Abstract
The essential C17 subunit of yeast RNA polymerase (Pol) III interacts with Brf1, a component of TFIIIB, suggesting a role for C17 in the initiation step of transcription. The protein sequence of C17 (encoded by RPC17) is conserved from yeasts to humans. However, mammalian homologues of C17 (named CGRP-RCP) are known to be involved in a signal transduction pathway related to G protein-coupled receptors, not in transcription. In the present work, we first establish that human CGRP-RCP is the genuine orthologue of C17. CGRP-RCP was found to functionally replace C17 in Δrpc17 yeast cells; the purified mutant Pol III contained CGRP-RCP and had a decreased specific activity but initiated faithfully. Furthermore, CGRP-RCP was identified by mass spectrometry in a highly purified human Pol III preparation. These results suggest that CGRP-RCP has a dual function in mammals. Next, we demonstrate by genetic and biochemical approaches that C17 forms with C25 (encoded by RPC25) a heterodimer akin to Rpb4/Rpb7 in Pol II. C17 and C25 were found to interact genetically in suppression screens and physically in coimmunopurification and two-hybrid experiments. Sequence analysis and molecular modeling indicated that the C17/C25 heterodimer likely adopts a structure similar to that of the archaeal RpoE/RpoF counterpart of the Rpb4/Rpb7 complex. These RNA polymerase subunits appear to have evolved to meet the distinct requirements of the multiple forms of RNA polymerases.
Transcription of eukaryotic genes is mediated by three related forms of nuclear RNA polymerases (Pol). These enzymes are large multisubunit proteins that require a distinct set of general transcription factors to transcribe their cognate genes (58, 72). In the well-studied case of the yeast Saccharomyces cerevisiae, the three enzymes contain 14 (Pol I), 12 (Pol II), and 17 (Pol III) different subunits and share a set of 5 conserved and 5 common subunits (9, 58). All but one of these subunits are represented in the archaeal enzyme (15, 36). In Pol II, this 10-subunit core enzyme is competent for the transcription of nonspecific DNA templates (16) but not for promoter-dependent transcription (18). The atomic structure of the yeast core Pol II enzyme has been recently determined (13).
Pol III makes many small nontranslated RNAs such as tRNAs, 5S rRNA, U6 snRNA, the 7S RNA of the signal recognition particle, and the RNA component of RNase P. Yeast Pol III contains six subunits (C82, C53, C37, C34, C31, and C17) that appear to have no counterparts in Pol I, Pol II, or archaeal RNA polymerase. All of these Pol III-specific subunits are essential for cell viability, like all the other Pol III components (9). Genetic and biochemical studies have shed light on the function of some Pol III-specific subunits. The triad of subunits C82-C34-C31 in yeast and their human counterparts, human RPC62 (hRPC62)-hRPC39-hRPC32, form a subcomplex that is required for promoter-dependent transcription initiation (66, 68, 71). One of these polypeptides, yeast C34, has been mapped the furthest upstream on the promoter (5) and shown to have a dual role in Pol III recruitment and in open complex formation via its interaction with the Brf1 component of TFIIIB (7, 30, 70). Similarly, its human counterpart, hRPC39, was found to physically interact with both human TATA-binding protein and hBrf1, two subunits of human TFIIIB (68). This subcomplex of Pol III subunits is therefore thought to play a role in preinitiation complex recognition. Recently, it was found that C31 interacts with another Pol III-specific subunit, C17, that also interacts with Brf1 (20). While the role of C17 as an essential subunit of yeast Pol III was well established on genetic and biochemical grounds, it was surprising that the putative mammalian homologue, based on sequence comparison with yeast C17, was known as a hormone receptor component (19, 41). Two other subunits, C37 and C53, specifically interact in a two-hybrid assay (21) and are essential for transcription in vivo (9) but their precise roles are unknown.
The present study deals with two components of Pol III, C17 and C25, that are essential for growth and structurally conserved from yeast to humans (20, 54). C25 is related in sequence to the Rpb7 subunit of Pol II, to the archaeal RpoE subunit, and to a lesser extent, to the A43 subunit of Pol I (36, 54, 62). Rpb7 forms with Rpb4 a stable complex that can be reversibly dissociated from the yeast Pol II enzyme (16, 18, 53). The Rpb7/Rpb4-depleted enzyme retains its transcription activity on nonspecific templates (18, 53), but the heterodimer Rpb7/Rpb4 is required for promoter-directed transcription in vitro (18, 50). Rpb4, which is dispensable in S. cerevisiae, is important to the stabilization of the Pol II-Rpb7 interaction under adverse conditions (43, 46, 59). Interestingly, Rpb4 is essential in Schizosaccharomyces pombe and was shown to interact with the phosphatase Fcp1 (32, 56). Recently, the crystal structure of the archaeal subunit heterodimer RpoE/RpoF, homologous to the Rpb7/Rpb4 complex, was determined at high resolution (67). Low-resolution three-dimensional maps of yeast Pol II, lacking or not lacking subunits Rpb4 and Rpb7, suggested that these two subunits form part of the floor of the DNA-binding cleft (27).
