is an intrinsic part of the assembly pathway (44). Because nuclear export of the RSV Gag protein is inhibited by treatment with LMB (44), Gag proteins that are capable of nuclear entry should accumulate within the nucleus when treated with LMB. To determine whether deletion of the fourth alpha-helix of the M domain disrupted nuclear entry of Gag, cells expressing Gag.GFP and B1c derivatives were treated with LMB for 2 h and examined by confocal microscopy (Fig. 7). Each of the Gag proteins with a deletion of the fourth alpha-helix was capable of nuclear entry and accumulated in the nucleus upon LMB treatment. While a portion of the Myr2.B1c.Gag and Myr2.T14K.B1c.Gag proteins remained associated with intracellular membranes following treatment, the majority of the protein was trapped in the nucleus. The efficient nuclear entry of B1c-deleted Gag proteins suggests that while the fourth alpha-helix is necessary for nuclear entry of MA, there might be additional downstream sequences that contribute to nuclear targeting of Gag. Furthermore, these results imply that deletion of the fourth alpha-helix of the M domain alters the terminal subcellular targeting step of assembly—the specific transport of Gag to the plasma membrane.
FIG. 7.
LMB treatment of Gag deletion proteins. To determine the ability of Gag proteins containing the B1c deletion to transit through the nucleus, live cells expressing each of the indicated GFP fusion proteins were treated for 2 h with LMB (10 ng/ml) and then examined by confocal microscopy.
DISCUSSION
The mechanism of RSV membrane targeting has remained elusive, as RSV Gag contains none of the membrane-targeting determinants found in other retroviral Gag proteins or cellular proteins, including amino-terminal fatty acid modifications or clustered basic residues. Association of cellular signal transduction proteins with the plasma membrane often involves modification of the penultimate glycine with a covalently attached myristate moiety added cotranslationally (54). Although myristatic acid is thought to insert into the lipid bilayer, this interaction appears to be transitory, as myristoylation is not sufficient for stable membrane association (28). Tight membrane association therefore requires an additional membrane-binding determinant (reviewed in references 24 and 39). For Src-related tyrosine kinases such as Fyn, Yes, and Lck, this signal is palmitate, a second fatty acid added posttranslationally in a reversible manner. Other proteins, such as Src itself, employ a polybasic stretch downstream of the myristate modification. These basic residues are believed to allow for electrostatic association with the negative phospholipids of the inner leaflet of the cell membrane.
This second signal for membrane binding, either palmitoylation or a polybasic cluster, not only stabilizes the association with the membrane but also provides specificity in plasma membrane targeting. Numerous examples demonstrate that elimination of this second signal results in relocation of the protein to intracellular sites. For example, mutations that prevent the palmitoylation of the cellular G protein α subunit (GαZ) result in relocalization of the mutant protein to intracellular membranes (11, 26), and the lack of palmitoylation results in the accumulation of the Src-family kinase p59Hck in lysosomes (4, 41). Mutation of the lysines which comprise the cellular kinase K-Ras(B) polybasic stretch redistributes the protein to the endoplasmic reticulum and Golgi apparatus (1, 5).
In a similar manner, membrane binding and the specificity of membrane targeting are separable in the HIV M domain. Numerous mutations and deletions affecting the polybasic stretch between amino acids 15 and 31 of the HIV M domain result in relocalization of the Gag precursor to intracellular membranes (10, 13, 19, 32, 48, 57). In some cases, rare budding structures of both immature (10, 48) and mature (32) viral particles can be observed at perinuclear, ER, and Golgi membranes. A similar phenotype has been observed for MLV, where mutations within the N terminus of MA redirect Gag to intracellular membranes (16). For MLV, intracellular aggregation of Gag proteins is seen along the ER membrane and incomplete budding structures are observed within the cytoplasm (46).
In this study, we report a similar phenotype for mutants of RSV Gag, a nonmyristoylated oncoviral Gag protein. Deletion of the fourth alpha-helix of the membrane-binding domain (ΔB1c) resulted in accumulation of the Gag precursor at intracellular membranes. Particle assembly was severely reduced in the B1c construct, yet overall membrane association was not impaired. Proper plasma membrane targeting was restored by stabilizing specific interactions with the plasma membrane either through the addition of basic residues or by the addition of a heterologous plasma membrane targeting signal. The fourth alpha-helix of the M domain was necessary but not sufficient for plasma membrane targeting, since this sequence was not able to redirect the soluble GFP protein to the plasma membrane (data not shown).
