Abstract
mi-er1 (previously called er1) was first isolated from Xenopus laevis embryonic cells as a novel fibroblast growth factor-regulated immediate-early gene. Xmi-er1 was shown to encode a nuclear protein with an N-terminal acidic transcription activation domain. The human orthologue of mi-er1 (hmi-er1) displays 91% similarity to the Xenopus sequence at the amino acid level and was shown to be upregulated in breast carcinoma cell lines and tumors. Alternative splicing at the 3′ end of hmi-er1 produces two major isoforms, hMI-ER1α and hMI-ER1β, which contain distinct C-terminal domains. In this study, we investigated the role of hMI-ER1α and hMI-ER1β in the regulation of transcription. Using fusion proteins of hMI-ER1α or hMI-ER1β tethered to the GAL4 DNA binding domain, we show that both isoforms, when recruited to the G5tkCAT minimal promoter, function to repress transcription. We demonstrate that this repressor activity is due to interaction and recruitment of a trichostatin A-sensitive histone deacetylase 1 (HDAC1). Furthermore, deletion analysis revealed that recruitment of HDAC1 to hMI-ER1α and hMI-ER1β occurs through their common ELM2 domain. The ELM2 domain was first described in the Caenorhabditis elegans Egl-27 protein and is present in a number of SANT domain-containing transcription factors. This is the first report of a function for the ELM2 domain, highlighting its role in the regulation of transcription.
mi-er1 (mesoderm induction early response 1), previously called er1, was first isolated as a novel fibroblast growth factor-regulated immediate-early gene from Xenopus embryonic cells induced to differentiate into mesoderm (22). Xmi-er1 encodes a nuclear protein that contains an N-terminal acidic domain with potent transcriptional activation activity (22).
The human orthologue of mi-er1 (hmi-er1) displays 91% similarity to the Xenopus sequence at the amino acid level (23) and has been shown to undergo tissue-specific alternative splicing to produce six protein isoforms that differ in their amino and carboxyl termini (24). The hmi-er1 isoform expression pattern is complex, but overall, expression of hmi-er1 isoforms is very low or undetectable in healthy adult tissues (23, 24). Human breast carcinoma cell lines and breast tumors, on the other hand, displayed elevated levels of hmi-er1 (23).
Alternate use of a facultative intron at the 3′ end of hmi-er1 produces two major isoforms, hMI-ER1α and hMI-ER1β, which contain distinct C-terminal domains (24). The α C terminus consists of 23 amino acids (aa) and includes the sequence LXXLL, a motif important for interaction with nuclear hormone receptors (15). This motif also bears some similarity to the Sin3A interaction domain of the MAD family of transcriptional repressors (4). The β C terminus contains 102 aa and includes the functional nuclear localization signal (NLS) (24, 25). The divergent amino acid sequences of the α and β C termini suggest that the two serve distinct functions.
Structurally, hMI-ER1 isoforms share a number of features with other transcriptional regulators. The common internal hMI-ER1 sequence contains the previously characterized acid activation domain (22), an ELM2 domain, and a signature SANT domain. The latter was first identified in the transcription factors SWI3, ADA2, N-CoR, and TFIIIB, from which the acronym “SANT” is derived (1). Other SANT domain-containing proteins include transcriptional regulatory molecules involved in nuclear hormone activity, such as N-CoR (1) and SMRT (21); molecules that are components of transcription- and chromatin-regulatory complexes, such as MTA-1 (33) and MTA-2 (37); and molecules that are important for regulating developmental events, such as Egl-27 (29) and CoREST (40). The SANT domain has been implicated in DNA binding as well as in protein-protein interactions (1), including interactions with complexes containing histone deacetylase (HDAC) (14, 40) and histone acetyltransferase (HAT) (30); however, the precise function(s) of the SANT domain in individual proteins remains to be determined. The ELM2 (EGL-27 and MTA1 homology) domain was first described in Egl-27, a Caenorhabditis elegans protein that plays a fundamental role in patterning during embryonic development (29). Interestingly, most ELM2 domain-containing proteins also possess a SANT domain, implying a structural and/or functional relationship between these two motifs. The ELM2 domain is conserved throughout evolution, but to date no function has been ascribed to this motif.
Given the potential role of hMI-ER1 as a novel transcription factor and its association with the neoplastic state, we investigated the role of the hMI-ER1α and hMI-ER1β isoforms in transcriptional regulation. In this paper, we demonstrate that both isoforms function as transcriptional repressors and that this repression involves recruitment of HDAC1 activity to the common ELM2 domain. This is the first report to characterize the function of the hMI-ER1 protein and to define a role for the ELM2 domain.
MATERIALS AND METHODS
Cells.
HeLa cervical carcinoma, C33A cervical carcinoma, HEK 293 human embryonic kidney, and NIH 3T3 fibroblast cell lines were obtained from the American Type Culture Collection and cultured as described previously (22).
Plasmids and constructs.
The plasmids used in this study include the G5tkCAT reporter plasmid (a kind gift from Diane Hayward, Johns Hopkins School of Medicine), which contains chloramphenicol acetyltransferase (CAT) linked to five GAL4 DNA binding sites and the herpes simplex virus minimal thymidine kinase (tk) promoter (16, 32, 38); the pM plasmid (Clontech), which contains the GAL4 DNA binding domain (DBD) and an NLS; CS3+MT, containing the Myc epitope tag (a kind gift from David Turner, University of Michigan); the pGBKT7 plasmid (Clontech), containing the Myc epitope tag; HA-Hdac3 (a kind gift from Mark Featherstone, McGill University); and a luciferase T7 control plasmid encoding full-length luciferase protein (Promega).
