Abstract
In a screen for Drosophila genes that interfere with transcriptional repression mediated by the Polycomb group of genes, we identified a dominant mutation affecting the Alhambra (Alh) gene, the fly homologue of the human AF10 gene. AF10 has been identified as a fusion partner of both MLL and CALM in infant leukemias. Both fusion proteins retain the leucine zipper domain of AF10 but not its PHD domain. We show here that, while the full-length ALH protein has no activity on Polycomb group-responsive elements (PREs), overexpression of the isolated ALH leucine zipper domain activates several PREs. Within the ALH full-length protein, the PHD domain inhibits the PRE deregulation mediated by the leucine zipper domain. This deregulation is conserved in the human AF10 leucine zipper domain, which confers the same activity on an oncogenic MLL-AF10 fusion protein expressed in Drosophila melanogaster. These data reveal new properties for the leucine zipper domain and thus might provide new clues to understanding the mechanisms by which AF10 fusion proteins in which the PHD domain is lost might trigger leukemias in humans.
The human MLL gene (also called HRX, ALL-1, and Htrx) is involved in translocations with up to 30 partner genes, creating fusion proteins in which the carboxy-terminal part of MLL is deleted and replaced in the correct reading frame by a part of the fusion partner. Such translocations are the most commonly observed cytogenetic alterations in infant acute leukemia and are also prevalent in secondary leukemia arising after treatment of neoplastic diseases with a topoisomerase II inhibitor. Several mammalian model systems have demonstrated that the generation of MLL fusion proteins can induce the transformation of hematopoïetic cells. For example, mice carrying a knock-in allele of a t(9;11) translocation, which fuses MLL with AF9, develop leukemia (14, 18). These experiments support the assumption that the generation of MLL fusions creates active oncogenes that are responsible for the development of leukemias in humans.
MLL is a very large protein (431 kDa) with homology to the Drosophila trithorax protein (TRX) in several domains (19, 27, 51). The trx gene is the prototype of a class of genes (the so-called trithorax group genes) which are required to positively maintain the proper expression of a number of Drosophila loci, including the homeotic genes. It is thought that TRX regulates homeotic expression at the level of chromatin organization, and it is likely that MLL serves the same role in vertebrates (54, 55).
The lack of functional information about the MLL partners cloned to date has made it difficult to generate testable hypotheses regarding their contribution to transformation. A specific function of the fusion partner, however, must be required, because in the knock-in studies, neither the N-terminal portion of MLL alone nor an MLL protein fused to a portion of the Myc protein showed transforming activity (14).
AF10 and its paralogous gene AF17 are two examples of human fusion partners of MLL (13, 44). Together with the Caenorhabditis elegans CEZF protein and the human BR140 protein, they belong to a small evolutionarily conserved family of proteins (12). Interestingly, the regions of homology between these proteins are restricted to two domains shared by known chromatin-associated proteins, the amino-terminal PHD finger and a carboxy-terminal leucine zipper domain. Leucine zippers are common in a number of transcriptional regulators but are also encountered in some chromatin-associated proteins such as the Drosophila Moira protein (MOR) (15). PHD domains are found in CBP, MLL, and TRX and in the Drosophila Polycomb group protein PCL (37). This suggests that PHD domains may be involved in chromatin-mediated gene expression mechanisms, yet this domain is lost in MLL fusions, and it thus might not be involved in the transformation process. Rather, the leucine zipper is the only region of homology between AF10 and AF17 that is retained in MLL fusions.
This observation points toward a crucial role of this domain in the transformation process. Further supporting this hypothesis, the C-terminal part of AF10 (including its leucine zipper domain) is consistently found fused to another partner gene, CALM, in leukemias (10). The central role played by the AF10 leucine zipper domain in MLL-AF10 fusions has recently been demonstrated by DiMartino et al. (17), who showed that the AF10 leucine zipper domain is required for leukemic transformation of myeloid progenitors by MLL-AF10. The murine AF10 homologue has been shown to be nuclear, consistent with a potential role of the whole protein in gene regulation (35). In agreement with this, a Drosophila AF10/17 gene homologue has recently been described and encodes a nuclear protein required to maintain proper even skipped expression in the central nervous system (3). However, the mechanism by which the AF10 or AF17 leucine zipper may trigger leukemia remains largely unknown.
Functional studies of several partners suggest a role in transcriptional regulation. For example, CBP, one of the fusion partners of MLL, possesses a histone acetyltransferase activity that has been shown to acetylate the lysine tails of histones and thus induces chromatin accessibility (for a review, see reference 9). In addition, it has been shown that AF9 interacts with MPc3, a Polycomb protein (29), and that the same MPc3 protein is recruited by the ENL moiety in the MLL-ENL fusion protein (24). Polycomb group gene (PC-G) proteins have a function opposite that of trithorax group gene (TRX-G) proteins, as they are chromatin-associated proteins involved in the maintenance of repressed transcriptional states of a number of genes, including homeotic loci (see below). Taken together, all these studies suggest that a unifying mechanism by which nuclear fusion partners “activate” MLL could be the gain of transcriptional effector potential. This is further suggested by a recent study which indicated that the AF10 leucine zipper domain binds GAS41, a protein that interacts with the human SWI/SNF complex, which acts to remodel chromatin and to modulate transcription (16). In all these cases, however, the in vivo activity of the fusion proteins on chromatin-dependent transcriptional regulation remains to be tested.
Mammalian and Drosophila Polycomb (Pc) group genes and trx group genes are structurally and functionally related (6), and a large part of our knowledge about Pc group and trx group gene function come from studies in Drosophila melanogaster. In flies, genetic and biochemical experiments have demonstrated that both PC-G group and TRX-G group proteins act on common DNA elements called Polycomb group-responsive elements (PREs) (49) and form different multiprotein complexes (31, 40, 42, 48) that have distinct activity upon transcriptional maintenance.
We previously demonstrated that the Drosophila polyhomeotic (ph) locus is transcriptionally controlled by PC-G and TRX-G proteins (21, 22). Here we report the identification of a neomorphic mutation affecting the Drosophila gene Alhambra (Alh). Alh is homologous to the human genes AF10 and AF17 and is identical to the previously described dalf/dAF10 gene (3, 34). Molecular genetic analysis indicates that while the Alh gene does not encode a PC-G protein, overexpression of the ALH leucine zipper alone is able to activate ph PRE transcription.
We tested the activity of the human AF10 leucine zipper domain and demonstrated that, like its Drosophila counterpart, it deregulates PRE activity in Drosophila cells. Furthermore, a human MLL-AF10 fusion has the same activity. Finally, we present evidence that the PHD domain within the full-length ALH protein inhibits this activity. Our results have implications not only for the role of the leucine zipper domain in oncogenic fusion proteins, but also for the involvement of PRE misregulation in malignant transformation.
MATERIALS AND METHODS
Fly strains and culture.
All strains were maintained on standard culture medium at 25°C. Except where otherwise stated, alleles have been described previously (22, 36). P{ry+ Δ2-3}(99B), a stable source of transposase, and Birm2, a second chromosome bearing 17 nonautonomous P elements, have been described (45). In order to assay eye color modification with the upstream activation sequence (UAS)/GAL4 binary system, we constructed a (UAS, yellow+) transformation vector (see section on plasmids and mutants constructions) and removed in situ, thanks to the action of the transposase, the white+ marker from P{daGAL4} to create P{daGAL4w−}.
Starting from 80 crosses between two dysgenic males and five balanced females, we were able to recover two bona fide P{daGAL4w−} events. P{GlassGAL4w− } was obtained in the same way and was a gift from Laurent Theodore. Flies carrying Fab7 containing the P{5F 24} and P{iab-7 860 bp} transgenes have been described (11, 28) and were obtained from G. Cavalli and P. Schedl, respectively. Flies carrying the Sex Combs reduced PRE P{Scr 8.2 XbaI} (26) were obtained from T. Kaufman, those with the engrailed PRE (P{en}) were from J. Kassis (32), and those carrying the 39C-4 and 39C-5 transgenes (52), allowing us to test position effect variegation and telomeric effect variegation, respectively, were obtained from L. Wallrath.
