Abstract
Previous findings that the vaccinia virus uracil DNA glycosylase is required for virus DNA replication, coupled with an inability to isolate a mutant with an active site substitution in the glycosylase gene, were surprising, as such enzymes function in DNA repair and bacterial, yeast, and mammalian null mutants are viable. To further study the role of the viral protein, we constructed recombinant vaccinia viruses with single or double mutations (D68N and H181L) in the uracil DNA glycosylase conserved catalytic site by using a complementing cell line that constitutively expresses the viral enzyme. Although these mutations abolished uracil DNA glycosylase activity, they did not prevent viral DNA replication or propagation on a variety of noncomplementing cell lines or human primary skin fibroblasts. In contrast, replication of a uracil DNA glycosylase deletion mutant occurred only in the complementing cell line. Therefore, the uracil DNA glycosylase has an essential role in DNA replication that is independent of its glycosylase activity. Nevertheless, the conservation of the catalytic site in all poxvirus orthologs suggested an important role in vivo. This idea was confirmed by the decreased virulence of catalytic-site mutants when administered by the intranasal route to mice.
Poxviruses, of which vaccinia virus is the prototype, are large, complex, double-stranded DNA viruses that replicate exclusively in the cytoplasm of host cells (18). Members of this large virus family encode numerous enzymes and factors for gene expression, genome replication, and virion assembly, which together provide considerable autonomy from host cell functions. A variety of genetic, biochemical, and molecular biological techniques have been used to identify proteins implicated in poxviral DNA replication. These viral proteins include a DNA polymerase, deoxynucleoside triphosphatase, protein kinase, DNA polymerase processivity factor, uracil DNA glycosylase, Holliday junction endonuclease, DNA topoisomerase, single-stranded DNA binding protein, DNA ligase, and enzymes involved in nucleotide metabolism, such as thymidine kinase, thymidylate kinase, ribonucleotide reductase, and dUTPase (18, 29). Only a subset of these proteins, however, is essential for viral DNA synthesis. One mutation abrogating viral DNA replication at the nonpermissive temperature was mapped to the D4R open reading frame (ORF), which encodes an active uracil DNA glycosylase (14, 27). This ubiquitous DNA repair enzyme is found in diverse organisms, and orthologs are present throughout the poxvirus family (31). Uracil arises in DNA through misincorporation of dUMP by DNA polymerase or through deamination of cytosine (11, 30). In eukaryotic and prokaryotic cells, uracil DNA glycosylase specifically recognizes uracil in DNA and initiates base excision repair by hydrolyzing the glycosylic bond linking uracil to a deoxyribose sugar. This activity creates an abasic site that is removed by a 5′-acting apurinic-apyrimidic (AP) endonuclease and a DNase, leaving a gap that is filled by DNA polymerase and sealed by ligase. However, no viral endonuclease that works in concert with the poxvirus uracil DNA glycosylase has been identified.
The finding that the vaccinia virus uracil DNA glycosylase is required for viral DNA replication (4, 14, 27) was surprising, since the excision of uracil residues from double-stranded DNA is generally not an essential process, as evidenced by the viability of null mutants of bacteria, yeast, and mammals (1, 3, 19). Two explanations were suggested for the importance of the D4R protein in viral DNA replication. One was that the D4R protein functions as an essential structural component of a multisubunit replication-repair complex in addition to its uracil excision activity (14, 27). Supporting this possibility, the D4R protein has been shown to interact with the A20R protein (12), an essential DNA replication factor (8, 10, 22). Alternatively, the removal of uracil residues might function in the initiation of DNA synthesis by allowing important protein-DNA interactions or participating in the production of a nick in the template (4). Similar mechanisms have been suggested for the corresponding herpesvirus enzymes under conditions in which the cellular uracil DNA glycosylase is absent or limiting (2, 5, 21).
One way to test the above hypotheses would be by producing recombinant viruses with uracil DNA glycosylase active-site mutations. The inability to isolate such vaccinia virus mutants by homologous recombination led to the conclusion that the viral glycosylase activity is essential (4). We considered that it would be useful to construct viruses with active-site mutations, even if they were not viable, for DNA replication studies. The availability of a D4R-expressing cell line which complements D4R deletion mutants (6) provided a tool for isolating nonviable D4R mutants. Using this cell line, we obtained vaccinia virus mutants with inactivating mutations in the glycosylase catalytic site of the D4R protein. Unexpectedly, these mutants had no defect in viral DNA replication and could propagate in a variety of cultured cells. Nevertheless, the mutations in the D4R protein decreased the virulence of vaccinia virus in a mouse infection model.
MATERIALS AND METHODS
Cells, virus, and plasmids.
