Abstract
Human papillomavirus type 1 (HPV1) E4 protein is associated with cytoplasmic and nuclear inclusions in productively infected keratinocytes. Here we have used transient expression of HPV1 E4 (also known as E1^E4) protein in keratinocytes to reproduce formation of E4 inclusions. Immunofluorescence analysis showed that progressive formation of inclusions correlated with diminished colocalization between E4 and keratin intermediate filaments (IFs). Our results support a model in which the HPV1 E4-keratin IF association is transient, occurring only at an early stage of inclusion formation. We also demonstrate that E4 induces relocation of the promyelocytic leukemia protein (PML) from multiple intranuclear speckles (ND10 bodies) to the periphery of nuclear E4 inclusions and that this activity is specific to full-length E4 protein. Analysis of HPV1-induced warts demonstrated that nuclear PML-E4 inclusions were present in productively infected keratinocytes, indicating that reorganization of PML occurs during the virus's replication cycle. It has been suggested that ND10 bodies are the sites for papillomavirus genome replication and virion assembly. Our finding that E4 induces reorganization of ND10 bodies in vitro and in vivo is further strong evidence that these domains play an important role in the papillomavirus life cycle. This study indicates that HPV1 is analogous to other DNA viruses that disrupt or reorganize ND10 domains, possibly to increase efficiency of virus infection. We hypothesize that HPV1 E4-induced reorganization of PML is necessary for efficient replication of the virus during the virus-producing phase.
Human papillomaviruses (HPVs) are double-stranded DNA viruses that induce benign or malignant tumors of both the skin and the mucosa. Despite differences in epithelial tropism and oncogenic potential, the life cycle of all HPV types (more than 80 types identified) is tightly coupled to the differentiation program of the infected epithelium. The virus infects cells of the proliferating basal layer, where the virus genome is established as a low-copy-number extrachromosomal plasmid and viral DNA replicates in synchrony with the host genome. Vegetative viral DNA replication initiates in infected cells that have moved up from the basal layer and begun to differentiate, and expression of structural proteins and assembly of new progeny occur in the uppermost and most differentiated regions of the epithelium (for a review see reference 37).
Abnormal cytological and histological features accompany HPV replication in epithelia (12, 26, 34). One feature that occurs in cutaneous warts is the presence of distinct inclusion bodies in the cytoplasm and nucleus of differentiating cells. The appearance and number of inclusion bodies present in infected cells vary between lesions induced by different HPV types. For instance, in HPV type 1 (HPV1)-induced warts the inclusions are small and numerous in cells of the parabasal layer and increase in size as the infected cell moves up toward the superficial layers, while in HPV4 infections, a single large fibrous inclusion is formed that almost fills the cytoplasm (12). Although the precise nature of these inclusion bodies is not known, HPV E4 proteins are associated with these structures (10, 14, 15, 49).
In HPV infections, E4 is the most abundant viral protein expressed and is derived from an E1^E4 spliced transcript initiated from a differentiation-inducible promoter that lies within the E7 open reading frame (11, 25, 30, 40, 42). Although no function has been assigned to this HPV protein, it is thought that E4 interacts with host cell structures and pathways that would otherwise inhibit efficient virion production and maturation in the differentiating keratinocyte (for a review of E4 see reference 44).
On the basis that transient expression of HPV16 E4 in epithelial cells induced the collapse of keratin intermediate filaments (IFs) (16, 46), it was proposed that E4 destroys the keratin matrix to compromise the strength of the keratinized squame in infected tissue and thereby promote efficient escape of the newly synthesized virions (16). However, expression of the HPV1 protein in epithelial cells did not collapse the keratin cytoskeleton even though the viral protein aligned along the keratin IFs (46). Neither was there disruption of the keratin matrix in cultured cells in which HPV1 E4 had formed in vivo-like inclusion bodies or in cells in naturally occurring lesions (49). The true nature of E4 inclusions and their role in E4 function therefore remain ones of conjecture. Here, we used transient expression of HPV1 E4 in human keratinocytes to reproduce the formation of in vivo-like cytoplasmic and nuclear E4 inclusions. We show that formation of E4 inclusions is associated with redistribution of the promyelocytic leukemia protein (PML), a component of ND10 bodies (also known as PML-oncogenic domains), which relocates from multiple intranuclear speckles to the periphery of nuclear E4 inclusions. We also show that nuclear PML-E4 inclusions are present in suprabasal keratinocytes of naturally occurring warts, indicating that reorganization of ND10 domains occurs during the replicative virus life cycle.
In human keratinocytes, HPV1 E4 protein accumulates into inclusion granules that are closely associated with E4-keratin filaments.