Genetic screen for new ph transcriptional regulators.
w1118; Birm2/CyO; Sb P{ry+ Δ2-3}(99B)/+ dysgenic males were crossed to y phlac+3 w1118 females at 18°C. A total of 5,330 Sb+ chromosomes were screened for either a pale eye color (males and females) or a red eye color (females only). We recovered one new ph allele (phrIIA) and one mutation on the third chromosome that we called Alhambra. (The Alhambra is a beautiful Moorish palace in Granada, Spain. Its name comes from the Arabic word hamra, which means red.) After 11 rounds of outcrossing over a y phlac+3 w1118 stock, polytene chromosome hybridization with the P element as a probe indicated the presence of two remaining insertions, at 79F and 84C. The defective P element of the Alh1 mutation was then replaced with a P{yellow+} transposon (8), allowing us to obtain a new Alh allele (Alhy+). Similar P replacements are described elsewhere (50).
The Alhy+ chromosome was then recombined over a Ki1 (83E) chromosome in order to make a Ki1 Alhy+ chromosome. Polytene chromosome hybridization demonstrated that this chromosome bore only one P insertion at 84C. In this strain, the loss of the yellow marker following mobilization of the P element was associated with reversion of the Alh phenotype, demonstrating that the P element insertion was responsible for this phenotype.
Molecular cloning of the Alhambra locus.
Genomic DNA flanking the Alhy+ P element insertion was recovered by plasmid rescue experiments (43) and used as a probe to screen a Lambda Zap Drosophila genomic library (Clontech) by standard procedures (47). Three overlapping clones, representing about 30 kb of genomic DNA, were recovered and sequenced. A search for Drosophila expressed sequence tags at the Berkeley Drosophila Genome Project (BDGP) (http://www.bdgp.org) identified several expressed sequence tags in the region. Four of them were analyzed in detail by restriction map analysis, and the longest was sequenced (GenBank accession no. AF217960). Later on, the sequence of the Drosophila genome (1) allowed us to look for new expressed sequence tags at BDGP and permitted the identification of a new class of Alh transcripts (Alh S), differing by having their transcriptional start located within the fifth intron of the Alh L transcript. These transcripts are identical to the one described previously (cDNA C1 [3]).
Northern blotting, reverse transcription-PCR, and molecular biology.
For developmental Northern blot analysis, total RNAs were extracted with Trizol (Life Technologies) and polyadenylated mRNA was purified with Oligotex (Qiagen) according to the manufacturer's conditions. Electrophoresis, transfer, and hybridization were done according to standard procedures (47).
For reverse transcription-PCR analysis of Alh transcripts, total RNAs were extracted with Trizol (Life Technologies) from 100 embryos, 10 third-instar larvae, 10 adult individuals, or 50 adult heads and resuspended in 10 μl of H2O. Then 1 μl of RNA was used in a 20-μl reverse transcription reaction with 200 U of Superscript Moloney murine leukemia virus reverse transcriptase (Life Technology) and random hexanucleotides. Then 1 μl of the resulting cDNAs was tested in a 50-μl PCR with 0.1 U of Taq polymerase (Promega).
Primers used to detect Alh L transcripts were Forward 3′Exon (1) (5′-ACTGCTCGCTCCCGATGTGC) and Reverse 5′Exon (4) (5′-GGGCGACGAACACGAATCGG). Alh S transcripts were identified with primer Forward Alh S (5′-GGACACCATGGACACCTCGC) and Reverse Exon6 (5′-GGCGGGCTGAGTTCGGTACG). For reverse transcription-PCR analysis of salivary glands and eye-antenna discs, reverse transcription was done on total RNA extracted from the corresponding tissues of 10 individuals and PCR was done on 5 μl of the resulting cDNAs.
Plasmid and mutant protein constructs.
To generate the pP{UAS, yellow+} vector, the mini-white gene was removed from pP{UAST} (5) by EcoRV digestion and replaced with a blunted SalI cassette from the Dint plasmid (25), encoding the yellow gene.
The UASy+-Alh L construct was generated by subcloning the 4,574-bp blunted BstEII fragment from the full-length Alh L cDNA in pP{UAS, yellow+} at the NotI restriction site (blunted). The UAS(y+)-Alh S construct was generated by subcloning the Pml1-EroRV fragment from the Alh S cDNA (cDNA C1 [3]) in pP{UAS, yellow+} at the NotI restriction site (blunted).
The Flag-tagged Alh L cDNA and the different mutant proteins were generated by PCR and cloned in pBlueScript KSII+, and the PCR products were completely sequenced to exclude errors introduced by Taq polymerase. They were then subcloned in pP{UAS, yellow+}.
The UAS(y+)-MLLNter construct was generated as follows. An amino-terminal Flag-tagged 5′ Mll encoding amino acids 1 to 1396 was excised by EcoRI and XhoI digestion from the pMSCV-5′Mll plasmid (kindly provided by J. F. DiMartino) (17) and cloned into pP{UAS, yellow+}. The UAS(y+)-MLL-AF10 fusion derived from pMSCV-5′MLL-AF10 (kindly provided by J. F. DiMartino) (17), encoding amino acids 1 to 1396 from MLL fused to the C-terminal portion of AF10 from amino acid 743. MLL-AF10 is amino-terminally Flag tagged and was excised by EcoRI and XhoI digestion before cloning in the pP{UAS, yellow+} plasmid. To generate the MLL-ALH fusion, an NdeI (blunted)-XhoI fragment from Alh L cDNA was subcloned in pMSCV-5′Mll and HpaI and XhoI digested, and then the fusion was cloned in pP{UAS, yellow+} as described above.
The glutathione S-transferase (GST)-LZ plasmid, corresponding to the GST-C-terminal ALH L fusion protein (including the leucine zipper domain, amino acids 938 to 1376 in the full-length protein) was generated by inserting the BamHI-XhoI fragment from the Alh L cDNA into the BamHI- and XhoI-digested pGEX-2T vector (Amersham Pharmacia Biotech). The plasmid encoding GST-ΔLZ, corresponding to amino acids 938 to 1280 from the ALH L protein fused to GST, was kindly provided by S. Bari.
Examination of eye pigmentation.
For all transgenic lines, flies of the same age were compared. Due to the localization of their transgenes on the X chromosome, transgenic lines phlac+3 and P{5F 24} are subject to dosage compensation. Consequently, for these transgenes, only heterozygous females were observed for eye color experiments. Flies were photographed under a Leica MZ12 dissecting microscope with a Nikon DXM 1200 digital camera.
Pigment assays were performed as described before (2). Heads from five flies (3 days old) were homogenized in 400 μl of 1:1 chloroform-ammonium hydroxide (0.1%) and spun for 2 min in a microcentrifuge. Then 100 μl of the aqueous layer was used to read absorbance at 485 nm. The experiment was repeated four times for each genotype and the average optical density at 485 nm (OD485) was calculated. For each experiment, the control was arbitrarily given a value of 1, and the values were calculated as a ratio of the corresponding control.
GST pulldown.
GST-LZ and GST-ΔLZ fusion proteins were isolated from Escherichia coli lysates (strain BL21; Novagen) by standard protocols. Resin-bound GST fusion proteins were pelleted, washed in lysis buffer (phosphate-buffered saline, 1 mM dithiothreitol, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 10 μg of pepstatin per ml, 10 μg of antipain per ml, 1 mM phenylmethylsulfonyl fluoride) and resuspended in phosphate-buffered saline containing 0.01% Triton X-100 and protease inhibitors (leupeptin, aprotinin, pepstatin, and antipain [all at 10 μg/ml], 1 mM phenylmethylsulfonyl fluoride). 35S-labeled ALH L full-length and mutant proteins were synthesized with a coupled in vitro transcription and translation kit (Promega) with the corresponding cDNAs cloned in pBlueScript KSII+ (see above) as the template in the presence of protease inhibitors. For the binding assay, again with protease inhibitors, 5 μg of fusion protein was incubated for 1 h at 4°C with 10 μl of [35S]methionine-labeled proteins in phosphate-buffered saline-0.01% Triton X-100. Following five washes with washing buffer (100 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl [pH 7.4], 0.05% Triton X-100), labeled proteins were eluted by boiling for 3 min in loading buffer. All bound eluate for each binding experiment was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Polytene chromosome immunostaining.
The protocol used for polytene chromosome immunostaining has been described (23) with antibody dilutions in blocking solution: 1:30 for anti-ALH (3), 1:30 for anti-Flag M2 (Sigma), 1:40 for anti-PH, 1:300 for anti-mouse immunoglobulin-fluorescein conjugate (Sanofi Diagnostics Pasteur), and 1:500 for anti-rabbit immunoglobulin G (heavy and light chain)-indocarbocyanine conjugate (Jackson).
RESULTS
Identification of Alhambra1, a new mutation that deregulates PRE activity.