Expression vectors were engineered to contain full-length or deletion mutants of hMI-ER1α or hMI-ER1β fused to the GAL4 DBD of the pM plasmid or to the Myc epitope tag of the CS3+MT plasmid. Specific primers incorporating 5′ and 3′ BamHI sites were used to amplify the entire coding sequence of either hMI-ER1α or hMI-ER1β, and the digested PCR fragments were inserted into the BglII site of either the pM or the CS3+MT plasmid. Fragments encoding the appropriate amino acid residues of hMI-ER1α or β deletion mutants were amplified by PCR using the primer pairs listed in Table 1. PCR products were cloned into pCR3.1 using the TA cloning kit (Invitrogen, Inc.), and EcoRI fragments were then inserted into the complementary sites of the pM or pGBKT7 plasmid. The deletion constructs were named according to the encoded amino acid residues of the hMI-ER1α or β protein. The GAL4-hMI-ER1(163-283) 213W→A and GAL4-hMI-ER1(163-283) 226FL→AA mutant constructs were generated by site-directed mutagenesis using two complementary primers (Oligos, Etc.) designed to contain the mutation and a QuikChange site-directed mutagenesis kit (Stratagene Inc.). All plasmids were sequenced to verify the junctions and the hMI-ER1 sequence.
TABLE 1.
PCR primers pairs used for preparing hmi-er1 constructs
| Constructa | Forward primer | Reverse primer |
|---|---|---|
| hmi-er1α aa 1-432 | 5′-CGGGATCCATATGGCGGAGCCATCTGTTG-3′ | 5′-CGGGATCCAAAACAAGACCACAGAAGC-3′ |
| hmi-er1β aa 1-512 | 5′-CGGGATCCATATGGCGGAGCCATCTGTTG-3′ | 5′-CGGGATCCTTAGTCATCTGTGTTTTCAAG-3′ |
| aa 1-283 | 5′-CACCATGGCGACATCTGTTGAATC-3′ | 5′-ATCCTCTCTAGCTGCTTTTACA-3′ |
| aa 1-155 | 5′-CACCATGGCGACATCTGTTGAATC-3′ | 5′-CAAAATATTTACATCGACGTGGGCG-3′ |
| aa 1-82 | 5′-CACCATGGCGACATCTGTTGAATC-3′ | 5′-TCTTCAGGTAGTCGAACAGTAC-3′ |
| aa 82-155 | 5′-CACCATGGAAGAAGATGAGGAAGAGGAAGAAGAG-3′ | 5′-CAAAATATTTACATCGACGTGGGCG-3′ |
| aa 82-432 | 5′-CACCATGGAAGAAGATGAGGAAGAGGAAGAAGAG-3′ | 5′-CGGGATCCAAAACAAGACCACAGAAGC-3′ |
| aa 163-432 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-CGGGATCCAAAACAAGACCACAGAAGC-3′ |
| α 286-432 | 5′-CACCATGGTTTGGACAGAGGAAGAGTGTA-3′ | 5′-CGGGATCCAAAACAAGACCACAGAAGC-3′ |
| β 286-512 | 5′-CACCATGGTTTGGACAGAGGAAGAGTGTA-3′ | 5′-CGGGATCCTTAGTCATCTGTGTTTTCAAG-3′ |
| aa 163-283 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-ATCCTCTCTAGCTGCTTTTACA-3′ |
| aa 179-283 | 5′-AAGGAGATTATGGTGGGCTCCATGTTTCAA-3′ | 5′-ATCCTCTCTAGCTGCTTTTACA-3′ |
| aa 163-238 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-CTTCTCATCACCTGTTCTTCTAGATGCATC-3′ |
| aa 239-283 | 5′-GGTGTAGAAGCAATTCCTGAAGGATCTCAC-3′ | 5′-ATCCTCTCTAGCTGCTTTTACA-3′ |
| aa 163-272 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-TCATCTTCTCAATGCTTCTTCTGTATCAAAATTGCA-3′ |
| aa 163-261 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-TCATTTAACCAATTCATATAAAGCCTGTTCATTGTC-3′ |
| aa 163-250 | 5′-CACCATGGAAGAATCTGAAGAAGATGAAGATT-3′ | 5′-TCATTTTATGTGAGATCCTTCAGGAATTGCTTC-3′ |
| 213W → A | 5′-GATCAGCTCCTGGCGGACCCTGAGTACTTACC-3′ | 5′-GGTAAGTACTCAGGGTCCGCCAGGAGCTGATC-3′ |
| 226FL → AA | 5′-GTGATTATAGCTGCTAAAGATGCATCTAGAAGAACAGGTGATGAGAAGG-3′ | 5′-CCTTCTCATCACCTGTTCTTCTAGATGCATCTTTAGCAGCTATAATCAC-3′ |
Deletion constructs were named according to the encoded amino acid residues of the hMI-ER1α or β protein.
Full-length human hdac1 cDNA was amplified by PCR from a testis library by using 5′-ACGGGAGGCGAGCAAGATGGCG-3′ and 5′-TCAGGCCAACTTGACCTCCTCCTTGAC-3′ as forward and reverse primers, respectively, and then cloned into pCR3.1 as described above.
Transfection and CAT assays.
All transfections were performed in duplicate in six-well plates with the indicated amount of plasmid DNA as previously described (22). A total of 1.5 × 105 cells/well was seeded 18 h prior to transfection, and cells were harvested after 48 h in culture. For trichostatin A (TSA) treatment, the medium was replaced 24 h after transfection with fresh medium with or without the indicated concentration of TSA, and the cells were cultured for an additional 24 h. Cell lysates were prepared and assayed for CAT protein by using a CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim) as described previously (22). The amount of CAT protein expressed in each sample was determined using a CAT standard curve supplied by the manufacturer, and this value was normalized to the amount of cellular protein in each sample.
Co-IP and Western blot analysis.