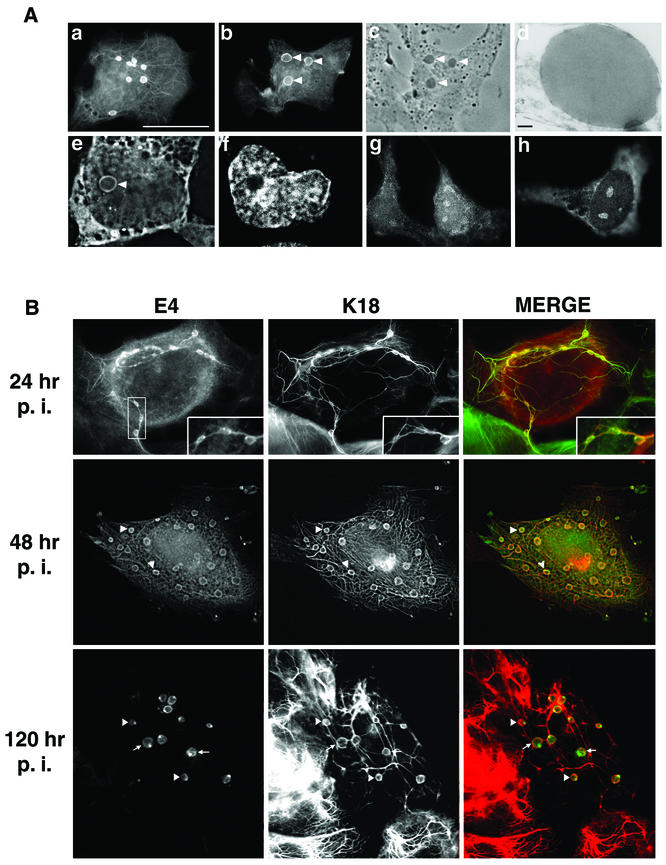
To recapitulate the formation of E4 inclusions in cultured cells, full-length HPV1 E4 protein (also known as E1^E4) was transiently expressed in a simian virus 40 (SV40)-immortalized human skin keratinocyte line, SVJD cells (46). HPV1 E1^E4 cDNA (46) was cloned into the BamHI site of the expression plasmid pcDNA3.0 (Invitrogen Life Technologies) to form pcDNA-1E4, and between 0.25 and 1 μg of DNA was transfected into 4 × 105 cells with PrimeFecta (Equibio, Ashford, United Kingdom) according to the manufacturer's instructions. In parallel experiments, SVJD cells were also infected with a recombinant adenovirus (Ad) expressing the HPV1 E1^E4 protein (Ad-1E4) constructed by methods described previously (7, 29, 51). After fixation with 4% paraformaldehyde for 5 min and permeabilization in cold (−20°C) acetone, pcDNA-1E4-transfected and Ad-1E4-infected cells were stained with the anti-HPV1 E4 monoclonal antibody (MAb) 4.37 (17) as previously described (46) and examined by immunofluorescence microscopy. In a majority (>80%) of pcDNA-1E4-transfected SVJD cells, HPV1 E4 localized to cytoplasmic filamentous networks (Fig. 1A, a), in agreement with our previous findings of E4 distribution in SVJD cells infected with a recombinant SV40-HPV1 E4 virus (46). However, in contrast to this previous study, multiple round E4 inclusion bodies were also visible in the cytoplasm and nuclei of up to 35% of cells (Fig. 1A, a, b, e, and f). Nuclear E4 was also associated with intranuclear foci that resembled nucleoli (Fig. 1A, g and h). These E4-stained foci were most notable in cells expressing low levels of cytoplasmic E4. HPV1 E4 expressed in Ad-1E4-infected SVJD cells also accumulated into inclusions, the size of these structures increasing at later times postinfection (p.i.) (Fig. 1B). Light and electron microscopic analysis showed the inclusions to be phase- and electron-dense structures (Fig. 1A, c and d, respectively), and with ultrastructures identical to those of the E4 inclusions detected in transfected VX2-R cells and in HPV1-induced warts (10, 49). In both pcDNA-1E4-transfected and Ad-1E4-infected cells, cytoplasmic E4 was not exclusively localized to filaments and/or inclusion granules but also formed a diffuse staining pattern that could be largely removed (or at least reduced) by decreasing the fixation time in 4% paraformaldehyde from 5 to 1 min (Fig. 1B and data not shown).
FIG. 1.
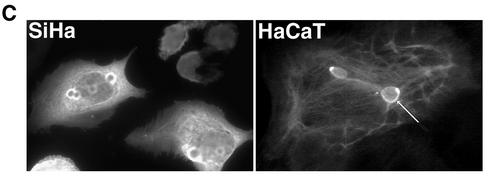
HPV1 E4 accumulates into in vivo-like inclusion bodies in human keratinocytes. (A) Localization of HPV1 E4 in pcDNA-1E4-transfected SVJD keratinocytes. At 48 h after transfection cells were fixed in 4% paraformaldehyde for 5 min prior to acetone permeabilization (10 min) and stained for E4 (MAb 4.37). Immune complexes were visualized with an anti-mouse Alexa Fluor 488 antibody. HPV1 E4 was associated with multiple inclusion bodies that were both phase dense (c, phase-contrast micrograph of cell shown in panel b; inclusions are indicated with arrowheads) and electron dense (d, electron micrograph showing one of the E4 inclusions formed in an Ad-1E4-infected SVJD cell; bar, 100 nm). Inclusions formed in the cytoplasm and nucleus. A deconvolved z section (0.3 μm) of a cell stained for E4 (e) and counterstained with nuclear stain 4′,6′-diamidino-2-phenylindole (Sigma Chemicals) (f) shows a large intranuclear E4 inclusion (arrowhead). In some cells, E4 was also localized to a cytoplasmic filamentous network (a) and small intranuclear foci that resemble nucleoli (g and h). Bar, 20 μm (6.5 μm in panels e and f). (B) Ad-1E4-infected SVJD keratinocytes at a multiplicity of infection of 25 to 50 were fixed at indicated times p.i. in 4% paraformaldehyde for 1 min prior to permeabilization with acetone. Cells were dual stained for E4 (MAb 4.37) and K18 (MAb CK5) and visualized with anti-mouse immunoglobulin G subclass-specific antibodies conjugated to Alexa 488 and Alexa 594 (Molecular Probes Inc.). Deconvolved z-section images are shown in gray and merged (E4 staining is green, K18 staining is red, and colocalization between red and green stains is shown as yellow). Insets in the top panels show enlargement of the marked area and highlight small E4 inclusions interconnected by E4-K18 filaments. In the middle and bottom panels, arrowheads indicate examples of inclusions that show colocalization between E4 and keratin at the periphery of the E4 inclusion. In the bottom panels arrows show E4 inclusions that are surrounded by keratin staining, but the two antigens do not appear to be colocalized. (C) Ad-1E4-infected SiHa and HaCaT keratinocytes (multiplicity of infection of 50) were fixed 48 and 120 h p.i., respectively. An arrow identifies one of the large inclusions formed in the HaCaT cell. HaCaT cells were a gift from N. Fusenig, University of Heidelberg.
To determine whether inclusion granule formation is a general feature of HPV1 E4 expression in keratinocyte lines, the spontaneously immortalized keratinocyte lines SCC 12F and HaCaT (9) and HPV DNA-positive cervical tumor keratinocytes (SiHa) were infected with Ad-1E4. HPV1 E4 accumulated into inclusion granules in all keratinocyte lines tested, although the number and size were variable between lines. In HaCaT cells for example, one or two very large cytoplasmic inclusions were associated with prominent E4 filaments, while in SiHa cells E4 filamentous staining was weak and many more inclusions (up to 10 in some cells) were detected that were most often located at the periphery of the nucleus (Fig. 1C).