Buffalo green monkey kidney (BGMK) cells were obtained from Y. Huang (Case Western Reserve University, Cleveland, Ohio), and human foreskin fibroblasts (HFF) were obtained from A. McBride (National Institutes of Health, Bethesda, Md.). In addition, a rabbit kidney cell line (RK-D4R-44.20) stably expressing the D4R protein, vaccinia virus vD4-ZG lacking a functional D4R gene, and plasmid pER, which contains D4R sequences flanked by D3 and D5 sequences, were gifts of F. G. Falkner (6). All cell lines, including HeLa, BS-C-1, and RK13, were maintained in Eagle's minimal essential medium (EMEM; Quality Biologicals, Inc. Gaithersburg, Md.) containing 10% fetal bovine serum (FBS). To construct plasmid pER-GFP, the enhanced green fluorescent protein (GFP) ORF under the control of the vaccinia virus P11 promoter was inserted into the intergenic region between the D4R and D5R sequences of SmaI- and HindIII-digested pER.
Site-directed mutagenesis.
Mutations in the D4R ORF of pER were created using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The following complementary oligonucleotides were synthesized to mutate active-site residues: D68N (5′ CGAGTATGTGTGTGTGGTATAAATCCGTATCCGAAAGATGG 3′) and H181L (5′ CCATAGTGGGATATCTTCCAGCGGCTAGAGACC 3′). The underlined bases represent mutated codons, and only the sense primers are shown. The generation of mutant plasmids by using oligonucleotides was carried out according to the manufacturer's instructions, and the mutations were confirmed by DNA sequencing.
Recombinant virus construction.
Approximately 106 RK-D4R-44.20 cells were infected with vD4-ZG at a multiplicity of 0.05 PFU per cell for 1 h at 37°C. The infected cells were washed twice with Opti-MEM (Invitrogen) and transfected with 2 μg of pER-GFP with or without D4R active-site mutations. After 5 h, the transfection mixture was replaced with EMEM containing 2.5% FBS, and the cells were harvested at 48 h in 0.5 ml of EMEM-2.5% FBS. Lysates were prepared by freezing and thawing the cells three times and sonicating them twice for 30 s. Recombinant viruses that expressed GFP were plaque purified five times on RK-D4R-44.20 cells. Recombinant GFP viruses with and without mutations were isolated, and their genetic purity was confirmed by PCR and sequencing.
Uracil DNA glycosylase assays.
Fluorescence assays for uracil DNA glycosylase activity were previously described (4, 17). Briefly, cells were infected with viruses at 10 PFU per cell or were mock infected. After a 1-h adsorption period, infection was allowed to proceed for 8 h at 37°C. The cells were washed twice with cold phosphate-buffered saline (140 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4, pH 7.4). The cells were then scraped into lysis buffer containing 50 mM NaCl, 50 mM Tris-HCl (pH 8.0), and 1% Nonidet P-40. Cell lysis was facilitated by rocking the mixture for 30 min at 4°C followed by centrifugation for 30 min at 12,000 × g. The supernatants were snap frozen in a dry ice-ethanol bath and stored at −80°C. The protein concentration was determined by using the Micro BCA Protein Assay Reagent kit (Pierce). Five micrograms of the total protein from each lysate was assayed for uracil DNA glycosylase activity.
To assay for uracil DNA glycosylase activity, protein from infected cell lysates was incubated with 1 μg of supercoiled pUC-19 either containing or lacking uracil residues. Uracil-containing pUC-19 was produced in Escherichia coli CJ236 (dut ung) cells (New England Biolabs). The reaction mixture also contained 50 mM Tris-HCl (pH 7.5), 20 mM EDTA, and 1 mg of heat-denatured gelatin/ml. AP sites induced in the substrate DNA were detected by removing aliquots of the reaction mixture at various times into a pH 12 buffer (20 mM K3PO4, 0.5 mM EDTA) containing 0.5 μg of ethidium bromide/ml and measuring the fluorescence. The mixture was then boiled for 4 min and quickly cooled, and fluorescence was measured again. Heat treatment causes the hydrolysis of AP sites, producing nicked and irreversibly denatured substrate DNA, resulting in a decrease in the amount of intercalated ethidium bromide and hence a loss of fluorescence. The percent fluorescence is proportional to the amount of supercoiled DNA remaining. A Turner (model 450) fluorometer was used for fluorescence measurements with filters that measure excitation at 520 nm and emissions at 605 nm.
Assays with the uracil-DNA glycosylase inhibitor protein (UGI) (New England Biolabs) were performed as described above except that 10 U of UGI were incubated with protein from each lysate in reaction buffer for 10 min at room temperature followed by 20 min on ice prior to the addition of substrate DNA.
