Abstract
Citrus tristeza virus (CTV) produces more than thirty 3′- or 5′-terminal subgenomic RNAs (sgRNAs) that accumulate to various extents during replication in protoplasts and plants. Among the most unusual species are two abundant populations of small 5′-terminal sgRNAs of approximately 800 nucleotides (nt) termed low-molecular-weight tristeza (LMT1 and LMT2) RNAs. Remarkably, CTV replicons with all 10 3′ genes deleted produce only the larger LMT1 RNAs. These 5′-terminal positive-sense sgRNAs do not have corresponding negative strands and were hypothesized to be produced by premature termination during plus-strand genomic RNA synthesis. We characterized a cis-acting element that controls the production of the LMT1 RNAs. Since manipulation of this cis-acting element in its native position (the L-ProI region of replicase) was not possible because the mutations negatively affect replication, a region (5′TR) surrounding the putative termination sites (nt ∼550 to 1000) was duplicated in the 3′ end of a CTV replicon to allow characterization. The duplicated sequence continued to produce a 5′-terminal plus-strand sgRNA, here much larger (∼11 kb), apparently by termination. Surprisingly, a new 3′-terminal sgRNA was observed from the duplicated 5′TR. A large 3′-terminal sgRNA resulting from the putative promoter activity of the native 5′TR was not observed, possibly because of the down-regulation of a promoter ∼19 kb from the 3′ terminus. However, we were able to observe a sgRNA produced from the native 5′TR of a small defective RNA, which placed the native 5′TR closer to the 3′ terminus, demonstrating sgRNA promoter activity of the native 5′TR. Deletion mutagenesis mapped the promoter and the terminator activities of the 5′TR (in the 3′ position in the CTV replicon) to a 57-nt region, which was folded by the MFOLD computer program into two stem-loops. Mutations in the putative stem-loop structures equally reduced or prevented production of both the 3′- and 5′-terminal sgRNAs. These mutations, when introduced in frame in the native 5′TR, similarly abolished the synthesis of the LMT1 RNAs and presumably the large 3′-terminal sgRNA while having no impact on replication, demonstrating that neither 5′- nor 3′-terminal sgRNA is necessary for replication of the replicon or full-length CTV in protoplasts. Differences between the 5′TR, which produced two plus-strand sgRNAs, and the cis-acting elements controlling the 3′ open reading frames, which produced additional minus-strand sgRNAs corresponding to the 3′-terminal mRNAs, suggest that the different sgRNA controller elements had different origins in the modular evolution of closteroviruses.
Citrus tristeza virus (CTV), a member of the genus Closterovirus of the family Closteroviridae, consists of a 19.3-kb single-stranded RNA genome of positive polarity organized into 12 open reading frames (ORFs) (9, 15). The virion RNA serves as a messenger to produce two polyproteins: one contains two papain-like proteases (L-Pro) plus methyltransferase-like (MTR) and helicase-like (HEL) domains, and the larger polyprotein has an additional RNA-dependent RNA polymerase-like domain that is thought to result from a +1 frameshift (9). The remaining 10 ORFs are expressed through a nested set of 3′-coterminal subgenomic RNAs (sgRNAs) (6). Only the 5′ ORFs 1a and 1b are required for replication in protoplasts (19), while the early-expressed 3′-most gene (p23) (14) is required for asymmetrical accumulation of plus and minus strands of both genomic and sgRNAs (20).
In infected cells, CTV produces an unusually large diversity of RNAs. In addition to the expected genomic RNA and its complementary copy, each 3′ ORF controller element produced three different RNAs: a 3′-terminal plus-strand RNA to serve as messenger for the adjacent ORF and a relatively large amount of a corresponding minus-strand sgRNA, plus a 5′-terminal plus-strand sgRNA, apparently produced by termination near the controller element during genomic RNA synthesis (4). Considering the 9 or 10 3′ controller elements (only 9 3′-terminal sgRNAs are visible from Northern blot hybridizations), this sums to 29 to 32 RNA species.
In addition, among the most abundant RNAs produced by CTV are two small 5′-coterminal plus-strand sgRNAs of ∼800 nucleotides (nt) (2, 12). These RNAs, designated low-molecular-weight tristeza (LMT) RNAs (12), are populations with heterogenous 3′ termini at nt 842, 854 (LMT1) and nt 744 to 746 (LMT2) for the VT strain of CTV (2). Since there were no corresponding minus-strand sgRNAs complementary to the LMT RNAs, it was proposed that the latter were produced by premature termination during genomic RNA synthesis (2, 12). All of the several different strains of wild-type CTV examined in protoplasts produced both populations of LMT RNAs, yet the CTV replicon with all of the 3′ ORFs deleted (19) produced only the larger LMT1 RNAs (2). Similar 5′-terminal plus-strand sgRNAs have been observed for other members of the Closteroviridae: a similarly sized RNA for Beet yellows virus (BYV), a related Closterovirus (5), and a larger RNA for Lettuce infectious yellows virus, a Crinivirus (18).
In the present study, we characterized a cis-acting element (the 5′ termination region [5′TR]) in the area corresponding to the 3′ termini of the LMT1 RNAs that effects the production of these sgRNAs. The sequences surrounding the putative LMT1 termination region were located within the replicase polypeptide coding region and could not be extensively manipulated without interfering with virus replication. We overcame this limitation by duplicating this region into the 3′ end of the genome of the CTV replicon, where the cis-acting element continued to function, producing a much longer 5′-terminal plus-strand RNA. Unexpectedly, this cis-acting element functioned as a strong sgRNA promoter when positioned near the 3′ terminus, inducing the accumulation of high levels of a new 3′-terminal plus-strand sgRNA. Mapping and characterization of the duplicated terminator/promoter element allowed precise mutagenesis of the 5′TR at its native location without affecting the replicase function. Remarkably, mutation of the native cis-acting element, such that neither the 5′-terminal nor the 3′-terminal sgRNAs were produced, failed to detectably reduce the replication of CTV in protoplasts.
MATERIALS AND METHODS
Plasmid construction.
The full-length cDNA clone of CTV, pCTV9, and CTV replicons, pCTV-ΔCla and pCTV-ΔCla333 (4, 19), were used as parent plasmids for DNA amplification and generation of mutants used in this study (Table 1). The nucleotide numbering and the sequence of the oligonucleotides designed were based on the sequence of the T36 isolate of CTV (9) (GenBank accession no. NC_001661). The mutants generated in pCTV-ΔCla333 were always ligated between unique XhoI and StuI restriction sites (see Fig. 1A).
TABLE 1.