Since we previously showed that HPV1 E4 colocalized to the keratin cytoskeleton in SV40-1E4-infected keratinocytes (46), cells were costained for E4 and keratin 18 (K18) (MAb CK5; Sigma Immunochemicals). Digital confocal analysis of these cells was performed by acquiring z-section images on a Zeiss Axioskop fluorescence microscope fitted with an Orca acquisition system and Openlab imaging system (Improvision, Coventry, United Kingdom). z images, acquired in different channels, were deconvolved by volume deconvolution prior to merger. The results showed coalignment between keratin and the E4 filaments as expected but also colocalization between keratin and E4 cytoplasmic inclusions (Fig. 1B). Colocalization between the two antigens was most notable at the periphery of the inclusions, where E4 and keratin staining was most intense (Fig. 1B; examples are indicated by arrowheads in middle and bottom panels). However, at later times p.i. some inclusions (indicated by arrows in Fig. 1B, bottom panels) do not show colocalization with keratin, even though keratin staining surrounds the inclusion granule. At early times in Ad-1E4 infection (24 h) the E4-keratin filaments interconnected the inclusions (Fig. 1B, top panel). However, as the infection proceeded, the inclusions increased in size and E4 localization to the keratin filaments diminished (Fig. 1B, middle panel) or was absent (Fig. 1B, bottom panel), even though the inclusions still appeared to be interconnected by the keratin filaments. There was no evidence that the cytoskeleton collapsed in the presence of HPV1 E4, supporting previous findings (46, 49).
From these experiments we conclude that localization of HPV1 E4 to keratin IFs may represent an early stage in the accumulation of E4 into inclusion structures. Inclusions appear to initialize at E4-IF complexes, but localization to the IFs is not maintained as inclusions increase in size (or mature). Keratins do, however, colocalize with E4 at the periphery of some of these more mature inclusions, although this does appear to diminish with increased maturity. These findings are in contrast to those of Rogel-Gaillard and colleagues, who found that, although keratin surrounded the HPV1 E4 inclusions, it did not colocalize with the inclusion (49). While this may reflect differences in the fixation of cells, we suggest that these inclusions represent more mature structures.
Efficiency of formation of HPV1 E4 inclusion bodies is reduced in fibroblasts.
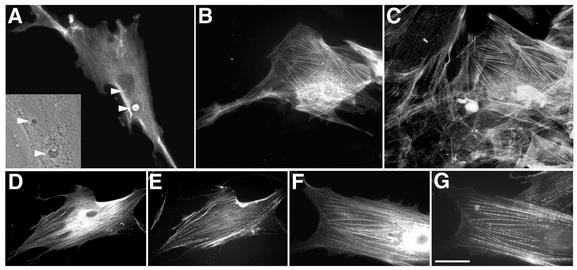
Since there is a close association between keratin IFs and E4 inclusion bodies, it is possible that an IF-E4 association is necessary in order for these structures to form. Therefore, to determine whether HPV1 E4 is able to form inclusions in cells that lack keratin IFs, human fibroblasts were infected with the recombinant Ad-1E4 virus and analyzed by immunofluorescence microscopy as described above for SVJD cells. At 48 h p.i., approximately 20% of E4-expressing fibroblasts contained E4 inclusion structures that appeared identical to those structures formed in keratinocytes (Fig. 2A). However, in contrast to the multiple inclusions formed in keratinocytes, in both fibroblast cell lines analyzed (WI-38 and a primary human fibroblast line) only a single small cytoplasmic inclusion and occasionally a single intranuclear inclusion were detected (Fig. 2A). These data indicate that the efficiency of E4 inclusion formation appears to be reduced compared to that for epithelial cells. While this may reflect differences in E4 expression between the cell types, it could indicate that an IF matrix contributes to efficient inclusion formation.
FIG. 2.
Subcellular distribution of HPV1 E4 in human fibroblasts. (A and B) Ad-1E4-infected human primary fibroblasts (multiplicity of infection of 50) were fixed 48 h p.i. Cells stained with an anti-HPV1 E4 MAb, 4.37, showed E4 localized to small cytoplasmic and intranuclear inclusions (indicated by arrowheads) that appeared identical to those formed in keratinocytes (inset, phase-contrast image of part of cell shown in panel A, which shows inclusions to be phase dense). E4 staining also formed a cytoplasmic network (B) that coaligned with actin stress fibers (C) . (D to G) Dual staining of cells infected with Ad-1E4Δ2-15 (D and E) or Ad-1E4Δ10-14 (F and G) showed that the mutant proteins associated with the actin cytoskeleton (D and F, anti-HPV1 MAb 4.37; E and G, phalloidin). Bar, 20 μm in panels A to E and 13 μm in panels F and G.
There was no evidence that E4 localized to vimentin IFs in fibroblasts, but in nearly 45% of E4-positive cells, E4 staining formed a cytoplasmic network that resembled the actin stress fiber network (Fig. 2B). This was confirmed by costaining the cells with Alexa 488-tagged phalloidin (Molecular Probes Inc.) that labels actin filaments (Fig. 2C). Cells infected with an Ad expressing β-galactosidase (a gift from Neil Blake, Institute for Cancer Studies, Birmingham, United Kingdom) did not show β-galactosidase localized to the actin filaments, indicating that this property is specific to the E4 protein (data not shown). We had previously shown that the leucine-rich motif (LLXLL) located near the N terminus of HPV1 E4 was important for localization of E4 to the keratin cytoskeleton (45). To determine whether this motif is also involved in mediating the interaction with the actin matrix, two recombinant Ads were generated that express HPV1 E4 mutant proteins that lack N-terminal amino acids (Δ2-15) or just the leucine cluster (Δ10-14) (45). Immunofluorescence analysis showed that both mutant proteins aligned with actin fibers (Fig. 2D to G), indicating distinct modes of interaction between E4 and the different cytoskeleton matrices.
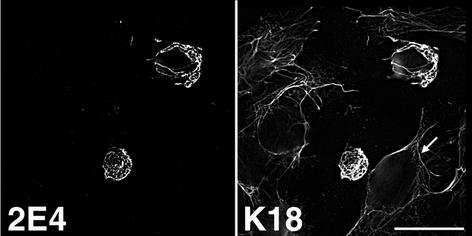
HPV2 E4 does not accumulate into inclusion structures in epithelial cells.