In vitro coupled transcription-translation reactions (TNTs; Promega) were performed as described previously (26). For in vitro coimmunoprecipitation (co-IP) assays, 10 μl each of 35S-labeled and unlabeled TNT mixtures programmed with the appropriate cDNAs were combined and incubated for 3 h at 4°C. Immunoprecipitation was performed as described previously (26) with either an anti-HDAC1 antibody (Santa Cruz Biotechnology, Inc.) or the anti-Myc monoclonal antibody 9E10 (a kind gift from K. Kao, Memorial University). For the input lanes, 0.5 μl of the indicated TNT mixture was used. All samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
For in vivo co-IP assays, either nontransfected HeLa cells or cells transfected with pCS3+MT, pCS3+MT-hmi-er1α, or pCS3+MT-hmi-er1β were used. Cells were lysed, and the insoluble material was removed by centrifugation at 12,000 × g for 10 min. For input lanes, one-third of the volume of the soluble fraction was used. Supernatants were subjected to immunoprecipitation with either an anti-hMI-ER1 antibody produced in our laboratory (24) or anti-HDAC1. Western blot analysis was performed as described previously (26) with anti-Myc or anti-HDAC1.
HDAC assays.
The [3H]acetate-labeled histone substrate for the HDAC enzyme assays was prepared from HeLa cells labeled with [3H]acetate in the presence of 300 nM TSA as described previously (39). For each sample, 1.5 × 105 HeLa cells were transfected with the appropriate construct, and after 48 h, the cells were lysed as described previously (17) and insoluble material was removed by centrifugation at 12,000 × g for 10 min. Supernatants were subjected to immunoprecipitation using anti-GAL4 (Upstate Biotechnology, Inc.), anti-Myc, or anti-HDAC1 antibody, and the immunoprecipitates were analyzed for HDAC activity as described previously (17) with 5,000 cpm of [3H]acetate-labeled histone substrate. Western blot analysis was performed as described above with anti-GAL4.
RESULTS
hMI-ER1α and hMI-ER1β isoforms act as transcriptional repressors.
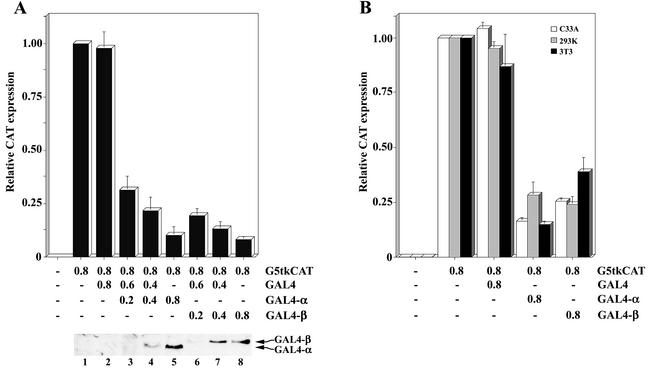
A previous study showed that XMI-ER1 is a nuclear protein and that the N-terminal acidic domain could function as a transcriptional activator (22). In this study, we set out to investigate the transcriptional regulatory activity of the hMI-ER1α and β isoforms by using the G5tkCAT reporter plasmid. This plasmid contains CAT linked to five GAL4 DNA binding sites along with the herpes simplex virus minimal tk promoter (16, 32, 38) to provide a constitutive level of CAT expression; thus, both activation and repression can be measured. HeLa cells were transfected with the reporter plasmid along with a plasmid expressing the GAL4 DBD alone or fused to hMI-ER1α (GAL4-α) or hMI-ER1β (GAL4-β). Transcriptional regulation was analyzed by determining the ability of such fusions to alter the level of CAT expression. Both hMI-ER1α and β significantly repressed transcription of G5tkCAT in a dose-dependent manner (P < 0.05) (Fig. 1A). This repression was not unique to HeLa cells, as similar results were also obtained with C33A cervical carcinoma cells, NIH 3T3 mouse fibroblasts, and HEK 293 human embryonic kidney cells (Fig. 1B).
FIG. 1.
Human MI-ER1α and MI-ER1β function as transcriptional repressors in vivo. Cells were transfected with the G5tkCAT reporter plasmid alone or with the pM plasmid containing an NLS and the GAL4 DBD (GAL4) alone or fused to hmi-er1α (GAL4-α) or hmi-er1β (GAL4-β); the amount of plasmid (in micrograms) used for transfection is indicated below each bar. Cells were harvested 48 h after transfection, and the amount of CAT protein (in nanograms per 100 μg of cellular protein) was determined as described in Materials and Methods. Expression values for all constructs were normalized to the CAT expression level obtained with G5tkCAT alone (relative CAT expression). Shown are the average values and standard deviations from at least three independent experiments. (A) Relative CAT expression in HeLa cells transfected with increasing amounts of GAL4-α or GAL4-β plasmid; in each case, the amount of GAL4 empty vector was adjusted so that the total amount of DNA used in each transfection was constant. The amount of GAL4-α or GAL4-β protein expressed in each sample was determined by Western blotting using an anti-GAL4 antibody. A representative blot is shown. (B) Relative CAT expression in C33A, HEK 293 (293K), and NIH 3T3 cells transfected with the G5tkCAT reporter plasmid alone or cotransfected with the indicated GAL4 construct. Shown are the average values and standard deviations from three independent experiments.
hMI-ER1α and hMI-ER1β repress transcription through a HDAC-dependent mechanism.
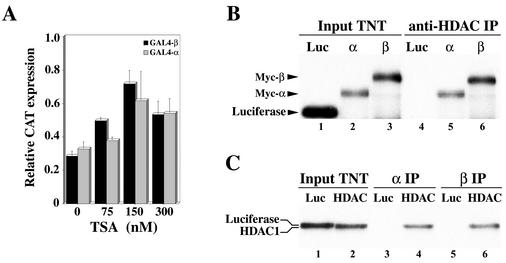
Recent studies have shown that transcriptional repression is often associated with recruitment of HDAC and/or chromatin-regulatory complexes containing HDAC activity (reviewed in reference 3). To test whether hMI-ER1α and β repress transcription through such a mechanism, we treated HeLa cells with TSA, a specific inhibitor of class I and class II HDACs (39), and examined the effect on hMI-ER1α- and β-mediated repression. Addition of TSA to the culture medium partially relieved repression of the tk promoter (Fig. 2A), suggesting that transcriptional repression by hMI-ER1α and β involves recruitment of HDAC activity.
