Abstract
Members of the DEAD-box family of helicases, distinguished by a core characteristic sequence of Asp-Glu-Ala-Asp, are expressed in a wide range of prokaryotes and eukaryotes and exhibit diverse cellular functions, including DNA transcription, recombination and repair, RNA processing, translation, and posttranslational regulation. Although ubiquitous, the function of most DEAD-box proteins is unknown. We and others have recently cloned DP103, which harbors conserved DEAD-box, helicase, and ATPase domains in its N terminus. DP103 (also termed Gemin3 and DDX20) interacts with SF-1, SMN, EBNA2, and EBNA3C in mammalian cells. Here we demonstrate that a discrete domain within the nonconserved C-terminal region of DP103 directly interacts with SF-1. This domain exhibits an autonomous repression function and is necessary and sufficient for repressing the transcriptional activity of SF-1. Furthermore, intact DP103 exhibits helicase activity. Importantly, the C-terminal domain is obligatory but not sufficient for this unwinding activity of DP103. Together, our results support a novel paradigm for transcriptional repression and demonstrate the bifunctional role of the C-terminal domain of DP103.
Members of the expanding family of DEAD-box proteins are found in diverse organisms, including viruses, bacteria, and eukaryotes from yeast to humans (30, 36). Proteins from this family are characterized by a core segment of similarly spaced conserved motifs. Among these motifs are ATPase motifs I (AXXGXGKS/T) and II (Asp-Glu-Ala-Asp, D-E-A-D; hence the family name), also known as Walker motifs A and B, respectively (66), which are responsible for binding the nucleotide triphosphate-Mg2+ complex (69). Additionally, motif III is defined as a helicase domain, and motif VI is thought to bind nucleic acids (reviewed in reference 33). Whereas ATPase motifs exhibit a similar biochemical function, the activity of other motifs, including helicase, remains conjectural for most family members. Besides the core segment region that contains the conserved motifs, the N- and C-terminal regions of individual DEAD-box proteins exhibit little or no sequence homology to each other. This modular organization suggests that the core region harbors conserved functions, whereas the N and C termini may play a role in protein-specific functions. These specialized functions may include discriminatory docking sites for proteins and/or additional RNA-binding motifs that confer substrate specificity or direct the DEAD-box protein to selective subcellular localization (13). Helicases are capable of unwinding duplex DNA or RNA in the presence of energy derived from nucleotide triphosphate hydrolysis. These enzymes are required for all aspects of nucleic acid metabolism in both prokaryotes and eukaryotes. They are involved in DNA repair, replication, recombination and transcription, RNA processing, transcript stability, and translation initiation (13, 16, 19, 52). DEAD-box proteins commonly display RNA helicase activity (12).
Several groups, including our own, have recently cloned DP103, a protein of 825 amino acids (aa) that contains the DEAD-box conserved region at the N terminus. Grundhoff et al. (20) identified DP103 by virtue of its interaction with Epstein-Barr virus nuclear antigens EBNA2 and EBNA3C and demonstrated its intrinsic ATPase activity. In addition, DP103 (also termed Gemin3 and DDX20) was found to directly interact with spinal muscular neuron (SMN) protein (5, 6, 65). Nevertheless, the function of DP103 in these contexts remained unclear. In the pursuit of mechanisms that modulate the transcriptional activity of the orphan nuclear receptor steroidogenic factor 1 (SF-1), we identified a previously unknown repression domain within SF-1 (42). Using this repression domain as bait in a yeast two-hybrid approach we cloned from a rat ovary library a novel regulator of SF-1, which is a murine homologue of DP103 (42). We found that DP103 is ubiquitously expressed at a low level, with higher expression predominantly in steroid-producing murine tissues, which also express SF-1 (42). Importantly, SF-1, EBNA2/EBNA3C, and SMN interact with DP103 via the nonconserved C-terminal region of DP103 (5, 20, 65), suggesting that this region is capable of forming protein complexes and potentially functions in a manner distinct from that of the conserved N-terminal DEAD-box core domain. Only one DEAD-box protein, p68, has been previously shown to interact with a member of the nuclear receptor superfamily (17). p68 binds the N-terminal AF-1 domain of an estrogen receptor and enhances its transcriptional activity. In contrast, DP103 interacts with the C-terminal repression domain of SF-1 and diminishes its transcriptional activity (42).
SF-1 is essential for endocrine and reproductive system development and function (22, 43, 48). Mice harboring a targeted disruption of the SF-1 gene lack gonads and adrenal glands, which consequently leads to early neonatal death from adrenal insufficiency (3, 37, 49, 53). Additionally, the ventromedial hypothalamic nucleus fails to fully develop in these mice, as do the pituitary gonadotropes (26, 71). Molecular analysis revealed that SF-1 regulates the expression of many steroidogenic enzymes (P450scc, P450c17, P450c21, P450c11, P450arom, and 3β-hydroxysteroid dehydrogenase) (39, 43, 48) and other pivotal regulators of endocrine and reproductive function, including steroidogenic acute regulatory protein (StAR), Mullerian inhibitory substance, gonadotropin-releasing hormone receptor, and luteinizing hormone-β (reviewed in references 22, 43, and 48). The well-synchronized expression of these proteins suggests that they are tightly modulated by SF-1 and possibly by additional regulatory proteins. Nevertheless, the mechanisms that govern SF-1 action are incompletely understood. SF-1 is an orphan receptor that binds its DNA response elements as a monomer and thus is not a target for modulation by a DNA binding heterodimerized partner (31, 38, 68). We and others have previously identified several SF-1 domains that modulate its activity through interactions with coregulators. These include the C-terminal AF-2 hexamer, interacting with SRC-1/CBP (12, 28); the distal repression domain, interacting with DAX-1 and the nuclear receptor corepressor (N-CoR) (11, 27); and the proximal interaction domain, interacting with both SRC-1 and DAX-1 (11, 12). Also vital for SF-1 function are serine203, which interacts with GRIP1 (23), and the proline-rich region near the DNA binding domain (35).
Repression of SF-1 activity by a DEAD-box protein constitutes a novel regulatory pathway, which may prove relevant to endocrine and reproductive homeostasis. To elucidate the biological action of DP103 we sought to analyze the repression of SF-1 by DP103. Here we mapped the domain that physically interacts with SF-1 to aa 721 to 825 within the nonconserved C-terminal region of DP103. Furthermore, we demonstrated that this domain is necessary and sufficient for the repression function of DP103. We also found that DP103 exhibits RNA helicase activity and that the nonconserved C-terminal region is obligatory for this activity.
MATERIALS AND METHODS
Plasmids.