We previously reported that transcription of the ph locus, encoding a PC-G protein, is itself under the control of both TRX-G and PC-G proteins, acting via PREs in the ph upstream regulatory region (21, 22). In order to recover new regulators of ph transcription, we made use of the phlac+3 strain described previously (22), in which a PlacW transgene (carrying the mini-white reporter gene) is inserted in the first intron of the ph-proximal transcription unit, thus bringing mini-white under the control of the endogenous ph PREs. In phlac+3 w/ph+ w flies, Pc group loss-of-function mutations induce an activation of the ph locus and lead to a darker (red) eye color of phlac+3 w/ph+ w females, while trx group mutations induce a repression of ph transcription and consequently a lighter (yellow) eye color of phlac+3 w/ph+ w females.
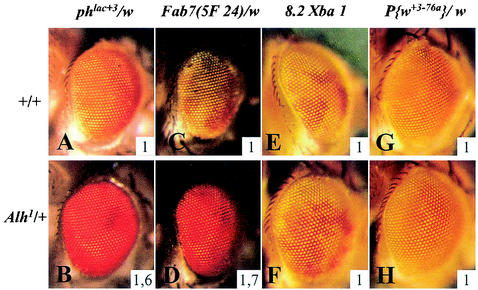
By screening P element-induced mutations for their ability to modify the eye color of phlac+3 w flies, we isolated the Alhambra1 (Alh1) mutation, which produces a red eye color instead of the regular orange eye color of phlac+3 w/ph+ w females (Fig. 1A and B). Furthermore, the Alh1 mutation also affected the transcription of the mini-white gene of a P{ph} transgene (21), indicating that it works at the level of the ph PRE. The Alh1 mutation has no effect on the transcriptional activity of a control mini-white gene present on the w, P{w+3-76a} chromosome (Fig. 1G and H), which carries a PlacW insertion insensitive to Pc group and trx group mutations (22). Therefore, Alh1 behaves as a strong Pc group mutation, since it activates ph transcription. Alh1 is homozygously viable, and both homozygous and heterozygous flies have the same phlac+3 eye color phenotype. This mutation also strongly deregulates the activity of Fab-7, a PRE from the bithorax homeotic complex (Fig. 1C and D). No effect was detected on the PRE of another homeotic gene, Sex comb reduced (Scr, Fig. 1E and F), or on an engrailed (en) PRE (not shown). Thus, Alh1 acts to disrupt silencing at specific PREs.
FIG. 1.
Alh1 mutation affects PRE regulation.The dominant Alh1 mutation has a strong effect upon phlac+3 (compare A and B) and on Fab-75F24 (compare C and D) eye color phenotypes. The lack of effect of Alh1 on Scr8.2XbaI eye color (compare E and F) shows that Alh1 has some PRE specificity. The Alh1 mutation does not modify the transcriptional activity of the control mini-white gene within the P{w+3-76a} transgene (compare G and H). The inserted numbers, which give the average OD485 values calculated from four experiments, help to quantify eye pigment differences (see Materials and Methods).
We further analyzed the effect of Alh1 on position effect variegation and on telomeric effect variegation (30, 53). Strains 39C-4 and 39 C-5 (52) allowed monitoring of the activity of centromeric and telomeric chromatin upon transcriptional regulation. Neither was sensitive to the Alh1 mutation (not shown), indicating that the mutation interacts specifically with Pc group and trx group transcriptional regulation.
Alh gene encodes an orthologue of human AF10 and AF17 proteins.
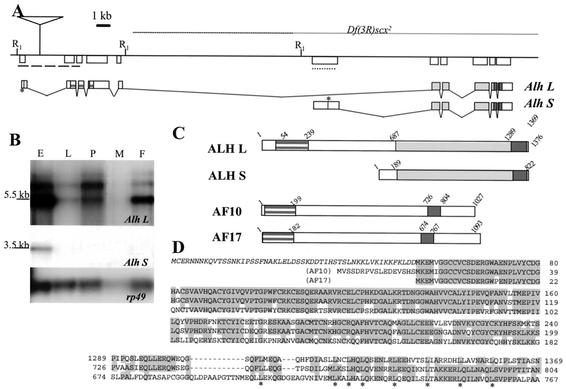
To clone the Alhambra locus, we took advantage of the P element insertion responsible for the Alh1 mutation (see Materials and Methods). Cloning and sequencing of the genomic DNA flanking the P element insertion site demonstrated that the Alh1 mutation is due to the insertion of a nonautonomous P element within the first intron of a transcriptional unit located at 84C on the right arm of chromosome 3 (Fig. 2A). This transcriptional unit produces two different transcripts visible on Northern blots (Alh L and Alh S, Fig. 2B). Alh L is expressed throughout development, while Alh S, which is produced from a different transcriptional start site (located into the fifth intron of Alh L), is detected only during embryogenesis. This transcriptional unit is identical to that of dalf, previously shown to be required for proper even skipped expression in RP2 neurons (3), and to dAF10 (34).
FIG. 2.
Alhambra locus. (A) Genomic organization of the Alhambra locus at cytological position 84C on 3R. EcoRI restriction sites are indicated (R1). A line represents genomic DNA. The localization of the P element P{Alh} responsible for the Alh1 mutation is indicated. It is inserted at nucleotide 26782 in the Drosophila genomic segment (GenBank no. AE003672). Genomic DNA deleted in the Df(3R)scx2 deficiency is represented by a thin line above the genomic map (according to reference 4). The two major transcripts Alh L and Alh S are represented below. Stars represent translational start sites. The regions encoding the PHD (striped grey block) and leucine zipper (dark grey box) domains are indicated. (B) Expression of Alh transcripts during development. A developmental Northern blot probed with a 4.5-kb genomic fragment adjacent to the P{Alh} insertion site (dashed line in A) was used to reveal Alh L transcripts. Alh-L is expressed at all stages during development. A probe corresponding to the sixth exon (dotted line in A) of Alh specifically reveals Alh S transcripts which are expressed only during the embryonic stage. Apparent sizes are indicated on the left. rp49 is shown as a loading control. E, embryos; L, third-instar larvae; P, pupae; M, males; F, females. (C) Schematic representation of ALH L, ALH S, AF10, and AF17 proteins. The PHD (striped grey block) and leucine zipper (dark grey box) are indicated. The predicted ALH L protein bears 53 more amino acids (amino acids 1 to 53) than the longest protein encoded by the class 3 cDNA described earlier (3). These amino acids are indicated in italic in D. ALH L and ALH L-∋1-53 behave exactly in the same way in vivo and are both able to rescue Alh loss of function (L. Perrin and J. M. Dura, unpublished data). Identical amino acid sequences between ALH L and ALH S are represented by a light grey box, while sequences specific to each protein are indicated by white boxes. (D) Sequence alignment of the PHD domain and leucine zipper domain of ALH and its human homologues AF10 and AF17. Italics indicate the 53 amino acids encoded by the first exon of Alh L that were not published previously (see panel C). Overall, the ALH PHD domain is 75% and 74% identical to the AF10 and AF17 PHD domains, respectively. The ALH leucine zipper displays 51% identity (68% similarity) with the AF10 leucine zipper. The conserved leucines are indicated (*).
The Alh L transcript that we have isolated bears one more 5′ exon and therefore potentially encodes a protein 53 amino acids longer than the longest protein encoded by the previously described class 3 Dalf cDNA (3). The ALH L protein is homologous to the human proteins AF10 and AF17 (encoded by the All1 fusion on chromosomes 10 and 17, respectively). Notably, ALH L bears one amino-terminal PHD and a C-terminal leucine zipper domain (Fig. 2C and D). As shown in Fig. 2D, the PHD domain displays strong homologies to both the AF10 and AF17 PHD domains (75% identity/85% similarity and 74% identity/85% similarity, respectively). The ALH leucine zipper domain displays more homology to the AF10 leucine zipper domain (51% identity and 68% similarity) than to the AF17 leucine zipper domain (34% identity and 54% similarity). Apart from these two domains, the central part of ALH L does not show homology with any known proteins. Computer analysis revealed that the ALH L protein is the only member of this family in the Drosophila genome. The ALH S protein retains only the leucine zipper domain in common with this family.
Alh1 mutation induces ectopic expression of Alh S transcripts.
