Abstract
Meltrin α (ADAM12) is a metalloprotease-disintegrin whose specific expression patterns during development suggest that it is involved in myogenesis and the development of other organs. To determine the roles Meltrin α plays in vivo, we generated Meltrin α-deficient mice by gene targeting. Although the number of homozygous embryos are close to the expected Mendelian ratio at embryonic days 17 to 18, ca. 30% of the null pups born die before weaning, mostly within 1 week of birth. The viable homozygous mutants appear normal and are fertile. Most of the muscles in the homozygous mutants appear normal, and regeneration in experimentally damaged skeletal muscle is unimpeded. In some Meltrin α-deficient pups, the interscapular brown adipose tissue is reduced, although the penetrance of this phenotype is low. Impaired formation of the neck and interscapular muscles is also seen in some homozygotes. These observations suggest Meltrin α may be involved in regulating adipogenesis and myogenesis through a linked developmental pathway. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) is a candidate substrate of Meltrin α, and we found that TPA (12-O-tetradecanoylphorbol-13-acetate)-induced ectodomain shedding of HB-EGF is markedly reduced in embryonic fibroblasts prepared from Meltrin α-deficient mice. We also report here the chromosomal locations of Meltrin α in the mouse and rat.
Meltrin α is a metalloprotease-disintegrin that belongs to the ADAM (for “a disintegrin and metalloprotease”) family. To date, more than 30 ADAMs in worms, flies, rodents, primates, and humans have been identified (3, 44). ADAMs are thought to be involved in various biological functions, including fertilization, myogenesis, neurogenesis, and the development of various epithelial tissues (3, 30, 35). For example, Fertilin α and Fertilin β, which were the first identified mammalian ADAMs, play important roles in fertilization (4, 8). Another example is Kuzbanian (ADAM10), which is known to be involved in neurogenesis by regulating Notch signaling (18, 28, 31, 40). Some ADAMs are catalytically active metalloproteases and participate in the proteolytic processing of the extracellular domains of membrane-anchored proteins (44). For example, TACE (ADAM17) was initially identified as the protease responsible for the processing of tumor necrosis factor alpha (2, 23). Studies on TACE-null mice then revealed that TACE is involved in the processing of the extracellular domains of several membrane-anchored proteins, including the tumor necrosis factor p75 receptor, the adhesion molecule L-selectin, the amyloid precursor protein, and transforming growth factor α (7). Another example is Meltrin β (ADAM19), which has been shown to be involved in the in vitro processing of Neuregulin β, another membrane-anchored growth factor (37). Other studies suggest that ADAMs are also involved in cell-cell or cell-extracellular matrix interactions through their interaction with integrins (6, 10, 27, 48) or proteoglycans (15).
We previously showed that Meltrin α promotes myotube formation in vitro (45). Furthermore, Meltrin α is specifically expressed in the muscles during the neonatal stages and in the bones of both neonates and adults. In addition, during embryogenesis, Meltrin α mRNA was found in the mesenchymes of the lungs and the intestines and in the placenta (17), which suggests that Meltrin α is involved in organogenesis. Based on its amino acid sequence, the metalloprotease domain of Meltrin α is presumed to be catalytically active. Supporting this notion are the reports that it has proteolytic activity in vitro. For example, human Meltrin α interacts with insulin-like growth factor-binding protein 3 (IGFBP-3) (36) and cleaves it in vitro (19). Furthermore, Meltrin α has also recently been implicated to act as a sheddase with heparin-binding epidermal growth factor-like growth factor (HB-EGF) (1). To further determine the function of Meltrin α in mouse development, we generated and analyzed Meltrin α gene-targeted mice.
Materials and Methods
Materials.
Chemicals were purchased from Sigma (St. Louis, Mo.) and nacalai tesque (Kyoto, Japan). Restriction enzymes and reagents for molecular biology were purchased from TaKaRa (Kyoto, Japan) and Toyobo (Osaka, Japan) unless otherwise indicated.
Construction of Meltrin α gene-targeting vector.
The Meltrin α gene isolated from a 129/SvJ genomic library (Stratagene) was used to construct the targeting vector. The PGKneobpA cassette from pPGKneobpA (39) was inserted between a 5′ homologous region (XhoI-BamHI [9 kb]) and a 3′ homologous region (SacII-SacI [1.5 kb]). The DT-A cassette (46) was ligated at the 3′ end of the targeting vector for negative selection. In this construct, the exon containing the initiation codon was deleted.
Generation of Meltrin α−/− mice.