Murine SF-1 cDNA was described previously (12). pBS-SF-1wt and pBSK-SF-1 mAAEY were generated as previously detailed (42). Full-length DP103 (aa 1 to 825) and a series of deletion mutants (aa 1 to 410, 411 to 825, 411 to 617, 411 to 521, 515 to 617, 456 to 547, 611 to 825, 611 to 727, and 721 to 825) were generated by PCR amplification with the mouse full-length DP103 plasmid pcDNA3-DP103 (42) as a template (see Table 1 for primers). For VP16 fusion proteins, PCR fragments were cloned downstream from the VP16 transactivation domain at the BamHI-PstI sites in a pVP16 vector (Clontech). For GAL4 fusion proteins, PCR fragments as described above as well as aa 1 to 617 and 1 to 727 were cloned downstream from GAL4 DNA binding domain (DBD) (amino acids 1 to 147) at the BamHI-PstI sites within the pM1 vector. Full-length p68 was cloned from human placental cDNA by using primers F-ATGTCGGGTTATTCGAGTGAC and R-TTATTGGGAATATCCTGTTGG and was cloned downstream from GAL4 DBD at the BamHI-HindIII sites within the pM2 vector. A similar approach was utilized for generation of His-tagged DP103 constructs, with in-frame subcloning at BamHI-PstI sites of pQE32 or at BamHI-BglI sites of PQE16 expression vectors (Qiagen Inc.). The activity of GAL4 fusion constructs was assessed by using either the reporter plasmid ΔGKI, which contains five GAL4 binding sites upstream of the E1B-TATA promoter, or GAL4 × 5-tkLuc, which contains five GAL4 binding sites upstream of the thymidine kinase minimal promoter; both were linked to luciferase. SF-1 activity was measured by using SF-1 luciferase reporter S25 as well as P450scc (a gift from J. S. Richards, Baylor College of Medicine, Houston, Tex.), as previously described (7, 42).
TABLE 1.
PCR primer sequences used to generate DP103 deletion mutants (5′ → 3′)a
| DP1031 | F GGATCCAAATGGCGGCGGCGACG |
| DP103410 | R CTGCAGTCATGTCAGTCCCAATGTAC |
| DP103411 | F GGATCCAAGTGACCTACTGTTGTAGA |
| DP103456 | F GGATCCAAGTTAAAGCTGCCATGCAT |
| DP103515 | F GGATCCAAAAGCTTCCTGTGAAAAGC |
| DP103521 | R CTGCAGTCAGTGGCTTTTCACAGGAA |
| DP103547 | R CTGCAGTCACTTAACTTGCTCTTCCA |
| DP103611 | F GGATCCAAGTGGAAATTATCCGGCAC |
| DP103617 | R CTGCAGTCAGTAGTGCCGGATAATTT |
| DP103721 | F GGATCCAAAAGGCAAGGCATAAAGAA |
| DP103727 | R CTGCAGTCAGCCTTCTTTATGCCTTG |
| DP103825 | R CTGCAGTCATTGGTTACCACGCATCA |
Numbers denote first (F) or last (R) amino acid in fragment.
Cell culture and transfections.
CV-1 and JEG3 cells were maintained and passaged as previously described (12). Adrenocortical Y1 cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and antibiotics at 37°C and 10% CO2. Cells were transfected by using the modified calcium-phosphate method described previously (8), which included 0.05 μg of cytomegalovirus (CMV)-β-galactosidase plasmid (to normalize for cell viability and transfection efficiency). Standard luciferase assays were performed 48 h after transfection as previously described (12) by using a Lumistar 96-well plate reader (BMG, Durham, N.C.). All experiments were performed in duplicate and were repeated at least three times. Results (means ± standard deviations [SD]), normalized to β-galactosidase activity, were expressed as relative luciferase units (cRLU). For Y1 RNA, cells were seeded in 6-well plates at a density of 150,000 cells/well. One day after transfection the cells were rinsed and cultured in medium containing 0.1 μM adrenocorticotropin (ACTH; Sigma) or water vehicle and were harvested for total RNA 24 h later. One day after transfection the cells were rinsed and cultured in medium containing 0.1 μM ACTH (Sigma) or water vehicle and were harvested for total RNA 24 h later.
Protein-protein interaction assay.
His-tagged DP103 fragments, described above, were expressed in Escherichia coli strain XL-1 Blue (Stratagene) and were induced for 4 h at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 0.5 M NaCl, 10 mM β-mercaptoethanol, 1% Tween 20, 10% glycerol, 15 mM imidazole) and then were incubated with 1 mg of lysozyme/ml for 30 min on ice, sonicated, and centrifuged at 15,000 × g at 4°C. The supernatant (soluble fraction) was saved for protein purification. The pellet was solubilized in a buffer containing 1.5% Sarkosyl, 25 mM triethanolamine, 1 mM EDTA, 2% Triton X-100, 1 mM CaCl2, and 15 mM imidazole. The pellet was then centrifuged and the supernatant was again stored. His-tagged proteins were purified with Ni-nitrilotriacetic acid (NTA) resin (Qiagen Inc.) under native conditions. The supernatant was applied to an appropriate volume of 50% slurry of Ni-NTA agarose resin and was gently mixed at 4°C for 1.5 h. The bound His-tagged proteins were washed three times by using gradients of 0.8, 8, and 20 mM imidazole in washing buffer (50 mM NaH2PO4 [pH 8.0], 0.5 M NaCl, 10 mM β-mercaptoethanol, 20% glycerol, and 2% Triton X-100) and were batch-eluted by increasing the imidazole concentration to 500 mM. Purified His-tagged DP103 fragments were concentrated by using a Centricon centrifugal filter (Millipore). SF-1 wild type and SF-1 mAAEY were in vitro translated and were labeled with [35S]methionine by using TNT (Promega). For the in vitro protein-protein interaction assay the His-tagged DP103 fragments bound to Ni-NTA agarose resin were equilibrated in 70 μl of binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40). Five microliters of [35S]Met-labeled SF-1 was added for 2 h at 4°C. The resin was washed four times with 800 μl of washing buffer supplemented with 40 mM imidazole. Bound proteins were released by boiling for 5 min, resolved on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE), and visualized by autoradiography. As controls, 5 μl of in vitro-translated SF-1 wild type or SF-1 mAAEY was reacted with Ni2+-NTA resin alone.
Expression analysis and hormone level.