One-step virus growth and plaque assays.
For one-step virus growth, confluent RK13, RK-D4R-44.20, or HFF cells in six-well plates were infected with 3 PFU of virus per cell and maintained at 37°C for 1 h. The inocula were removed, and the cells were washed twice and overlaid with 2 ml of EMEM containing 2.5% FBS. The cells were maintained at 37°C and harvested at various times after infection, collected by centrifugation, and resuspended in EMEM containing 2.5% FBS. The cells were disrupted by three cycles of freezing and thawing and two 30-s bursts of sonication. Virus yields were determined by titration on RK-D4R-44.20 cells at 37°C. One-step virus growth in serum-starved HFF was performed as described above except that confluent cells were grown in EMEM supplemented with 0.2% FBS for 72 h prior to, during, and following infection.
For plaque assays, RK-D4R-44.20 monolayers in six-well plates were infected with 10-fold serial dilutions of virus for 1 h. After adsorption, the inocula were removed and the cells were washed twice, overlaid with EMEM containing 0.5% methylcellulose-5% FBS, and incubated at 37°C for 2 days. The cell monolayers were stained with crystal violet, and the plaques were counted.
Analysis of viral DNA synthesis by slot blot hybridization.
Confluent RK-D4R-44.20 and RK13 cells in six-well plates were infected with 3 PFU of virus per cell as described above. The cells were harvested at various times by scraping, collected by centrifugation, washed once with phosphate-buffered saline, and resuspended in 0.3 ml of 1.5 M NaCl-0.15 M sodium citrate (pH 7.0)-1 M ammonium acetate. The cells were disrupted by three cycles of freezing and thawing. After the cells were vortexed, 50 μl of each sample was spotted in duplicate onto an Immobilon-Ny+ transfer membrane in a slot blot apparatus. The membrane was treated sequentially with 0.5 M NaOH-1 M Tris-HCl (pH 7.5) and 0.3 M NaCl-0.03 M sodium citrate (pH 7.0) and cross-linked by UV irradiation. Viral DNA was detected using the Quick-Hyb protocol (Stratagene) with vaccinia virus G7L and G8L genes that were 32P labeled using a random-priming kit (Invitrogen) and autoradiography. The radiolabeled DNA was quantified with a Storm 860 PhosphorImager (Molecular Dynamics) and ImageQuant software.
Measurement of virulence.
Purified virus, used for virulence studies, was prepared in the following manner. RK-D4R-44.20 cells (2.5 × 108) were infected with 3 PFU of virus per cell and harvested 3 days after infection. The cells were resuspended in swelling buffer (10 mM Tris, pH 9.0) and Dounce homogenized. The cytoplasm was separated from the nuclei by low-speed centrifugation and layered on a 36% sucrose cushion. After centrifugation at 13,500 rpm in an SW 28.1 rotor for 80 min at 4°C, the virus pellet was suspended in 10 mM Tris (pH 9.0). Plaque assays were performed as described above to determine the virus titers. The virulence of vaccinia virus for mice was ascertained as previously described (9, 13, 20). Essentially, groups of four 6-week-old female BALB/c mice were anesthetized and inoculated intranasally with 103 to 107 PFU of wild-type or mutant virus in 20 μl of 10 mM Tris (pH 9.0). Each mouse was weighed daily, and animals whose body weight was ≤70% of the initial value were sacrificed to avoid further stress and suffering.
RESULTS
Construction of vaccinia viruses with mutations in the catalytic site of uracil DNA glycosylase.
The conserved active-site residues of human, herpes simplex virus type 1, and E. coli uracil DNA glycosylases have been visualized by crystal structure analyses (15, 16, 23, 25). Despite the large overall sequence differences between the poxviral and other uracil DNA glycosylases, the active-site residues are retained (Fig. 1). Tyr-70, Phe-79, and Asn-120 of vaccinia virus were predicted to form the DNA recognition pocket, and Asp-68 and His-181 are likely to be involved in cleavage of the glycosylic bond. A previous study of human uracil DNA glycosylase demonstrated that mutations of the corresponding amino acids involved in cleavage inhibited enzyme activity by >99% (15). Moreover, either of these mutations in the vaccinia virus uracil DNA glycosylase completely eliminated enzyme activity (4).
FIG. 1.