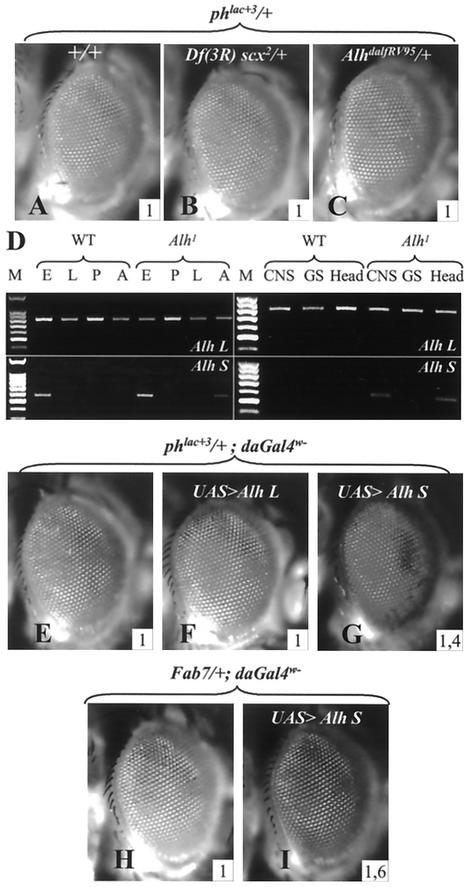
The cloning of the Alh locus allowed us to analyze the Alh1 mutation at both the genetic and molecular levels. Alh1 appears not to cause a lack of function of the gene. Indeed, the Df(3R)scx2 deficiency, which uncovered the Alh locus (4), and the AlhDalfRV95 mutation, an Alh null allele (3), have no effect on phlac+3 eye color (Fig. 3A to C). Furthermore, Alh loss-of-function mutations do not change the phenotypes of the Pc group mutants (Psc1, PscArp1, PcK, ph0, and dMi-24) or of the trx group mutants [mor1, Df (3)XS (ash-2−), trxE2, TrlR85, and z1] (22) we have tested, as would be the case for a classical Pc group or trx group mutation (data not shown). These results indicate that the Alh gene does not belong to either of these groups. Localization of ALH L protein on polytene chromosomes further confirms this proposal, since it does not localize on PREs (see below).
FIG. 3.
Functional study of Alh1 mutation. (A, B, and C) Alh loss of function does not affect PRE regulation. Neither Df(3R) scx2 (B) nor AlhdalfRV95 (C) mutations modify the eye color of phlac+3/w females. (D) Alh L and Alh S expression in wild-type and Alh1 individuals revealed by reverse transcription-PCR analysis. Reverse transcription-PCR was performed with oligonucleotides specific for Alh L and Alh S transcripts on RNAs extracted from wild-type (WT) and Alh1 mutant individuals at different stages (left) or from different tissues (right). M, molecular size markers, E, embryos; L, larvae; P, pupae; A, adults; CNS, central nervous system plus eye antenna disc; GS, salivary glands. The long transcript (Alh L, top) is expressed ubiquitously at all developmental stages in wild-type and mutant Alh1 flies. The short transcript (Alh S, bottom), which is expressed only during embryogenesis in the wild type, is found in Alh1 adults and more specifically in the head (line labeled head). At the third-instar larval stage, Alh S is expressed in the eye antenna disc and/or central nervous system but not in salivary glands. (E to G). Effects of Alh L and Alh S overexpression on phlac+3 eye color. The corresponding cDNAs were inserted in the P{y+UAS} vector (see Materials and Methods), and their expression was induced with P{daGAL4w−}, allowing expression of the different cDNAs without interfering with the phlac+3 white phenotype. Alh L overexpression has no effect on phlac+3 eye color (compare E and F), while that of Alh S strongly activates the ph PRE and resembles the mutant Alh1 phenotype (compare E and G). In addition, Alh S overexpression also activates an Fab7 PRE present in the 18.8.6 transgene (H and I). The inserted numbers give the average OD485 values calculated from four experiments (see Materials and Methods).
Reverse transcription-PCR analysis showed that, in Alh1 flies as well as in wild-type flies, the Alh L transcript is expressed throughout development (Fig. 3D). On the other hand, and contrary to the wild-type situation, the Alh S transcript is weakly but reproducibly detected in Alh1 adults (Fig. 3D, lane F). This ectopic expression is more readily detected if RNAs are extracted from fly heads rather than from whole flies (Fig. 3D, lane head). Although this short transcript is not detected in whole mutant third-instar larvae (Fig. 3D, lane L), reverse transcription-PCR of RNA extracted from the eye-antenna disc attached to the central nervous system shows specific expression of the Alh S transcript in the Alh1 mutant (Fig. 3D, lane CNS). Moreover, this expression seems to be tissue specific, since no expression is detected in salivary glands (Fig. 3D, lane SG).
Overexpression of Alh S transcript induces PRE deregulation.
In order to assay the in vivo roles of Alh transcript products upon ph transcriptional regulation, we ectopically expressed Alh L and Alh S in adults. Although the overexpression of Alh L has no effect on ph transcription (Fig. 3E and F), ectopic expression of Alh S induces a strong disruption of ph silencing (Fig. 3G). This level of ph transcription is similar to that seen in the original Alh1 mutant. The same results are obtained when Alh S is induced in eye cells only, with a GlassGAL4 driver, suggesting that it acts cell autonomously on PRE silencing. Furthermore, as for the Alh1 mutation, Alh S overexpression deregulates a Fab7 PRE (Fig. 3H and I) but has no effect on the P{w+3-76a} transgene (not shown). These results strongly suggest that the original Alh1 mutation is a neomorph (gain-of-function) type of mutation which leads to ectopic expression of the Alh S transcript in the head, leading to deregulation of several PREs.
Overexpression of the ALH leucine zipper domain alone activates PREs, a property conserved in the human AF10 leucine zipper domain.
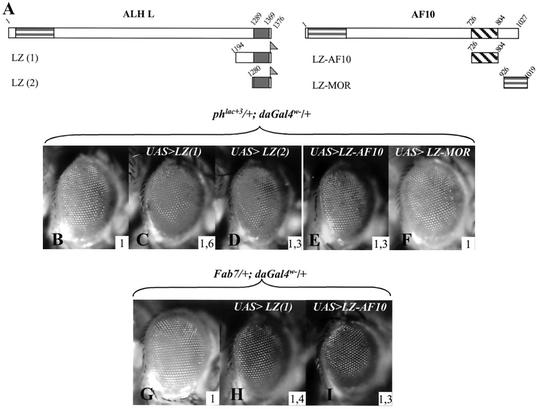
As stated above, the sole characteristic region in the ALH S protein is the leucine zipper domain. In order to test the involvement of this domain in PRE deregulation, we constructed transgenic fly lines expressing the carboxy-terminal part of ALH proteins, which includes the leucine zipper domain (LZ-1 construct, Fig. 4A). Overexpression of this truncated protein induces a strong derepression activity that is comparable to that obtained with the Alh1 mutation (Fig. 4B and C). A comparable effect is observed when a variant encompassing only the leucine zipper domain is assayed (LZ-2, Fig. 4D). This construct, however, has a weaker effect, which could be due to a defect in folding of the leucine zipper domain in this shortest construct. Nevertheless, these results clearly indicate a gain-of-function action of the ALH leucine zipper towards ph PRE regulation. Both LZ-1 and LZ-2 also activate a Fab-7 PRE (the LZ-1 effect is given as an example in Fig. 4H), further supporting the fact that overexpression of the leucine zipper domain alone is sufficient to deregulate PREs.
FIG. 4.
ALH and AF10 leucine zipper domains deregulate PRE activity. (A) Schematic representation of the different constructs used. Amino acids present in these constructs are indicated and refer to amino acid positions in the full-length ALH L and AF10 proteins (represented above). The leucine zipper domain of the Drosophila moira-encoded protein MOR (amino acids 926 to 1019) was also analyzed. The corresponding cDNAs were inserted in the P{y+UAS} vector, and their expression was induced with P{daGAL4 w−}. The carboxy-terminal Flag tags are indicated (shaded flag). (B to F) Effects of overexpression of leucine zipper domains on phlac+3 eye color phenotype. Overexpression of the carboxy-terminal region of ALH containing the leucine zipper domain, LZ(1), strongly deregulates ph silencing (C). A comparable but lighter effect is obtained with the isolated leucine zipper domain LZ(2) (D). (E) Expression of the AF10 leucine zipper domain alone also relieves ph silencing. (F) The MOR leucine zipper domain has no effect on ph silencing when overexpressed. (G to I) Overexpression of LZ(1) (H) and AF10-LZ(I) also activates an Fab7 PRE present in the 18.8.6 transgene. The inserted numbers give the average OD485 values calculated from four experiments (see Materials and Methods).