Total cellular RNA was isolated from Y1 cells by using TRIzol (Life Technologies, Inc.) following the manufacturer's instructions and was incubated with DNase I (1 U/10 μg of RNA; Ambion, Inc.) at 37°C for 30 min and was inactivated with DNase Inactivation Reagent (Ambion) at room temperature for 2 min. The RNA was ethanol precipitated and quantified. For quantitative PCR we initially performed reverse transcription (RT) with 0.5 μg of total RNA in a 50-μl RT reaction (Applied Biosystems, Inc.) which included 5 μl of Taqman RT buffer, 11 μl of 25 mM MgCl2, 10 μl of 10 mM deoxynucleoside triphosphate mixture, 2.5 μl of 50 μM random primer, 20 U of RNase inhibitor, and 1.25 μl of murine leukemia virus reverse transcriptase. Transcript level was determined by quantitative PCR with a 5700 Sequence Detector System (Applied Biosystems, Inc.). Each 50 μl of PCR included 3 μl of RT products, forward primers, and reverse primers (Table 2) and 25 μl of SYBR Green master mixture (Applied Biosystems, Inc.). Transcript level was determined by using GeneAmp 5700 SDS software. For Western immunoblotting, transfected CV-1 cells were lysed and protein concentration was determined as previously described (50). Total cell lysate (30 μg) was subjected to SDS-10% PAGE, transferred to Immobilon-P transfer membrane (Millipore), and blotted with primary mouse anti-GAL4DBD antibody (sc510; Santa Cruz) and horseradish peroxidase-conjugated goat anti-mouse secondary antibody. The signal was visualized by chemiluminescence (Amersham Pharmacia Biotech). A rabbit polyclonal antibody and a monoclonal antibody (Chemicon) were used for detection of DP103 and P450scc, respectively. Protein levels were quantified with densitometry (PhosphorImager; Molecular Dynamics, Inc.).
TABLE 2.
Primer sequences used for quantitative RT-PCR (5′→3′)
| mRNA | Primer sequencea
|
|
|---|---|---|
| F′ | R′ | |
| P450scc | CCAGTGTCCCCATGCTCAA | CAGCTGCATGGTCCTTCCA |
| P450c21 | CCCGGGTTTTCTGCACTTC | TGTAGATGGGCCCGAGTTTC |
| StAR | GGAGATGCCGGAGCAGAGT | GCCAGTGGATGAAGCACCAT |
| SF-1 | CGCACCATCAAGTCTGAGTATCC | GCTGCAATATGAGCTCTGGTACA |
F′, forward sequence; R′, reverse sequence.
Progesterone levels (in nanograms per milliliter) in media from Y1 cells transfected with the different DP103 plasmids were determined by using enzyme immunoassay (Oxford Biomedical Research, Oxford, Mich.). Measurements were performed in duplicate, each from a pair of wells identically transfected 48 h prior to analysis of hormone concentration.
Helicase assays.
For generation of RNA strands, a pSV72 vector (Promega) was first linearized with BamHI and was transcribed with SP6 polymerase in the presence of [α-32P]CTP (150 μCi) as recommended by the manufacturer (Promega), yielding a 50-nucleotide transcript from the polylinker of pSV72 (GAACTCGAGCAGCTGA AGCTTGCATGCCTGCAGGTCGACTCTAGAGGATC; the 15-bp duplex region is underlined). For the complementary strand, pSV72 was linearized with AccI and was transcribed with T7 polymerase in the presence of unlabeled nucleotides, yielding a 66-nucleotide transcript (GGGAGACCGGCAGATCTGATATCATCGATGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCG; the 15-bp duplex region is underlined). The two strands were separated by using a denaturing 8 M urea-6% PAGE. The bands were excised and eluted overnight in 0.5 M ammonium acetate, 1 mM EDTA, and 0.1% SDS on a 4°C rotator. The DNA oligomers, which correspond to the RNA oligomers described above, were purchased from Integrated DNA Technologies. The short DNA strand was [γ-32P]ATP labeled by using T4 kinase (Life Technologies, Inc.). RNA and DNA strands were phenol-chloroform extracted and ethanol precipitated. Unlabeled long strands were annealed to labeled short strands by gradual cooling from 95 to 37°C over 3 h in a mixture of 20 mM HEPES, KOH (pH 7.2), 250 mM NaCl, and 1 mM EDTA. The annealed substrates were resolved on a native 8% PAGE, excised, eluted, and ethanol precipitated as described above and then were dissolved in diethyl pyrocarbonate-treated H2O. The helicase assays were performed in 20 μl of buffer (20 mM Tris-HCl [pH 8.0], 70 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 3 mM ATP, 20 U of RNasin, 200 μg of bovine serum albumin/ml) by using partially duplexed substrates (50 fmol) and 50 ng of purified His-tagged DP103. After 15 min of incubation at 37°C the reaction was quenched with 5 μl of stop solution (3% SDS, 30% glycerol, and 150 mM EDTA). Reaction products were separated on SDS-10% PAGE in 1× TBE (45 mM Tris-borate-1 mM EDTA) at 4°C and 160 V for 1.5 h. The gel was then dried and analyzed by using a PhosphorImager. Double-stranded substrates, incubated at 37°C or boiled, served as negative and positive controls, respectively.
RESULTS
The C-terminal domain of DP103 (aa 721 to 825) physically interacts with SF-1.
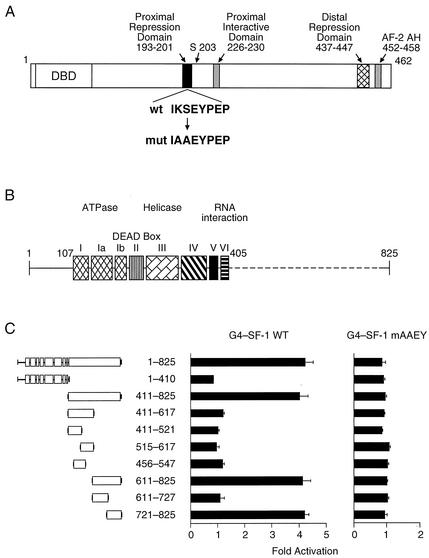
We have previously shown that DP103 directly interacts with the proximal repression domain (PRD) of SF-1 (aa 193 to 201, Fig. 1A) and that mutations in PRD significantly diminish this interaction (42). In addition, we have demonstrated that the nonconserved C-terminal domain of DP103 (aa 405 to 825, Fig. 1B) mediates the interaction with SF-1 (42). To identify the specific region within DP103 that mediates the interaction with SF-1 we generated a series of DP103 C-terminal fragments, fused to the activation domain of VP16, and tested for their interaction with GAL4-SF-1120-462 by employing a mammalian two-hybrid approach. As shown in Fig. 1C, GAL4-SF-1120-462 chimeric protein interacted with intact DP103 as well as with each DP103 fragment that harbored the C-terminal DP103721-825 region. Importantly, the conserved N-terminal (aa 1 to 410) fragments of DP103 failed to interact with SF-1. As expected, we found no interaction between any DP103 construct and GAL4-SF-1120-462 that was mutated in PRD (Fig. 1C). To obtain further support for the direct interaction of DP103721-825 with SF-1 we examined the interaction of the two proteins in vitro. For this purpose, we tested the ability of resin-bound His-tagged DP103 fragments to pull down wild-type or mutant 35S-labeled SF-1. As shown in Fig. 2, only fragments 411 to 825 and 721 to 825 of DP103 interacted with 35S-labeled SF-1. The interaction was not observed in the presence of mutant SF-1. Taken together, DP103721-825 is required for the direct association of DP103 with SF-1.