Sequence alignment of uracil DNA glycosylase proteins of viruses and cells. The shaded segments indicate identical amino acids. The circles indicate conserved active-site residues. The solid circles indicate the conserved amino acids of the vaccinia virus uracil DNA glycosylase that were mutated in this study. The numbers of the amino acid residues of each protein are indicated at the left and right of the lines. HSV-1, herpes simplex virus type 1; VAC, vaccinia virus; MPXV, monkeypox virus; VAR, variola major virus; EVM, ectromelia virus; CMPV, camelpox virus; CPXV, cowpox virus; YAB, Yaba-like disease virus; SWPV, swinepox virus; MYX, myxoma virus; RFB, rabbit fibroma virus; LSD, lumpy skin disease virus; SPPV, sheeppox virus; MCV, molluscum contagiosum virus; FPV, fowlpox virus.
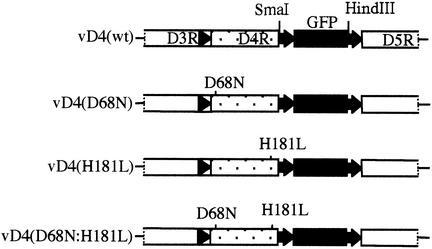
Vaccinia virus mutants containing a single D68N or H181L mutation or double mutations in the catalytic site were made by homologous recombination between vD4-ZG (6), a mutant referred to here as vΔD4 that has the central 300 bp of the D4R ORF replaced with a cassette expressing E. coli β-galactosidase and xanthine-guanine phosphoribosyltransferase, and plasmids with mutated D4R ORFs. Recombination was carried out in RK-D4R-44.20 cells, an RK13 cell line that stably expresses the D4R protein and can support the replication of D4R deletion mutants (6). To facilitate the isolation of recombinant viruses, the plasmid vectors contained a GFP expression cassette downstream of the D4R ORF (Fig. 2). Plaques comprised of recombinant viruses were identified by fluorescence microscopy and were picked for further purification, amplification, and analysis. Recombinant GFP viruses with a single mutation, vD4(D68N) or vD4(H181L), a double mutation, vD4(D68N:H181L), or no mutation, vD4(wt), were isolated, and their genetic purity was confirmed by PCR analysis and sequencing (data not shown).
FIG. 2.
Diagram of portions of recombinant viral genomes. The names of the recombinant viruses are indicated on the left. The region between the D3R and D5R ORFs is shown. The D4R promoter (the arrowhead upstream of D4R) is located within the D3R ORF. The GFP ORF preceded by the vaccinia virus P11 late promoter (the arrow immediately upstream of GFP ORF) was inserted into the intergenic region upstream of the D5R ORF and its promoter. Amino acid substitutions in the catalytic site of D4R (D68N and H181L) are indicated.
Replication of D4R mutants in noncomplementing cell lines.
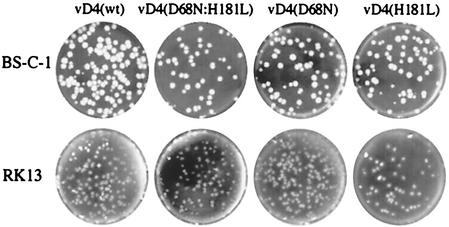
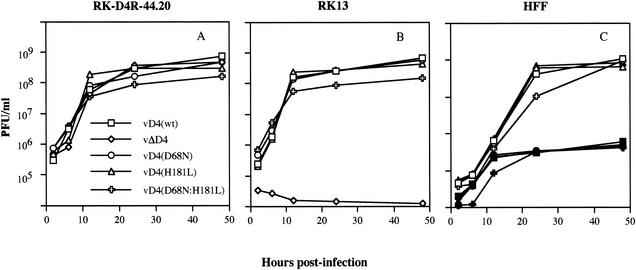
To our surprise, we found that the D4R active-site mutants formed plaques in noncomplementing BS-C-1 and RK13 cell lines (Fig. 3). In contrast, vΔD4R formed plaques only in the complementing cell line (not shown), as previously reported (6). To measure virus replication quantitatively, one-step growth experiments were carried out. At appropriate times, cells were harvested, and virus yields were determined on the complementing RK-D4R-44.20 cell line to avoid any preference for the wild-type virus. As expected, wild-type and D4R mutant viruses all replicated in the complementing RK-D4R-44.20 cells (Fig. 4A). In addition, each of the viruses except vΔD4R replicated in noncomplementing RK13 cells (Fig. 4B). Indeed, the growth curves of the wild type and single active-site mutants were almost superimposed. The double mutant grew to slightly lower levels than the others, but this was also true in the complementing cell line. Human HeLa and monkey BGMK cells also supported replication of the D4R active-site mutants but not the D4R deletion mutant (data not shown).
FIG. 3.
Plaques of recombinant viruses in noncomplementing cell lines. Recombinant viruses with wild-type or mutated D4R ORFs were plated on BS-C-1 or RK13 cell monolayers and covered with a semisolid methylcellulose overlay. After 2 days at 37°C, the monolayers were stained with crystal violet.