HPV2 is a major cause of skin warts, and HPV2 E4 is associated with multiple cytoplasmic granules in cells of the suprabasal epithelial layers (15). Therefore, to determine whether the in vivo intracellular distribution of HPV2 E4 could be recapitulated in cultured cells, a DNA fragment containing the HPV2 E1^E4 cDNA (6) was cloned into the BamHI site of pcDNA3.0 to form pcDNA-2E4 and used to transfect SVJD cells. Cells were fixed and dual stained with an anti-HPV2 E4 MAb, 2C05 or 1F10 (6), and the K18 MAb CK5. At 24 h posttransfection, HPV2 E4 was aligned with the keratin cytoskeleton and the E4-keratin IFs collapsed to form a single fibrous body (Fig. 3 and data not shown). There was no evidence that HPV2 E4 formed multiple inclusion granules resembling those formed in naturally occurring HPV2-induced warts, even at later times posttransfection (72 h). It is possible, therefore, that HPV1 and HPV2 E4 proteins assemble into inclusion structures by different mechanisms. The collapse of keratin IFs induced by HPV2 E4 indicates that this cutaneous E4 behaves more like the E4 protein of HPV16 that infects mucosal epithelia than like the cutaneous HPV1 protein (16, 46). We suggest that this behavior can be explained by the fact that HPV2 E4 has greater sequence homology with mucosal E4 proteins and notably contains a mucosal E4-specific, C-terminal motif (VXV/LXLH/RL, where X is any amino acid) that has been shown to be necessary for the collapse of HPV16 E4-IFs in epithelial cells (45, 47, 53). The physiological significance of HPV2 and HPV16 E4's rearrangement of the keratin cytoskeleton is unclear. There is no evidence, as yet, that indicates that this function occurs in vivo, and it might reflect an anomaly of E4 overexpression in cultured epithelial cells that do not recapitulate the phenotype of the differentiating keratinocyte (44).
FIG. 3.
HPV2 E4 colocalizes to keratin IFs and induces collapse of the keratin matrix into a fibrous bundle. SVJD cells were transfected with pcDNA-2E4, fixed 48 h p.i., and dual stained with anti-HPV2 E4 MAb IF10 and K18 MAb. Note that the keratin cytoskeleton in neighboring E4-negative cells remains intact; one is shown by the arrow. Bar, 20 μm.
PML is associated with nuclear HPV1 E4 inclusion bodies.
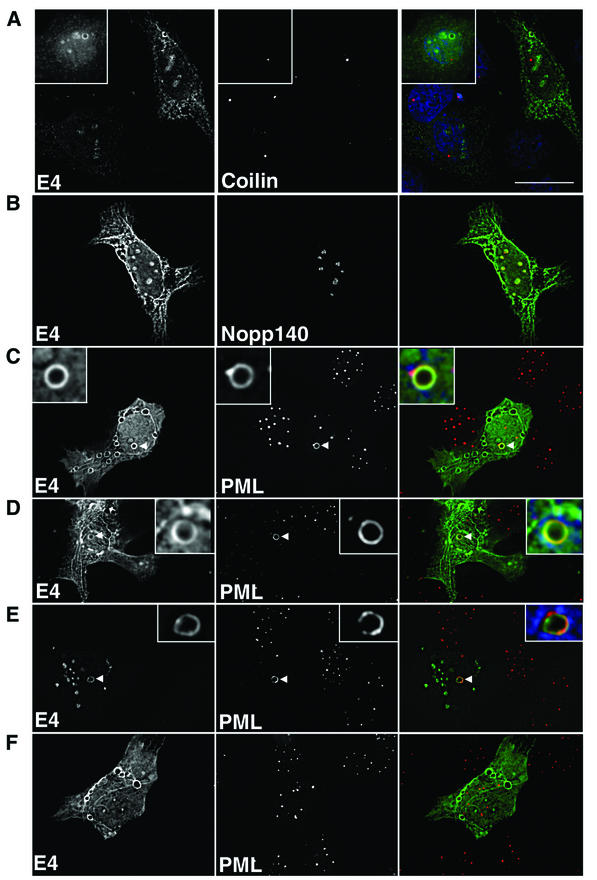
While cytoplasmic HPV1 E4 inclusions are closely associated with keratin IFs in cultured epithelial cells and in warts, nuclear E4 inclusions are devoid of such structures (12, 49). To characterize the subnuclear topology of E4 inclusions, pcDNA-1E4-transfected SVJD cells were costained with antibodies to HPV1 E4 and components of various nuclear structures: coiled bodies, nucleoli, and ND10 bodies. The results of digital confocal analysis of these cells are shown in Fig. 4. Coiled bodies were visualized by using a MAb to coilin (3) (a gift from Maria Carmo-Fonseca, University of Lisbon) and appeared as several bright spots in SVJD cells. Nuclear E4 was not associated with coiled bodies, nor was the number of coiled bodies in these cells different from those in untransfected cells (Fig. 4A). Antibodies specific to nucleolar components Nopp140 (a gift from Thomas Meier, Yeshiva University) and C23 (MAb MS-3; Santa Cruz Biotechnology) costained the small E4-stained foci (Fig. 4B and data not shown), confirming that, in a fraction of E4-expressing cells, the viral protein localized to the nucleolus. ND10 bodies were identified by using a rabbit antibody to PML (Medical and Biological Laboratories, Nagoya, Japan), the defining component of these nuclear domains. In mock-transfected cells, PML staining appeared as multiple bright speckles located throughout the nucleus (Fig. 4G and H). However, in cells containing nuclear E4 inclusions the number of PML speckles was markedly reduced and the majority of PML localized to the periphery of the nuclear E4 inclusion (Fig. 4C and D). In fact, between 5 and 8.5% of E4-positive cells contained nuclear inclusions, and in all these cells, PML was relocated to the E4 inclusion. The redistribution of PML was not an artifact of the fixation treatment, as a similar PML staining pattern formed in cells treated for the shorter fixation time (Fig. 4E). PML-E4 inclusions were also recognized by a different antibody to PML (MAb PG-M3; Santa Cruz Biotechnology) and to E4 (MAb 9.95) (Fig. 4E and data not shown). In the majority of cells showing abnormal PML distribution, PML staining surrounded the E4 inclusion (Fig. 4C and D) but was also observed to accumulate predominantly to one side of the granule, sometimes forming a bright spot at the edge (Fig. 4C and E). PML and E4 were colocalized, but sometimes only partially, at the periphery of inclusions, suggesting a close association between these two molecules (Fig. 4C to E). Redistribution of PML was dependent on the formation of intranuclear E4 inclusions, because PML had a normal distribution in cells containing only cytoplasmic E4 inclusion bodies (Fig. 4F).