FIG. 1.
DP103 interacts with the PRD in SF-1 (aa 193 to 201) through aa 721 to 825. (A) Schematic diagram depicting the main transcriptional regulatory domains of SF-1. A mutated PRD, known to abrogate SF-1 repression, is shown. (B) A diagram of DP103, denoting eight highly conserved motifs within the N-terminal region of DEAD-box family proteins as well as the nonconserved C-terminal region. (C) DP103721-825 interacts with wild type SF-1 but not with PRD mutant SF-1. Interaction was detected by using a mammalian two-hybrid assay, with SF-1120-462 fused downstream from GAL4 and DP103 fragments (as shown) fused to the VP16 activation domain. Plasmids were transiently transfected into CV-1 cells along with the GAL4 reporter plasmid ΔGKI. Results represent three independent experiments performed in duplicate, expressed as fold activation over control in which the empty VP16 plasmid was used and normalized to β-galactosidase activity.
FIG. 2.
DP103721-825 physically interacts with SF-1's PRD (aa 193 to 201) in vitro. Wild-type SF-1 (W) and PRD mutant SF-1 (M) cloned in pBSK were expressed and labeled with [35S]methionine in vitro by using a TNT transcription-translation system. Labeled SF-1 was incubated with His-tagged DP103 fragments bound to Ni2+-NTA agarose resin. Bound SF-1 was detected by using SDS-10% PAGE and autoradiography. Input included 20% SF-1 that was used for each assay. The empty Ni2+-NTA agarose resin, reacted with wild-type or mutant SF-1, served as negative control.
The C-terminal domain of DP103 (aa 721 to 825) represses SF-1 activity.
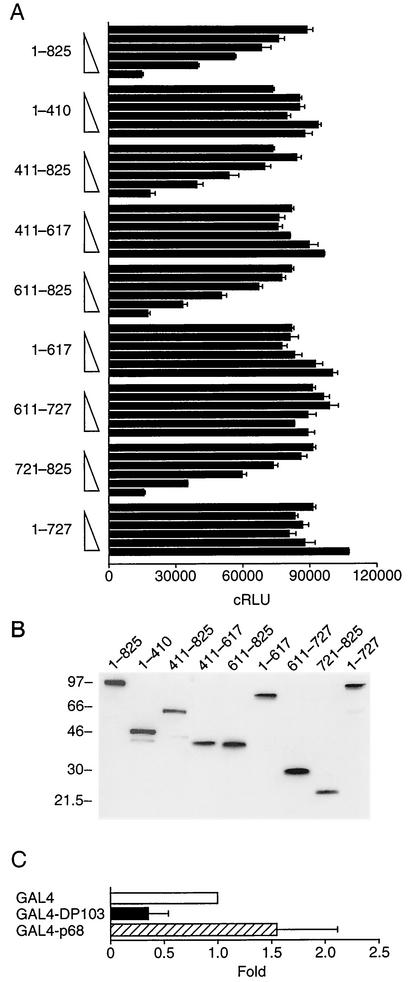
We have previously demonstrated that DP103 represses the transcriptional activity of SF-1. We therefore sought to determine if DP103 contains an intrinsic repression domain. For this purpose we fused full-length DP103 or a series of DP103 fragments to GAL4 and determined the ability of these chimeric proteins to regulate the transcriptional activity of a GAL4 reporter plasmid. As shown in Fig. 3A, only GAL4-DP103 chimeric constructs that contained aa 721 to 825 (411 to 825, 611 to 825, and 721 to 825) repressed reporter activity. The repression was concentration dependent, and the degree of repression was similar to that of GAL4 fused to full-length DP103. In contrast, GAL4-DP103 constructs spanning aa 1 to 410, 411 to 617, 1 to 617, 611 to 727, or 1 to 727 did not repress the GAL4 reporter. To confirm an equivalent expression of transfected GAL4-DP103 constructs we used Western immunoblotting to analyze the expression of these chimeric proteins in CV-1 cells. The expression of GAL4-DP103 constructs was uniform (Fig. 3B), indicating that the differences in transcriptional activity represent transcriptional repression function of DP103 fragments. As a control we also demonstrated that a chimeric protein of GAL4 DBD fused to the DEAD-box protein p68, which has been shown to enhance the activity of the estrogen receptor (17), did not repress the transcriptional activity of a GAL4 reporter plasmid (Fig. 3C). Taken together these findings indicate that aa 721 to 825 of DP103, which directly interact with SF-1, harbor a transcriptional repression domain.
FIG. 3.
DP103 harbors a repression domain at aa 721 to 825. (A) CV-1 cells were cotransfected with increasing amounts (0, 0.03, 0.1, 0.3, 1.0, and 3.0 μg) of either full-length DP103 or DP103 fragments, fused to GAL4, along with the reporter plasmid GAL4 × 5-tkLuc. Results (means ± SD) are expressed as cRLU normalized to β-galactosidase activity and represent three independent experiments, each performed in duplicate. (B) Western immunoblotting of the GAL4-DP103 fusion proteins analyzed in panel A, demonstrating equal cellular expression of transfected plasmids. Lysates (30 μg) were prepared from CV-1 cells that were transfected with 3 μg of each DP103 plasmid, separated by SDS-PAGE, and immunodetected by using anti-GAL4 antibody as described in Materials and Methods. (C) The DEAD-box protein p68 (3 μg, expressed as chimeric protein GAL4-p68) does not repress the reporter plasmid GAL4 × 5-tkLuc in CV-1 cells. Results are means of three independent experiments, each performed in duplicate, and are expressed as fold over control in which the empty GAL4 plasmid was used.