FIG. 4.
One-step growth of recombinant viruses in complementing and noncomplementing cells. Confluent RK-D4R-44.20 (A), RK13 (B), and HFF (C) cells were infected with 3 PFU of recombinant virus per cell. HFF were infected under normal (open symbols) and serum-starved (solid symbols) conditions. The infected cells were harvested at the indicated times, and their titers were determined on RK-D4R-44.20 cells. Virus yields were plotted as PFU per milliliter.
Previous work with herpesviruses suggested that uracil DNA glycosylase mutants replicated well in actively growing cells because of complementation by cellular uracil DNA glycosylase but less well in resting or other cells that are deficient in the enzyme (2, 5, 21). To test for a similar effect with vaccinia virus, we infected HFF that had been held in low serum to induce a resting state. We found that the wild type and active-site mutants produced similar virus titers in the serum-deprived cells, though the titers of all were reduced by approximately 2 log units compared to cells kept at normal serum concentrations (Fig. 4C).
Prichard et al. (21) demonstrated that a human cytomegalovirus mutant lacking the uracil DNA glycosylase gene is hypersensitive to the nucleoside analog 5-bromodeoxyuridine. We performed similar experiments using the D4R catalytic-site mutants. Primary HFF were infected with D4R mutant viruses and treated with different concentrations of the nucleoside analog for 2 days. After the infected cells were harvested, the virus titers were determined on the complementing RK-D4R-44.20 cells. The wild-type and mutant viruses were equally sensitive to 5-bromodeoxyuridine (data not shown).
Absence of virus-induced uracil DNA glycosylase activity.
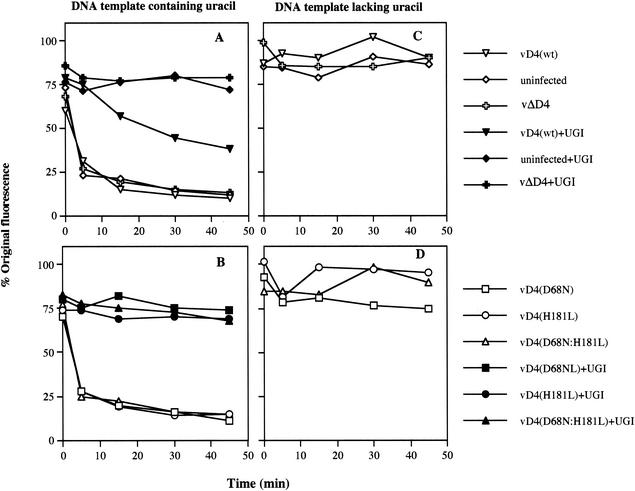
Previous studies using a rabbit reticulocyte lysate to express the vaccinia virus uracil DNA glycosylase with D68N or H181L mutations demonstrated complete loss of enzymatic activity as measured by a fluorescence-based assay with uracil-containing DNA as a substrate (4). To distinguish between viral and cellular activities, Ellison et al. (4) added UGI (28), which irreversibly binds to and inactivates cellular but not viral uracil DNA glycosylase. Without UGI treatment, uracil DNA glycosylase activity was detected in extracts of both mock- and vD4(wt)- or vΔD4-infected cells (Fig. 5A). Indeed, under these conditions, the viral activity was not above the cellular level as previously found by Ellison et al. (4). The specificity of the assay was demonstrated by its dependence on uracil residues in the DNA substrate (Fig. 5C and D). After UGI treatment, the activities of uninfected and vΔD4-infected cell extracts were eliminated, whereas that of vD4(wt) was only reduced due to a mixture of cellular and viral activities (Fig. 5A). The uracil DNA glycosylase activity of extracts of cells infected with vD4(D68N), vD4(H181L), or vD4(D68N:H181L) was entirely eliminated by UGI, indicating that there was no contribution from the viral enzyme (Fig. 5B). Absence of activity in UGI-treated extracts of mutant virus-infected cells was also found using assay conditions in which the UGI-resistant activity in parallel extracts of wild-type virus-infected cells caused the fluorescence to decrease to baseline values within 15 min (data not shown). Western blot analysis of the extracts from the virus-infected cells used in the assay showed similar amounts of D4R protein (data not shown).
FIG. 5.
Fluorescence assay of uracil DNA glycosylase activity. RK13 cells were mock infected or infected with recombinant virus at 10 PFU/cell. After 8 h at 37°C, the cells were harvested and lysed. Fluorescence assays were performed using 5 μg of cell extract protein and 1 μg of supercoiled pUC-19 DNA containing (A and B) or lacking (C and D) uracil residues. At the indicated times, aliquots of the reaction mixture were removed into pH 12 buffer, and fluorescence was measured before and after the mixtures were boiled. The percent fluorescence represents the amount of supercoiled pUC-19 remaining. The presence of nicks in the uracil containing pUC 19, prepared from E. coli CJ236 (dut ung) cells, accounted for the values of <100% at zero time.