Mutants and their parent plasmids used in this studya
| Mutant | Description of mutation |
|---|---|
| pCTV-ΔCla333 | Parent plasmid, replicon with no sgRNA (4) (Fig. 1) |
| pCTV:524/956 | nt 524-956 inserted into pCTV-ΔCla333 |
| pCTV:524/1164 | nt 524-1164 inserted into pCTV-ΔCla333 |
| pCTV:1497/2124 | nt 524-2124 inserted into pCTV-ΔCla333 |
| pCTV:635/956 | nt 635-956 inserted into pCTV-ΔCla333 |
| pCTV:705/956 | nt 705-956 inserted into pCTV-ΔCla333 |
| pCTV:524/789 | nt 524-789 inserted into pCTV-ΔCla333 |
| pCTV:524/775 | nt 524-775 inserted into pCTV-ΔCla333 |
| pCTV:524/705 | nt 524-705 inserted into pCTV-ΔCla333 |
| pCTV:789/956 | nt 789-956 inserted into pCTV-ΔCla333 |
| pCTV:719/775 | nt 719-775 inserted into pCTV-ΔCla333 |
| pCTV:524/956 | Parent plasmid, mutant pCTV:524/956 above (Fig. 1) |
| pCTV:524/956 M1 | Substitution in SL2 sequence |
| pCTV:524/956 M2 | Deletion of loop of SL2 sequence |
| pCTV:524/956 M3 | Deletion of loop of SL1 sequence |
| pCTV:524/956 M4 | Substitution of loop of SL1 sequence |
| pCTV:524/956 M5 | Substitution of loop of SL1 sequence |
| pCTV:524/956 M6 | Substitution of stem of SL1 sequence |
| pCTV:524/956 M7 | Substitution of stem of SL1 sequence |
| pCTV:524/956 M8 | Compensatory change of SL1 sequence |
| pCTV:524/956 M9 | Site-specific changes in SL1 sequence |
| pCTV:524/956 M10 | Site-specific changes in SL1 sequence |
| pCTV:524/956 M11 | Site-specific changes in SL1 sequence |
| pCTV-ΔCla | Parent plasmid, replicon with one hybrid sgRNA (19) |
| pCTV-ΔCla M12 | Substitution in SL2 sequence |
| pCTV-ΔCla M13 | Substitution in SL2 sequence |
| pCTV-ΔCla M14 | Substitution in SL1 sequence |
| pCTV-ΔCla M15 | Substitution in SL1 sequence |
| pCTV-ΔCla M16 | Substitution in SL1 sequence |
| pCTV-ΔCla M17 | Site-specific changes in SL1 sequence |
| pCTV-ΔCla M18 | Site-specific changes in SL1 sequence |
| pCTV-ΔCla M19 | Site-specific changes in SL1 sequence |
| pCTV dRNA 2892/p23 | |
| pCTV9 | Parent plasmid, wild-type CTV (19) |
| pCTV9 M13 | Substitution in SL2 sequence |
| pCTV9-M14 | Substitution in SL1 sequence |
| pCTV9-M15 | Substitution in SL1 sequence |
| pCTV9-M16 | Substitution in SL1 sequence |
Mutants in the first two groups contained 5′TR duplicated in the 3′ end of pCTV-ΔCla333 between XhoI and StuI sites (Fig. 1, 2, and 4). Mutants in the third group contained 5′TR duplicated in the 3′ end of pCTV-ΔCla333. The mutation was introduced by overlap extension PCR and ligated between XhoI and StuI sites (Fig. 6). Mutants in the fourth group contained changes in the 5′TR in its native position in pCTV-ΔCla. The mutations were generated by overlap extension PCR and ligated between unique StuI and BglII restriction sites (Fig. 7). Mutant pCTV dRNA 2892/p23 contained a large internal deletion (nt 2893 to 18393) in CTV9 (Fig. 3). Mutants in the final group contained changes in 5′TR in its native position in CTV9 (Fig. 8).
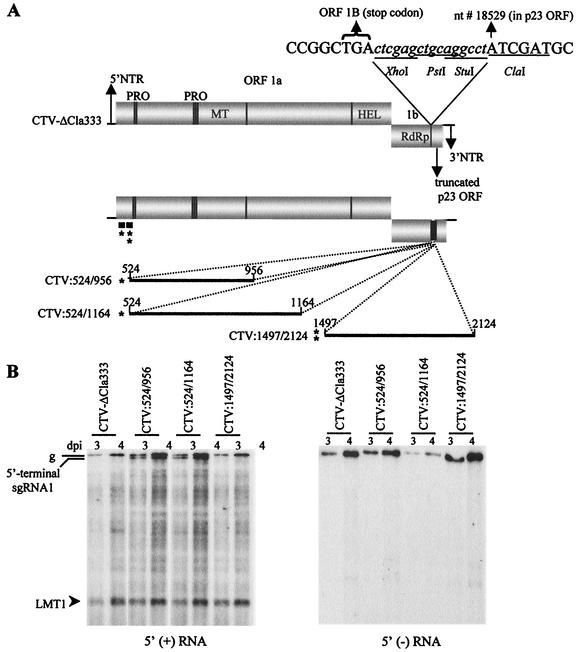
FIG. 1.
Synthesis of 5′-terminal sgRNA1 by the 5′TR. (A) Schematic representation of the organization of the CTV replicon, CTV-ΔCla333, and CTV replicons containing portions of CTV, nt 524 to 956 (CTV:524/956), nt 524 to 1164 (CTV:524/1164) or nt 1497 to 2124 (CTV:1497/2124) placed between the end of ORF 1a/1b and the truncated p23 ORF. Nucleotide numbers of the 5′ regions (solid black lines) ligated into CTV-ΔCla333 are also shown. The open boxes represent ORFs 1a and 1b and the truncated p23 ORF. The two papain-like proteases (PRO), methyltransferase-like (MT), helicase-like (HEL), and RNA-dependent RNA polymerase (RdRp) domains of ORF 1a/1b in the genome of CTV-ΔCla333 are indicated. The relative positions of the 5′TR and non-5′TR elements (black boxes with one or two asterisks, respectively) in CTV-ΔCla333 are indicated below the L-ProI domain. (B) Northern blot analysis of the total RNA isolated 3 and 4 days postinoculation (dpi) of N. benthamiana mesophyll protoplasts and hybridized with the 5′ plus- and minus-strand (left and right panels, respectively) RNA-specific probes. Positions of the genomic (g), 5′-terminal sgRNA1, and LMT1 RNAs are indicated.
A positive oligonucleotide (5′-3′ nt 524 to 543) that contained an XhoI restriction enzyme site at the 5′ end and two negative primers (3′-5′ nt 956 to 943 and 1164 to 1147) were used to amplify DNA fragments corresponding to the LMT region to construct pCTV:524/956 and pCTV:524/1164, respectively (Table 1). Similarly, a plus-strand primer (5′-3′ nt 1497 to 1516) with an XhoI site at the 5′ end and a minus-strand primer (3′-5′ nt 2124 to 2092) were used to construct pCTV:1497/2124.
The defective RNA (dRNA) was generated by a large internal deletion in CTV9 (19) created by overlap extension PCR (7). The plus-strand overlap PCR primer contained sequence that corresponded to 5′-3′ nt 2872 to 2892, followed by 5′-3′ nt 18394 to 18414. The minus-strand primer represented the complementary sequence of the plus-strand primer. The outer 5′ PCR primer corresponded to 5′-3′ nt 773 to 793, while the extreme 3′ PCR primer was complementary to the 3′ end of CTV and contained a NotI restriction site. The final amplified product was digested with BglII (at nt 1026) and NotI and ligated between similarly digested CTV-ΔCla333. The dRNA construct, CTVdRNA 2892/p23, thus contained up to 2,892 nt at the 5′ end, followed by the entire coding region of p23 ORF and the 3′ nontranslated region (3′NTR) towards the 3′ end.
Deletion mutants were generated to identify minimal sequences of the 5′TR. Mutants CTV:635/956, CTV:705/956, and CTV:789/956 were generated using DNA fragments amplified by plus-strand primers, with the 5′ end starting at nt 635, 705, and 789, respectively, and with an XhoI site at the 5′ end, and a constant minus-strand primer (3′-5′ nt 956 to 943). The 3′ deletion mutants CTV:524/789, CTV:524/775, and CTV:524/705 were generated using a constant plus-strand primer (5′-3′ nt 524 to 543) that contained an XhoI site and minus-strand primers (3′-5′ nt 789 to 769, 775 to 750, or 705 to 683, respectively). Construct pCTV:719/775 was generated using a plus-strand primer (nt 719 to 736 with an XhoI site at the 5′ end) and a minus-strand primer complementary to the 3′ end of the CTV genome (3′- 5′ nt 19296 to 19269 with a NotI restriction site at the 5′ end). DNA was amplified using pCTV:524/775 as template, digested with XhoI and NotI, and ligated between similarly digested pCTV:524/775. Mutants CTVM1-CTVM11 were generated by overlap extension PCR using a set of positive and negative primers that contained the intended mutation. The 5′- and 3′-end primers (5′-3′ nt 524 to 543 and 3′-5′ nt 956 to 943, respectively) used for the final overlap DNA amplification step corresponded to the primers that were used to construct mutant CTV:524/956.