Our results suggest that Gag is able to associate nonspecifically with intracellular membranes until plasma membrane specificity is achieved. Many cellular proteins achieve membrane specificity as a late event in membrane binding (56). Proteins containing a single signal for membrane binding sample both intracytoplasmic membranes and the plasma membrane through reversible associations. Addition of the second modification, often palmitoylation at a specific subcellular location, increases membrane affinity, thereby tethering the protein at the site of modification. RSV Gag might also temporally separate membrane binding and specific plasma membrane association; the fourth alpha-helix of the M domain might enable Gag to achieve the appropriate conformation for plasma membrane association after sampling intracellular membranes. Alternatively, the fourth alpha-helix may lock RSV Gag into the final conformation for specific plasma membrane targeting, thereby bypassing prior association of Gag with intracellular membranes.
However, rather than affecting the conformation of Gag, the fourth alpha-helix of the M domain might encode sequence determinants that promote stable association with the plasma membrane. It is possible that the Myr0.B1c.Gag protein does not contain enough basic residues to be stably maintained at the plasma membrane; the additional three basic residues in the fourth alpha-helix could therefore enable Gag to achieve the threshold for interaction with the negatively charged cytoplasmic face of the plasma membrane. Alternatively, the fourth alpha-helix of RSV Gag might not associate directly with membrane phospholipids but may instead provide a protein interaction domain that anchors RSV Gag to another plasma membrane associated protein.
Our results are also consistent with a model whereby the MA domain adopts different conformations in its mature form and in the context of the Gag polyprotein. It has been proposed that HIV Gag utilizes a myristyl-switch mechanism in its interaction with the plasma membrane (33, 59). For HIV Gag, the myristate moiety and a polybasic motif facilitate plasma membrane association; upon proteolysis of Gag, the mature MA protein sequesters the myristate moiety to increase its solubility, thereby becoming available to participate in postentry events of infection within the cytoplasm. This hypothesis is supported by biochemical data indicating that the membrane association of MA is much weaker than Gag (19, 32, 47, 59) and that MA and Gag bind to the membrane in distinct conformations (9, 43). We found that the mature MA protein of RSV also binds membranes much more weakly than does the full-length Gag protein. Deletion of the fourth alpha-helix increases nonspecific membrane association, perhaps by altering the conformation of MA to resemble that present in the context of the Gag polyprotein. In an analogous fashion, the last alpha-helix of HIV-1 MA is postulated to regulate the switch between the conformations of the mature, soluble MA protein and the immature membrane-bound form (18).
Both the association of Gag with intracellular membranes, which prevents the efficient release of viral particles, and the association of the mature MA protein with inappropriate cellular membranes may be deleterious to viral replication. Viruses containing the Myr2.T14K.B1c and Myr2.HB12 Gag mutants manifest postassembly blocks in the viral life cycle (14, 34). Despite efficient particle assembly, these viruses contain monomeric genomic RNA. This finding suggests that while an altered membrane-binding domain may allow Gag to transit to the plasma membrane and release particles, formation of an infectious virion appears to be more complex than simply assembling particles at the plasma membrane. The wild-type membrane-targeting domain of Gag might be necessary for association of Gag with a specific plasma membrane microenvironment, a cellular cofactor, or a subcellular trafficking pathway that is required for genomic RNA dimerization. In addition, these viral mutants show postentry defects, suggesting that alteration of the M domain might have additional effects required early in the establishment of infection. Thus, the signals present in Gag that confer specific plasma membrane targeting may provide additional essential functions in the viral life cycle.
Acknowledgments
We thank Rachel Garbitt for critical review of the manuscript, Carol Wilson and Karen Bone for technical assistance, and John Wills for reagents and insightful discussions.
This work was supported by a grant from the NIH to L.J.P. (R01 CA76534). L.Z.S. is supported by a predoctoral fellowship from the National Science Foundation.