FIG. 2.
Repression by hMI-ER1α and hMI-ER1β occurs through a HDAC-dependent mechanism. (A) HeLa cells were cotransfected with 0.8 μg of the G5tkCAT reporter plasmid and 0.8 μg of the GAL4, GAL4-α, or GAL4-β plasmid and cultured in the presence or absence of TSA. Cells were harvested 48 h after transfection, and the amount of CAT protein (in nanograms per 100 μg of cellular protein) was determined as described in Materials and Methods. The values for GAL4-α- and GAL4-β-transfected cells are presented as a proportion of the value obtained with the GAL4 empty vector (relative CAT expression). Shown are the average values and standard deviations from three independent experiments. (B) 35S-labeled TNT mixtures programmed with cDNA encoding luciferase (Luc), Myc-α (α), or Myc-β (β) were loaded directly on the gel (lanes 1 to 3) or incubated with unlabeled TNT mixtures programmed with hdac1 cDNA and subjected to immunoprecipitation (IP) with anti-HDAC1 (lanes 4 to 6) as described in Materials and Methods. Proteins were visualized by SDS-PAGE and autoradiography. (C) 35S-labeled TNT mixtures programmed with cDNA encoding luciferase or HDAC1 were loaded directly on the gel (lanes 1 and 2) or incubated with unlabeled TNT mixtures programmed with cDNA encoding Myc-α (lanes 3 and 4) or Myc-β (lanes 5 and 6) and subjected to IP with anti-Myc; proteins were visualized as described for panel B. In panels B and C, the positions of luciferase, HDAC1, Myc-α, and Myc-β proteins are indicated.
To further investigate the nature of the TSA-sensitive repression, we utilized co-IP analysis to examine the ability of hMI-ER1α and β to physically associate with HDACs both in vitro and in vivo. We began our investigation with HDAC1. Myc-tagged hMI-ER1α (Myc-α) and hMI-ER1β (Myc-β) were synthesized in vitro and mixed with in vitro-translated HDAC1 and then subjected to immunoprecipitation with anti-HDAC1; both hMI-ER1α and hMI-ER1β were detected in HDAC1 immunoprecipitates, while luciferase, a control protein, was not (Fig. 2B). Reciprocal experiments using anti-Myc for immunoprecipitation confirmed that HDAC1 could associate with either hMI-ER1 isoform (Fig. 2C). Similar experiments were performed with HDAC3 but failed to reveal an interaction with hMI-ER1α or hMI-ER1β (data not shown).
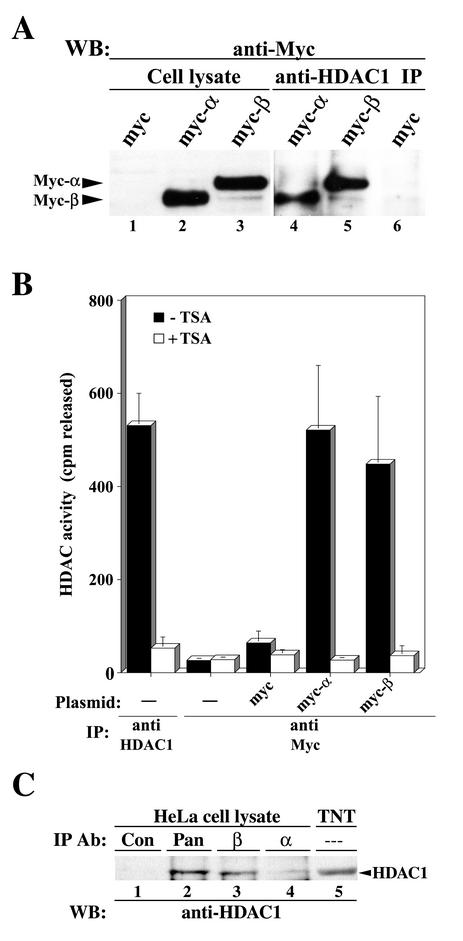
In vivo analysis involved transient expression of the Myc tag empty vector, Myc-α, or Myc-β in HeLa cells. Cell extracts were subjected to immunoprecipitation with anti-HDAC1, followed by Western blotting with anti-Myc. As shown in Fig. 3A, both hMI-ER1α and β isoforms coimmunoprecipitated with endogenous HDAC1. Next, we tested whether HDAC activity was recovered in hMI-ER1α and β immunoprecipitates. Cell extracts from HeLa cells transfected with the Myc tag empty vector, Myc-α, or Myc-β were incubated with anti-Myc, and the immunoprecipitates were assayed for HDAC activity. Positive and negative controls consisted of extracts from mock-transfected cells immunoprecipitated with anti-HDAC1 and anti-Myc, respectively. As shown in Fig. 3B, both Myc-α and Myc-β immunoprecipitates contained significant levels of HDAC activity and this activity was inhibited by TSA. On the other hand, control samples from mock-transfected cells or cells transfected with the Myc tag empty vector did not contain significant HDAC activity.
FIG. 3.