The site-specific mutations used to examine putative secondary structures in the native 5′TR were constructed in a modified pCTV-ΔCla that contained a unique StuI site at the 5′ end upstream of the SP6 RNA polymerase sequence (19). The 5′ primer was designed upstream of the SP6 RNA polymerase sequence, while the 3′ primer (3′-5′ nt 1164 to 1147) was located at the 3′ end of the unique BglII site in CTV-ΔCla. The overlap extension PCR products were digested with BglII and ligated between StuI- and BglII-digested pCTV-ΔCla. Site-specific mutations in the native 5′TR of full-length CTV (CTV9) were introduced by the exchange of the ∼15.6-kb DNA fragment between NcoI and NotI sites from CTV9 (19) into CTV-ΔCla mutants.
The homology of the 5′TR of different isolates of CTV was determined using Clustal W (23).
Transfection of Nicotiana benthamiana protoplasts and Northern blot hybridization analysis.
Synthesis of the transcripts from NotI-linearized templates, polyethylene glycol-mediated transfection of the N. benthamiana mesophyll protoplasts, subsequent isolation of the total RNA, and Northern hybridization analyses were performed as described by Satyanarayana et al. (19). The 5′ and 3′ digoxigenin-labeled riboprobes used in this study have been described earlier (19).
RESULTS
Sequences surrounding the 3′ terminus of the LMT1 RNAs produce 5′-terminal sgRNAs.
The ∼800-nt 5′-terminal sgRNAs (LMT1 and LMT2) that were found associated with CTV were suggested to be products of premature termination during synthesis of genomic RNAs (2). One possibility was that a cis-acting element of the minus-strand template RNA near the 3′ termini of the LMT RNAs effects termination. For simplicity, we refer to this region as the 5′ termination region (5′TR). To identify and examine a possible termination element in this region, we would need to be able to delete adjacent areas and to mutate it. However, this area, located ∼700 to 900 nt from the 5′ end of the genome, encodes the N terminus of the leader protease I (L-ProI) (9). Mutations made in this region interfere with replication of the virus (references 16 and 19 and unpublished data). To overcome this limitation, we chose to duplicate the 5′TR to a position within the genome that would not interfere with replication and would allow examination of whether these sequences function as a transcription terminator. Previously we developed a CTV replicon, CTV-ΔCla333, which retains none of the 3′ genes but replicates efficiently in protoplasts without producing any 3′ sgRNAs (4). This replicon contains the 5′NTR, ORF 1a/1b (nt 1 to 10857), followed by a few unique restriction sites and ∼800 nt of the 3′ end of the CTV genome (nt 18529 to 19296 [Fig. 1A]) (4). We inserted a second set of the 5′TR sequences into CTV-ΔCla333 between ORF1a/1b and the 3′ end of CTV-ΔCla333 to examine whether the inserted sequences in this context produced a new, correspondingly longer 5′-terminal sgRNA.
We ligated relatively large portions of the 5′TR, including the putative termination sites (∼nt 800) plus ∼200 to 400 nt in both directions, into CTV-ΔCla333 to generate CTV:524/956 (nt 524 to 956) and CTV:524/1164 (nt 524 to 1164) (Fig. 1A). As a control, a region of similar size not near the putative termination sites, but immediately 3′ of the catalytic His464 residue of L-ProI (nt 1497 to 2124) (9), was similarly ligated into CTV-ΔCla333 to generate CTV:1497/2124 (Fig. 1A). RNA transcripts of these constructs were transfected into N. benthamiana mesophyll protoplasts, and the total RNA isolated 3 and 4 days postinoculation was analyzed by Northern blot hybridizations using 5′ plus- or minus-strand RNA-specific riboprobes (Fig. 1B). All three constructs replicated and produced plus- and minus-strand genomic RNAs plus the 5′-terminal plus-strand LMT1 sgRNA expected for the replicon (Fig. 1B) (2). In addition, constructs CTV:524/956 and CTV:524/1164 produced a new plus-strand RNA of ∼11.0 kb (referred to as sgRNA1) that was slightly smaller than the genomic RNA (Fig. 1B). The smaller size of this RNA and its detection by the 5′-terminal probe but not the 3′-terminal plus-strand RNA-specific probe (see below) suggested that it was a 5′-terminal sgRNA. A corresponding minus-strand RNA was not detected (Fig. 1B, right panel). Accumulation of sgRNA1 was not observed from RNAs extracted from protoplasts infected with CTV-ΔCla333 or CTV:1497/2124 (Fig. 1B). Thus, the insertions containing the putative termination region in the new context near the 3′ end of the CTV replicon resulted in the production of an additional 5′-terminal sgRNA.
Synthesis of a new 3′-terminal sgRNA from the duplicated 5′TR.
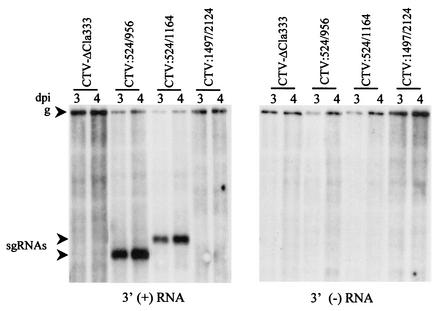
Previously, when examining the elements controlling mRNA production by the 3′ genes, we found that each controller element produced a large 5′-terminal plus-strand sgRNA in addition to the 3′-coterminal mRNA (4), which appears to be similar to that shown in Fig. 1B. Thus, if the 5′TR functioned similarly to the 3′ controller elements, it might produce a 3′-terminal sgRNA in addition to the 5′-terminal sgRNA. To examine this possibility, we hybridized the total RNA derived from protoplasts infected with mutant CTV-ΔCla333, CTV:524/956, CTV:524/1164, or CTV:1497/2124 with 3′ plus- or minus-strand RNA-specific riboprobes (Fig. 2). Constructs CTV:524/956 and CTV:524/1164 produced new 3′-terminal plus-strand sgRNAs of ∼1.0 and 1.2 kb, respectively (Fig. 2). The increase in the size of the 3′-terminal sgRNA in CTV:524/1164 was correlated with the larger 5′TR ligated into CTV-ΔCla333 compared to that of CTV:524/956 (Fig. 1A). The accumulation levels of the 3′-terminal sgRNAs were much greater than those of the genomic RNA and of the corresponding 5′-terminal sgRNAs. In contrast to the controller elements of 3′ ORFs, no minus-strand sgRNA corresponding to the 3′-terminal sgRNA was detected (Fig. 2). The new 3′-terminal sgRNAs were not observed from CTV-ΔCla333- or CTV:1497/2124-infected protoplasts, but normal levels of the genomic plus- and minus-strand RNAs (Fig. 2) and the small 5′-terminal LMT1 RNAs (Fig. 1) were observed. These results suggested that when duplicated into the 3′ end of the CTV replicon, the 5′TR functioned in the production of both 5′- and 3′-terminal plus-strand sgRNAs and that the smaller 5′TR subset, nt 524 to 956, was sufficient for the production of both RNAs.