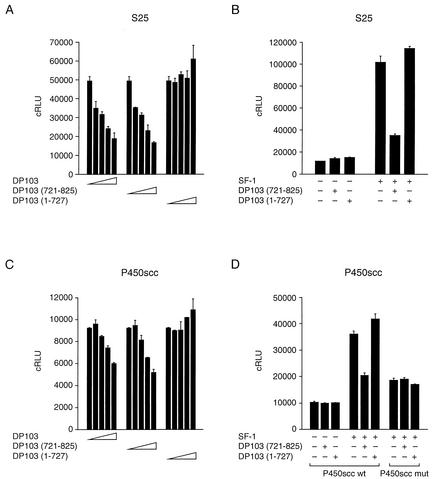
Having localized the repression domain of DP103 to aa 721 to 825, a domain that interacts with SF-1, we hypothesized that DP103721-825 is necessary and sufficient for repression of SF-1. We tested this hypothesis by utilizing two approaches. We initially analyzed the effect of DP103721-825 on reporter gene activation by SF-1. For this purpose we cotransfected JEG3 cells with an SF-1 expression plasmid, along with increasing concentrations of either GAL4-DP1031-825, GAL4-DP103721-825, or GAL4-DP1031-727 expression plasmids and the SF-1 synthetic reporter S25, which contains two SF-1 binding elements (TCAAGGTCA) upstream from the luciferase gene (12). As shown in Fig. 4A, the repression domain at DP103721-825 repressed SF-1 activity in a concentration-dependent manner, reproducing the repression by intact DP103. There was no effect on basal promoter activity in the absence of SF-1 (Fig. 4B). The estrogen receptor coactivator GAL4-p68 did not repress SF-1 activity (data not shown). We repeated the experiment with the rat P450scc-Luc promoter. This promoter binds SF-1 at positions −79 (SCC2) and −51 (SCC1) and is activated by coexpressed SF-1 (7, 9). As expected, coexpression of either GAL4-DP1031-825 or GAL4-DP103721-825, but not GAL4-DP1031-727, diminished P450scc-Luc activity (Fig. 4C). In the absence of SF-1 there was no repression of basal promoter activity. Similarly, there was no effect of DP103 constructs on the P450scc promoter that was mutated at both SF-1 binding elements (Fig. 4D).
FIG. 4.
DP103721-825 represses the transcription of SF-1 target promoters. (A) The concentration-dependent influence of GAL4-fused full-length or truncated DP103 on the transcriptional activity of SF-1. JEG3 cells were cotransfected with 0.05 μg of CMV-SF-1 and either GAL4-DP1031-825, GAL4-DP103721-825, or GAL4-DP1031-727 (0, 0.1, 0.3, 1.0, and 3.0 μg), along with 0.5 μg of the SF-1 luciferase reporter plasmid S25. (B) The repression effect of DP103 is observed only in the presence of SF-1. Transfection was performed as described above, with 3 μg of GAL4-DP103721-825 or GAL4-DP1031-727. The empty expression vector CMV-neo or GAL4 was used as control for SF-1 and DP103, respectively. (C) Concentration-dependent influence of GAL4-fused full-length or truncated DP103 on the transcriptional activity of an SF-1-responsive rat P450scc reporter. Transfection was performed as described above, with 0.5 μg of the SF-1 luciferase reporter plasmid P450scc. (D) SF-1 is required for the repression effect of DP103. The repression was abrogated when the two SF-1 binding elements in the P450scc promoter were mutated. Transfection was performed as described above. Results (means ± SD) are expressed as cRLU normalized to β-galactosidase activity and represent three independent experiments, each performed in duplicate.
In a second approach, we analyzed the influence of DP103721-825 on physiological targets for SF-1 in Y1 adrenocortical cells. These cells express SF-1 and DP103 endogenously (42), and steroid production by these cells is inducible by ACTH (44, 59). We determined the influence of overexpressed DP103 on the expression of P450scc, P450c21, and StAR, measured by quantitative real-time PCR. Transfection of GAL4-DP1031-825 (3 μg) led to a 2.4-fold increase in DP103 protein and resulted in a concentration-dependent reduction in basal and ACTH-induced expression of P450scc and P450c21 (Fig. 5A and B). A similar effect was observed with overexpression of GAL4-DP103721-825 but not with GAL4-DP1031-727, which is devoid of the repression domain. The reduced expression of P450scc protein (twofold) was confirmed by Western analysis (data not shown). Interestingly, there was no repression of StAR by any of the DP103 constructs (Fig. 5C). DP103 constructs did not affect SF-1 expression (Fig. 5D), indicating that the down-regulation of SF-1 targets could not be attributed to altered SF-1 expression. To confirm the repressive effect of GAL4-DP1031-825 and GAL4-DP103721-825 on steroidogenesis we measured progesterone production by Y1 cells, transfected with the three GAL4-DP103 expression plasmids. We found that the medium progesterone level was significantly reduced in Y1 cells transfected with GAL4-DP1031-825 or GAL4-DP103721-825 but not GAL4-DP1031-727 (Fig. 5E). The expression of DP103 was unchanged by ACTH (data not shown). Together, our data clearly demonstrate that DP103 represses the activity of SF-1 and that the DP103721-825 domain is necessary and sufficient for this effect.
FIG. 5.
DP103721-825 diminishes the expression of SF-1 target genes in the adrenocortical line. Y1 cells were transfected with increasing amounts (0, 1.0, and 3.0 μg) of GAL4-DP1031-825, GAL4-DP103721-825, or GAL4-DP1031-727. Steroidogenesis was stimulated with 0.1 μM of ACTH or water vehicles 18 h after transfection. Total RNA was isolated and processed for quantitative RT-PCR as described in Materials and Methods. (A) Expression of P450scc mRNA. (B) Expression of P450c21 mRNA. (C) Expression of StAR mRNA. (D) GAL4-DP103 fusion proteins do not diminish the expression of SF-1. Results (means ± SD) are expressed as fold expression over control (no ACTH, no GAL4-DP103) and represent three independent experiments, each performed in duplicate. (E) Progesterone levels in media from cultured Y1 cells transfected with GAL4-DP103 plasmids (3 μg) and collected over 24 h prior to hormone determination. Results (means ± SD) are expressed as fold over control (GAL4) and represent three to five independent experiments, each performed in duplicate. An asterisk denotes a P value of <0.05 (Student's t test).
DP103 exhibits RNA helicase activity that requires its C-terminal repression domain.