REFERENCES
- 1.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carreno, S., M.-E. Gouze, S. Schaak, L. J. Emorine, and I. Maridonneau-Parini. 2000. Lack of palmitoylation redirects p59Hck from the plasma membrane to p61Hck-positive lysosomes. J. Biol. Chem. 275:36223-36229. [DOI] [PubMed] [Google Scholar]
- 5.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 6.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 8.Eastman, S. W., and M. L. Linial. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich, L. S., S. Fong, S. Scarlata, G. Zybarth, and C. Carter. 1996. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry 35:3933-3943. [DOI] [PubMed] [Google Scholar]
- 10.Facke, M., A. Janetzko, R. L. Shoeman, and H.-G. Krausslich. 1993. A large deletion in the Matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 67:4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishburn, C. S., P. Herzmark, J. Morales, and H. R. Bourne. 1999. Gbg and palmitate target newly synthesized Gaz to the plasma membrane. J. Biol. Chem. 274:18793-18800. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara, T., K. Oda, S. Yokota, A. Takatsuki, and Y. Ikehara. 1988. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263:18545-18552. [PubMed] [Google Scholar]
- 13.Gallina, A., G. Mantoan, G. Rindi, and G. Milanesi. 1994. Influence of MA internal sequences, but not of the myristylated N-terminus sequence, on the budding site of HIV-1 Gag protein. Biochem. Biophys. Res. Comm. 204:1031-1038. [DOI] [PubMed] [Google Scholar]
- 14.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, M., L. Jelinek, S. Whiting, and E. Barklis. 1990. Transport and assembly of gag proteins into Moloney murine leukemia virus. J. Virol. 64:5306-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatanaka, H., O. Iourin, Z. Rao, E. Fry, A. Kingsman, and D. I. Stuart. 2002. Structure of equine infectious anemia virus matrix protein. J. Virol. 76:1876-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermida-Matsumoto, L., and M. D. Resh. 1999. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55gag and p17MA. J. Virol. 73:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishna, N. K., S. Campbell, V. M. Vogt, and J. W. Wills. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishna, N. K., R. A. Weldon, Jr., and J. W. Wills. 1996. Transport and processing of the Rous sarcoma virus Gag protein in the endoplasmic reticulum. J. Virol. 70:1570-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 23.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCabe, J. B., and L. G. Berthiaume. 1999. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell 10:3771-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonnell, J. M., D. Fushman, S. M. Cahill, W. Zhou, A. Wolven, C. B. Wilson, T. D. Nelle, M. D. Resh, J. Wills, and D. Cowburn. 1998. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 279:921-928. [DOI] [PubMed] [Google Scholar]
- 26.Morales, J., C. S. Fishburn, P. T. Wilson, and P. T. Bourne. 1998. Plasma membrane localization of Gaz requires two signals. Mol. Biol. Cell 9:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscovici, C., M. G. Moscovici, J. Jimenez, M. M. C. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95-103. [DOI] [PubMed] [Google Scholar]
- 28.Murray, D., N. Ben-Tal, B. Honig, and S. McLaughlin. 1997. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure 5:985-989. [DOI] [PubMed] [Google Scholar]
- 29.Nelle, T. D., and J. W. Wills. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paillart, J.-C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parent, L. J., C. B. Wilson, M. D. Resh, and J. W. Wills. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provitera, P., F. Bouamr, D. Murray, C. Carter, and S. Scarlata. 2000. Binding of equine infectious anemia virus matrix protein to membrane bilayers involves multiple interactions. J. Mol. Biol. 296:887-898. [DOI] [PubMed] [Google Scholar]
- 38.Resh, M. D. 1994. Myristoylation and palmitoylation of Src family members: the fats of the matter. Cell 76:411-413. [DOI] [PubMed] [Google Scholar]
- 39.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Rhee, S. R., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 41.Robbins, S. M., N. A. Quintrell, and J. M. Bishop. 1995. Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and their association with caveolae. Mol. Cell. Biol. 15:3507-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarlata, S., L. S. Ehrlich, and C. A. Carter. 1998. Membrane-induced alterations in HIV-1 Gag and matrix protein-protein interactions. J. Mol. Biol. 277:161-169. [DOI] [PubMed] [Google Scholar]
- 44.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM-1 dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz, A. M., and A. Rein. 1989. Unmyristoylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J. Virol. 63:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soneoka, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Mutagenesis analysis of the murine leukemia virus matrix protein: identification of regions important for membrane localization and intracellular transport. J. Virol. 71:5549-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of retroviral proteins., p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 50.van't Hof, W., and M. D. Resh. 2000. Targeting proteins to the plasma membrane and membrane microdomains by N-terminal myristoylation and palmitoylation. Methods Enzymol. 327:317-330. [DOI] [PubMed] [Google Scholar]
- 51.Verderame, M. F., T. D. Nelle, and J. W. Wills. 1996. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 70:2664-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welker, R., A. Janetzko, and H.-G. Krausslich. 1997. Plasma membrane targeting of chimeric intracisternal A-type particle proteins leads to particle release and specific activation of the viral proteinase. J. Virol. 71:5209-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilcox, C., J.-S. Hu, and E. N. Olson. 1987. Acylation of proteins with myristic acid occurs cotranslationally. Science 238:1275-1278. [DOI] [PubMed] [Google Scholar]
- 55.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolven, A., H. Okamura, Y. Rosenblatt, and M. D. Resh. 1997. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol. Biol. Cell 8:1159-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, X., X. Yu, T.-H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix block intracellular transport of the Gag precursor. J. Virol. 67:6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70:8540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]