FIG. 2.
Synthesis of 3′-terminal sgRNAs by the 5′TR in the 3′ position. A Northern blot analysis of the total RNA isolated 3 and 4 days postinoculation (dpi) of N. benthamiana protoplasts transfected with the constructs outlined in Fig. 1 is shown. The blots were hybridized with 3′ plus- and 3′ minus-strand (left and right panels, respectively) RNA-specific probes. Positions of the genomic (g) and sgRNAs are indicated.
The native 5′TR element produces a 3′ terminal sgRNA.
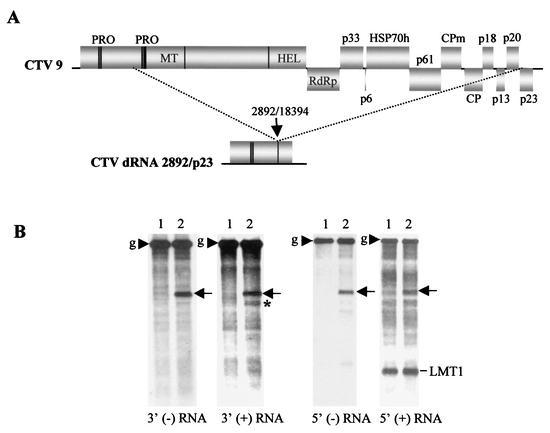
Synthesis of a 3′-terminal sgRNA by the 5′TR element when present in the 3′ position in CTV:524/956 suggested that the native 5′TR in the 5′ end of the genome probably also synthesized a 3′-terminal sgRNA. We did not detect such an sgRNA during replication of CTV-ΔCla333 (Fig. 2). However, if it were produced, it would be only ∼800 nt smaller than the full-length replicon RNA and might not be resolved as a distinct band during Northern blot hybridization analysis. Also, the abundance of this RNA would probably be low based on the observation that other sgRNAs tend to be produced in smaller amounts when positioned farther from the 3′ terminus (4, 19). To overcome this, we constructed a dRNA (CTVdRNA:2892/p23) with 2,892 nt of the 5′ end ligated to the p23 ORF plus the 3′NTR such that a potential 3′-terminal sgRNA from the native 5′TR would be resolved (Fig. 3A). CTV-ΔCla333 was used as the helper replicon because it produces no 3′-terminal sgRNAs to interfere with the examination of a new 3′-terminal sgRNA. The dRNA and CTV-ΔCla333 were cotransfected into N. benthamiana protoplasts, and the total RNA was analyzed with the 3′ and 5′ plus- or minus-strand RNA-specific probes. The dRNA and CTV-ΔCla333 replicated in protoplasts and produced full (dRNA and replicon)-length plus- and minus-strand RNAs (Fig. 3B). In addition, a smaller 3′ terminal plus-strand sgRNA was produced (Fig. 3B). However, no complementary minus-strand sgRNA was observed (Fig. 3B). The size of the new 3′-terminal sgRNA, ∼750 nt smaller than the dRNA, and the lack of hybridization with the 5′ plus- and minus-strand RNA-specific probes (Fig. 3B) are consistent with its being produced by the 5′TR.
FIG. 3.
Synthesis of a 3′-terminal sgRNA from the 5′TR element in a dRNA. (A) Schematic diagram of the CTV dRNA generated by a large internal deletion between nt 2892 and 18394 in CTV9. The different domains of ORF 1a/1b are as outlined in the legend to Fig. 1A. (B) Northern blot hybridizations showing the accumulation of the 3′ and 5′ plus- and minus-strand RNAs from the total RNA isolated from protoplasts transfected with the helper replicon (CTV-ΔCla333) (lanes 1) and the helper replicon plus the dRNA (lanes 2). The positions of the genomic (g) RNA of the helper replicon (arrowheads) and the dRNA (arrows) are indicated. The 3′-terminal sgRNA (indicated by an asterisk, observed only with the 3′ plus-strand RNA-specific probe) and LMT1 RNAs (observed only with the 5′ plus-strand RNA-specific probe) are also indicated.
Determination of a minimal region required for production of the 3′- and 5′-terminal sgRNAs.
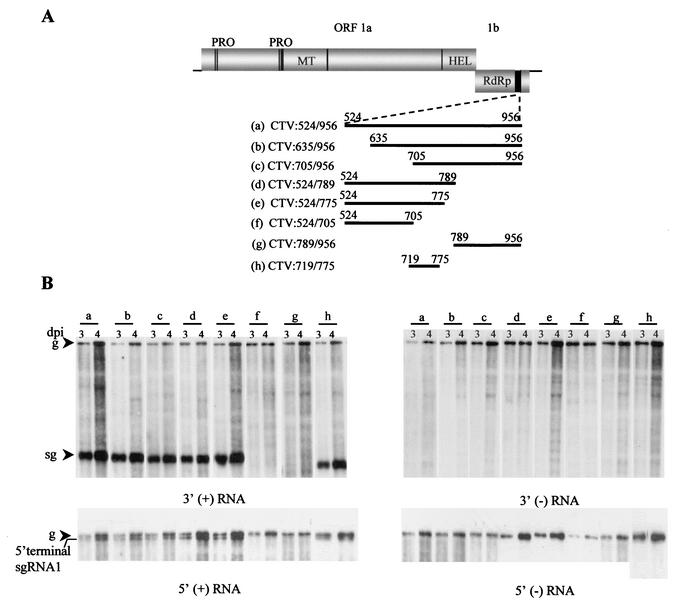
The sizes of the LMT1 RNAs suggested that the population of 3′ termini would correspond to ∼800 nt. To delineate a minimal set of sequences required for the production of the 3′-terminal sgRNA and the 5′-terminal sgRNA1, a series of progressive deletions were made from the 5′ end of the insertion in CTV:524/956 to generate mutants CTV:635/956, CTV:705/956, and CTV:789/956; these mutants contained 322, 252, or 168 nt of the 5′TR, respectively (Fig. 4A, b, c, and g). Similarly, progressive deletions were introduced from the 3′ end of the insertion in CTV:524-956 to generate CTV:524/789, CTV:524/775, and CTV:524/705 containing 266, 252 or 182 nt of the 5′TR, respectively (Fig. 4A, d to f).
FIG. 4.
Deletion analysis of the 5′TR in the 3′ position in CTV-ΔCla333. (A) Schematic diagram of CTV-ΔCla333 containing progressive deletions from the 5′ and 3′ ends in the 5′TR with their corresponding nucleotide endpoints. (B) Results of the accumulation of 3′ plus- and 3′ minus-strand (left and right panels, respectively) genomic (g) and sgRNA in Northern blot hybridization analysis of the total RNA isolated 3 and 4 days postinoculation (dpi) of the protoplasts transfected with the mutants outlined in panel A. Positions of the genomic and sgRNAs (sg) are indicated. In the bottom panels, only the top portion of the Northern blot hybridizations is presented, showing the accumulation of RNAs that reacted with 5′-terminal plus-strand RNA-specific (left) or 5′-terminal minus-strand-specific (right) riboprobes.