FIG. 4.
Subnuclear topology of HPV1 E4 in SVJD keratinocytes. pcDNA-1E4-transfected SVJD cells were fixed at 48 h in 4% paraformaldehyde for 5 min (A to D and F) or 1 min (E) and permeabilized in acetone. Cells were dual stained for E4 (MAb 4.37) and cellular factors of coilin bodies (A, coilin), nucleoli (B, Nopp140), and ND10 bodies (C to F; PML, rabbit anti-PML antibody). Deconvolved z sections are shown as individual images (gray) and merged (right panels). In merged images, E4 is green; coilin, Nopp140, and PML are red; and where shown 4′,6′-diamidino-2-phenylindole staining of nuclei is blue. Yellow in merged images indicates colocalization between green and red colors. (C to E) Note that in cells containing E4 inclusion bodies PML was reorganized to the periphery of the intranuclear E4 inclusion. The insets in panels C to E are enlargements of the inclusion bodies indicated by arrowheads. PML was partly colocalized with E4. (E) PML-E4 inclusions were also observed in cells that had been fixed differently and stained with a different anti-PML antibody (MAb PG-M3). (F) Note that reorganization of PML did not occur in cells that contain only cytoplasmic E4 inclusions. (G and H) Mock-transfected cells dual stained with 4.37 (G) or 9.95 (H) and PML. Bar, 20 μm (insets, 5.5 μm).
Redistribution of PML is dependent on expression of the full-length E4 protein.
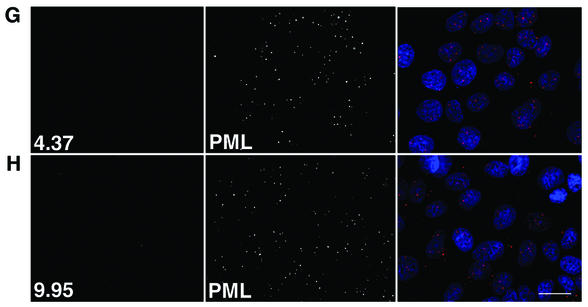
During the HPV1 replication cycle, the E4 protein is subjected to a series of posttranslational modifications, including proteolytic processing. The full-length E4 protein (17K) is cleaved to form several smaller species (16K and 10/11K) (10, 14). These share C-terminal sequences with the full-length protein but vary in the extent of N-terminal sequence retained (17, 45). 16K E4 lacks only the first 15 amino acids, while almost half the protein (58 amino acids) has been cleaved off to form the 11K species (45, 48). These modifications of 17K E4 may result in alteration of E4 function, as the processed polypeptides lack sequences that are important for E4's association with the keratin cytoskeleton (45). Since the N terminus of E4 may have an important role in E4 function, we next examined whether the expression of these modified forms of HPV1 E4 also affected the intranuclear distribution of PML. HPV1 E4 N-terminal deletion mutants Δ2-15 and Δ2-58 have been described in previous studies (4, 45) and closely resemble the modified E4 proteins 16K and 11K, respectively, found in HPV1-induced warts (45). The mutant cDNAs were cloned into pcDNA3.0 to form pcDNA-1Δ2-15 and pcDNA-1Δ2-58. Following transfection into SVJD keratinocytes, cells were costained for the E4 proteins and PML and examined by immunofluorescence microscopy. Both mutant proteins localized primarily to the nucleus (Fig. 5) in agreement with our previous findings (45). Interestingly, the 16K protein located to the nucleolus (Fig. 5, top panel, and data not shown), while the 11K protein was excluded from this subnuclear structure (Fig. 5, bottom panel, and data not shown), indicating that sequences between amino acids 16 and 59 are involved in nucleolar localization of E4. Neither protein, however, was observed to accumulate into inclusions, and PML remained localized to multiple speckles throughout the nucleus, similar to neighboring nontransfected cells (Fig. 5). We noted that the number of ND10 bodies was low (five bodies or fewer) in 42% of 16K-expressing cells (Fig. 5, middle panel). However, a similar number of non-16K-expressing SVJD cells also contained a low number of ND10 bodies, indicating that this may not be significant.
FIG. 5.
HPV1 E4 proteins that lack N-terminal sequences do not induce reorganization of PML in keratinocytes. SVJD keratinocytes were transfected for 48 h with pcDNA-1E4Δ2-15 (top and middle panels) and pcDNA-1E4Δ2-58 (bottom panels). These plasmids express proteins that closely resemble the truncated E4 proteins, 16K and 11K, expressed in HPV1-induced warts (45). Cells were dual stained for E4 (MAb 4.37, top and middle panels; p1p7 antibody, bottom panels) and PML (rabbit anti-PML antibody). Note that the Δ2-58 protein lacks epitopes of anti-HPV1 E4 MAbs and therefore was recognized with a rat polyclonal antibody, p1p7 (18). In merged images, E4 is green and PML is red. Truncated proteins do not assemble into inclusions in SVJD cells, and PML is localized to multiple speckles, a similar distribution as that of PML in adjacent nontransfected cells. However, cells expressing high levels of 16K protein often contain a low number of PML-stained speckles (a typical example is shown in the middle panels). Note that 16K, but not 11K, E4 is localized to nucleoli (compare top with bottom panels). The inset in the bottom panel shows a 16K-expressing cell stained with p1p7 to show that this antibody is able to recognize nucleolar E4. Bar, 20 μm (inset, 25 μm).
PML is associated with intranuclear E4 inclusion granules in HPV1-induced skin warts.