We show here that C17 is akin to Rpb4 and RpoF and specifically interacts with C25, thus underscoring the remarkable conservation of this heterodimeric structure throughout archaeal and eukaryotic evolution. Sequence analysis, further supported by molecular modeling, indicated a structural relationship between the C25/C17 and RpoE/RpoF pairs of subunits. We also identified the human counterpart of C17 and found that it replaces C17 in vivo and supports transcription when incorporated in yeast Pol III. Unexpectedly, the human C17 is identical to a protein called CGRP-RCP, previously described as a component of a signal transduction cascade related to membrane-bound G proteins.
MATERIALS AND METHODS
Yeast strains and methods.
Standard yeast media, growth conditions, and genetic techniques were used as described previously (3). The yeast strains used in this study were constructed by using genetic techniques based on the transformation of lithium acetate-treated cells and plasmid shuffling assays. The products of the YJL011C and YKL144C open reading frames (ORF) are designated C17 and C25, respectively. The human homologue of the C17 protein is referred to as CGRP-RCP or HsC17.
Construction of plasmids.
Two oligonucleotides harboring BamHI and SalI restriction sites, respectively, were used to amplify by PCR the ORF encoding the HsC17 protein from a Homo sapiens cDNA library (a gift from M. Minet). The amplified DNA fragment was cloned into the yeast vector pGEN (61) or pBFG1 (a gift from J. H. Camonis), creating pGEN-HsRPC17 (2μm TRP1 pPGK1::HsRPC17::tCYC1) and pBFG1-HsRPC17 (2μm LEU2 pPGK1::2HA-HsRPC17::tPGK1), respectively. The untagged (HsC17, 148 amino acids) or N-terminally hemagglutinin (HA)-tagged (HA-HsC17, 173 amino acids) forms of HsC17 were expressed from these plasmids. The BamHI-SalI DNA fragment from pCMc17 (20) was cloned into pGEN, creating pGEN-RPC17. The RPC17 ORF was amplified by PCR from S. cerevisiae genomic DNA and inserted into the BamHI and SalI sites of pBFG1 to give pBFG1-RPC17, which produces the N-terminal HA-tagged form of C17 (185 amino acids). The BamHI-SalI DNA fragment from pYc17 (20) was cloned between the BamHI-XhoI sites of the multicopy pFL44L (2μm URA3) vector (6), creating pFL44L-RPC17. Two oligonucleotides harboring the BamHI restriction site and DNA sequences hybridizing 360 bp downstream or upstream of the RPC25 ORF were used to amplify an S. cerevisiae genomic DNA fragment of 1,380 bp that was cloned into pFL44L to give pFL44L-RPC25. The RPC25 ORF was also amplified by PCR with primers bearing NotI or BamHI sites and encoding a single copy of the HA epitope. The resulting DNA fragment was inserted between the NotI-BamHI sites of pACYC184-11b (a gift from S. Fribourg) to give pACYC184-RPC25 (ori p15A1 TcR CmR pT7::RPC25-HA::tT7), expressing a C-terminal HA-tagged version of C25. All PCR constructions were sequence verified.
In vivo visualization of C17-GFP fusion.
C-terminal fusion of green fluorescent protein (GFP) (S65T) to C17 was introduced into the RPC17 chromosomal locus of strain YPH500, as described previously (40). The correct chromosomal integration of the PCR-amplified DNA cassette was verified by PCR. In vivo visualization of C17-GFP fusion (or C160-GFP fusion, a generous gift from A. Chevalier) was performed as described previously (38) with cells grown in yeast extract-peptone-dextrose (YPD) medium to an A600 of 0.5.
Heterocomplementation with HsRPC17 ORF.
Plasmid pGEN-HsRPC17 or pBFG1-HsRPC17 was used to transform strain YMLF1 (20), in which the null mutant Δrpc17::HIS3 is complemented by plasmid pYc17 (CEN URA3 RPC17). Plasmid pGEN-RPC17, pGEN-HsRPC17, pBFG1-RPC17, or pBFG1-HsRPC17 was substituted for pYc17 by plasmid shuffling on plates containing 5-fluoroorotic acid. The resulting strains were named YMS1 to YMS4.
Dosage-dependent suppression.
High-copy suppressors were isolated after transformation of the YMS4 (HA-HsC17) and DS3-6b (MATa ura3-52 trp1-Δ63 his3-Δ200 leu2 rpc25-S100P; a gift from O. Gadal) strains with a yeast genomic library containing Sau3A fragments inserted at the BamHI sites of the pFL44L vector (63). Suppressor candidates were identified by restriction mapping and sequencing.
Two-hybrid assays.
Two-hybrid assays were performed essentially as described previously (70). After transformation of the Y190 yeast strain with a combination of plasmids, six independent transformants were grown as patches for 2 days at 30°C on selective medium. Transcriptional activation of the lacZ reporter gene was assayed by overlaying the cells with 10 ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) agar and incubating the plates for 24 h at 30°C.