All mutants replicated in protoplasts and accumulated plus- and minus-strand genomic RNAs (Fig. 4B). However, production of the new 3′- and 5′-terminal sgRNAs was observed only with constructs CTV:524/956, CTV:635/956, CTV:705/956, CTV:524/789, and CTV:524/775 (Fig. 4B). These results suggested that the region between nt 705 to 775 was involved in the production of both 5′- and 3′-terminal sgRNAs. We generated another mutant, CTV:719/775 (Fig. 4A, h), with a smaller subset of the 5′TR (57 nt) inserted into CTV-ΔCla333. This mutant also replicated normally and still produced both sgRNAs (Fig. 4B, lanes h). None of these mutants accumulated detectable levels of a minus-strand sgRNA complementary to the 3′- or 5′-terminal plus-strand sgRNA (Fig. 4B). Thus, the 57-nt segment of the 5′TR element, when duplicated in the 3′ end of the replicon, was sufficient to produce both 5′- and 3′-terminal plus-strand sgRNAs.
Predicted secondary structure of the minimal 5′TR element.
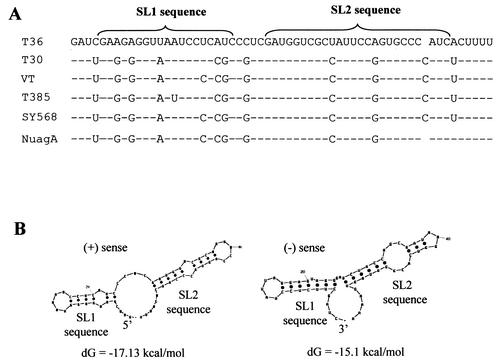
The sequences of 5′TR encode a part of the leader protease of ORF 1a (9) that has an unusually high level of sequence conservation (>82%) among different isolates of CTV (Fig. 5A), while surrounding sequences are substantially less highly conserved. The smallest subset of sequences (57 nt) of the 5′TR that was active for the synthesis of both 3′- and 5′-terminal sgRNAs was predicted to fold into two stem-loops separated by a small spacer region (Fig. 5B) (MFOLD program [28]). Both plus and minus strands gave approximately similar patterns of folding, with free energies of −17.13 and −15.1 kcal/mol, respectively. Since this cis-acting element is involved in the production of plus-strand sgRNAs, it is expected that the minus-strand sequence would be functional. However, since the convention of nucleotide numbering corresponds to the plus-strand, we use plus-strand numbering, in which the putative stem-loop 1 (SL1) and 2 (SL2) sequences encompass nt 723 to 741 and 746 to 770, respectively, separated by a 4-nt spacer (nt 742 to 745).
FIG. 5.
(A) Alignment of the minimal size of the 5′TR element of T36 isolate (identified by mutational analysis) with the corresponding region of other isolates of CTV. The GenBank accession numbers of the sequences of different CTV isolates are as follows: T36, NC_001661; T30, AF260651; VT, U56902; T385, Y18420; SY568, AF001623; and NUagA, AB046398. Only the nucleotide differences from T36 are shown. Hyphens represent the conserved nucleotides in all isolates, and the gaps were introduced for optimal alignment. Positions of the predicted stem-loop 1 (SL1) and stem-loop 2 (SL2) are indicated. (B) The predicted secondary structure of plus (left) and minus (right) strands of the 5′TR element and their respective free energies obtained using the MFOLD program (28). The putative stem-loops (SL1 and SL2) are indicated.
Mutagenesis of the predicted stem-loops of the 5′TR element.
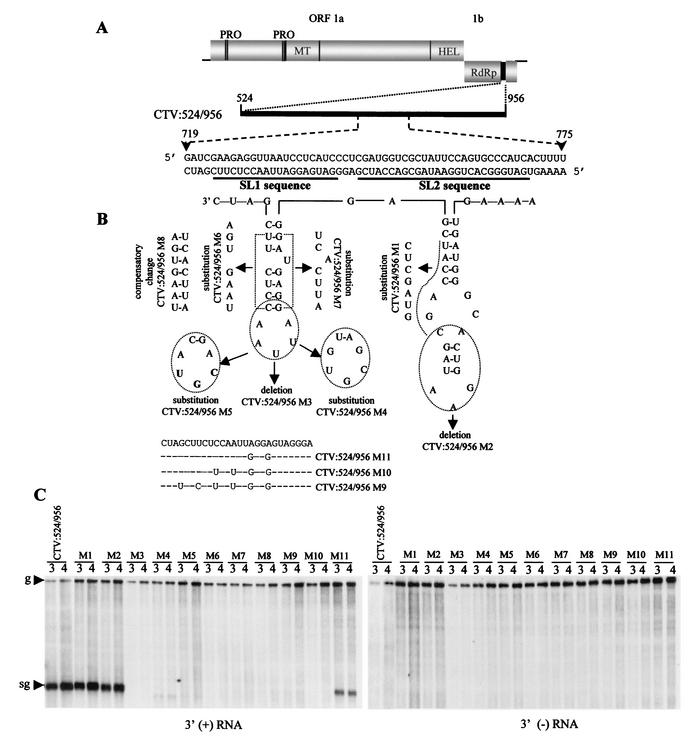
In an attempt to determine whether the predicted secondary structures correlate with biological activity, we introduced mutations designed to alter both the primary and secondary structures or to alter the primary structure while attempting to preserve similar secondary structures (Fig. 6). The mutations, including deletions, substitutions with nonviral sequences, compensatory changes, and site-specific substitutions (Fig. 6B), were introduced into CTV:524/956.
FIG. 6.
Mutational analysis of the 5′TR element. (A) Schematic representation of the mutant CTV:524/956, which contained nt 524 to 956 in the 3′ region of CTV-ΔCla333. The nucleotide sequence of the plus and minus strands of nt 719 to 776 in CTV:524/956 is shown as RNA sequence. Positions of stem-loops 1 and 2 (SL1 and SL2) are shown as dark lines below the negative strand sequence. (B) Diagrammatic representation of the putative stem-loop structures (SL1 and SL2) of the minus-strand RNA sequence as predicted by MFOLD and schematic outline of different mutations (substitutions, deletions, compensatory changes, and site-specific changes) introduced in SL1 and SL2. See the text for the details of mutations in SL1 and SL2. (C) Northern blot hybridization analysis of the total RNA isolated 3 and 4 days postinoculation from protoplasts transfected with the mutants outlined above. Accumulation of the genomic (g) and sgRNAs hybridized with 3′-plus- and minus-strand (left and right panels, respectively) RNA-specific riboprobes are shown.
(i) Mutations in SL2 sequences.
Mutant CTV:524/956 M1 contained a replacement of nt 747 to 753 (UACCAGC) by nonviral nucleotides (CUCGAUG) to disrupt the predicted stem of SL2. CTV:524/956 M2 had a deletion of nt 753 to 763 to remove a portion of stem and putative loop region of SL2 (Fig. 6B). These mutants replicated and produced 3′-terminal sgRNAs (Fig. 6C) and 5′-terminal sgRNA1 (data not shown) at levels similar to those produced by the unmodified control, CTV:524/956, suggesting that SL2 played no more than a minor role in the production of these RNAs.
(ii) Mutations in SL1 sequences.
Mutant CTV:524/956 M3 contained a deletion of the putative loop (nt 729 to 735), which should destroy the secondary structure of SL1. Mutant CTV:524/956 M4 contained a replacement of the putative loop (nt CAAUUAG) by nonviral nucleotides (UGUGCGA), while CTV:524/956 M5 contained a substitution of the central nucleotides of the loop (AUU to UGC) (Fig. 6B). All three mutants replicated efficiently but did not produce the 3′-terminal (Fig. 6C) or 5′-terminal (data not shown) sgRNAs. The next set of mutations were introduced in the putative stem region of SL1. Mutant CTV:524/956 M6 contained a replacement of nt 723 to 729 by nonviral nucleotides (AGUGAAU), and CTV:524/956 M7 contained a replacement of nt 735 to 741 of the other part of the stem by nonviral nucleotides (AUUCACU) to disrupt the predicted stem of SL1; while CTV:524/956 M8 had compensatory mutations with nonviral sequences of both CTV:524/956 M6 and CTV:524/956 M7 to restore the predicted stem structure of SL1 (Fig. 6B). These mutants replicated normally in protoplasts but produced neither the 3′-terminal sgRNA (Fig. 6C) nor the 5-terminal sgRNA1 (data not shown). Thus, all changes examined within the SL1 area destroyed the function of this cis-acting element.