A functional HDAC1 coimmunoprecipitates with hMI-ER1α and hMI-ER1β in vivo. (A) Cell lysates from HeLa cells transiently transfected with myc tag empty vector or myc-α or myc-β plasmid were prepared, and equivalent amounts of protein from each sample were either added directly to sample buffer (lanes 1 to 3) or subjected to immunoprecipitation (IP) with anti-HDAC1 (lanes 4 to 6); Western blot (WB) analysis was performed using anti-Myc. The positions of the Myc-α and Myc-β proteins are indicated. (B) HeLa cells (1.5 × 105 cells per sample) were transfected with myc tag empty vector or myc-α or myc-β plasmid and lysed, and the supernatants were subjected to IP with anti-Myc. Additional controls consisted of mock-transfected HeLa cell extracts immunoprecipitated with anti-HDAC1 or anti-Myc. Immunoprecipitates were assayed for HDAC activity in the presence or absence of 300 nM TSA as described in Materials and Methods. Shown are the average values and standard deviations from three independent experiments. (C) HeLa cell lysates were subjected to IP with nonimmune serum (lane 1) or anti-pan hMI-ER1 (lane 2), anti-hMI-ER1β-specific (lane 3), or anti-hMI-ER1α-specific (lane 4) antiserum. HDAC1 protein from an in vitro TNT mixture was loaded in lane 5. Western blot (WB) analysis was performed using anti-HDAC1. The position of the HDAC1 protein is indicated.
The next set of experiments was designed to investigate whether endogenous complexes containing hMI-ER1 and HDAC1 exist in the cell and thus to rule out the possibility that the observed interaction was an artifact of overexpression. Co-IP analysis of extracts from nontransfected HeLa cells was performed using hMI-ER1α-specific, hMI-ER1β-specific, or pan-hMI-ER1 antibodies. As shown in Fig. 3C, HDAC1 protein was detected in all hMI-ER1 immunoprecipitates but not in the control.
Taken together, these results demonstrate that hMI-ER1α and hMI-ER1β isoforms are physically associated in vivo with a functional HDAC1 protein.
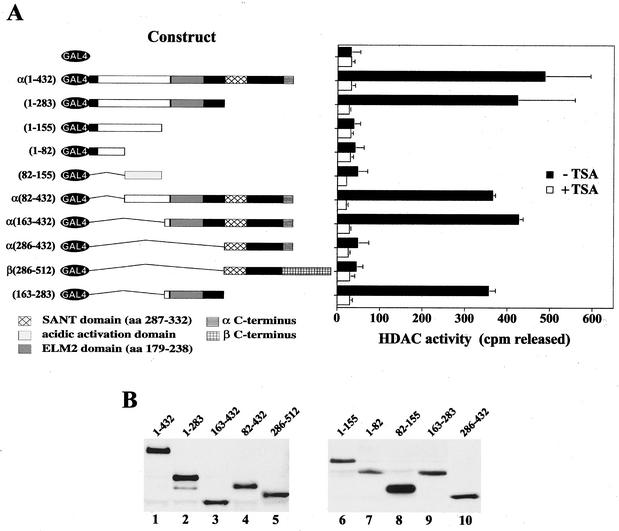
hMI-ER1α and β recruit HDAC activity through a region containing the ELM2 domain.
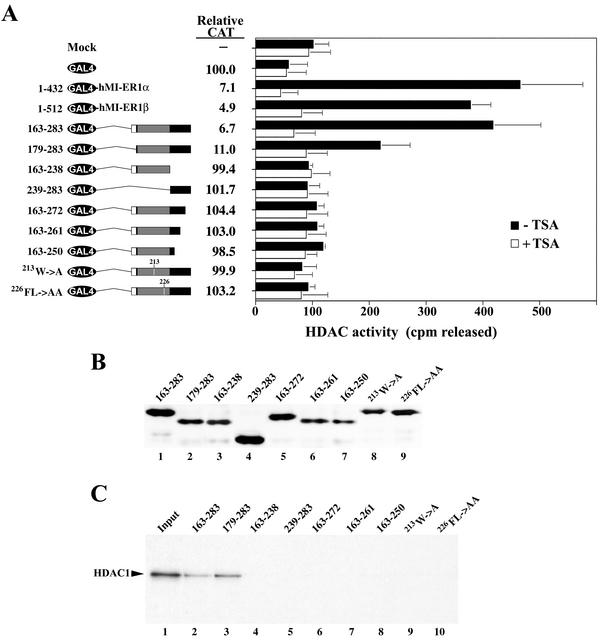
To determine which region of hMI-ER1 protein was responsible for recruitment of HDAC activity, a series of GAL4 DBD-hMI-ER1α and GAL4 DBD-hMI-ER1β deletion mutants were constructed and transiently expressed in HeLa cells. Expression of all constructs was confirmed by Western blotting (Fig. 4B), and anti-GAL4 immunoprecipitates were tested for HDAC activity in the presence or absence of TSA (Fig. 4A). This deletion analysis revealed that a single 120-aa region common to both hMI-ER1α and β was sufficient for recruitment of HDAC activity. This region is located between aa 163 and 283 and contains the conserved ELM2 domain of hMI-ER1.
FIG. 4.
hMI-ER1 associates with HDAC activity through a region containing the ELM2 domain. (A) Deletion mutants of hMI-ER1α or β fused to GAL4 were transfected into HeLa cells (1.5 × 105 cells per sample). The schematic on the left illustrates the constructs used and shows a scaled representation of the hMI-ER1 sequence and each of its domains. The individual domains are identified in the legend below the schematic, and the hMI-ER1 amino acid residues encoded by each construct are listed on the left. Cell extracts were prepared 48 h after transfection and subjected to immunoprecipitation with anti-GAL4. Immunoprecipitates were assayed for HDAC activity in the presence or absence of 300 nM TSA as described in Materials and Methods. The histogram shows the average values and standard deviations from three independent experiments. (B) The expression of the GAL4-hMI-ER1 fusion protein in each sample used in panel A was examined by Western blotting using an anti-GAL4 antibody. Indicated above each lane are the hMI-ER1 residues encoded by the construct.
The ELM2 domain functions as a transcriptional repression domain through recruitment of HDAC1.