The conservation between the AF10 and ALH leucine zipper domains (see Fig. 2) led us to test, in flies, the effect of overexpressing the human AF10 leucine zipper domain on ph silencing. Like the ALH leucine zipper, the AF10 leucine zipper has a reproducible deregulating effect on ph transcription in vivo (Fig. 4E) that is also observed on the Fab7 PRE (Fig. 4I). In order to test whether the observed effect is specific for the ALH and AF10 leucine zippers or whether it is due to a general activity of leucine zipper domains when overexpressed, we tested the effect of overexpression of the leucine zipper domain of the Drosophila protein MOR on ph transcription. mor encodes a TRX group protein and is thus involved in transcriptional maintenance (15). The MOR leucine zipper domain overexpressed in vivo has no effect on ph transcription (Fig. 4F), indicating that PRE derepression is specific for the AF10 and ALH leucine zipper domains.
AF10 leucine zipper confers PRE activation activity on a human oncogenic MLL-AF10 fusion.
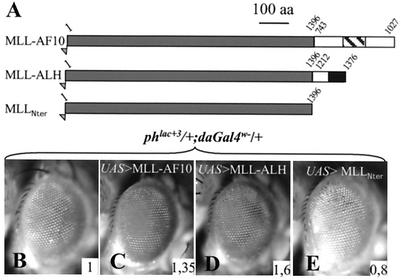
In humans, all the different oncogenic fusion proteins involving AF10 retain the leucine zipper domain. These include MLL-AF10 and CALM-AF10, suggesting a particular role for the AF10 leucine zipper in the transformation process (13, 39). In order to test if the PRE deregulation potential of the AF10 leucine zipper domain has an effect on the activity of an MLL-AF10 fusion in Drosophila cells, we generated transgenic lines bearing a P{UAS> MLL-AF10} transgene which allowed the expression of a human oncogenic MLL-AF10 fusion protein (generous gift of G. F. DiMartino and M. L. Cleary). This fusion encompasses the N terminus of MLL and the C terminus of AF10, including its leucine zipper domain (see Fig. 5).
FIG. 5.
MLL-AF10 and MLL-ALH fusion proteins dominantly suppress ph silencing. (A) Schematic representation of fusion proteins. An oncogenic human MLL-AF10 fusion was inserted into the P{y+UAS} vector in order to allow its expression in flies. A chimeric protein containing the N-terminal part of human MLL (amino acids 1 to 1396) fused to the C-terminal part of ALH (amino acids 1212 to 1376) was cloned into the same vector. The N-terminal part of MLL alone was also analyzed. (B to E) Effects of expression of these proteins on ph silencing. The expressions of the oncogenic fusion MLL-AF10 (C) and of MLL-ALH (D) relieve ph silencing to different degrees. Contrarily, the N-terminal domain of MLL alone (E) produces slightly more ph silencing. The inserted numbers give the average OD485 values calculated from four experiments (see Materials and Methods).
When expressed in Drosophila cells, MLL-AF10 induces a reduction of ph PRE silencing (Fig. 5C). Interestingly, a chimeric MLL-ALH protein made of the N-terminal part of human MLL and the C-terminal part of Drosophila ALH also removed ph silencing (Fig. 5D). The effects of MLL-AF10 and MLL-ALH fusions are not due to the MLL Nter region (MLLNter), since this protein fragment does not relieve ph silencing when expressed alone (Fig. 5E). In fact, a weak increase in ph silencing is even observed, which may be correlated to the presence of a potential repressive domain in this part of the protein (46). In all cases, the same effects are observed with the Fab7 PRE (not shown). The leucine zipper domains are the sole homologous domains in the portions of ALH and AF10 that were fused to MLL. Given the above-mentioned effects of these leucine zipper domains on PRE activity, these results indicate that, in Drosophila cells, the AF10 leucine zipper domain as well as that of ALH confers PRE deregulation activity on the human oncogenic MLL-AF10 fusion.
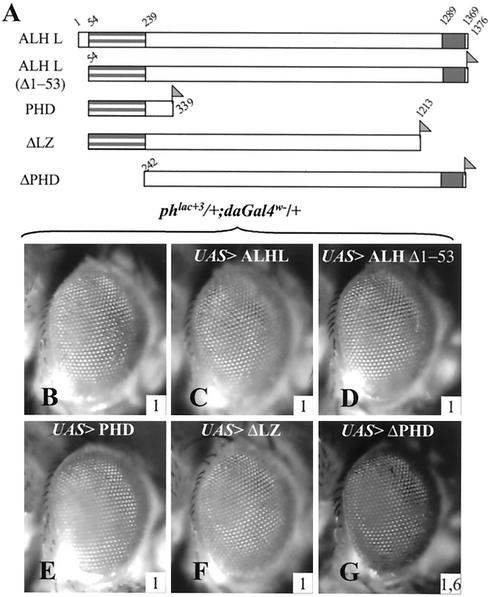
Structure-function analysis of ALH L protein reveals antagonism between PHD and leucine zipper domains in PRE regulation.
A noticeable feature of the ALH S protein is that, compared to ALH L, it is devoid of the PHD domain. In order to characterize the functions of the PHD and leucine zipper domains with respect to PRE regulation, we constructed transgenic lines that allowed the specific expression of different parts of the ALH L protein. As shown in Fig. 6, none of the protein variants that encompass the PHD domain have an effect on ph PRE silencing (Fig. 6B to G). These protein variants include, apart from the full-length proteins (ALH L and ALH LΔ1-53), a deletion of the leucine zipper (ΔLZ, Fig. 6F) and a variant corresponding essentially to the PHD domain (PHD, Fig. 6E). These data indicate that the PHD domain is unable to deregulate the ph PRE. Conversely, expression of a variant corresponding to the ALH L protein with its PHD domain deleted induces a strong deregulation of ph silencing (ΔPHD, Fig. 6G), in a manner similar to that observed when the leucine zipper domain or the ALH S protein is overexpressed. This clearly indicates that, within ALH L, the PHD domain inhibits the effect of the leucine zipper domain activity on PRE regulation.
FIG. 6.
Within ALH L, the PHD domain inhibits the activity of the leucine zipper domain. (A) Schematic representation of the different constructs used. The amino acids present in the different proteins are indicated. The corresponding cDNAs were inserted in the P{y+UAS} vector, and their expression was induced with P{daGAL4 w−}. The PHD domain is shown as a striped grey block (at the left), and the leucine zipper domain is shown as a uniform grey block (on the right). The ALH L Δ1-53 construct has a deletion of the first 53 amino acids and is thus identical to the class 3 protein described earlier (3). The carboxy-terminal Flag tag is indicated (shaded flag). (B to G) Effects of overexpression of truncated ALH L proteins on phlac+3 eye color phenotype. (C to F) All the constructs that bear the PHD domain of ALH are unable to modify ph silencing (ALH L, ALH L Δ1-53, PHD, and ΔLZ). (G) Only the construct that does not contain the PHD (ΔPHD) displays a disruption of ph silencing. The inserted numbers give the average OD485 values calculated from four experiments (see Materials and Methods).
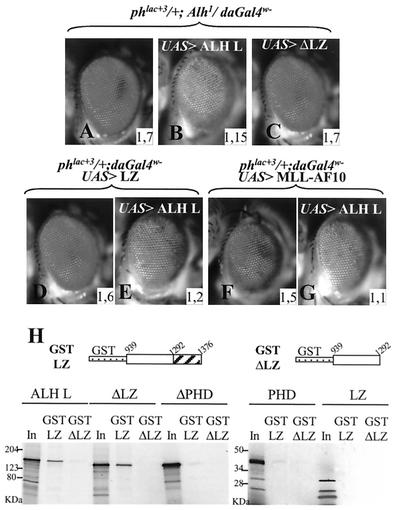
Moreover, coexpression of the full-length protein ALH L, which shows no effect by itself, together with the leucine zipper domain leads to inhibition of the effect of the leucine zipper domain (Fig. 7D and E). ALH L overexpression therefore seems to be able to titrate the action of the isolated leucine zipper domain. ALH L overexpression also reduces the effect of the Alh1 mutation (Fig. 7A and B) and of ΔPHD overexpression (not shown). In contrast, ALH L-ΔLZ has no effect in this assay, indicating that, within the ALH L protein, the leucine zipper domain is necessary for this activity (Fig. 7C). We next tested the ability of ALH L overexpression to inhibit PRE deregulation mediated by the MLL-AF10 fusion protein. As shown in Fig. 7F and G, ALH L is able to reduce the effect of the MLL-AF10 fusion protein, suggesting that conserved residues in both proteins mediate this titration effect. Not surprisingly, the MLL-ALH effect was also reduced when it was overexpressed together with ALH L (not shown).