We next examined a series of site-specific mutations in the SL1 area that could be useful later in examination of the native 5′TR in the 5′ end of the genome. These mutants were designed to cause minimal disruption of the N terminus of L-ProI encoded by the 5′TR while attempting to disrupt sgRNA production. Mutants CTV:524/956 M9, CTV:524/956 M10, and CTV:524/956 M11 contained replacements by nonviral nucleotides, as shown in Fig. 6B. These mutants replicated and produced normal amounts of genomic RNAs, but the accumulation of the 3′-terminal sgRNA was not observed with CTV:524/956 M9 and CTV:524/956 M10, while a reduced level of accumulation of this sgRNA was observed with CTV:524/956 M11 (Fig. 6C). In addition, the LMT RNAs accumulated normally from the native 5′TR, suggesting that none of the 3′ duplications or mutations affected the native cis-acting element (data not shown).
LMT RNAs are not required for the replication of CTV in protoplasts.
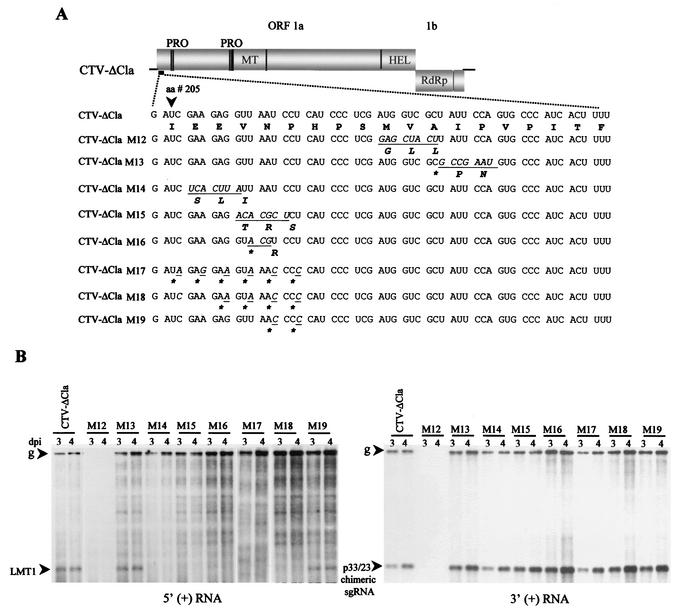
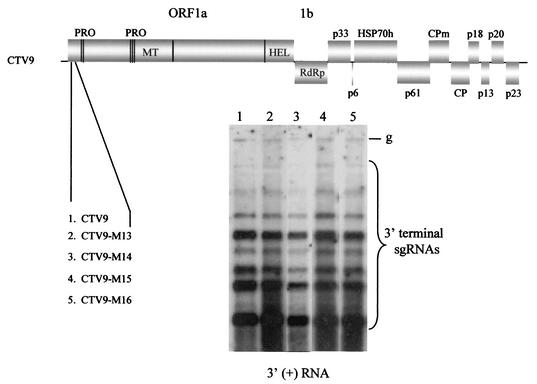
To examine the function of the LMT1 RNAs and the undetected but expected 3′-terminal sgRNA produced from the native 5′TR, mutations that were shown to alter the function of the duplicated 5′TR in the 3′ end of the replicon were incorporated into the native 5′TR and examined for genomic RNA replication and production of LMT1 RNAs. For this purpose, we used a CTV replicon, CTV-ΔCla (19). Use of this replicon allowed us to monitor whether the function of the 5′TR effected replication and/or the synthesis of the sgRNA from the 3′ ORF (p33) controller element. Total RNAs from protoplasts transfected with the transcripts of mutants were analyzed by Northern blot hybridization analysis using 3′ or 5′ plus-strand RNA-specific riboprobes. Mutant CTV-ΔClaM12 (Fig. 7A; similar mutation to M1 in Fig. 6B) contained nonviral nucleotides GAGCUACUU encoding Glu212, Leu213, and Leu214 in place of AUGGUCGCU, which encoded Met212, Val213, and Ala214, resulting in three amino acid substitutions in L-ProI. This change completely abolished the ability of the mutant to replicate in protoplasts (Fig. 7B). Other mutants (CTV-ΔClaM13, CTV-ΔClaM14, CTV-ΔClaM15, and CTV-ΔClaM16) also contained amino acid alterations in L-ProI as shown in Fig. 7A. CTV-ΔClaM13, with two amino acids altered, resulted in continued accumulation of the LMT1 RNAs. The other mutations (CTV-ΔClaM14 to CTV-ΔClaM16) that prevented production of the 5′TR-induced 3′-terminal sgRNA when positioned in the 3′ region of the replicon (Fig. 6C; mutants M6, M4, and M5) similarly prevented the production of the LMT1 RNAs but allowed normal production of genomic and subgenomic plus- and minus-strand RNAs. It is possible that the prevention of accumulation of the LMT1 RNAs could have resulted from changes in the amino acid sequence of L-ProI rather than from a change in the primary structure of the 5′TR. To examine this possibility, the “wobble” bases of codons were mutated in CTV-ΔClaM17 to CTV-ΔClaM19 to avoid changes in the amino acid sequence of L-ProI (Fig. 7A). These mutants replicated in protoplasts producing normal amounts of genomic plus-strand (Fig. 7B) and minus-strand (data not shown) RNAs. The accumulation of the LMT1 RNAs was not observed in mutants CTV-ΔClaM17 (the nucleotides changed were nt 722, 725, 728, 731, 734, and 737) and CTV-ΔClaM18 (nt 728, 731, 734, and 737). Small amounts of LMT1 RNAs accumulated in cells infected with mutant CTV-ΔClaM19, which had the same mutation as mutant M11 (Fig. 6B) (which also accumulated the 3′-terminal sgRNA, albeit at reduced levels). The synthesis of the p33/p23 chimeric sgRNA controlled by the native p33 controller was not affected by any of the mutations (Fig. 7B, right panel). Similarly, mutations were introduced into the 5′TR in the full-length CTV, CTV9. The mutants CTV9-M13 to CTV9-M16 (Fig. 8) (corresponding to CTV-ΔClaM13 to CTV-ΔClaM16) replicated in protoplats and produced genomic RNAs and sgRNAs approximately similar to those of CTV9 (Fig. 8). These results suggested that the 5′TR functions (synthesis of LMT RNAs and the large 3′-terminal sgRNA) were not needed in protoplasts for replication of the genomic RNAs or production of 3′ ORF sgRNAs.
FIG. 7.
Mutational analysis of the 5′TR at its native position in CTV-ΔCla. (A) Diagrammatic representation of the organization of the CTV replicon, CTV-ΔCla (19). The nucleotide sequence nt 719 to 776 (in the 5′TR) showing triplets (codons) and their corresponding amino acids (below the nucleotide sequence) are shown. The underlined sequences in italics indicate changes introduced into the primary sequence, and the resultant amino acid changes are presented below the RNA sequence. A change in the nucleotide sequence that did not result in an amino acid change is indicated by an asterisk. (B) Accumulation of the 5′ plus-strand and 3′ plus-strand (left and right panels, respectively) RNAs as demonstrated by Northern blot hybridization analysis of the total RNA isolated 3 and 4 days postinoculation (dpi) of the protoplasts transfected with the mutants. Positions of the genomic (g) and the LMT1 RNAs in the left panel and the genomic (g) and the chimeric p33/p23 sgRNA in the right panel are indicated by arrowheads.