The linearized targeting vector (20 μg) was electroporated (250 V, 500 μF) into 107 R1 embryonic stem (ES) cells (26) and selected with 180 μg (active form) of G418 (Gibco-BRL)/ml for 7 to 10 days. Homologous recombinants were selected by PCR and confirmed by Southern blot analysis. The forward primer in the PGKneobpA cassette was 5′-TGGATGTGGAATGTGTGCGAGG-3′, and the reverse primer outside the targeting vector was 5′-TTCCATTGCTCAGCGGTGCTGTC-3′. PCR was performed with LA Taq DNA polymerase (TaKaRa, Kyoto, Japan) for 30 cycles at 98°C for 20 s and at 68°C for 10 min in a volume of 50 μl. Two ES clones that yielded hybridization bands of the correct size gave rise to germ line chimeras by the aggregation method (26). The resulting chimeras were backcrossed to C57BL/6J, and N8 mice were used in the following experiments. The genotypes were determined by either PCR (for embryos) or Southern blot analysis. Genotyping PCR was performed as follows: the primer set in the PGKneobpA cassette (5′-TGGAGAGGCTATTCGGCTATGACTGGG-3′ and 5′-ATGCAGCCGCCGCATTGCAT-3′) was used to detect targeted alleles. PCR was performed with AmpliTaq Gold DNA polymerase (PE Applied Biosystems) for 40 cycles at 96°C for 15 s and at 70°C for 30 s in a volume of 15 μl. The primer set in the exon containing the initiation codon (5′-GCGCTCTGCCATTGTCGCCG-3′ and 5′-GGCAGACTCAGGGCAGTAGGACTTCCC-3′) was used to detect wild-type alleles. PCR conditions used were the same as those for the targeted allele detection PCR, except that the annealing and extension temperatures used were both 64°C. Southern blot and reverse transcription-PCR (RT-PCR) analyses were performed as follows: genomic DNA from ES cells or mouse tail was digested with restriction enzymes, electrophoresed through a 0.8% agarose gel and transferred to Hybond-XL membrane (Amersham Pharmacia Biotech). Hybridization was performed according to standard methods (33) with a 32P-labeled DNA probe made by using a Megaprime DNA labeling kit (Amersham Pharmacia Biotech). A 1.2-kb SacI-HindIII fragment immediately downstream of the 3′ end of the 3′ homology region was used as the probe. RT-PCR was performed as follows. mRNA was extracted from embryos by using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech). cDNA was synthesized from the mRNA by using the SuperScript first-strand synthesis system (Invitrogen). PCR was carried out with AmpliTaq Gold DNA polymerase. The primers used to detect mRNA of Meltrin α were 5′-GATGACCAAGTACGTAGAGCTGG-3′ and 5′-TCATGGAGCCTGGTGAATGGG-3′ (for the metalloprotease domain), 5′-GAGTGTGACTGCGGAGAACCGGAGGAA-3′ and 5′-ATTTTCCCACACTTGGCATCTCTCA-3′ (for the disintegrin domain), 5′-GTCAAGGTGGTGCAAGCCGA-3′ and 5′-TGATGGGACCACTGTCTGTGC-3′ (for the cysteine-rich domain), and 5′-GACGTTGATGCGGCTGCTGTTC-3′ and 5′-GCGTCGAGGGGCCTGCTGATG-3′ (for the cytoplasmic domain). The glyceradehyde-3-phosphate dehydrogenase 0.45-kb control amplimer set (Clontech) was used as a positive control. Mice were maintained under specific-pathogen-free conditions in environmentally controlled clean rooms at the Laboratory Animal Research Center, Institute of Medical Science, University of Tokyo, the Animal Facility, Tokyo Institute of Medical Science, and at the Institute for Frontier Medical Sciences, Kyoto University. The experiments were conducted according to institutional ethical guidelines for animal experimentation and safety guidelines for gene manipulation experiments.
Histology.
Embryos were fixed in 4% paraformaldehyde-phosphate-buffered saline (PBS). Fixed samples were dehydrated by sequentially increased ethanol concentrations, cleared in xylene, and then embedded in paraffin. The embedded samples were sectioned into 4.5-μm-thick slices and stained with hematoxylin and eosin (HE).
In situ hybridization.
In situ hybridization was performed with digoxigenin-labeled antisense and sense riboprobes prepared by in vitro transcription according to the manufacturer's protocol (Roche Molecular Biochemicals). The antisense probe for Meltrin α consisted of two 1.3-kb fragments that spanned nucleotides 93 to 1417 and nucleotides 1997 to 3338.
Mouse embryonic fibroblasts.
Embryonic day 13.5 (E13.5) embryos from Meltrin α knockout and wild-type mice were used to generate mouse embryonic fibroblasts (14). Briefly, the head, limbs, and viscera were removed from the embryos, and the carcasses were minced and then trypsinized in 0.05% trypsin 0.02% EDTA in PBS for 10 min at 37°C. Cells were collected and grown in 10% fetal calf serum in Dulbecco’s modified Eagle medium (DMEM).
HB-EGF ectodomain shedding assay.