Further analysis of the region containing residues 163 to 283 was performed to determine the minimum sequence required for recruitment of HDAC1 and to determine whether this region was important for the transcriptional repression activity of hMI-ER1. We constructed a series of GAL4-hMI-ER1(163-283) deletion mutants and employed a site-directed mutagenesis approach to specifically examine the role of the ELM2 domain. For this purpose, a highly conserved tryptophan (W) at position 213 and phenylalanine-leucine (FL) at positions 226 and 227 were changed to alanines (A) to produce two mutant GAL4-hMI-ER1(163-283) constructs, 213W→A and 226FL→AA, respectively.
All constructs were transfected into HeLa cells, and expression of the GAL4 fusion proteins was verified by Western blotting (Fig. 5B). Extracts from these cells were assayed for CAT expression and HDAC activity (Fig. 5A). Our analysis revealed that deletion of the N-terminal 16 aa in GAL4-hMI-ER1(179-283) reduced the associated HDAC activity and had a slight effect on repression. Further deletion of N-terminal residues reduced HDAC activity and transcriptional repression to control levels (Fig. 5A). At the C-terminal end, all deletions, even one as small as 11 aa [GAL4-hMI-ER1(163-272)], completely eliminated HDAC activity and transcriptional repression. Mutant proteins 213W→A and 226FL→AA were also unable to recruit HDAC activity or to function in transcriptional repression (Fig. 5A), confirming the role of the ELM2 domain in mediating these activities. Thus, only GAL4-hMI-ER1(163-283) and GAL4-hMI-ER1(179-283) showed significant levels of associated HDAC activity and transcriptional repression. Moreover, these were the only two constructs that were able to coimmunoprecipitate HDAC1 (Fig. 5C). These data demonstrate that the minimum sequence for recruitment of HDAC1 activity and repression of transcription is contained in aa 179 to 283.
FIG. 5.
The ELM2 domain of hMI-ER1 can recruit HDAC1 activity and repress transcription in vivo. (A) Deletion mutants of hMI-ER1(163-283) fused to GAL4 were transfected into HeLa cells for HDAC activity measurements or were cotransfected with the G5tkCAT reporter plasmid for transcriptional repression assays. The schematic illustrates the constructs used, and the hMI-ER1 amino acid residues encoded by each construct are listed on the left. 213W→A and 226FL→AA constructs contain residues 163 to 283 with alanine substitutions at 213W and 226FL, respectively. HDAC activity measurements were performed as described in the legend to Fig. 3B, and the average values and standard deviations from three independent experiments are shown. Repression was determined by measuring CAT expression levels as described in the legend to Fig. 2. (B) The expression of the GAL4-hMI-ER1 fusion protein in each sample used in panel A was examined by Western blotting using an anti-GAL4 antibody. Indicated above each lane are the hMI-ER1 residues encoded by each construct. (C) 35S-labeled TNT mixtures programmed with cDNA encoding HDAC1 were loaded directly on the gel (Input; lane 1) or incubated with unlabeled TNT mixtures programmed with Myc-tagged constructs of the regions listed in panel A and then subjected to immunoprecipitation using anti-Myc (lanes 2 to 10). Proteins were visualized by SDS-PAGE and autoradiography. The position of the HDAC1 protein is indicated.
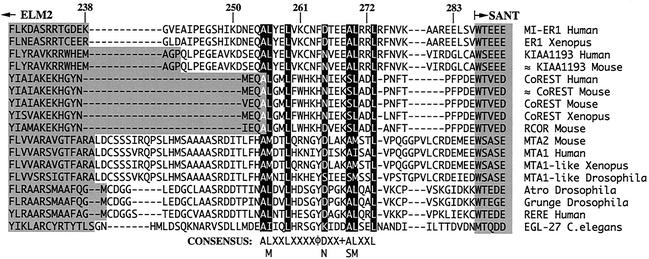
While our analysis showed that the ELM2 domain (aa 179 to 238) is required for HDAC1 binding and transcriptional repression, it was clear that an additional sequence C terminal to this domain is essential. The ELM2 domain was originally defined on the basis of sequence conservation in a small number of available protein sequences (29). A reexamination of the alignment by using a larger number of ELM2-containing proteins revealed conservation of an additional sequence C terminal to the defined ELM2 domain (Fig. 6). This highly conserved sequence encompasses aa 255 to 273 of hMI-ER1 and contains the consensus ALXXLX5DX3ALXXL, where the second and last leucines are invariant.
FIG. 6.
Alignment of ELM2 domains reveals additional conserved sequence. The ELM2 regions of proteins from the Pfam and GenBank databases were aligned using ClustalW. Shown is the amino acid sequence from the C-terminal end of the ELM2 domain to the beginning of the SANT domain in each protein. Residues belonging to these two domains are shaded. Highly conserved residues in the region C terminal to the ELM2 domain are shown with white lettering and highlighted in black. The consensus sequence is listed below the alignment; X represents any amino acid; φ represents Y, F, or H; and + represents a charged residue. The numbers listed above the alignment correspond to amino acid positions in the hMI-ER1 protein sequence. The accession numbers for the sequences used in this alignment (from top to bottom) are as follows: AF515447, O42194, AB033019, XM_125783, Q9UKL0, XM_127140, XM_127140, AJ311849, Q9JMK4, Q9R190, Q13330, O94776, Q9VNF6, Q9VNF6, Q9NHX6, Q9P2R6, and Q09228.
The data presented here illustrate that the previously delineated ELM2 domain is not sufficient for activity but requires an additional C-terminal sequence. We show that this additional sequence is highly conserved among ELM2-containing proteins, indicating that the functional ELM2 domain extends further downstream than previously described and in hMI-ER1 includes aa 179 to 283.
DISCUSSION
In this report, we investigated the transcriptional regulatory function of the hMI-ER1α and β isoforms. We have shown that both can function equally well as transcriptional repressors through recruitment of HDAC1 and that the evolutionarily conserved ELM2 domain, which is common to both, is required for these activities. Thus, the two alternate C-terminal domains appear to serve distinct functions.