Effects of D4R mutations on viral DNA synthesis.
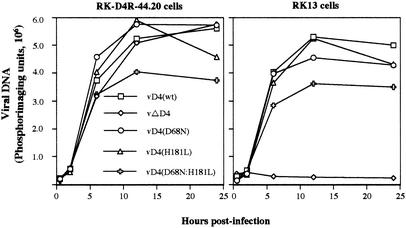
Although the uracil DNA glycosylase activity of the D4R protein was not required for virus replication, we decided to analyze viral DNA synthesis directly to see if there were any detectable differences. To measure viral DNA synthesis, RK13 and RK-D4R-44.20 cells were infected with wild-type and mutant viruses. At various times, infected cells were harvested and total DNA was prepared from the infected cell lysates. The samples were spotted onto membranes, and the DNA was then denatured and hybridized in situ with a 32P-labeled viral DNA probe. In the complementing RK-D4R-44.20 cell line, viral DNA synthesis largely reached plateau values by 12 h after infection in each case, although the level of DNA was slightly lower for the double mutant (Fig. 6). A similar result was obtained with noncomplementing RK13 cells for each of the viruses except vΔD4, which was unable to replicate (Fig. 6). These data confirmed the importance of the D4R protein for viral DNA replication but indicated that the uracil DNA glycosylase activity was not essential.
FIG. 6.
Accumulation of viral DNA in complementing and noncomplementing cells infected with recombinant viruses. RK-D4R-44.20 and RK13 cells were infected with 3 PFU of recombinant virus with wild-type or mutated D4R ORFs. At the indicated times, the cells were harvested and the total DNA was purified. The quantity of viral DNA was determined by dot blot hybridization and quantified with a PhosphorImager.
D4R active-site mutants are less pathogenic than wild-type virus.
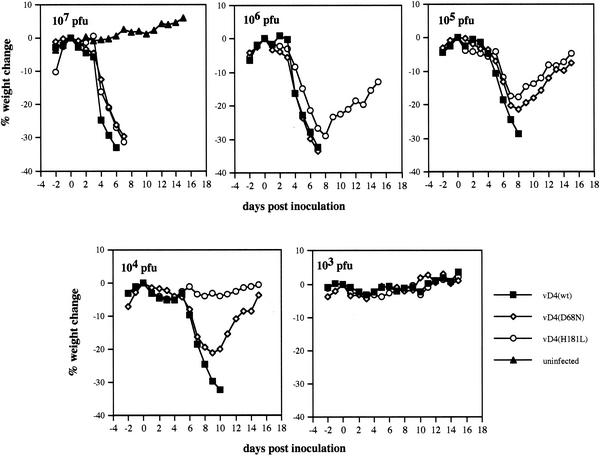
The conservation of the uracil DNA glycosylase active site in the D4R orthologs of all poxviruses (Fig. 1) strongly suggested that it has an important role in the virus life cycle. One way of examining this was to compare the pathogenicities of the recombinant viruses with wild-type and mutated D4R genes in a mouse intranasal-infection model. All animals infected with ≥104 PFU of virus expressing wild-type D4R suffered severe illness and were sacrificed within 6 to 10 days when their weight loss equaled or exceeded 30% (Fig. 7). In contrast, animals infected with 104 to 105 PFU of virus with mutated D4R suffered temporary weight loss, and none had to be sacrificed (Fig. 7). Only when 106 to 107 PFU of mutant virus was inoculated did the weight losses reach 30%. vD4(H181L) appeared to be slightly less pathogenic than vD4(D68N).
FIG. 7.
Determination of viral pathogenesis. Groups of four 6-week-old female BALB/c mice were intranasally infected with 103 to 107 PFU of purified recombinant virus. Each day, the mice were weighed, and animals that lost ≥30% of their initial weight were sacrificed. The average weight for each group at the indicated times was plotted.