Embryonic fibroblast cells prepared from wild-type and knockout mice were seeded in 6-cm dishes at a density of 2 × 105 cells/dish and cultured for 48 h with DMEM-10% fetal calf serum. Cells were incubated for 1 h at 37°C with 100 nM TPA (12-O-tetradecanoylphorbol-13-acetate). Cells were washed three times with ice-cold Hanks buffer and biotinylated with 0.1 mg of sulfo-NHS-biotin/ml in 50 mM HEPES (pH 7.5)-0.15 M NaCl for 10 min on ice. Excess reagent was quenched and removed by washing with ice-cold DMEM-fetal calf serum. Cells were lysed with a buffer containing 1% Triton X-100, 1 mM EDTA, 1 mM (p-amidinophenyl)methanesulfonyl fluoride HCl, 1 μg of aprotinin/ml, and 0.4 M NaCl in 20 mM HEPES (pH 7.2). After centrifugation of the lysates at 15,000 rpm for 10 min, supernatants were collected and incubated with 2 μg of HB-EGF antibody H1 for 2 h at 4°C, followed by incubation with 10 μl of protein G-Sepharose (50% suspension) for 2 h at 4°C. After centrifugation of the mixes, the pellets were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting as described previously (13).
FISH.
The chromosomal assignment of the murine and rat Meltrin α genes was made by direct R-banding fluorescence in situ hybridization (FISH) with a mouse cDNA fragment as a probe (nucleotides 93 to 1417). Chromosome preparation and FISH detection were performed as described previously (21, 22).
RESULTS
Generation of Meltrin α-deficient mice.
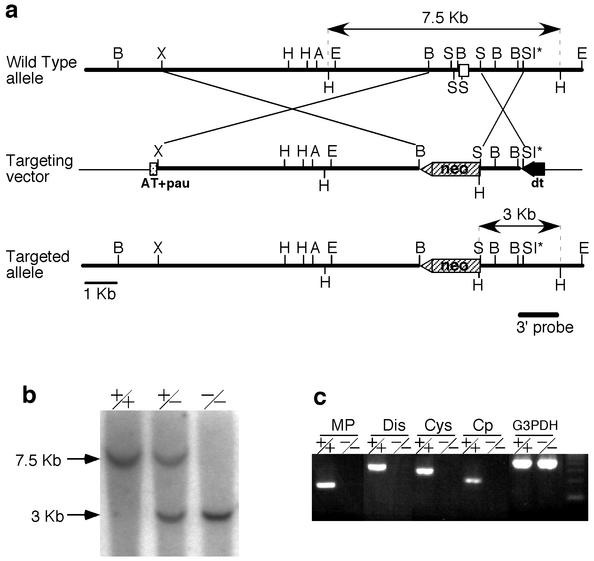
Meltrin α-deficient mice were generated by homologous recombination. The exon containing the initiation codon was replaced with the neomycin-resistant cassette (Fig. 1a). Homologous recombination was confirmed by Southern blot analysis (Fig. 1b). The hetero- and homozygotes had a normal appearance, and the males and females were both fertile. To confirm that homozygotes do not express Meltrin α, RT-PCR was performed. PCR primer sets for each of the metalloprotease, disintegrin, cysteine-rich, and cytoplasmic domains of Meltrin α did not detect Meltrin α mRNA in the homozygous mutant embryos (E14.5) (Fig. 1c). Despite the normal appearance of the null homozygotes, the number of these homozygotes at weaning was lower than the expected Mendelian ratio in the heterozygous crosses, although the ratio of wild types, heterozygotes, and homozygotes was Mendelian prior to birth (E16.5 to E18.5) (Table 1). That is, during the perinatal period, especially P1-2, the number of homozygous mutants declined, with ca. 30% of the homozygotes dying before weaning. The cause of death is still unclear. Similar results were observed for both of our lines of Meltrin α-deficient mice, which were obtained from independent ES clones.
FIG. 1.
Generation of Meltrin α-deficient mice. (a) Targeted disruption of the Meltrin α gene. Exon 1 containing the first methionine codon (open box) was replaced by a neocassette. Neo (hatched box) is the neomycin resistance gene, AT+pau (dotted box) indicates the AT-rich RNA polymerase II destabilizing signal and pausing signal, and DT (closed box) is the diphtheria toxin A fragment cassette. The 3′ probe represents the position of the external probe used for Southern blot analysis, and the expected HindIII fragments are indicated by arrows. Abbreviations: B, BamHI; X, XhoI; H, HindIII; A, Asp718; E, EcoRI; S, SacII; SI, SacI. The asterisk indicates that the SacI sites in the targeting vector are not unique. (b) Southern blot analysis of mouse tail genomic DNA. The expected DNA fragments for the targeted allele and the wild-type allele are 3 and 7 kb, respectively. +/+, wild-type; +/−, heterozygote; −/−, homozygote. (c) RT-PCR analysis of mRNA from embryonic fibroblasts isolated from E13.5 embryos. MP, primer detecting the metalloprotease domain; Dis, primer detecting the disintegrin domain; Cys, primer detecting the cysteine-rich domain; Cp, primer detecting the cytoplasmic domain. +/−, heterozygote; −/−, homozygote.