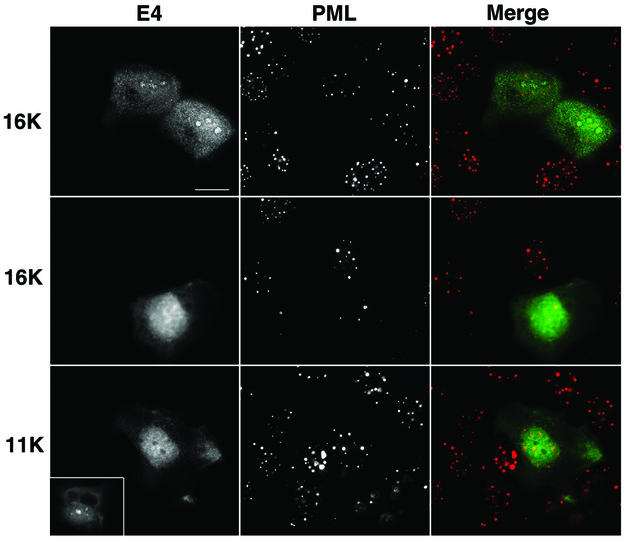
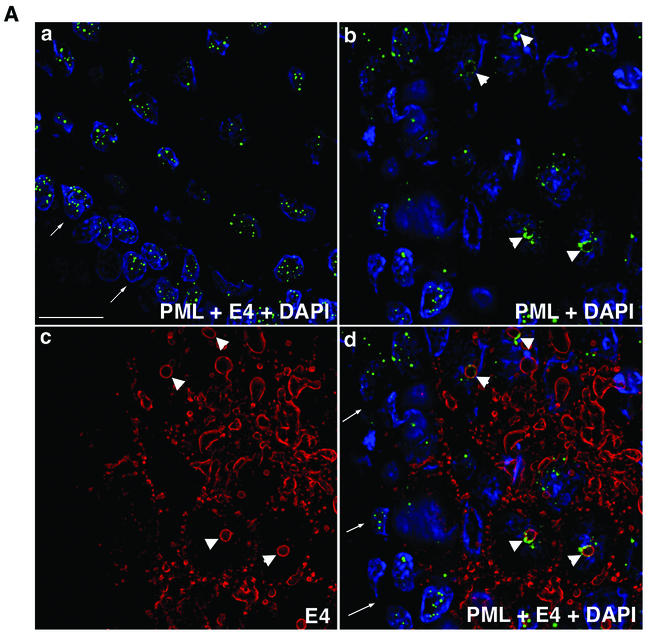
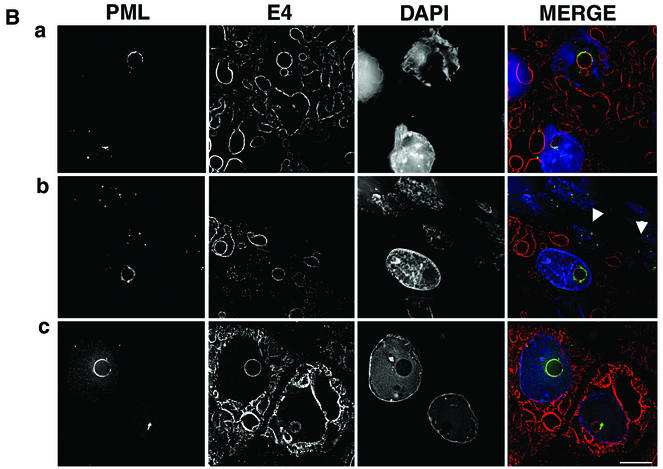
The results described above indicate that expression of full-length HPV1 E4 protein induces the relocation of PML to the periphery of intranuclear E4 inclusion bodies formed in monolayer cultures of human keratinocytes. To determine whether a similar redistribution of PML occurs during the natural productive cycle of the virus, we examined PML and E4 localization in HPV1-induced skin warts. Frozen tissue sections (4 μm) of three HPV1-induced skin warts (collected locally and also kindly provided by Thomas Iftner, University of Tuebingen, Tuebingen, Germany) were fixed in 4% paraformaldehyde for 8 min and permeabilized for 10 min in 0.2% Triton X-100. Sections were dual stained with anti-HPV1 E4 MAbs and the anti-PML rabbit antibody, and the distribution of the two antigens was examined by digital confocal microscopy (Fig. 6). E4 localized to multiple inclusions in cells immediately above the basal cell layer and persisted up through the epithelial layers, as previously reported (10, 14). In E4-positive cells, PML relocated to E4-stained intranuclear inclusion bodies (Fig. 6A, b, c, and d; examples are indicated by arrowheads). In the underlying basal cells that are negative for E4 expression PML staining was normal, forming multiple speckles (Fig. 6A, d). The sequestration of PML to E4 inclusions was specific to productively infected cells because, in regions of the wart tissue that are E4 negative, PML is localized to multiple intranuclear speckles in both basal and differentiated cells (Fig. 6A, a). The redistribution of PML occurred in E4-positive cells immediately above the basal layer (Fig. 6B, b), indicating that this is an early event in the productive phase of HPV1 replication. In the transfection experiments (Fig. 4 and 5) only the full-length E4 protein (17K) induced sequestration of PML to nuclear inclusions. To confirm that the PML-E4 inclusions formed in the warts also contained 17K protein, sections were stained with an anti-E4 MAb (1D11) that recognizes the leucine-rich epitope present at the N terminus of the protein and which is lost during processing of the protein. Thus, this antibody detects the full-length HPV1 E4 protein but not the processed forms (S. Roberts, unpublished data). The 1D11 MAb recognized the PML-E4 inclusions, confirming that these structures contained full-length E4 protein (Fig. 6B, b and c). Like PML localization at E4 inclusions formed in pcDNA-1E4-transfected SVJD keratinocytes, in HPV1-infected cells, PML was partially colocalized with E4 at the periphery of the inclusion (Fig. 6B). In some cases, PML nearly surrounded the E4 structures (Fig. 6B, a and c), but in others it accumulated predominantly to one side (Fig. 6B, b).
FIG. 6.