DISCUSSION
Many large DNA viruses, including members of the poxvirus and herpesvirus families, encode the DNA repair enzyme uracil DNA glycosylase. Poxviruses are unique, however, in that the enzyme is required for replication in actively growing cultured cells (14, 27). The inability to isolate viable vaccinia virus mutants with active-site mutations in the uracil DNA glycosylase seemed to confirm the hypothesis that the enzyme activity is required for DNA replication (4). In the study by Ellison et al. (4), the isolation of active-site mutants depended on the rescue of a temperature-sensitive virus that expressed an unstable D4R protein under nonpermissive conditions. We took an alternative approach, based on the availability of a cell line that expresses the wild-type D4R protein, and succeeded in isolating such mutants. It was not surprising that the mutants replicated in the complementing cell line. However, these mutants also replicated in all tested noncomplementing cell lines, including the one used by Ellison et al. (4), as well as in quiescent serum-deprived primary HFF. Since our mutations were identical to ones designed in the previous study, we attribute the different outcomes to the methods used for their isolation.
Further characterization of the single and double catalytic-site mutations confirmed the absence of detectable uracil DNA glycosylase activity. Nevertheless, viral DNA replication was unimpeded. Since viral DNA synthesis does not occur in cells infected with D4R deletion mutants, the D4R protein must have a role in DNA replication that is independent of the uracil DNA glycosylase activity. A recent global yeast two-hybrid analysis of vaccinia virus ORFs showed that the D4R protein interacts with the A20R DNA replication protein (12). Moreover, the A20R protein interacted with two additional proteins implicated in viral DNA replication. A more recent study (7) demonstrated that the A20R protein has nonoverlapping sites for interactions with the three proteins, suggesting the existence of a replication complex that includes the D4R protein. Human uracil DNA glycosylase is physically associated with DNA polymerase α, suggesting that here too the DNA repair enzyme is associated with a replication complex (26). It is possible that the D4R protein participates in initiation or elongation steps of DNA replication by recruiting other viral proteins in addition to serving as a repair enzyme. Efforts to test this hypothesis are planned but will require purified replication proteins and the development of an appropriate in vitro system.
It seems unlikely that the cellular uracil DNA glycosylase could operate on poxvirus DNA because the latter is replicated in specialized cytoplasmic factory areas. Herpesviruses replicate in the nucleus, however, and it has been suggested that cellular uracil DNA glycosylase can complement viral uracil DNA glycosylase mutants in tissue culture cell lines (2, 5, 21). The delayed replication of human cytomegalovirus uracil DNA glycosylase deletion mutants in serum-deprived primary human fibroblasts was attributed to decreased levels of the cellular enzyme (2, 21). Similarly, the decreased neurovirulence of herpes simplex virus mutants was attributed to the absence of uracil DNA glycosylase in neurons (5). Roles for viral or cellular uracil DNA glycosylase in producing nicks for the initiation of DNA replication or removal of uracil at origins of DNA replication have been suggested (2, 5).
Vaccinia virus also differed from cytomegalovirus in another way. Although cytomegalovirus mutants unable to express uracil DNA glycosylase are hypersensitive to 5-bromodeoxyuridine (21), the vaccinia virus catalytic-site mutants had the same sensitivity as wild-type virus. This similarity could reflect the absence of a cytoplasmic AP endonuclease activity or the inability of the viral uracil DNA glycosylase to excise halogenated uracil.
Although the uracil DNA glycosylase catalytic-site mutants replicated normally in vitro, decreased pathogenicity was demonstrated by intranasal infection of mice. We were not surprised by this result, as the uracil DNA glycosylase active site is conserved in poxviruses and must therefore provide a selective advantage. A similar decrease in pathogenicity of herpes simplex virus uracil DNA glycosylase deletion mutants was attributed to decreased replication in neurons, which lack this enzyme activity (24). However, because of the specialized site of vaccinia virus DNA replication, it seems unlikely that differences in cellular uracil DNA glycosylase content would matter. Further studies with animal models may help to explain the loss of virulence and determine whether mutation of the active site would be useful in generating attenuated vaccinia virus strains for use as recombinant vaccines.
Several orthopoxvirus enzymes are being considered as targets for antiviral drugs. Although the catalytic site of the uracil DNA glycosylase is not essential for vaccinia virus DNA replication in vitro, drugs could provide therapeutic benefit by decreasing pathogenicity. In addition, the protein is highly conserved among orthopoxviruses; there are only five amino acid differences between the proteins of vaccinia virus and variola major virus, suggesting that an inhibitor could be broad enough to inhibit all orthopoxvirus orthologs. The cellular enzyme, though sharing the active-site residues, is quite divergent: there are 185 amino acid differences between the human uracil DNA glycosylase and the variola major virus homolog, making selectivity for the viral enzyme a possibility.
In summary, we have found that the poxvirus uracil DNA glycosylase is a multifunctional protein, and future work will be directed toward understanding its role in DNA replication, which is not dependent on glycosylase activity, and its role in virulence, which is.
Acknowledgments
We thank L. Wyatt and E. Katz for helpful discussions, N. Cooper for maintaining and providing tissue culture cells, and G. McFadden for antiserum to the vaccinia virus DNA glycosylase.