TABLE 1.
Genotypic analysis of mice from intercrossesa
| Mating (female vs male) | Age | No. (%) of mice with genotype:
|
||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| +/− vs +/− | 3 wk | 66 (30.8) | 101 (47.2) | 47 (22.0) |
| P1-P2 | 51 (29.0) | 92 (52.3) | 33 (18.8) | |
| E16.5-E18.5 | 25 (23.6) | 46 (43.4) | 35 (33.0) | |
| +/− vs −/− | 3 wk | 80 (58.4) | 57 (41.6)b | |
+/+, wild type; +/−, heterozygote; −/−, homozygote.
P < 0.03.
Histological analysis of Meltrin α−/− embryos.
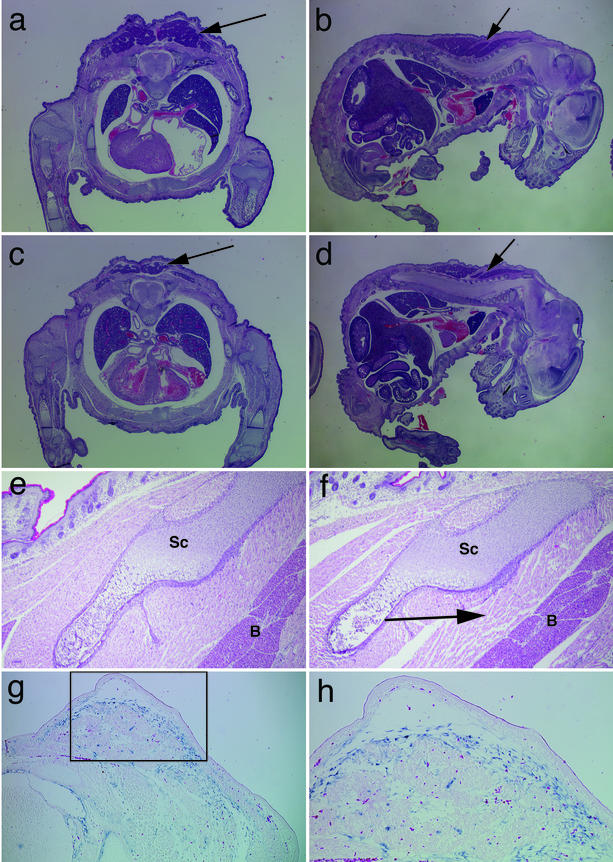
The Meltrin α-deficient mice appeared grossly normal at weaning. However, detailed anatomical analysis of neonatal mice revealed that the interscapular brown adipose tissue (BAT) was reduced in ca. 30% of the Meltrin α-deficient mice (Fig. 2). Although the number and morphology of the adipose lobes were similar to those of wild-type mice, the lobes were significantly smaller. Some of the homozygotes exhibited looser condensation of adipocytes than the wild-type or heterozygous littermates at ca. E16.5, probably due to the lower cell number (data not shown). In addition, some of the perinatal and newborn homozygotes (E17.5-P1) showed impaired formation of the neck and interscapular muscles (Fig. 2f). Although these muscles occupy areas between the bone and skin that are similar in size to that in the wild type or heterozygotes, the muscle fibers close to the adipose tissues were found not to be as tightly packed in some homozygotes as in the non-null littermates. Although the number of homozygous neonates with impaired BAT or interscapular muscle formation was low (ca. 30%), such defects were never observed in wild-type neonates. However, at 8 weeks of age, all homozygous mice had normal-sized BATs, although some of the interscapular muscles remained slightly reduced in size (data not shown). Since it was not possible to monitor individual mice throughout development, it is not clear whether the size of the BAT recovers after birth or whether mutants with small BATs die.
FIG.2.
Histological analysis of Meltrin α−/− embryos. Heterozygous (a, b, and e) and homozygous (c, d, and f) neonates (P1) were fixed, embedded in paraffin, sectioned, and stained with HE. The frontal section (a and c) and saggital section (b and d) are shown. BAT is indicatedby arrows in panels a to d. The arrow in panel f shows impaired muscle development. Sc, scapular bone; B, BAT. (g and h) The expression of Meltrin α mRNA in heterozygous embryos was detected by in situ hybridization. At E14.5, Meltrin α is expressed in bones, muscles, and the peripheral region of condensing interscapular mesenchymal cells. (Panel h shows an enlarged region of panel g [boxed].)
Muscle regeneration in Meltrin-α-deficient mice.
Meltrin-α is activated both in vitro and in vivo during the differentiation of muscle satellite cells (5). Thus, we evaluated the effect Meltrin-α deficiency has on muscle regeneration. When the anterior tibial muscles of adult Meltrin α-deficient mice were experimentally damaged by cardiotoxin treatment, muscle regeneration comparable to that of wild-type mice was observed (data not shown). When we crossed Meltrin α-deficient mice with mdx mice, which are muscular dystrophy mutants that show enhanced muscle degeneration and muscle regeneration (9, 41), no enhancement of muscle degeneration or decreased regeneration was observed in the Meltrin-α mdx double mutants (data not shown).