PML is redistributed to intranuclear E4 inclusions in HPV1-induced warts. (A) Tissue sections of wart were dual stained for E4 (MAb 9.95; E4 staining is now shown in red) and PML (rabbit antibody, green), and nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue). (a) Merged deconvolved z sections of area of wart negative for E4 staining. Note that PML was localized to multiple intranuclear speckles in basal cells (arrows indicate basal layer) and suprabasal cells. (b to d) PML distribution in area of wart expressing E4. Deconvolved z-section images are shown as PML and DAPI merged (b); E4 alone (c); and E4, PML, and DAPI merged (d). In suprabasal cells, PML was reorganized from multiple speckles to ring structures (b) that are localized to intranuclear E4 inclusions (d; arrows indicate basal cell layer). Arrowheads indicate PML-E4 inclusions. Bar, 20 μm. (B) Reorganization of PML to E4 inclusions was identical to that observed for E4-expressing cultured keratinocytes. Deconvolved z sections are shown as individual images (gray) and merged (E4, red; PML, green; DAPI, blue). PML either nearly completely surrounds the E4 inclusion (a and c) or accumulates predominantly to one side (b). PML shows partial colocalization with E4 (depicted as yellow color in merged images). Note that in panel b the E4-positive cell immediately above the basal layer (indicated by arrowheads) contains a PML-E4 inclusion and that PML distribution is normal in basal cells. Note that the wart section in panel a is stained with MAb 9.95 and that wart sections in panels b and c are stained with anti-E4 MAb 1D11. MAb 1D11 recognizes only full-length E4 (E1^E4) protein. Bar, 10 μm.
To conclude, our study shows for the first time that full-length HPV1 E4 induces reorganization of PML, an essential component of ND10 domains, to intranuclear E4 inclusion bodies (Fig. 4 and 5) and that sequestration of PML to E4 structures occurs during the productive cycle of the virus (Fig. 6). The function of ND10 bodies is an enigma (for reviews of ND10 function, see references 8 and 36), but they have been connected to the replication strategies of several DNA and RNA viruses (reviewed in references 21 and 43) and have also been linked to the replicative cycle of PVs (13, 24, 27, 28, 41, 50). It has been suggested previously that, in parallel with other DNA viruses (31), these ND10 domains may be sites of HPV genome replication (50). In a transient replication assay, replication centers containing the HPV11 viral replication proteins E1 and E2 and replication origin-containing DNA localized (partially and completely) to ND10 domains (50). Late stages of the PV life cycle could also be linked to ND10 domains. The minor capsid protein L2 of both bovine PV1 (BPV1) and HPV16 is able to recruit E2 to ND10 bodies (13, 28), and BPV1 L2 also directed the other coat protein L1 to ND10 bodies (13). Therefore, L2 protein may mediate the assembly and packaging of newly synthesized virions at ND10 bodies by inducing E2-virus genome complexes and the major capsid protein to accumulate at these subnuclear domains (13, 41). Although these interactions did not result in ND10 body disruption, HPV33 L2 induced the release of Sp100 from these domains, and a concomitant recruitment of the transcriptional repressor Daxx and such changes to ND10 composition could facilitate packaging of the viral genome (24). Our finding that PML is associated with E4-containing nuclear structures in keratinocytes is further strong evidence that ND10 bodies play an important role in the replicative cycle of these small DNA viruses. Since PML is the defining component of ND10 bodies and is essential for their proper formation and integrity (32), it is reasonable to assume that E4 disrupts the normal nuclear organization of these cellular bodies. In this respect, E4 is analogous to herpes simplex virus type 1 ICP0 (22, 23), Epstein-Barr virus (EBV) BLZF-1 (1), cytomegalovirus IE1 (2, 33, 39), and Ad5 E4orf3 (20, 35), viral early proteins that induce ND10 body disruption and/or reorganization.
Redistribution of PML to nuclear E4 inclusions occurred in suprabasal keratinocytes of naturally occurring lesions, as early as the parabasal cells, but there was no obvious disruption of ND10 bodies in the underlying basal cells (Fig. 6). Initiation of E4 expression has been shown to correlate with the onset of vegetative genome replication in naturally occurring lesions including those induced by HPV1 (10, 18). We suggest, therefore, that E4-induced reorganization of PML represents a switch in the HPV life cycle, from the nonproductive maintenance stage of replication in the basal cells to the virus-producing productive phase in differentiating cells. Reorganization of the ND10 domains may be necessary only when HPV changes its replication strategy. Interestingly, such a scenario does have parallels with the replication strategies of some lytic viruses (5, 21, 52). For example, EBV does not appear to require an association with ND10 bodies during latent infections; however, upon induction of lytic replication ND10 components are dispersed sequentially, and EBV replication compartments develop at PML-containing remnants (5). The effect of E4-induced redistribution of PML on the replicative life cycle of PVs is the subject of further investigation. However, overexpression of PML has been linked elsewhere to induction of both G1 arrest of the cell cycle and apoptosis (38), and both effects are at odds with efficient virus production in suprabasal keratinocytes. Furthermore, there is evidence that ND10 bodies have a role in interferon's antiviral functions, and PML may have an important role in this function (reviewed in reference 43). It is, therefore, tempting to speculate that E4-induced PML sequestration relieves repressive activities of PML and ND10 bodies that would otherwise inhibit or limit the productive phase of the HPV life cycle in the differentiating keratinocytes.
PML's redistribution was directly linked to the formation of nuclear E4 inclusions, since no significant change in PML location was apparent in cells containing only cytoplasmic inclusions (Fig. 4). While this may indicate that cytoplasmic inclusions have a different function from that of the nuclear structures, it does not rule out the possibility that this may be related to modification of ND10 domains. Whatever the function of these cytoplasmic structures, our data imply that they form in close association with IFs but that this association is diminished or lost as inclusions increase in size (Fig. 1). Our results support a model in which the HPV1 E4-keratin IF association is transient, occurring only at an early stage of inclusion formation. Maturation of the inclusions correlates with loss of E4 localization to keratins (Fig. 1). In natural infections, therefore, only a small population of E4 molecules may associate with the keratin IFs for a short time in the infection process. The transient nature of the E4-IF interaction may explain the lack of detection of a significant E4-keratin interaction in HPV1 warts (19, 49).
The reorganization of ND10 domains is a novel function for E4, and analysis of the molecular interactions between the E4 inclusions and ND10 components should shed new light on the role of this protein in the PV life cycle. Given the emerging link between these cellular structures and viral genome replication and encapsidation, we hypothesize that E4-induced reorganization of ND10 bodies is necessary for efficient replication of the virus during the virus-producing phase.
Acknowledgments
This work was supported by a program grant from Cancer Research UK.