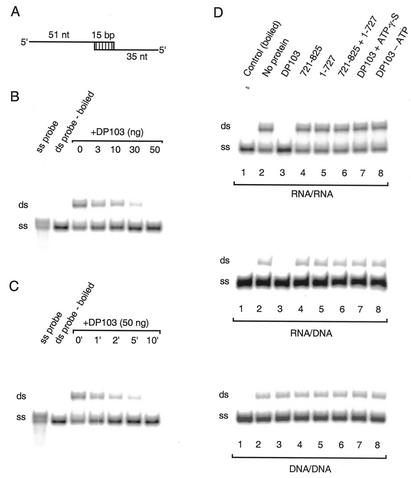
The N-terminal region of DP103 (see Fig. 1B) is highly homologous with DEAD-box proteins (6, 20, 42). This homologous region harbors a helicase domain known to exhibit unwinding activity in other members of this family (4, 25, 47, 54, 57, 70). We have thus far demonstrated that the nonconserved C-terminal domain of DP103 interacts with SF-1 and is necessary and sufficient for repression of SF-1's activity. We therefore questioned whether or not the C-terminal domain is necessary for the helicase function of DP103. To address this question we first sought to determine whether or not DP103 exhibits helicase activity. For that purpose we generated several duplex oligonucleotides of RNA/RNA, RNA/DNA, and DNA/DNA sequences, designed to serve as DP103 substrates. We used bacterially expressed, His-tagged purified DP103 as well as fragments aa 721 to 825 and aa 1 to 727 to determine helicase activity by coincubation with labeled duplex substrates (Fig. 6A). Using the RNA/RNA duplex substrate we initially determined optimal reaction conditions. We found that full-length DP103 exhibited unwinding activity, evident at a protein level as low as 3 ng. The reaction was concentration dependent with complete unwinding using 50 ng of DP103 (Fig. 6B). The reaction was time dependent and was maximal after 10 min (Fig. 6C). All subsequent reactions were therefore performed over 15 min with 50 ng of each protein. We next examined the helicase activity of the C-terminal repression domain of DP103. As shown in Fig. 6D, upper panel, while wild-type DP103 exhibited complete unwinding of the RNA/RNA double-stranded substrate, there was no such activity with DP103721-825 or DP1031-727 fragments (Fig. 6D, lanes 3 to 5). Moreover, we were unable to reproduce unwinding activity even when DP103721-825 and DP1031-727 fragments were coincubated in trans with the labeled duplex substrate (Fig. 6D, lane 6). The RNA unwinding activity was ATP dependent, as it was not observed in the absence of ATP or in the presence of the nonhydrolyzable ATP analog ATP-γ-S (Fig. 6D, lanes 7 and 8). We reproduced these results with RNA/DNA duplexes, demonstrating unwinding activity with full-length DP103 but not with the fragments that span aa 721 to 825 or aa 1 to 727 (Fig. 6D, middle panel). In contrast, we observed no DNA/DNA unwinding activity with any of the proteins (compare lane 3 in the lower panel of Fig. 6D to the same lane on the two other panels). We conclude that the DEAD-box protein DP103 is capable of RNA helicase activity and that the C-terminal domain at aa 721 to 825 is obligatory but not sufficient for this activity of DP103.
FIG. 6.
DP103 exhibits RNA helicase activity in vitro. Helicase assays (5′→3′) were performed under standard conditions as described in Materials and Methods, with 50 fmol of either 32P-labeled RNA/RNA, RNA/DNA, or DNA/DNA duplex substrates, along with His-tagged DP103 in the presence of ATP (3 mM) at 37°C. (A) The structure of the artificial RNA or DNA duplex substrates. (B) RNA helicase activity of DP103 is concentration dependent. A concentration range of full-length His-tagged DP103 (0, 3, 10, 30, and 50 ng) was examined. (C) A time course for the RNA helicase activity of full-length DP103 at the time points indicated. (D) Helicase assay with 50 ng of proteins as indicated, along with RNA/RNA (upper panel), RNA/DNA (middle panel), or DNA/DNA (lower panel) substrates. ATP (3 mM) was present unless indicated as being absent or replaced with ATP-γ-S (3 mM). All the helicase assays were performed for 15 min. Boiled double-stranded (ds) substrates or substrate incubated without protein at 37°C was used as control, indicating the position of released single-stranded (ss) or ds probe, respectively. 32P-labeled short ss probe was also included in the experiments depicted in panels B and C to highlight the position of released ssRNA or ssDNA.
DISCUSSION
Members of the DEAD-box family of putative RNA helicases exhibit diverse cellular functions, including DNA transcription, recombination and repair, RNA processing, translation, and posttranslational gene regulation, in eukaryotes and prokaryotes (2, 10, 13, 21, 30, 58). Newly discovered proteins of unknown function are added to this family on the basis of homology with the DEAD-box, ATPase, or helicase domains. Several of these proteins contain additional domains of unknown function. DP103 was cloned by us and others on the basis of its physical interaction with transcriptional regulators. DP103 contains conserved DEAD-box, helicase, and ATPase domains. In contrast, the C-terminal region of DP103 is not conserved in other members of the DEAD-box family. We have previously determined that DP103 binds a repression domain within SF-1 and represses its activity (42). Our data and the degree of repression are consistent with the findings of Voss et al., who have recently demonstrated that DP103 represses the transcriptional activation of the viral latent membrane protein 1 promoter by the EBNA2/SMN complex in B-lymphocytes (65). The repression was attenuated when a DP103 binding-deficient SMN mutant was used (65). Here we examined the repression function of DP103 and located the interaction domain to aa 721 to 825 within the C terminus of DP103. Consistent with our results, DP103 interacts with the viral proteins EBNA2 and EBNA3C through the region spanning aa 666 to 824 (5, 20, 65). This region was also determined to mediate the repression function of DP103 in a recent analysis of DP103 interaction with the Ets repressor METS (32). Together, our data lead to the unexpected conclusion that the C-terminal domain is necessary and sufficient for SF-1 interaction and repression of transcription, and hence the transcriptional repression of SF-1 by DP103 does not require helicase activity.
Having demonstrated DP103's helicase activity we found that the C-terminal domain is obligatory for RNA unwinding activity of DP103, thereby establishing the pivotal role of this domain in the dual function of DP103. Unlike the repression function, the C-terminal domain is not sufficient for unwinding activity, which requires additional domains within the conserved N terminus. Interestingly, a similar role for a nonconserved C-terminal domain was observed for the bacterial DEAD-box helicases DbpA and YxiN, in which a unique C-terminal domain binds to 23S rRNA, thus bestowing sequence specificity and stimulating the nonspecific helicase activity of the N-terminal conserved domains (14, 34, 60).
The regulation of SF-1 by the C-terminal domain of a DEAD-box helicase constitutes a previously unidentified mechanism for regulation of a nuclear receptor. This C-terminal domain physically interacts with the proximal repression domain of SF-1 and exhibits an autonomous repression function. This conclusion is supported by the fact that GAL4-DP103721-825 is sufficient to repress a GAL4 reporter gene, and DP103721-825 represses SF-1. Consistent with these findings, we have previously demonstrated that DP103 represses the transcriptional activity of an estradiol-stimulated estrogen receptor fused to SF-1's PRD (42). Importantly, the repression function of DP103721-825 is observed by using relevant SF-1 reporter constructs as well as P450scc and P450c21, known transcriptional targets for SF-1 in adrenocortical cells. These results cannot be explained by altered SF-1 expression, as its level is unchanged by overexpressed DP103. Interestingly, although SF-1 was shown to up-regulate the StAR promoter in vitro (56), we found that DP103 failed to repress StAR expression. The reason for this nonuniform influence of DP103 on SF-1 targets is unclear and may suggest that the interaction of SF-1 with DP103 is target selective. Interestingly, Bland et al. demonstrated that StAR expression was not reduced in haploinsufficient SF-1 heterozygous mice (3), suggesting that SF-1 may not play a dominant role in the regulation of StAR.