Expression of Meltrin α in the BAT-forming region during embryogenesis.
In situ hybridization was performed to analyze the expression pattern of Meltrin α during BAT development in the heterozygotes. At E14.5, Meltrin α transcripts were detected in mesenchymal cells in the BAT-forming region beneath the skin (Fig. 2g and h). These Meltrin-α-expressing mesenchymal cells extended to the region of the developing neck muscles adjacent to the BAT tissues, suggesting that these Meltrin α-positive mesenchymal cells are involved in the embryonic development of BAT and adjacent muscle tissues.
HB-EGF ectodomain shedding.
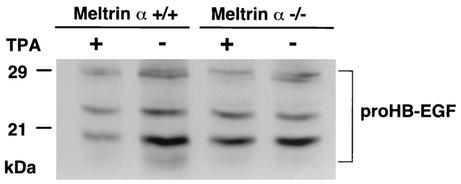
Recent studies with metalloprotease inhibitors have implicated metalloproteases in the ectodomain shedding of HB-EGF (1). Therefore, we examined whether Meltrin α can act as a sheddase of HB-EGF in mouse embryonic fibroblasts prepared from E13.5 Meltrin α-deficient or wild-type embryos. Expression of Meltrin α in embryonic fibroblasts was confirmed by RT-PCR (Fig. 1c). Endogenous HB-EGF was detected by cell surface biotinylation, immunoprecipitation, and Western blotting. HB-EGF shedding was evaluated in untreated cells and in TPA-treated cells (Fig. 3). In wild-type embryonic fibroblasts, TPA induced the processing of proHB-EGF (the membrane-anchored form), resulting in the loss of the cell surface proHB-EGF. In contrast, TPA did not induce processing of proHB-EGF in Meltrin α-deficient embryonic fibroblasts. This suggests that Meltrin α contributes to the ectodomain shedding of HB-EGF.
FIG. 3.
HB-EGF ectodomain shedding assay. Detection of surface proHB-EGF in mouse embryonic fibroblasts treated with or without TPA. The multiple bands of proHB-EGF are derived from various N-terminal truncations and heterogeneous glycosylations.
Chromosomal mapping of the murine and rat meltrin α genes.
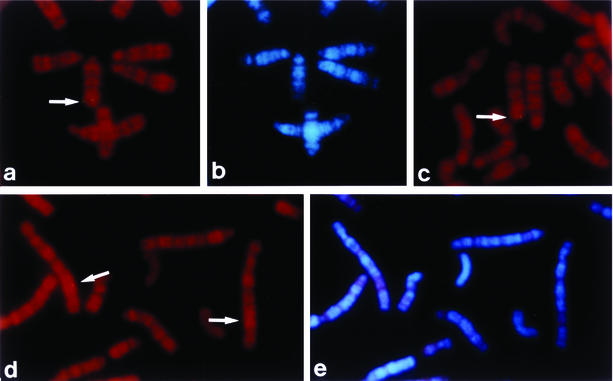
The chromosomal locations of the murine and rat meltrin α genes were determined by direct R-banding FISH with a murine cDNA fragment as a probe (Fig. 4). The Meltrin α genes were localized to the F3 distal −F4 band of mouse chromosome 7 and the q43 proximal band of rat chromosome 1. Conserved linkage homology between these species has been identified in these areas (20, 21, 34, 38, 47). To date, suggestive mutations have not been mapped to Meltrin α. We attempted to linkage map the Meltrin α gene by interspecific backcross analysis with progeny derived from the mating of (C57BL6 3 Mus spretus)F1 × M. spretus mice. However, this attempt failed because of the presence of an additional Meltrin α-like gene in M. spretus chromosome 2.
FIG. 4.
Chromosomal localization of the Meltrin α gene. Chromosomal localization of the Meltrin α gene on R-banded murine (a to c) and rat (d and e) chromosomes. The chromosomal locations of the murine and rat Meltrin α genes were determined by using a murine cDNA fragment as a biotinylated probe. The hybridization signals are indicated by arrows. The signals are localized to murine chromosome 7F3 distal −F4 and to the rat chromosome 1q43 (proximal). The metaphase spreads were photographed with Nikon B-2A (a, c, and d) and UV-2A (b and e) filters. R-band and G-band patterns are demonstrated in panels a, c, and d and in panels b and e, respectively.
DISCUSSION
Meltrin α (ADAM12) was originally cloned from myogenic cells. Its expression pattern suggests that it is involved in various organogenic processes, particularly myogenesis (12, 17, 45). In the present study, we examined the function of Meltrin α during mouse development by generating mice lacking the Meltrin α gene. Unexpectedly, no major morphological abnormalities were observed in Meltrin α−/− mice, and the homozygous mutants were viable and fertile. However, ca. 30% of homozygotes did die within 1 week of birth. The cause of death is unknown. In addition, some homozygous mutants showed impaired formation of interscapular BAT (E17.5-P1).