FIG. 7.
ALH L overexpression reduces PRE deregulation mediated by ALH leucine zipper and MLL-AF10 fusion. (A to E) The ALH L protein overexpressed in Alh1 mutants reduces the effect of the Alh1 mutation (compare A and B). If a ΔLZ protein is expressed, no effect is detected, indicating the requirement of the leucine zipper domain for this titration effect (C). The same titration effect is observed when ALH L is coexpressed together with the isolated leucine zipper domain (D and E). ALH L also reduces PRE activation mediated by MLL-AF10 overexpression (F and G). The inserted numbers give the average OD485 values calculated from four experiments (see Materials and Methods). (H) GST pulldown experiments. The C-terminal part of ALH L (amino acids 938 to 1376; GST-LZ) containing the leucine zipper domain was fused to GST. As a control, the same region with the leucine zipper domain deleted (amino acids 938 to 1280; GST-ΔLZ) was fused to GST. Different truncated proteins (the same that were expressed in vivo in Fig. 4 and 6) were produced in vitro and tested for their affinity for the two GST fusion proteins. Only the complete protein ALH L and the ΔLZ protein interact in vitro with the leucine zipper domain. The PHD domain alone and the ΔPHD protein have a very weak affinity for the leucine zipper domain. We note that, in vitro, the leucine zipper domain does not interact with itself. None of the proteins tested interacts with the GST-ΔLZ fusion protein. “In” corresponds to 5% of the input labeled proteins. Apparent molecular masses are indicated on the left.
Leucine zipper domains are known for forming homodimers or heterodimers (7, 38). One hypothesis that would explain the titration effect of ALH L is that it interacts with the isolated leucine zipper domain via its own leucine zipper domain. We tested a potential oligomerization of the ALH leucine zipper domain in vitro with the GST pulldown technique. The different variants of the ALH L protein described previously (Fig. 5 and 6) were in vitro transcribed and translated, and their affinity for the C-terminal part of the ALH protein (which includes the leucine zipper domain) fused to GST was tested (GST-LZ protein, Fig. 7H). The specificity of the interaction with the leucine zipper domain was controlled by use of the GST-ΔLZ fusion protein, which corresponds to the same part of the ALH protein without the leucine zipper domain.
The full-length protein ALH L interacts with the leucine zipper domain in a specific way, since it binds to the GST-LZ protein but not to the GST-ΔLZ protein (Fig. 7H). However, the ALH L-ΔLZ protein is also able to interact with the leucine zipper domain in a specific way, which seems to rule out a homophilic interaction with this domain. This is confirmed by the inability of the leucine zipper domain alone to interact with the GST-LZ protein. The analysis of the other protein variants indicates that the PHD domain is involved in the interaction with the leucine zipper domain. This is shown by the very weak interaction between the ALH L-ΔPHD and GST-LZ fusion proteins. Nevertheless, the PHD domain appears not to be sufficient for this interaction because, when assayed alone, it binds the GST-LZ fusion protein only very weakly (Fig. 7H).
In summary, the GST pulldown results indicate that the ALH leucine zipper domain does not interact with itself and argue in favor of an interaction between the PHD and leucine zipper domains, perhaps with additional involvement of other parts of the protein. Therefore, the titration of the leucine zipper domain by the ALH L protein observed in vivo might be mediated by interaction of the isolated leucine zipper domain with the complete protein via the PHD domain and a region in the central part of the protein. The titration effect observed with MLL-AF10 suggests that ALH L could interact with the AF10 leucine zipper domain through conserved residues within the AF10 leucine zipper domain. The lack of titration effect by the ALH L-ΔLZ protein, which also interacts in vitro with the leucine zipper domain, suggests that the inhibition observed in vivo also requires the presence of the leucine zipper domain in the complete protein, for instance, by allowing its interaction with other factors.
ALH is a chromosomal protein independent of the PH/PC complex.
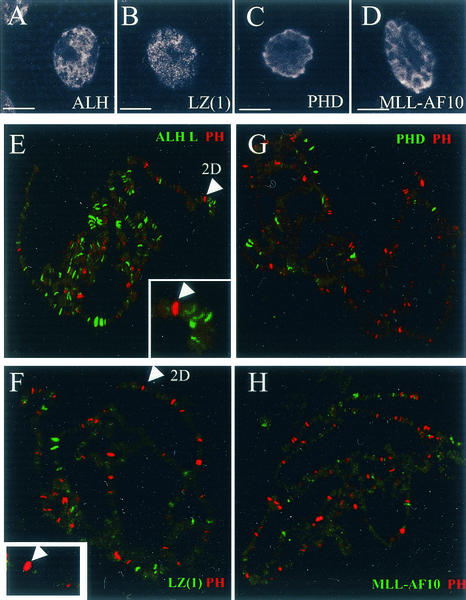
With a polyclonal anti-ALH antibody (anti-DALF [3]), we examined the distribution of the wild-type ALH L protein on salivary gland polytene chromosomes. Since Alh S is not transcribed in this tissue (see above), this allowed the specific detection of ALH L. As published previously (3), ALH localization is nuclear. This is also the case in this tissue, as revealed by immunostaining of whole salivary glands (Fig. 8A). Examination of ALH L on squashed nuclei reveals that the protein is associated with chromatin and binds more than 100 discrete sites in euchromatin (Fig. 8E). We also addressed whether ALH L and PH colocalize on salivary gland chromosomes. Double immunostaining of ALH and PH proteins reveals mutually exclusive staining patterns. This indicates that ALH L does not localize on PREs and is consistent with the lack of activity of the whole protein on PRE-mediated silencing.
FIG. 8.
Immunofluorescence localization of ALH, ALH-LZ, ALH-PHD, MLL-AF10, and PH proteins on salivary gland cells and polytene chromosomes. Whole-mount salivary glands (A to D) and polytene chromosomes (E to H) of third-instar larvae were treated with mouse polyclonal anti-ALH (A and E), monoclonal anti-Flag M2 (B to D and G to H), and rabbit polyclonal PH antibodies (E to H). The bound antibodies were detected with fluorescein-labeled anti-mouse immunoglobulin antibody and indocarbocyanine-labeled anti-rabbit immunoglobulin antibody. (A to D) All the proteins tested present a nuclear localization. These include endogenous ALH, ALH-LZ, ALH-PHD, and MLL-AF10. Bars, 5 μm. (E to H) Merge of PH staining and ALH (E), leucine zipper (F), PHD (G), and MLL-AF10 (H) staining. (E) ALH protein (green) binds over 100 euchromatic sites but never overlaps those of PH (red). We can particularly note the absence of ALH at 2D, corresponding to the ph locus, which is a PH binding site. (F and G) The isolated leucine zipper and PHD domains of the ALH L protein and the MLL-AF10 fusion protein present only faint association on polytene chromosomes that varies depending on the chromosome set examined. No consistent overlapping with PH localization is observed, particularly at 2D (magnified view in F).
To test the subcellular localization of the different protein variants used in this study, we used a Flag epitope tag. In these experiments, the cDNAs were ubiquitously expressed under the control of the daGAL4 driver. Anti-Flag immunodetection indicates that, whatever the tissue and the construct tested, all the proteins are nuclear. Immunodection of ALH leucine zipper, ALH PHD, and the MLL-AF10 fusion in salivary gland cells is given as examples (Fig. 8B to D). Next, we tested the distribution of the ALH leucine zipper, ALH PHD, and MLL-AF10 proteins on polytene chromosomes. As shown in Fig. 8F to H, these proteins only weakly bind polytene chromosomes. Indeed, they are detected on 5 to 30 euchromatic chromosomal bands, depending on the chromosome set examined, and in most cases these bands were very weakly stained.
On whole-mount salivary glands, however, these proteins were detected at levels comparable to those of the endogenous ALH L protein (Fig. 8A to D). Furthermore, this faint staining was not due to a failure of the anti-Flag antibody to stain polytene chromosomes, as this antibody was able to detect a Flag-tagged ALH L protein on salivary gland polytene chromosomes at a level comparable to that obtained with the polyclonal anti-ALH antibody (not shown). Therefore, these results might indicate that most of the proteins are not associated with chromatin. In addition, no consistent colocalization with PH was noticed, indicating that these proteins might not act directly at the level of PREs. Indeed, the ph locus at chromosomal band 2D on the X chromosome, while labeled by PH protein, is not labeled by the ALH leucine zipper, ALH PHD, or MLL-AF10.