It is interesting that the α isoform does not contain a functional NLS and that its subcellular localization in NIH 3T3 cells is predominately cytoplasmic (24). We postulated that controlling the subcellular localization of hMI-ER1 by alternate splicing provides a mechanism for regulation of its nuclear activities. It is also possible that the α isoform may be transported to the nucleus through regulated interactions with another nuclear protein(s), such as HDAC1. Cotransport to the nucleus through such a piggyback mechanism has been reported for a number of proteins, including the retinoblastoma gene product (Rb) and Hsp90 (18, 41). Interestingly, the C terminus of the α isoform possesses a class III LXXLL motif (NR box), a nuclear hormone receptor interaction domain found in a number of transcriptional coactivators (6, 15; reviewed in reference 2) that may be important in regulating the differential subcellular localizations of hMI-ER1 in steroid hormone responsive tissues.
We found that hMI-ER1 functions in transcriptional repression in several cell lines; however, we cannot exclude the possibility that in other cell lines or under other conditions, it could function in transcriptional activation. Indeed, the N-terminal region of Xenopus MI-ER1 was shown to function as a powerful transcriptional activator (22). Furthermore, the ability to both repress and activate transcription has been reported for a number of other transcription factors, including the nuclear hormone receptors (reviewed in references 2 and 35), MTA2 (20), and the HOX-PBX complex (27). In most cases, differential recruitment of coactivator or corepressor complexes determines the net effect on transcription. We are currently investigating whether hMI-ER1 can also function as a transcriptional activator and whether the α isoform is in fact involved in nuclear receptor transactivation.
Although hMI-ER1 possesses a SANT domain, which is structurally related to the DBD of the Myb protein, we have not been able to show that hMI-ER1 possesses specific DNA binding ability (unpublished data). Indeed, proteins like hMI-ER1 that contain a single SANT domain do not normally possess DNA binding activity (1). Thus, hMI-ER1 most likely functions as a corepressor in a manner similar to other SANT-containing corepressors, such as N-CoR, SMRT, and CoREST (14, 40).
Eleven human HDACs have been identified (7, 10, 11, 13, 17, 19, 34, 42) and have been divided into three classes based on their homology to Saccharomyces cerevisiae proteins (5, 8, 9, 12, 28, 31, 36), and it is believed that the different HDACs may have unique temporal and spatial expression patterns that contribute to tissue-specific regulation of transcription- and chromatin-regulatory complexes. We have shown that both hMI-ER1α and β interact with HDAC1 and that this is a native interaction, not just an artifact of overexpression by transfection. We could not, however, detect any interaction with HDAC3, suggesting some specificity in the interaction of hMI-ER1 with HDAC family members. Whether other HDAC family members can interact specifically with hMI-ER1 to regulate transcription in distinct cell types is currently being investigated.
We mapped the domain responsible for recruiting HDAC1 activity and mediating transcriptional repression to aa 179 to 283, a region common to all hMI-ER1 isoforms and containing the ELM2 domain. Our mutational analysis revealed that an intact ELM2 domain is required for these activities but that additional sequences, not previously included in the ELM2 domain by sequence comparisons (29) or Pfam alignments, are also critical for these activities. A more detailed analysis of the sequences carboxy terminal to the ELM2 domain in hMI-ER1 and other ELM2 domain-containing proteins revealed conservation of additional sequences, including the consensus sequence ALXXLX5DX3ALXXL. Secondary-structure analysis of this region with the SOPMA and COILS algorithms revealed that this region has a propensity to form α-helical structures and that the region from aa 263 to 290 has a high probability of forming a coiled coil. These data suggest that the structural integrity of this region is critical for recruitment of HDAC1 activity, with the region being involved either directly by associating with HDAC1 or indirectly by recruitment of an HDAC1-containing complex.
Our data suggest that other ELM2-containing proteins, including CoREST, MTA1, and MTA2, may interact with HDACs in a manner similar to that of hMI-ER1. While all of these are known components of characterized transcription- and chromatin-regulatory complexes containing one or more HDACs, the interaction domain(s) for most has not been determined. You et al. (40) concluded that the SANT domain of the corepressor CoREST was required for association with HDAC1; however, we note that the construct used in this study also contained the ELM2 domain. Our constructs were designed to separate the two domains and clearly showed that the SANT domain of hMI-ER1 does not interact with HDAC1. We do, however, acknowledge that different molecules may utilize these domains differently and/or may cooperate in binding HDAC-containing complexes. Indeed, the SANT domain of the related corepressor SMRT, which does not contain an ELM2 domain, has been implicated in HDAC3 binding (14).
It is interesting that virtually all proteins containing an ELM2 domain also contain a SANT domain. Recently, the SANT domain of ADA2, a component of the SAGA complex in S. cerevisiae, has been shown to recruit HAT activity (30). We are currently investigating whether the SANT domain of hMI-ER1 can recruit HAT activity; positive results would support a model in which the ELM2 and SANT domains function in opposing directions to provide precise regulation of transcriptional activity.
Acknowledgments
This work was supported by a grant from the Canadian Institutes for Health Research to L.L.G. and G.D.P. The hybridoma producing the monoclonal antibody 9E10 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa Department of Biological Sciences, Iowa City, Ia.