REFERENCES
- 1.Burgers, P. M., and M. B. Klein. 1986. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J. Bacteriol. 166:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courcelle, C. T., J. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Requirement of uracil-DNA glycosylase during transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan, B. K., and B. Weiss. 1982. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherchia coli. J. Bacteriol. 151:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellison, K. S., W. Peng, and G. McFadden. 1996. Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J. Virol. 70:7965-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Focher, F., A. Verri, P. Verzeletti, P. Mazzarello, and S. Spadari. 1992. Uracil in oriS of herpes simplex 1 alters its specificity recognition by origin binding protein (OBP): does virus induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma 102:S67-S71. [DOI] [PubMed] [Google Scholar]
- 6.Holzer, G. W., and F. G. Falkner. 1997. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 71:4997-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishii, K., and B. Moss. Mapping interaction sites of the A20R protein component of the vaccinia virus DNA replication complex. Virology, in press. [DOI] [PubMed]
- 8.Ishii, K., and B. Moss. 2001. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J. Virol. 75:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettle, S., N. W. Blake, K. M. Law, and G. Smith. 1995. Vaccinia virus serpins B13R (SPI-2) and B22R (SPI-1) encode Mr 38.5 and 40K, intracellular polypeptides that do not affect virus virulence in a murine intranasal model. Virology 206:136-147. [DOI] [PubMed] [Google Scholar]
- 10.Klemperer, N., W. McDonald, K. Boyle, B. Unger, and P. Traktman. 2001. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J. Virol. 75:12298-12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl, T. 1994. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 12.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntosh, A. A., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millns, A. K., M. S. Carpenter, and A. M. DeLange. 1994. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology 198:504-513. [DOI] [PubMed] [Google Scholar]
- 15.Mol, C. D., A. S. Arvai, R. J. Sanderson, G. Slupphaug, B. Kavli, H. E. Krokan, D. W. Mosbaugh, and J. A. Tainer. 1995. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell 82:701-708. [DOI] [PubMed] [Google Scholar]
- 16.Mol, C. D., A. S. Arvai, G. Slupphaug, B. Kavli, I. Alseth, H. E. Krokan, and J. A. Tainer. 1995. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell 80:869-878. [DOI] [PubMed] [Google Scholar]
- 17.Morgan, A. R., and J. Chlebek. 1988. Quantitative assays for uracil-DNA glycosylase of high sensitivity. Biochem. Cell Biol. 66:157-160. [DOI] [PubMed] [Google Scholar]
- 18.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Nilsen, H., I. Rosewell, P. Robins, C. F. Skjelbred, S. Andersen, G. Slupphaug, G. Daly, H. Krokan, T. Lindahl, and D. E. Barnes. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5:1059-1065. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson, J. E., and G. L. Smith. 1994. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology 204:376-390. [DOI] [PubMed] [Google Scholar]
- 21.Prichard, M. N., G. M. Duke, and E. S. Mocarski. 1996. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 70:3018-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punjabi, A., K. Boyle, J. DeMasi, O. Grubisha, B. Unger, M. Khanna, and P. Traktman. 2001. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J. Virol. 75:12308-12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam, C. D., M. J. Shroyer, A. J. Lundquist, C. D. Mol, A. S. Arvai, D. W. Mosbaugh, and J. A. Tainer. 1999. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 287:331-346. [DOI] [PubMed] [Google Scholar]
- 24.Pyles, R. B., and R. L. Thompson. 1994. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 68:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savva, R., K. McAuley-Hecht, T. Brown, and L. Pearl. 1995. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature 373:487-493. [DOI] [PubMed] [Google Scholar]
- 26.Seal, G., and M. A. Sirover. 1986. Physical association of the human base excision repair enzyme uracil DNA glycosylase with the 70,000-dalton catalytic subunit of DNA polymerase alpha. Proc. Natl. Acad. Sci. USA 83:7608-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart, D. T., C. Upton, M. A. Higman, E. G. Niles, and G. McFadden. 1993. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J. Virol. 67:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi, I., and J. Marmur. 1963. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature 197:794-795. [DOI] [PubMed] [Google Scholar]
- 29.Traktman, P. 1996. Poxvirus DNA replication, p. 775-798. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Tye, B. K., P. O. Nyman, I. R. Lehman, S. Hochhauser, and B. Weiss. 1977. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc. Natl. Acad. Sci. USA 74:154-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upton, C., D. T. Stuart, and G. McFadden. 1993. Identification of a poxvirus gene encoding a uracil DNA-glycosylase. Proc. Natl. Acad. Sci. USA 90:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]