Whereas the biological activity of putative RNA helicases has been demonstrated (12, 16, 28, 36, 54), ATP-dependent RNA helicase activity has been characterized for only some of these proteins (13, 14, 45, 46, 62, 63). Our data clearly indicate that, similar to other RNA helicases (15, 45, 63), DP103 exhibits RNA helicase activity in an ATP-dependent fashion and in a 5′ to 3′ direction with a 5′ single-strand overhang and is thus distinguished from activity of DNA helicases (51, 54, 61). Whether or not DP103 is capable of bidirectional unwinding activity, as shown for several other helicases, is presently unknown.
Our studies do not elucidate the mechanism of transcriptional repression by DP103. Repression of unliganded nuclear receptors is commonly mediated by N-CoR (SMRT)/mSin3/histone deacetylase (HDAC) complexes (24, 40). Similarly, we and others have demonstrated that DAX-1 can serve as an adapter molecule that recruits the nuclear receptor corepressor N-CoR to SF-1 (11, 27). A recent publication by Klappacher et al. provides evidence that DP103 interacts with METS to repress Ets by assembling a complex of N-CoR, Sin3A, HDAC-2, and HDAC-5 (32). We could not demonstrate an interaction between DP103 and DAX-1 (data not shown). Whether or not this mechanism underlies the repression of SF-1 by DP103 is unclear. Although other DEAD-box proteins regulate gene transcription, the involvement of helicase function in this process is inconsistent (1, 18, 41, 55, 64). One example is RHII/Gu, in which RNA helicase activity is involved in c-Jun-activated transcription during neuronal differentiation of PC12 cells (67). Because residues between aa 721 to 825 in DP103 are pivotal in conferring specificity to DP103 tethering, it is also conceivable that this region may sequester a transactivating cofactor or may disrupt a transcriptionally active nucleic acid-protein complex (29), resulting in diminished transcriptional activation. Moreover, the fact that helicase activity was not recapitulated when both DP1031-727 and DP103721-825 were coexpressed in trans suggests that the C-terminal domain modulates intramolecular conformation of DP103, paramount for interaction with additional protein complexes and unwinding activity. Dissecting these mechanisms in vitro and in vivo is imperative for our understanding of the physiological role of helicase activity and chromatin remodeling.
Acknowledgments
This investigation was supported by grants HD-34110 and HD-37571 from the National Institutes of Health (to Y.S.).
We thank J. S. Richards (Baylor College of Medicine) for the P450scc promoter; Tim Lohman, Jeff Milbrandt, and Peter Crawford (Washington University School of Medicine) for discussions; and Elena Sadovsky and Lori Rideout for technical assistance.
REFERENCES
- 1.Aratani, S., R. Fujii, T. Oishi, H. Fujita, T. Amano, T. Ohshima, M. Hagiwara, A. Fukamizu, and T. Nakajima. 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol. 21:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bland, M. L., C. A. Jamieson, S. F. Akana, S. R. Bornstein, G. Eisenhofer, M. F. Dallman, and H. A. Ingraham. 2000. Haploinsufficiency of steroidogenic factor-1 in mice disrupts adrenal development leading to an impaired stress response. Proc. Natl. Acad. Sci. USA 97:14488-14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddeker, N., K. Stade, and F. Franceschi. 1997. Characterization of DbpA, an Escherichia coli DEAD box protein with ATP independent RNA unwinding activity. Nucleic Acids Res. 25:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, L., K. M. Hunter, P. Mohaghegh, J. M. Tinsley, M. A. Brasch, and K. E. Davies. 2000. Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum. Mol. Genet. 9:1093-1100. [DOI] [PubMed] [Google Scholar]
- 6.Charroux, B., L. Pellizzoni, R. A. Perkinson, A. Shevchenko, M. Mann, and G. Dreyfuss. 1999. Gemin3. A novel dead box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chau, Y. M., P. A. Crawford, K. G. Woodson, J. A. Polish, L. M. Olson, and Y. Sadovsky. 1997. The role of steroidogenic Factor 1 in basal and 3′,5′-cAMP-mediated regulation of cytochrome P450scc enzyme in the mouse. Biol. Reprod. 57:765-771. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, J. W., D. S. Lala, K. L. Parker, and J. S. Richards. 1994. Steroidogenic factor-1 binding and transcriptional activity of the cholesterol side-chain cleavage promoter in rat granulosa cells. Endocrinology 134:1499-1508. [DOI] [PubMed] [Google Scholar]
- 10.Company, M., J. Arenas, and J. Abelson. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349:487-493. [DOI] [PubMed] [Google Scholar]
- 11.Crawford, P. A., C. Dorn, Y. Sadovsky, and J. Milbrandt. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 18:2949-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford, P. A., J. A. Polish, G. Ganpule, and Y. Sadovsky. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient, for potentiation by SRC-1. Mol. Endocrinol. 11:1626-1635. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 14.Diges, C. M., and O. C. Uhlenbeck. 2001. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 20:5503-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, M. X., R. B. Johnson, X. L. Sun, K. A. Staschke, J. Colacino, and Q. M. Wang. 2002. Comparative characterization of two DEAD-box RNA helicases in superfamily II: human translation-initiation factor 4A and hepatitis C virus non-structural protein 3 (NS3) helicase. Biochem. J. 363:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen, A., M. Sattah, T. Gazitt, K. Neal, P. Szauter, and J. Lucchesi. 1998. A novel DEAD-box RNA helicase exhibits high sequence conservation from yeast to humans. Biochim. Biophys. Acta 1397:131-136. [DOI] [PubMed] [Google Scholar]
- 17.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Fujii, R., M. Okamoto, S. Aratani, T. Oishi, T. Ohshima, K. Taira, M. Baba, A. Fukamizu, and T. Nakajima. 2001. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 276:5445-5451. [DOI] [PubMed] [Google Scholar]
- 19.Fuller-Pace, F. V. 1994. RNA helicases: modulators of RNA structure. Trends Cell Biol. 4:271-274. [DOI] [PubMed] [Google Scholar]
- 20.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 21.Hamm, J., and A. I. Lamond. 1998. Spliceosome assembly: the unwinding role of DEAD-box proteins. Curr. Biol. 8:R532-534. [DOI] [PubMed] [Google Scholar]
- 22.Hammer, G. D., and H. A. Ingraham. 1999. Steroidogenic factor-1: its role in endocrine organ development and differentiation. Front. Neuroendocrinol. 20:199-223. [DOI] [PubMed] [Google Scholar]
- 23.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 25.Hirling, H., M. Scheffner, T. Restle, and H. Stahl. 1989. RNA helicase activity associated with the human p68 protein. Nature 339:562-564. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, Y., X. Luo, R. Abbud, J. H. Nilson, and K. L. Parker. 1995. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol. Endocrinol. 9:478-486. [DOI] [PubMed] [Google Scholar]