We are grateful to A. Pater for helpful discussion. We thank Paula Ryan, Corinne Mercer, and Yuan Lew for excellent technical assistance.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1270-1304. [DOI] [PubMed] [Google Scholar]
- 3.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 4.Brubaker, K., S. M. Cowley, K. Huang, L. Loo, G. S. Yochum, D. E. Ayer, R. N. Eisenman, and I. Radhakrishnan. 2000. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell 103:655-665. [DOI] [PubMed] [Google Scholar]
- 5.Buggy, J. J., M. L. Sideris, P. Mak, D. D. Lorimer, B. McIntosh, and J. M. Clark. 2000. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 350:199-205. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C., J. D. Norris, H. Gron, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol. Cell. Biol. 19:8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer, D. D., R. Cai, U. Bhatia, F. A. Asselbergs, C. Song, R. Terry, N. Trogani, R. Widmer, P. Atadja, and D. Cohen. 2002. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 277:6656-6666. [DOI] [PubMed] [Google Scholar]
- 8.Fischle, W., S. Emiliani, M. J. Hendzel, T. Nagase, N. Nomura, W. Voelter, and E. Verdin. 1999. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274:11713-11720. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa, Y., T. Kawakami, K. Sudo, J. Inazawa, A. Matsumine, T. Akiyama, and Y. Nakamura. 1996. Isolation and mapping of a human gene (RPD3L1) that is homologous to RPD3, a transcription factor in Saccharomyces cerevisiae. Cytogenet. Cell Genet. 73:130-133. [DOI] [PubMed] [Google Scholar]
- 10.Gao, L., M. A. Cueto, F. Asselbergs, and P. Atadja. 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 277:25748-25755. [DOI] [PubMed] [Google Scholar]
- 11.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 12.Grozinger, C. M., C. A. Hassig, and S. L. Schreiber. 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guardiola, A. R., and T. P. Yao. 2002. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 277:3350-3356. [DOI] [PubMed] [Google Scholar]
- 14.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh, J. J.-D., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, E., Z. Chen, T. Fredrickson, Y. Zhu, R. Kirkpatrick, G. F. Zhang, K. Johanson, C. M. Sung, R. Liu, and J. Winkler. 2000. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 275:15254-15264. [DOI] [PubMed] [Google Scholar]
- 18.Kang, K. I., J. Devin, F. Cadepond, N. Jibard, A. Guiochon-Mantel, E.-E. Baulieu, and M.-G. Catelli. 1994. In vivo functional protein-protein interaction: nuclear targeted hsp90 shifts cytoplasmic steroid receptor mutants into the nucleus. Proc. Natl. Acad. Sci. USA 91:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, H. Y., C. H. Lee, A. Komarov, C. C. Han, and R. M. Evans. 2002. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 277:187-193. [DOI] [PubMed] [Google Scholar]
- 20.Matsusue, K., S. Takiguchi, Y. Toh, and A. Kono. 2001. Characterization of mouse metastasis-associated gene 2: genomic structure, nuclear localization signal, and alternative potentials as transcriptional activator and repressor. DNA Cell. Biol. 20:603-611. [DOI] [PubMed] [Google Scholar]
- 21.Ordentlich, P., M. Downes, W. Xie, A. Genin, N. B. Spinner, and R. M. Evans. 1999. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl. Acad. Sci. USA 96:2639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paterno, G. D., Y. Li, H. A. Luchman, P. J. Ryan, and L. L. Gillespie. 1997. cDNA cloning of a novel, developmentally regulated immediate early gene activated by fibroblast growth factor and encoding a nuclear protein. J. Biol. Chem. 272:25591-25595. [DOI] [PubMed] [Google Scholar]
- 23.Paterno, G. D., F. C. Mercer, J. J. Chayter, X. Yang, J. D. Robb, and L. L. Gillespie. 1998. Molecular cloning of human er1 cDNA and its differential expression in breast tumours and tumour-derived cell lines. Gene 222:77-82. [DOI] [PubMed] [Google Scholar]
- 24.Paterno, G. D., Z. Ding, Y.-Y. Lew, G. N. Nash, F. C. Mercer, and L. L. Gillespie. 2002. Genomic organization of the human mi-er1 gene and characterization of alternatively spliced isoforms: regulated use of a facultative intron determines subcellular localization. Gene 295:79-88. [DOI] [PubMed] [Google Scholar]
- 25.Post, J. N., L. L. Gillespie, and G. D. Paterno. 2001. Nuclear localization signals in the Xenopus FGF embryonic early response 1 protein. FEBS Lett. 502:41-45. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, P. J., and L. L. Gillespie. 1994. Phosphorylation of phospholipase C gamma 1 and its association with the FGF receptor is developmentally regulated and occurs during mesoderm induction in Xenopus laevis. Dev. Biol. 166:101-111. [DOI] [PubMed] [Google Scholar]
- 27.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shore, D. 2000. The Sir2 protein family: a novel deacetylase for gene silencing and more. Proc. Natl. Acad. Sci. USA 97:14030-14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solari, F., A. Bateman, and J. Ahringer. 1999. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development 126:2483-2494. [DOI] [PubMed] [Google Scholar]
- 30.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 31.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 32.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toh, Y., S. Kuninaka, K. Endo, T. Oshiro, Y. Ikeda, H. Nakashima, H. Baba, S. Kohnoe, T. Okamura, G. L. Nicolson, and K. Sugimachi. 2000. Molecular analysis of a candidate metastasis-associated gene, MTA1: possible interaction with histone deacetylase 1. J. Exp. Clin. Cancer Res. 19:105-111. [PubMed] [Google Scholar]
- 34.Tong, J. J., J. Liu, N. R. Bertos, and X. J. Yang. 2002. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 30:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torchia, J., C. Glass, and M. G. Rosenfeld. 1998. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 10:373-383. [DOI] [PubMed] [Google Scholar]
- 36.Verdel, A., and S. Khochbin. 1999. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem. 274:2440-2445. [DOI] [PubMed] [Google Scholar]
- 37.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 38.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 40.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zacksenhaus, E., Z. Jiang, Y. J. Hei, R. A. Phillips, and B. L. Gallie. 1999. Nuclear localization conferred by the pocket domain of the retinoblastoma gene product. Biochim. Biophys. Acta 1451:288-296. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, X., P. A. Marks, R. A. Rifkin, and V. M. Richon. 2001. Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. USA 98:10572-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]