DISCUSSION
We have shown that the loss of the conserved PHD domain in the ALH L protein, the Drosophila AF10/17 homologue, confers PRE derepression activity. This activity is also shared by the leucine zipper domain alone (when overexpressed) and is conserved in the corresponding domain of human AF10. Furthermore, our study demonstrates that, in Drosophila cells, the AF10 leucine zipper domain confers PRE regulation properties on the human MLL-AF10 oncogenic fusion.
The AF10 and AF17 regions that are present in oncogenic fusion proteins include their leucine zipper domains, the only region of homology between the two proteins (12). This is the case not only in the MLL-AF10 and MLL-AF17 fusions but also in t(10;11)(p13;q14), which fuses AF10 to CALM. Indeed, CALM-AF10 chimeric transcripts but not the AF10-CALM reciprocal message have been consistently detected in patients bearing such translocations (10, 39). This observation clearly suggests a particular role of the leucine zipper domain in leukemias. Indeed, DiMartino et al. (17) recently demonstrated that the AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. In mammals, however, functional studies of either the AF10 or AF17 leucine zipper domain are lacking. With D. melanogaster, and thanks to several amenable genetic tools, we were able to decipher some important properties of the Drosophila AF10/AF17 homologue that can be extended to the AF10 leucine zipper domain.
Our observation that the ALH L protein, although bound to polytene chromosomes, does not overlap PH binding sites might indicate that the ALH L protein is involved in transcriptional maintenance in complexes that are different from PC-G/TRX-G complexes. Indeed, Alh is required for the maintenance but not the initiation of eve expression in RP2 neurons of the Drosophila central nervous system (3). In this respect, its function resembles that of trx group genes. It might be pointed out, however, that, contrary to Pc and trx group genes, we show here that Alh loss of function does not interact with any Pc or trx group mutation tested. In addition, we were unable to detect any effect of Alh L loss or gain of function on PRE activity, whatever the PRE tested. While known Pc group and trx group genes do not exhibit functional tissue specificity, Alh loss of function has been rescued by neuron-specific expression of Alh L transcripts (3), indicating that its function is required in the nervous system only.
In addition to its association with polytene chromosomes, a chromatin function for ALH L is further reinforced by the observation that the PHD domain is a common feature of numerous chromatin-associated proteins such as TRX, CBP, and the PC-G protein PCL. In a recent study, it was shown that the PCL PHD domain mediates interaction with an Enhancer of zeste-encoded protein, another PC-G protein (41). We show here that the ALH L PHD domain contacts the ALH leucine zipper domain in vitro. PHD domains thus appear to mediate protein-protein interactions. Identification of the direct interactors with the ALH PHD domain is currently under investigation in our laboratory.
Our data clearly demonstrate that, within ALH L, the PHD domain behaves as a repressor of the leucine zipper domain with respect to PRE deregulation. Indeed, while in all our assays the ALH L protein has no effect on PRE activity, the ALH S protein, which lacks the PHD domain, induces a strong disruption of PRE silencing when overexpressed. The same activity was observed when an ALH L protein with its PHD domain deleted was used and is also caused by overexpression of the isolated leucine zipper domain. The loss of the PHD domain thus confers novel properties on the ALH L protein. This is further supported by our in vivo competition experiments. ALH L overexpression is able to inhibit the activity of ALH S, ALH L-Δ-PHD, and the ALH leucine zipper with respect to PRE regulation. This suggests that the inhibition effect of the PHD domain over the leucine zipper domain that occurs in cis within ALH L can also take place in trans between the ALH L protein and the leucine zipper domain.
This genetic interaction between the PHD and leucine zipper domains with respect to PRE regulation is further supported by our in vitro experiments. Indeed, we show that the leucine zipper domain interacts directly with the ALH L protein and that the PHD domain, while not sufficient, is necessary for this interaction. The absence of the PHD domain, which reveals the PRE regulation properties of the leucine zipper domain, might allow the latter domain to contact other proteins within the nucleus, which in turn may induce PRE transcriptional activation (see below). Given the strong homology between the PHD domain of ALH L and its human homologues, and given the functional homology of the leucine zipper of both ALH and AF10 regarding PRE regulation, the same situation is likely to prevail in the human proteins. In support of this hypothesis, we show that ALH L overexpression is able to reduce MLL-AF10-mediated PRE deregulation. Importantly, the PHD domain is lost in all oncogenic fusions affecting AF10 and AF17. Thus, the oncogenic properties of the human fusions could, at least in part, originate from loss of the PHD domain, which, in the full-length AF10 and AF17 proteins, might inhibit some properties of the leucine zipper domain.
In a recent study, Dobson et al. (20) documented that an MLL-β-galactosidase fusion has oncogenic potential in mice. This suggested that truncation and fusion of MLL can be sufficient for tumorigenesis and that the fusion partner may not have an active role in the fusion. It may rather have a permissive function, such as dimerization, a property of β-galactosidase itself. However, the MLL-β-galactosidase fusion induced leukemia with a lower frequency and a longer latency than other MLL fusions tested. The observation that AF10 is involved in leukemias independently of MLL in CALM-AF10 fusions might rule out the possibility that AF10 has only a passive role in leukemias. Indeed, we demonstrate that the AF10 leucine zipper domain, like its Drosophila counterpart, is able to activate several PREs when overexpressed alone. Furthermore, while the MLL amino-terminal portion has very weak silencing activity on Drosophila PREs, an MLL-AF10 fusion protein behaves as a strong PRE activator in our system. This demonstrates that the C-terminal part of AF10 (and most probably its leucine zipper domain) confers PRE activation potential on the MLL-AF10 fusion. This is confirmed by the observation that the MLL-ALH leucine zipper has the same activity and points to an active function of the AF10 leucine zipper in the capacity of MLL-AF10 fusion to activate Drosophila PREs.
The ALH and AF10 leucine zipper domains are specifically involved in PC-G- and TRX-G-mediated transcriptional regulation. We failed to detect any effect of these domains (or of the MLL fusion proteins) on the hsp70 minimal promoter carried by the PlacW transgene in the P{w+3-76a} chromosome or on position effect variegation (53) or telomeric position effect (30). However, none of the proteins tested (ALH-LZ, ALH L-ΔPHD, and MLL-AF10) were detected associated with PREs on polytene chromosomes from larval salivary glands. This observation suggests an indirect action of the leucine zipper domains on PRE activity.
As discussed above, the activity of the ALH leucine zipper domain on PRE regulation is observed only when it is disconnected from the PHD domain. Since the leucine zipper and PHD domains are able to interact directly in vitro, the dissociation of both domains could allow the leucine zipper domain to contact new factors that are unable to interact with it in the normal context of ALH L. Given the above-mentioned specificity for PRE-mediated transcriptional regulation, members of the PC-G and/or TRX-G complexes are likely candidates for such interactions. These interactions would titrate and inhibit the activity of PC-G proteins or enhance that of TRX-G proteins. We are currently searching for interactors with the isolated leucine zipper domain that do not interact with the full-length ALH L protein.
Importantly, we show that the leucine zipper domain activity is conserved from flies to humans. This may indicate that this activity requires conserved residues in these domains. Thus, the interactions that the AF10 leucine zipper domain is able to make in Drosophila cells could be conserved in humans. Indeed, mammals have PC-G and TRX-G homologues that are conserved both structurally and functionally, and as it is in Drosophila, their activity is required to maintain homeotic gene expression (6). Furthermore, several studies have documented a role of different homeotic genes in leukemias. For example, engineered coexpression of HOX a-9 and of its coactivator MEIS 1 in murine hematopoietic cells leads to rapid development of acute myeloid leukemias (33). Hox deregulation could thus be a unifying theme of the development of leukemia diseases. Consistent with this idea, it has recently been reported that two MLL fusion partners, AF9 and ENL, interact directly with PC3, a mammalian Polycomb homologue. In these cases, however, a direct role of the oncogenic fusions in PC-G-mediated regulation has not been tested.
Our study thus provides the first evidence that an MLL fusion can play on PC-G/TRX-G activity and furthermore demonstrates that this activity is conferred by the fusion partner and not by the MLL moiety. With Drosophila melanogaster, biochemical and genetic analysis will allow us to understand the mechanisms by which the AF10 leucine zipper domain and the MLL-AF10 fusion interfere with PC-G- and TRX-G-mediated transcriptional regulation.
Acknowledgments
We are particularly indebted to S. Bahri for fly stocks, plasmids, and antibody, to G. F. DiMartino and M. L. Cleary for MLLNter and MLL-AF10 cDNAs, and to L. Ringrose for comments on the manuscript. We thank G. Cavalli, J. Kassis, T. Kaufman, P. Schedl, and L. Theodore for fly stocks. We thank Siegfried Bozza for precious help in polytene chromosome immunostaining. We also thank M. Sémériva for helpful discussions.