Mammals have two types of adipose tissue, namely, white adipose tissue (WAT) and BAT (32). Although WAT stores excess energy as triglycerides and releases free fatty acids in response to energy requirements, BAT dissipates energy in the form of heat through the uncoupling of oxidative phosphorylation. It functions as a thermogenerator to maintain the body temperature and protects against obesity. Thus, the impaired formation of BAT can cause hypothermia in neonates. Parents sometimes do not nurse neonates with low body temperatures, which might explain the semilethality of the homozygous mutant neonates. Nevertheless, all surviving adult homozygous mutants had apparently normal BAT and WAT (perigonadal).
It is noteworthy that some Meltrin α-deficient mice also show hypotrophy of muscles adjacent to BAT and that others show a reduction of both tissues. In vitro studies have revealed that both muscle and fat cells can be induced from pluripotent stem cell lines or marrow-derived stromal cells (29, 42). Although the identity and mechanisms of differentiation of such presumptive mesenchymal precursor cells remains unknown, the embryonic expression of Meltrin α and the effect of its absence on BAT and adjacent muscle formation shown in the present study support the idea that the development of these tissues are intimately related. Furthermore, recent studies demonstrated that transgenic mice overexpressing Meltrin α have an upregulated formation of adipose tissues (16). Together, these results suggest that Meltrin-α plays regulatory roles in the formation of muscle and adipose tissues.
Recently, experiments with metalloprotease inhibitors have also implicated Meltrin α in cardiac hypertrophy and in the ectodomain shedding of HB-EGF (1). Although the hearts of the Meltrin α−/− mice appear normal, it will be of interest to test whether hypertrophy occurs under conditions of pressure overload. However, we did find that Meltrin α indeed contributes to TPA-stimulated HB-EGF ectodomain shedding in embryonic fibroblasts. The importance of HB-EGF in adipogenesis has not been examined, although its expression in interscapular mesenchymal cells has been observed (unpublished data). On the other hand, insulin-like growth factor I (IGF-I) (32) is one of the cytokines that plays an essential role in adipogenesis and stimulates myogenesis (11, 24, 25). Meltrin α interacts with IGFBP-3 and cleaves it in vitro (19, 36), suggesting that Meltrin α may also regulate the release of active IGFs in BAT and muscle cell lineages by cleaving IGFBPs.
It is not clear at this stage why the levels of penetrance of neonatal lethality, BAT malformation, and hypotrophic muscle formation are so low in the Meltrin α-null mutants. It is possible that other ADAMs may compensate for the loss of Meltrin α due to functional redundancy between Meltrin α and other ADAMs.
Acknowledgments
This work was supported in part by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology and by research grants from the Japanese Health Science Foundation, the National Center of Neurology and Psychiatry of the Ministry of Health and Welfare of Japan, and CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation.
We thank T. Obata for technical assistance, T. Fujimori for helpful comments, and R. T. Yu for critically reading the manuscript.
REFERENCES
- 1.Asakura, M., M. Kitakaze, S. Takashima, Y. Liao, F. Ishikura, T. Yoshinaka, H. Ohmoto, K. Node, K. Yoshino, H. Ishiguro, H. Asanuma, S. Sanada, Y. Matsumura, H. Takeda, S. Beppu, M. Tada, M. Hori, and S. Higashiyama. 2002. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat. Med. 8:35-40. [DOI] [PubMed] [Google Scholar]
- 2.Black, R. A., C. T. Rauch, C. J. Kozlosky, J. J. Peschon, J. L. Slack, M. F. Wolfson, B. J. Castner, K. L. Stocking, P. Reddy, S. Srinivasan, N. Nelson, N. Boiani, K. A. Schooley, M. Gerhart, R. Davis, J. N. Fitzner, R. S. Johnson, R. J. Paxton, C. J. March, and D. P. Cerretti. 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 385:729-733. [DOI] [PubMed] [Google Scholar]
- 3.Black, R. A., and J. M. White. 1998. ADAMs: focus on the protease domain. Curr. Opin. Cell Biol. 10:654-659. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, C. P., T. G. Wolfsberg, C. W. Turck, D. G. Myles, P. Primakoff, and J. M. White. 1992. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature 356:248-252. [DOI] [PubMed] [Google Scholar]