FIG. 8.
Replication of full-length CTV containing mutations in the 5′TR. A diagrammatic representation of mutations in CTV is shown. A Northern blot hybridization analysis of the total RNA isolated 4 days after inoculation of protoplasts inoculated with the wild type (CTV9) or mutants CTV9-M13 to CTV9-M16 (mutations in the 5′TR of CTV9-M13 to CTV9-M16 are as in CTV-ΔCla M13 to CTV-ΔClaM16 in Fig. 7A, respectively) and probed with the 3′ plus-strand RNA-specific riboprobe. Positions of the genomic (g) and sgRNAs are indicated.
DISCUSSION
Analysis of a putative cis-acting element such as that involved in the production of the LMT RNAs would generally require extensive mutations, deletions, or substitutions. However, this cis-acting element probably was positioned within the ORF 1a region encoding L-ProI. Previous mutations within the L-ProI domain of CTV (reference 19 and unpublished data) or BYV (17) greatly debilitated or prevented replication. Thus, traditional mapping in this region was not possible. However, the continued function of the duplicated 5′TR element inserted near the 3′ terminus facilitated its examination. The insertion of a second 5′TR did not affect the activity of the native element, which continued to produce normal amounts of LMT1 RNAs. The second 5′TR element produced a much larger 5′-terminal sgRNA at a lower level of accumulation than the LMT RNAs. The initial analysis of the duplicate 5′TR in the 3′ end of the replicon permitted precise mutagenesis of the native element without disrupting the replicase proteins of ORF 1a.
Remarkably, the duplicate 5′TR element located near the 3′ end caused the production of large amounts of a newly observed 3′-terminal sgRNA, suggesting that the native 5′TR also produces a much larger 3′-terminal sgRNA that is below our level of detection. This supposition was supported by the visible 3′-terminal sgRNA produced by the native 5′TR from the dRNA.
Since only plus-strand sgRNAs accumulated due to the 5′TR, it appears that the 5′-terminal sgRNAs were produced via termination and the 3′-terminal sgRNA was produced by classical promotion: the replicase initiating internally on the complementary minus strand of the genomic RNA. It could be argued that production of a 5′-terminal sgRNA and a 3′-terminal sgRNA which additively are approximately the same size as the genomic RNA is reminiscent of mRNA synthesis by minus-strand RNA viruses (22). However, CTV did not express its sgRNAs in the expected polar ratio. In fact, CTV sgRNA production was directly related to the position of the 5′TR within the genome. The 5′TR elements placed at two different positions in the genome appeared to produce their sgRNAs in opposite ratios; in each case, the smaller sgRNA accumulated to the higher level. As previously shown for CTV 3′ ORF controller elements, alteration of position did not equally affect the accumulation of 5′- and 3′-terminal sgRNAs (4). Distance from the 3′ terminus appears to decrease 3′-terminal sgRNA accumulation considerably more than distance from the 5′ terminus affects the accumulation of 5′-terminal sgRNAs.
CTV has 10 internal ORFs expressed via 9 or 10 3′-coterminal sgRNAs. We previously characterized some of the elements for the production of these sgRNAs (4). The 5′TR and the 3′ controller elements appear to have some similarities. The 5′TR, along with the controller elements of the major and minor coat protein genes, mapped to minimal elements of approximately 50 nt which were folded by the computer into two stem-loop structures. However, mutagenesis provided little support for the secondary structures, perhaps because primary structures also were important. Both types of cis-acting elements produce 5′- and 3′-terminal plus-strand sgRNAs that, when added together, approximate the size of the genomic RNA. Otherwise, the 5′TR element differed considerably from the 3′ controller elements. A unique characteristic of the 3′ controller elements is that each controls the production of a third sgRNA: relatively large amounts of a minus-strand sgRNA complementary to the 3′-terminal mRNA. The 5′TR produced only plus strands of the 3′-terminal sgRNA. The production of the minus-strand sgRNAs was repressed by the p23 gene (19), whereas the p23 gene did not appear to affect the function of the 5′TR. Also, even though the 5′TR and the 3′ ORF controller elements were each folded into a pair of stem-loop structures by the MFOLD program, the sensitivities to mutagenesis were opposite: the 5′-most region of the minimal active 5′TR and the 3′-most region of the minimal 3′ ORF controller elements tolerated little mutagenesis, whereas their opposite regions tolerated substantial mutation. These differences between the sgRNA controllers at opposite ends of the CTV genome suggest that during modular evolution of this virus, the different controllers might have originated from different sources, perhaps from recombination between different viruses, as has been suggested previously for CTV (2, 11).
Although the 5′TR element produced only plus strands of the 3′-terminal sgRNA, suggesting internal initiation on the minus strand, a dilemma is that we have not been able to distinguish how the 3′-ORF controller elements work; which sgRNA, the plus or minus strand, is produced from the full-length RNA? Are the 3′-terminal plus-strand mRNAs produced via internal promotion from the full-length cRNA (classic promotion) (13), or are the subgenomic minus strands produced by termination at the controller region, followed by transcription of mRNAs from the minus subgenomics as demonstrated for arteriviruses (24)? Production first of the minus-strand sgRNAs followed by transcription of the plus-strand sgRNAs from the minus-sense sgRNAs would suggest a cis-acting element on the 3′ ends of the minus-strand sgRNAs for initiation of mRNA transcription. However, deletions or insertions to completely alter the expected cis-acting element, the 3′ regions of minus-strand sgRNAs, had no effect on either plus- or minus-strand sgRNA synthesis, casting doubt on this mechanism (4). Synthesis first of the plus-strand sgRNAs via promotion leaves the question of how and why the minus-strand sgRNAs are produced. An obvious possibility would be that the replicase complex initiates minus-strand RNA synthesis at the 3′ terminus of all plus strands (the minus-strand promoter), copying the mRNAs to produce minus-strand sgRNAs. This strategy would appear to be supported by the fact that the p23 protein reduces the synthesis of both genomic and subgenomic minus strands (20). However, the 5′TR produces 3′-terminal sgRNAs which apparently have 3′-terminal sequence identical to that of the 3′ORF mRNAs, and thus the identical promoter for minus-strand synthesis. Yet, the sgRNAs from the 5′TR were not copied to produce minus strands, suggesting that minus-strand sgRNAs also are not copied from the 3′-ORF mRNAs. Thus, we appear to be left with a scheme in which the 3′-ORF controller elements produce 3′-terminal plus-strand sgRNAs by promotion and minus-strand sgRNA independently by termination.
Both LMT1 and LMT2 RNAs are produced by wild-type CTV of several strains (2, 10), while only LMT1 RNAs are produced by the replicons with all or most of the 3′ genes deleted. This demonstrates that none of the 3′ genes are necessary for production of the longer LMT1 RNAs, but it might suggest an involvement of these genes in production of the shorter 5′-terminal LMT2 RNAs. Alternatively, some 3′ gene functions, such as assembly or movement, might process the larger LMT RNAs into the population of smaller RNAs.