We thank Susan Rookes and Darren Parkin for excellent technical assistance and Lesley Tompkins and Peter Whittle (Life Sciences Electron Microscopy Centre, University of Birmingham) for invaluable assistance with electron microscopy. We also acknowledge the continued cooperation and support by local dermatologists and chiropodists without whom this research would not be possible.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida, F., R. Saffrich, W. Ansorge, and M. Carmo-Fonseca. 1998. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142:899-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashmole, I., P. H. Gallimore, and S. Roberts. 1998. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1^E4 protein necessary for E4 oligomerisation in vivo. Virology 240:221-231. [DOI] [PubMed] [Google Scholar]
- 5.Bell, P., P. M. Lieberman, and G. G. Maul. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800-11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthoud, T. J. 1997. Ph.D. thesis. University of Birmingham, Birmingham, United Kingdom.
- 7.Blake, N., S. Lee, I. Redchenko, W. Thomas, N. Steven, A. Leese, P. Steigerwald-Mullen, M. G. Kurilla, L. Frappier, and A. Rickinson. 1997. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 7:791-802. [DOI] [PubMed] [Google Scholar]
- 8.Borden, K. L. B. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markam, and N. E. Fusenig. 1988. Normal keratinisation in a spontaneously immortalized human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitburd, F., O. Croissant, and G. Orth. 1987. Expression of human papillomavirus type-1 E4 gene products in warts. Cancer Cells (Cold Spring Harbor) 5:115-122. [Google Scholar]
- 11.Chow, L. T., S. S. Reilly, T. R. Broker, and L. B. Taichman. 1987. Identification and mapping of human papillomavirus type 1 RNA transcripts recovered from plantar warts and infected epithelial cell cultures. J. Virol. 61:2581-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croissant, O., F. Breitburd, and G. Orth. 1985. Specificity of cytopathic effect of cutaneous human papillomaviruses. Clin. Dermatol. 3:43-55. [DOI] [PubMed] [Google Scholar]
- 13.Day, P. M., R. B. S. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorbar, J., D. Campbell, R. J. A. Grand, and P. H. Gallimore. 1986. Identification of the human papillomavirus-1a E4 gene products. EMBO J. 5:355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doorbar, J., I. Coneron, and P. H. Gallimore. 1989. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology 172:51-62. [DOI] [PubMed] [Google Scholar]
- 16.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 17.Doorbar, J., H. S. Evans, I. Coneron, L. V. Crawford, and P. H. Gallimore. 1988. Analysis of HPV1 E4 gene expression using epitope-defined antibodies. EMBO J. 7:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 19.Doorbar, J., E. Medcalf, and S. Napthine. 1996. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology 218:114-126. [DOI] [PubMed] [Google Scholar]
- 20.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:197-207. [DOI] [PubMed] [Google Scholar]
- 21.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 22.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110 and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO. J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Florin, L., F. Schafer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein L2. Virology 295:97-107. [DOI] [PubMed] [Google Scholar]
- 25.Grassman, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 70:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross, G., H. Pfister, M. Hagedorn, and L. Gissman. 1982. Correlation between human papillomavirus (HPV) type and histology of warts. J. Investig. Dermatol. 78:160-164. [DOI] [PubMed] [Google Scholar]
- 27.Guccione, E., P. Massimi, A. Bernat, and L. Banks. 2002. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology 293:20-25. [DOI] [PubMed] [Google Scholar]
- 28.Heino, P., J. Zhou, and P. F. Lambert. 2000. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304-314. [DOI] [PubMed] [Google Scholar]
- 29.Hitch, A. J. L. 1999. Ph.D. thesis. University of Birmingham, Birmingham, United Kingdom.
- 30.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daax to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 34.Koss, L. G., and G. R. Durfee. 1956. Unusual patterns of squamous epithelium of the uterine cervix; cytologic and pathologic study of koilocytotic atypia. Ann. N. Y. Acad. Sci. 63:1245-1261. [DOI] [PubMed] [Google Scholar]
- 35.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 36.Maul, G. G., E. Yu, A. M. Ishov, and A. L. Epstein. 2000. Review: properties and assembly mechanisms of ND10, PML, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 37.McMurray, H. R., D. Nguyen, T. F. Westbrook, and D. J. McCance. 2001. Biology of human papillomaviruses. Int. J. Exp. Pathol. 82:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenicity of acute promyelocytic leukaemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 39.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasseri, M., R. Hirochika, T. R. Broker, and L. T. Chow. 1987. A human papilloma virus type 11 transcript encoding an E1^E4 protein. Virology 159:433-439. [DOI] [PubMed] [Google Scholar]
- 41.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. S. Roden. 2001. L1 interaction domains of papillomavirus L2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palermo-Dilts, D. A., T. R. Broker, and L. T. Chow. 1990. Human papillomavirus type 1 produces redundant as well as polycistronic mRNAs in plantar warts. J. Virol. 64:3144-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, S. 2002. Biology of the E4 protein, p. 119-142. In D. J. McCance (ed.), Human papillomaviruses, in press. Elsevier Science B.V., Amsterdam, The Netherlands.
- 45.Roberts, S., I. Ashmole, L. J. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 68:6432-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, S., I. Ashmole, S. R. Rookes, and P. H. Gallimore. 1997. Mutational analysis of the human papillomavirus type 16 E1^E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J. Virol. 71:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, S., I. Ashmole, T. M. T. Sheehan, A. H. Davies, and P. H. Gallimore. 1994. Human papillomavirus type 1 E4 protein is a zinc-binding protein. Virology 202:865-874. [DOI] [PubMed] [Google Scholar]
- 49.Rogel-Gaillard, C., G. Pehau-Arnaudet, F. Breitburd, and G. Orth. 1993. Cytopathic effect in human papillomavirus type 1-induced inclusion warts: in vitro analysis of the contribution of two forms of the viral E4 protein. J. Investig. Dermatol. 101:843-851. [DOI] [PubMed] [Google Scholar]
- 50.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson, G. W. G., and A. Akrigg. 1992. Constitutive and enhanced expression from the CMV major IE promoter in a defective adenovirus vector. Nucleic Acids Res. 20:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, Q.-L., T. F. Smith, E. J. Lefkowitz, L. T. Chow, and T. R. Broker. 1994. Nucleic acid and protein sequence alignments of human and animal papillomaviruses constrained by functional sites. The University of Alabama at Birmingham Press, Birmingham.