- 5.Borneman, A., R. Kuschel, and A. Fujisawa-Sehara. 2000. Analysis for transcript expression of meltrin alpha in normal, regenerating, and denervated rat muscle. J. Muscle Res. Cell Motil. 21:475-480. [DOI] [PubMed] [Google Scholar]
- 6.Bridges, L. C., P. H. Tani, K. R. Hanson, C. M. Roberts, M. B. Judkins, and R. D. Bowditch. 2002. The lymphocyte metalloprotease MDC-L (ADAM 28) is a ligand for the integrin α4β1. J. Biol. Chem. 277:3784-3792. [DOI] [PubMed] [Google Scholar]
- 7.Buxbaum, J. D., K. N. Liu, Y. Luo, J. L. Slack, K. L. Stocking, J. J. Peschon, R. S. Johnson, B. J. Castner, D. P. Cerretti, and R. A. Black. 1998. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 273:27765-27767. [DOI] [PubMed] [Google Scholar]
- 8.Cho, C., D. O. Bunch, J. E. Faure, E. H. Goulding, E. M. Eddy, P. Primakoff, and D. G. Myles. 1998. Fertilization defects in sperm from mice lacking Fertilin β. Science 281:1857-1859. [DOI] [PubMed] [Google Scholar]
- 9.Dangain, J., and G. Vrbova. 1984. Muscle development in mdx mutant mice. Muscle Nerve 7:700-704. [DOI] [PubMed] [Google Scholar]
- 10.Eto, K., W. Puzon-McLaughlin, D. Sheppard, A. Sehara-Fujisawa, X. P. Zhang, and Y. Takada. 2000. RGD-independent binding of integrin alpha9beta1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J. Biol. Chem. 275:34922-34930. [DOI] [PubMed] [Google Scholar]
- 11.Florini, J. R., D. Z. Ewton, and S. A. Coolican. 1996. Growth hormone and the insulin-like growth factor system in myogenesis. Endocrinol. Rev. 17:481-517. [DOI] [PubMed] [Google Scholar]
- 12.Gilpin, B. J., F. Loechel, M. G. Mattei, E. Engvall, R. Albrechtsen, and U. M. Wewer. 1998. A novel, secreted form of human ADAM 12 (Meltrin α) provokes myogenesis in vivo. J. Biol. Chem. 273:157-166. [DOI] [PubMed] [Google Scholar]
- 13.Goishi, K., S. Higashiyama, M. Klagsbrun, M. Ishikawa, E. Mekada, and N. Taniguchi. 1995. Phorbol ester induces the rapid processing of cell surface heparin-binding EGF-like growth factor: conversion from juxtacrine to paracrine growth factor activity. Mol. Biol. Cell 6:967-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Iba, K., R. Albrechtsen, B. Gilpin, C. Frohlich, F. Loechel, A. Zolkiewska, K. Ishiguro, T. Kojima, W. Liu, J. K. Langford, R. D. Sanderson, C. Brakebusch, R. Fassler, and U. M. Wewer. 2000. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J. Cell Biol. 149:1143-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi, N., X. Xu, R. tajima, P. Kronqvist, C. Sundberg, F. Loechel, R. Albrechtsen, and U. M. Wewer. 2002. ADAM 12 protease induces adipogenesis in transgenic mice. Am. J. Pathol. 160:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurisaki, T., A. Masuda, N. Osumi, Y. Nabeshima, and A. Fujisawa-Sehara. 1998. Spatially- and temporally-restricted expression of meltrin α (ADAM12) and β (ADAM19) in mouse embryo. Mech. Dev. 73:211-215. [DOI] [PubMed] [Google Scholar]
- 18.Lieber, T., S. Kidd, and M. W. Young. 2002. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16:209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loechel, F., J. W. Fox, G. Murphy, R. Albrechtsen, and U. M. Wewer. 2000. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem. Biophys. Res. Commun. 278:511-515. [DOI] [PubMed] [Google Scholar]
- 20.Lyon, M. F., Y. Cocking, and X. Gao. 1996. Mouse chromosome atlas. Mouse Genome 95:731-798. [Google Scholar]
- 21.Matsuda, Y., Y. N. Harada, S. Natsuume-Sakai, K. Lee, T. Shiomi, and V. M. Chapman. 1992. Location of the mouse complement factor H gene (cfh) by FISH analysis and replication R-banding. Cytogenet. Cell Genet. 61:282-285. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda, Y., T. Imai, T. Shiomi, T. Saito, M. Yamauchi, T. Fukao, Y. Akao, N. Seki, H. Ito, and T. A. Hori. 1996. Comparative genome mapping of the ataxia-telangiectasia region in mouse, rat, and Syrian hamster. Genomics 34:347-352. [DOI] [PubMed] [Google Scholar]
- 23.Moss, M. L., S. L. Jin, M. E. Milla, D. M. Bickett, W. Burkhart, H. L. Carter, W. J. Chen, W. C. Clay, J. R. Didsbury, D. Hassler, C. R. Hoffman, T. A. Kost, M. H. Lambert, M. A. Leesnitzer, P. McCauley, G. McGeehan, J. Mitchell, M. Moyer, G. Pahel, W. Rocque, L. K. Overton, F. Schoenen, T. Seaton, J. L. Su, J. D. Becherer, et al. 1997. Cloning of a disintegrin metalloproteinase that processes precursor tumour necrosis factor-alpha. Nature 385:733-736. [DOI] [PubMed] [Google Scholar]