So the final dilemma is defining the functions of the RNAs produced by the 5′TR. It is possible that only one of the RNAs is functional and the other is “accidental.” There is considerable information suggesting that specific sequences or structures cause pausing and termination of polymerases (3, 8, 25, 27). It is possible that the 5′-terminal sgRNAs of CTV are produced by accidental termination at promoter structures. A normal function of the 3′ ORF controller elements (4), as observed with Sindbis virus (26) and Tobacco mosaic virus (TMV) (unpublished data from our laboratory), is to produce 5′-terminal sgRNAs, perhaps “accidentally,” in addition to the subgenomic mRNAs. But why would the virus have a sgRNA promoter ∼800 nt from the 5′ terminus? One possible function for a near-full-length RNA missing the 5′ terminus could be to serve as a dedicated messenger for replicase-associated proteins. The genomic RNA of plus-strand RNA viruses is thought to function as the mRNA for replicase, as the template for minus-strand synthesis, and as the substrate for assembly. Additionally, CTV has to make nine or ten different 3′-ORF mRNAs. Thus, the competition for the genomic RNA might increase with size and complexity in ssRNA viruses. Since both ends of the viral RNAs can function in the production of both plus- and minus-strand RNAs (1), an RNA with the absence of the 5′ cis-acting elements could be free to function only as an mRNA. However, the fact that mutations that prevented the promoter function of the 5′TR had no negative effect on the replication of CTV discredits this hypothesis. It also is possible that the LMT RNAs are functional and the promotion is accidental. The psuedoknots in the 3′NTR of TMV function effectively as an apparent accidental promoter (21). However, when the native 5′TR was mutated to produce neither the LMT1 RNAs nor the 3′-terminal sgRNA, we saw no effect on the replication of CTV in protoplasts. Is it possible that neither sgRNA is functional?
Acknowledgments
We thank John Cook and Cecile Robertson for their technical assistance.
This research was supported by an endowment from the J. R. and Addie Graves family and grants from the Florida Agricultural Experiment Station, the Florida Citrus Production Research Advisory Board, US-Israel BARD, and USDA/ARS cooperative agreement 58-6617-4-018. M. A. Ayllón was the recipient of a postdoctoral fellowship from the Ministerio de Educación y Ciencia (Spain).
Footnotes
University of Florida Agricultural Experiment Station journal series R-09057.
REFERENCES
- 1.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ clover-leaf in poliovirus RNA is a cis acting replication element required for negative-strand synthesis. EMBO J. 15:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Che, X., D. Piestun, M. Mawassi, T. Satyanarayana, S. Gowda, W. O. Dawson, and M. Bar-Joseph. 2001. 5′-coterminal subgenomic RNAs in citrus tristeza virus-infected cells. Virology 283:374-381. [DOI] [PubMed] [Google Scholar]
- 3.Deng, L., and S. Shuman. 1997. Elongation properties of vaccinia virus RNA polymerase: pausing, slippage, 3′ end addition, and termination site choice. Biochemistry 36:15892-15899. [DOI] [PubMed] [Google Scholar]
- 4.Gowda, S., T. Satyanarayana, M. A. Ayllón, M. R. Albiach-Martí, M. Mawassi, S. Rabindran, S. M. Garnsey, and W. O. Dawson. 2001. Characterization of the cis-acting elements controlling subgenomic mRNAs of Citrus tristeza virus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology 286:134-151. [DOI] [PubMed] [Google Scholar]
- 5.He, X-H., A. L. N. Rao, and R. Creamer. 1997. Characterization of beet yellows closterovirus-specific RNAs in infected plants and protoplasts. Phytopathology 87:347-352. [DOI] [PubMed] [Google Scholar]
- 6.Hilf, M. E., A. V. Karasev, H. R. Pappu, D. J. Gumpf, C. L. Niblett, and S. M. Garnsey. 1995. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology 208:576-582. [DOI] [PubMed] [Google Scholar]
- 7.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Puller, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Jager, J., and J. D. Pata. 1999. Getting a grip: polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 9:21-28. [DOI] [PubMed] [Google Scholar]
- 9.Karasev, A. V., V. P. Boyko, S. Gowda, O. V. Nikolaeva, M. E. Hilf, E. V. Koonin, C. L. Niblett, K. Cline, D. J. Gumpf, R. F. Lee, S. M. Garnsey, D. J. Lewandowski, and W. O. Dawson. 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511-520. [DOI] [PubMed] [Google Scholar]
- 10.Mawassi, M., T. Satyanarayana, S. Gowda, M. R. Albiach-Marti, C. Robertson, and W. O. Dawson. 2000. Replication of heterologous combinations of helper and defective RNA of citrus tristeza virus. Virology 267:360-369. [DOI] [PubMed] [Google Scholar]
- 11.Mawassi, M., E. Mietkiewska, R. Goffman, G. Yang, and M. Bar-Joseph. 1996. Unusual sequence relationships between two isolates of citrus tristeza virus. J. Gen. Virol. 77:2359-2364. [DOI] [PubMed] [Google Scholar]
- 12.Mawassi, M., E. Mietkiewska, M. E. Hilf, L. Ashoulin, A. V. Karasev, R. Gafny, R. F. Lee, S. M. Garnsey, W. O. Dawson, and M. Bar-Joseph. 1995. Multiple species of defective RNAs in plants infected with citrus tristeza virus. Virology 214:264-268. [DOI] [PubMed] [Google Scholar]
- 13.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Navas-Castillo, J., M. R. Albiach-Martí, S. Gowda, M. E. Hilf, S. M. Garnsey, and W. O. Dawson. 1997. Kinetics of accumulation of citrus tristeza virus RNAs. Virology 228:92-97. [DOI] [PubMed] [Google Scholar]
- 15.Pappu, H. R., A. V. Karasev, E. J. Anderson, S. S. Pappu, M. E. Hilf, V. J. Febres, R. M. G. Eckloff, M. McCaffery, V. P. Boyko, S. Gowda, V. V. Dolja, E. V. Koonin, D. J. Gumpf, K. C. Cline, S. M. Garnsey, W. O. Dawson, R. Lee, and C. L. Niblett. 1994. Nucleotide sequence and organization of eight 3′ open reading frames of the citrus tristeza closterovirus genome. Virology 199:35-46. [DOI] [PubMed] [Google Scholar]
- 16.Peng, C.-W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of the leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1998. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J. Virol. 72:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio, L., H.-H. Yeh, T. Tian, and B. W. Falk. 2000. Heterogeneous population of defective RNAs associated with Lettuce infectious yellows virus. Virology 271:205-212. [DOI] [PubMed] [Google Scholar]
- 19.Satyanarayana, T., S. Gowda, V. P. Boyko, M. R. Albiach-Martí, M. Mawassi, J. Navas-Castillo, A. V. Karasev, V. Dolja, M. E. Hilf, D. J. Lewandowski, P. Moreno, M. Bar-Joseph, S. M. Garnsey, and W. O. Dawson. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. USA 96:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satyanarayana, T., S. Gowda, M. A. Ayllón, M. R. Albiach-Martí, S. Rabindran, and W. O. Dawson. 2002. The p23 protein of Citrus tristeza virus controls asymmetrical RNA accumulation. J. Virol. 76:473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivprasad, S., G. P. Pogue, D. J. Lewandowski, J. Hidalgo, J. Donson, L. K. Grill, and W. O. Dawson. 1999. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255:312-323. [DOI] [PubMed] [Google Scholar]
- 22.Shuman, S. 1997. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology 227:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Hippel, P. H. 1998. An integrated model of the transcription complex in elongation, termination, and editing. Science 281:660-665. [DOI] [PubMed] [Google Scholar]
- 26.Wielgosz, M., and H. Huang. 1997. Novel viral RNA species in Sindbis virus-infected cells. J. Virol. 71:9108-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, K. S., and P. H. von Hippel. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 92:8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]