Postdoctoral training from the Indo-French Centre for the Promotion of Advanced Research (IFCPAR) and the Société de Secours des Amis des Sciences supported L.P. S.B. was supported by a predoctoral fellowship from the Ministère de l'Education Nationale and by a grant from the Association pour la Recherche sur le Cancer. This work was supported by grants from the IFCPAR (project no. 1603-1), the Centre National de la Recherche Scientifique (ATIPE no. 7), the Association pour la Recherche sur le Cancer (no. 5658), and also the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, et al.. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, M. 1989. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Bahri, S. M., W. Chia, and X. Yang. 2001. The Drosophila homolog of human AF10/AF17 leukemia fusion genes (Dalf) encodes a zinc finger/leucine zipper nuclear protein required in the nervous system for maintaining EVE expression and normal growth. Mech. Dev. 100:291-301. [DOI] [PubMed] [Google Scholar]
- 4.Baker, B. S., and M. F. Wolfner. 1988. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 2:477-489. [DOI] [PubMed] [Google Scholar]
- 5.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 6.Brock, H. W., and M. van Lohuizen. 2001. The Polycomb group-no longer an exclusive club? Curr. Opin. Genet. Dev. 11:175-181. [DOI] [PubMed] [Google Scholar]
- 7.Busch, S. J., and P. Sassone-Corsi. 1990. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6:36-40. [DOI] [PubMed] [Google Scholar]
- 8.Busseau, I., and A. Bucheton. 1999. A Drosophila enhancer detector transposon marked with the yellow gene. Dros. Infect. Serv. 82:94-95. [Google Scholar]
- 9.Cairns, B. R. 2001. Emerging roles for chromatin remodeling in cancer biology. Trends Cell Biol. 11:S15-21. [DOI] [PubMed] [Google Scholar]
- 10.Carlson, K. M., C. Vignon, S. Bohlander, J. A. Martinez-Climent, M. M. Le Beau, and J. D. Rowley. 2000. Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia 14:100-104. [DOI] [PubMed] [Google Scholar]
- 11.Cavalli, G., and R. Paro. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93:505-518. [DOI] [PubMed] [Google Scholar]
- 12.Chaplin, T., P. Ayton, O. A. Bernard, V. Saha, V. Della Valle, J. Hillion, A. Gregorini, D. Lillington, R. Berger, and B. D. Young. 1995. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood 85:1435-1441. [PubMed] [Google Scholar]
- 13.Chaplin, T., O. Bernard, H. B. Beverloo, V. Saha, A. Hagemeijer, R. Berger, and B. D. Young. 1995. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood 86:2073-2076. [PubMed] [Google Scholar]
- 14.Corral, J., I. Lavenir, H. Impey, A. J. Warren, A. Forster, T. A. Larson, S. Bell, A. N. McKenzie, G. King, and T. H. Rabbitts. 1996. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 85:853-861. [DOI] [PubMed] [Google Scholar]
- 15.Crosby, M. A., C. Miller, T. Alon, K. L. Watson, C. P. Verrijzer, R. Goldman-Levi, and N. B. Zak. 1999. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debernardi, S., A. Bassini, L. K. Jones, T. Chaplin, B. Linder, D. R. de Bruijn, E. Meese, and B. D. Young. 2002. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99:275-281. [DOI] [PubMed] [Google Scholar]
- 17.DiMartino, J. F., P. M. Ayton, E. H. Chen, C. C. Naftzger, B. D. Young, and M. L. Cleary. 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99:3780-3785. [DOI] [PubMed] [Google Scholar]
- 18.DiMartino, J. F., and M. L. Cleary. 1999. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol. 106:614-626. [DOI] [PubMed] [Google Scholar]
- 19.Djabali, M., L. Selleri, P. Parry, M. Bower, B. D. Young, and G. A. Evans. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukemias. Nat. Genet. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 20.Dobson, C. L., A. J. Warren, R. Pannell, A. Forster, and T. H. Rabbitts. 2000. Tumorigenesis in mice with a fusion of the leukemia oncogene Mll and the bacterial lacZ gene. EMBO J. 19:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fauvarque, M. O., and J. M. Dura. 1993. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 7:1508-1520. [DOI] [PubMed] [Google Scholar]
- 22.Fauvarque, M. O., V. Zuber, and J. M. Dura. 1995. Regulation of polyhomeotic transcription may involve local changes in chromatin activity in Drosophila. Mech. Dev. 52:343-355. [DOI] [PubMed] [Google Scholar]
- 23.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Cuellar, M. P., O. Zilles, S. A. Schreiner, M. Birke, T. H. Winkler, and R. K. Slany. 2001. The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene 20:411-419. [DOI] [PubMed] [Google Scholar]
- 25.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1:996-1004. [DOI] [PubMed] [Google Scholar]
- 26.Gindhart, J. G., Jr., and T. C. Kaufman. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701-708. [DOI] [PubMed] [Google Scholar]
- 28.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemenway, C. S., A. C. de Erkenez, and G. C. Gould. 2001. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene 20:3798-3805. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff, S. 1992. Position effect and related phenomena. Curr. Opin. Genet. Dev. 2:907-912. [DOI] [PubMed] [Google Scholar]
- 31.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 32.Kassis, J. A. 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136:1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroon, E., J. Krosl, U. Thorsteinsdottir, S. Baban, A. M. Buchberg, and G. Sauvageau. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linder, B., N. Gerlach, and H. Jackle. 2001. The Drosophila homolog of the human AF10 is an HP1-interacting suppressor of position effect variegation. EMBO Rep. 2:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linder, B., L. K. Jones, T. Chaplin, A. Mohd-Sarip, U. A. Heinlein, B. D. Young, and V. Saha. 1998. Expression pattern and cellular distribution of the murine homologue of AF10. Biochim. Biophys. Acta 1443:285-296. [DOI] [PubMed] [Google Scholar]
- 36.Lindsley, D. T., and G. G. Zimm. 1992. The genome of Drosophila melanogaster. Academic Press, Inc., San Diego, Calif.
- 37.Lonie, A., R. D'Andrea, R. Paro, and R. Saint. 1994. Molecular characterisation of the Polycomblike gene of Drosophila melanogaster, a trans-acting negative regulator of homeotic gene expression. Development 120:2629-2636. [DOI] [PubMed] [Google Scholar]
- 38.Motohashi, H., J. A. Shavit, K. Igarashi, M. Yamamoto, and J. D. Engel. 1997. The world according to Maf. Nucleic Acids Res. 25:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narita, M., K. Shimizu, Y. Hayashi, T. Taki, M. Taniwaki, F. Hosoda, H. Kobayashi, H. Nakamura, N. Sadamori, H. Ohnishi, F. Bessho, M. Yanagisawa, and M. Ohki. 1999. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br. J. Haematol. 105:928-937. [DOI] [PubMed] [Google Scholar]
- 40.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connell, S., L. Wang, S. Robert, C. A. Jones, R. Saint, and R. S. Jones. 2001. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J. Biol. Chem. 276:43065-43073. [DOI] [PubMed] [Google Scholar]
- 42.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 43.Pirrotta, V. 1988. Vectors for P-mediated transformation in Drosophila. Bio/Technology 10:437-456. [DOI] [PubMed] [Google Scholar]
- 44.Prasad, R., D. Leshkowitz, Y. Gu, H. Alder, T. Nakamura, H. Saito, K. Huebner, R. Berger, C. M. Croce, and E. Canaani. 1994. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc. Natl. Acad. Sci. USA 91:8107-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowley, J. D. 1998. The critical role of chromosome translocations in human leukemias. Annu. Rev. Genet. 32:495-519. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 49.Simon, J., A. Chiang, W. Bender, M. J. Shimell, and M. O'Connor. 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158:131-144. [DOI] [PubMed] [Google Scholar]
- 50.Taillebourg, E., and J. M. Dura. 2000. P element replacement at the linotte/derailed locus in Drosophila: presence of the wild-type region in the homologous chromosome increases the efficiency. Dros. Infect. Serv. 83:91-93. [Google Scholar]
- 51.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 52.Wallrath, L. L., and S. C. Elgin. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9:1263-1277. [DOI] [PubMed] [Google Scholar]
- 53.Weiler, K. S., and B. T. Wakimoto. 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29:577-605. [DOI] [PubMed] [Google Scholar]
- 54.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]