- 27.Ito, M., R. Yu, and J. L. Jameson. 1997. DAX-1 inhibits SF-1 mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 17:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, M., R. N. Yu, and J. L. Jameson. 1998. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol. 12:290-301. [DOI] [PubMed] [Google Scholar]
- 29.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121-125. [DOI] [PubMed] [Google Scholar]
- 30.Jankowsky, E., and A. Jankowsky. 2000. The DExH/D protein family database. Nucleic Acids Res. 28:333-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastner, P., M. Mark, and P. Chambon. 1995. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell 83:859-869. [DOI] [PubMed] [Google Scholar]
- 32.Klappacher, G. W., V. V. Lunyak, D. B. Sykes, D. Sawka-Verhelle, J. Sage, G. Brard, S. D. Ngo, D. Gangadharan, T. Jacks, M. P. Kamps, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 2002. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 109:169-180. [DOI] [PubMed] [Google Scholar]
- 33.Korolev, S., N. Yao, T. M. Lohman, P. C. Weber, and G. Waksman. 1998. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 7:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kossen, K., and O. C. Uhlenbeck. 1999. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, L. A., E. F. Chiang, J. C. Chen, N. C. Hsu, Y. J. Chen, and B. C. Chung. 1999. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol. Endocrinol. 13:1588-1598. [DOI] [PubMed] [Google Scholar]
- 36.Linder, P., and P. P. Slonimski. 1989. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc. Natl. Acad. Sci. USA 86:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, X., Y. Ikeda, and K. L. Parker. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77:481-490. [DOI] [PubMed] [Google Scholar]
- 38.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 39.Morohashi, K. I., and T. Omura. 1996. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 10:1569-1577. [DOI] [PubMed] [Google Scholar]
- 40.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima, T., C. Uchida, S. F. Anderson, C. G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 42.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 43.Parker, K. L., and B. P. Schimmer. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocrine Rev. 18:361-377. [DOI] [PubMed] [Google Scholar]
- 44.Rainey, W. E., K. Oka, R. R. Magness, and J. I. Mason. 1991. Ovine fetal adrenal synthesis of cortisol: regulation by adrenocorticotropin, angiotensin II and transforming growth factor-beta. Endocrinology 129:1784-1790. [DOI] [PubMed] [Google Scholar]
- 45.Rogers, G. W., Jr., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236-12244. [DOI] [PubMed] [Google Scholar]
- 46.Rossler, O. G., A. Straka, and H. Stahl. 2001. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 29:2088-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadovsky, Y., and C. Dorn. 2000. Function of steroidogenic factor 1 during development and differentiation of the reproductive system. Rev. Reprod. 5:136-142. [DOI] [PubMed] [Google Scholar]
- 49.Sadovsky, Y., P. Webb, G. Lopez, J. Baxter, V. Cavailles, M. Parker, P. Fitzpatrick, E. Gizeng-Ginsberg, and P. Kushner. 1995. Transcriptional activators differ in their response to overexpression of TBP. Mol. Cell. Biol. 15:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaiff, W. T., M. G. Carlson, S. D. Smith, R. Levy, D. M. Nelson, and Y. Sadovsky. 2000. Peroxisome proliferator-activated receptor-γ modulates differentiation of human trophoblast in a ligand-specific manner. J. Clin. Endocrinol. Metab. 85:3874-3881. [DOI] [PubMed] [Google Scholar]
- 51.Scheffner, M., R. Wessel, and H. Stahl. 1989. SV40 T antigen catalyzed duplex DNA unwinding. Curr. Top. Microbiol. Immunol. 144:37-45. [DOI] [PubMed] [Google Scholar]
- 52.Schwer, B., and T. Meszaros. 2000. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 19:6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinoda, K., H. Lei, H. Yoshii, N. Nomura, M. Nagano, H. Shiba, H. Sasaki, Y. Osawa, Y. Ninomiya, O. Niwa, K. I. Morohashi, and E. Li. 1995. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev. Dyn. 204:22-29. [DOI] [PubMed] [Google Scholar]
- 54.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugawara, T., J. A. Holt, M. Kiriakidou, and J. F. Strauss III. 1996. Steroidogenic factor 1-dependent promoter activity of the human steroidogenic acute regulatory protein (StAR) gene. Biochemistry 35:9052-9059. [DOI] [PubMed] [Google Scholar]
- 57.Tang, P. Z., C. H. Tsai-Morris, and M. L. Dufau. 1999. A novel gonadotropin-regulated testicular RNA helicase. A new member of the dead-box family. J. Biol. Chem. 274:37932-37940. [DOI] [PubMed] [Google Scholar]
- 58.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 59.Tremblay, Y., and A. Belanger. 1984. Effect of acute ACTH administration on plasma steroid levels in the dog. Steroids 44:57-66. [DOI] [PubMed] [Google Scholar]
- 60.Tsu, C. A., K. Kossen, and O. C. Uhlenbeck. 2001. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 7:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tuteja, N., and R. Tuteja. 1996. DNA helicases: the long unwinding road. Nat. Genet. 13:11-12. [DOI] [PubMed] [Google Scholar]
- 62.Uhlmann-Schiffler, H., O. G. Rossler, and H. Stahl. 2002. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J. Biol. Chem. 277:1066-1075. [DOI] [PubMed] [Google Scholar]
- 63.Valdez, B. C., D. Henning, K. Perumal, and H. Busch. 1997. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu—two activities in separate domains of the same protein. Eur. J. Biochem. 250:800-807. [DOI] [PubMed] [Google Scholar]
- 64.von Hippel, P. H., and E. Delagoutte. 2001. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104:177-190. [DOI] [PubMed] [Google Scholar]
- 65.Voss, M. D., A. Hille, S. Barth, A. Spurk, F. Hennrich, D. Holzer, N. Mueller-Lantzsch, E. Kremmer, and F. A. Grasser. 2001. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 75:11781-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westermarck, J., C. Weiss, R. Saffrich, J. Kast, A. M. Musti, M. Wessely, W. Ansorge, B. Seraphin, M. Wilm, B. C. Valdez, and D. Bohmann. 2002. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 21:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson, T., T. Fahrner, and J. Milbrandt. 1993. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol. Cell. Biol. 13:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wittinghofer, A., and E. F. Pai. 1991. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 16:382-387. [DOI] [PubMed] [Google Scholar]
- 70.Yu, E., and G. W. Owttrim. 2000. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 28:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, L., M. Bakke, Y. Krimkevich, L. J. Cushman, A. F. Parlow, S. A. Camper, and K. L. Parker. 2001. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development 128:147-154. [DOI] [PubMed] [Google Scholar]