- 24.Musaro, A., K. McCullagh, A. Paul, L. Houghton, G. Dobrowolny, M. Molinaro, E. R. Barton, H. L. Sweeney, and N. Rosenthal. 2001. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 27:195-200. [DOI] [PubMed] [Google Scholar]
- 25.Musaro, A., K. J. McCullagh, F. J. Naya, E. N. Olson, and N. Rosenthal. 1999. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400:581-585. [DOI] [PubMed] [Google Scholar]
- 26.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nath, D., P. M. Slocombe, A. Webster, P. E. Stephens, A. J. Docherty, and G. Murphy. 2000. Meltrin γ (ADAM-9) mediates cellular adhesion through α6β1 integrin, leading to a marked induction of fibroblast cell motility. J. Cell Sci. 113(Pt. 12):2319-2328. [DOI] [PubMed] [Google Scholar]
- 28.Pan, D., and G. M. Rubin. 1997. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90:271-280. [DOI] [PubMed] [Google Scholar]
- 29.Pittenger, M. F., A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig, and D. R. Marshak. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284:143-147. [DOI] [PubMed] [Google Scholar]
- 30.Primakoff, P., and D. G. Myles. 2000. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 16:83-87. [DOI] [PubMed] [Google Scholar]
- 31.Qi, H., M. D. Rand, X. Wu, N. Sestan, W. Wang, P. Rakic, T. Xu, and S. Artavanis-Tsakonas. 1999. Processing of the Notch ligand delta by the metalloprotease Kuzbanian. Science 283:91-94. [DOI] [PubMed] [Google Scholar]
- 32.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Satoh, H., M. C. Yoshida, and M. Sasaki. 1989. High resolution chromosome banding in the Norway rat, Rattus norvegicus. Cytogenet. Cell Genet. 50:151-154. [DOI] [PubMed] [Google Scholar]
- 35.Schlondorff, J., and C. P. Blobel. 1999. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 112(Pt. 21):3603-3617. [DOI] [PubMed] [Google Scholar]
- 36.Shi, Z., W. Xu, F. Loechel, U. M. Wewer, and L. J. Murphy. 2000. ADAM 12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J. Biol. Chem. 275:18574-18580. [DOI] [PubMed] [Google Scholar]
- 37.Shirakabe, K., S. Wakatsuki, T. Kurisaki, and A. Fujisawa-Sehara. 2001. Roles of Meltrin β/ADAM19 in the processing of neuregulin. J. Biol. Chem. 276:9352-9358. [DOI] [PubMed] [Google Scholar]
- 38.Somssich, I. E., and H. Hameister. 1996. Standard karyotype of early replicating bands (RBG-banding), p. 1450-1451. In M. F. Lyon, S. Rastan, and S. D. M. Brown (ed.), Genetic variants and strains of the laboratory mouse. Oxford University Press, Oxford, England.
- 39.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 40.Sotillos, S., F. Roch, and S. Campuzano. 1997. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124:4769-4779. [DOI] [PubMed] [Google Scholar]
- 41.Stedman, H. H., H. L. Sweeney, J. B. Shrager, H. C. Maguire, R. A. Panettieri, B. Petrof, M. Narusawa, J. M. Leferovich, J. T. Sladky, and A. M. Kelly. 1991. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature 352:536-539. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, S. M., and P. A. Jones. 1979. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17:771-779. [DOI] [PubMed] [Google Scholar]
- 43.Turner, A. J., and N. M. Hooper. 1999. Role for ADAM-family proteinases as membrane protein secretases. Biochem. Soc. Trans. 27:255-259. [DOI] [PubMed] [Google Scholar]
- 44.Wolfsberg, T. G., P. Primakoff, D. G. Myles, and J. M. White. 1995. ADAM, a novel family of membrane proteins containing a disintegrin and metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J. Cell Biol. 131:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagami-Hiromasa, T., T. Sato, T. Kurisaki, K. Kamijo, Y. Nabeshima, and A. Fujisawa-Sehara. 1995. A metalloprotease-disintegrin participating in myoblast fusion. Nature 377:652-656. [DOI] [PubMed] [Google Scholar]
- 46.Yagi, T., S. Nada, N. Watanabe, H. Tamemoto, N. Kohmura, Y. Ikawa, and S. Aizawa. 1993. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal. Biochem. 214:77-86. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, J., T. Kuramoto, and T. Serikawa. 1994. A rat genetic linkage map and comparative maps for mouse or human homologous rat genes. Mamm. Genome 5:63-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, M., R. Graham, G. Russell, and P. I. Croucher. 2001. MDC-9 (ADAM-9/Meltrin γ) functions as an adhesion molecule by binding the αvβ5 integrin. Biochem. Biophys. Res. Commun. 280:574-580. [DOI] [PubMed] [Google Scholar]