Purification of RNA Pol III and in vitro transcription.
Whole-cell extracts from strains YMS1 (C17), YMS2 (HsC17), YMS3 (HA-C17), and YMS4 (HA-HsC17) were prepared from 6-liter cultures grown at 30°C on YPD to an A600 of 2. Micropurifications of wild-type or mutant RNA Pol III were performed as described previously (25). RNA polymerase activity during purification was monitored by nonspecific transcription assays on a poly(dA-dT) template. The subunit composition of the Pol III preparations (1 μg) was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on polyacrylamide gels. Proteins were revealed by silver staining or by Western blotting with monoclonal 16B12 antibodies (anti-HA) or anti-Pol III polyclonal antibodies by using an Amersham enhanced chemiluminescence kit. Transcription reactions were performed as described previously (25). Transcription mixtures (40 μl) contained transcription buffer (20 mM HEPES-KOH [pH 8], 5 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, 10% glycerol), 8 U of RNasin (Amersham), 0.6 mM (each) ATP, GTP, and CTP, 0.03 mM UTP, 10 μCi of [32P]UTP, TFIIIB, TFIIIC, RNA Pol III (50 ng), and 160 ng of plasmid DNA harboring the SUP4 tRNATyr gene. The final KCl concentration was 100 mM. The transcription reaction was allowed to proceed for 45 min at 25°C. Transcripts were analyzed by electrophoresis on an 8 M urea gel (6% polyacrylamide).
Primer extension.
Yeast strains YMS1 and YMS2 were grown in YPD to an A600 of 0.4 at 30°C and then shifted at 37°C. Cultures (maintained at an A600 of 0.4 by dilution with prewarmed YPD) were incubated at 37°C for 1, 10, 24, or 33 h. Cells were collected, and RNAs were prepared as described previously (24). Primer extension analysis was performed as described previously (2). Three micrograms of RNAs was used for annealing with a 5′-end-labeled oligonucleotide complementary to the sequence from +61 to +86 (located within the intron) of the tRNAIle gene. DNAs were recovered by ethanol precipitation, resuspended in 50% formamide, and denatured for 3 min at 90°C before loading onto a 7% polyacrylamide sequencing gel.
Expression and purification of recombinant proteins.
The Escherichia coli BL21(DE3) strain was transformed with plasmid pET-C17 encoding an N-terminal His6-T7-tagged form of C17 (20), plasmid pACYC184-RPC25 encoding a C-terminal HA-tagged version of C25, or a combination of both plasmids. Cells were grown at 37°C to A600 of 0.8 in Luria-Bertani medium supplemented with kanamycin (60 μg/ml) or chloramphenicol (34 μg/ml). The expression of recombinant proteins was induced with 50 μM isopropyl-1-thio-β-d-galactopyranoside. Cells were incubated for 3 h at 24°C with shaking, harvested by centrifugation, and resuspended in transcription buffer supplemented with 1 mM phenylmethylsulfonyl fluoride and a set of protease inhibitors (Complete; Boehringer). After cell lysis by sonication at 4°C, crude extracts containing recombinant T7-C17, C25-HA, or both proteins were recovered by centrifugation for 45 min at 20,000 × g and 4°C.
Immunopurification experiments.
Immunopurifications were performed by using 2 μg of purified 12CA5 monoclonal antibodies coupled to 40 μl of magnetic beads (8 × 108 beads/ml in phosphate-buffered saline supplemented with 0.5% serum albumin bovine and 10% glycerol coated with rat monoclonal antibodies directed against mouse immunoglobulin G2b [Dynal M450]). After 1 h of incubation at 10°C, the beads were extensively washed with phosphate-buffered saline and then with transcription buffer. The beads were then incubated overnight with gentle shaking at 10°C with 240 μg of crude extracts containing T7-C17, C25-HA, or both recombinant proteins, washed three times with 500 μl of transcription buffer, and transferred to another tube. Immunopurified proteins were eluted with Laemmli buffer, separated by SDS-PAGE, and revealed by Western blotting with anti-T7 (Novagen) or 16B12 anti-HA antibodies. The reciprocal experiment which used beads coated with anti-T7 antibodies could not be performed due to a nonspecific binding of C25-HA to the beads.
Protein identification by mass spectrometry.
Highly purified human Pol III (10 μg; a generous gift from Martin Teichmann of R. G. Roeder's laboratory) was prepared as described previously (68) and subjected to SDS-PAGE on a 4 to 12% Bis-Tris gel (Invitrogen). After staining with SimplyBlue (Invitrogen), protein bands migrating with an apparent mass between 6 and 20 kDa were isolated and subjected to in-gel tryptic digestion (60). Matrix-assisted laser desorption ionization mass spectra of peptide mixtures were obtained with an Autoflex mass spectrometer (Bruker Daltonik, Bremen, Germany). Monoisotopic peptide masses were assigned and used for database searching with the MS-Fit program (http://prospector.ucsf.edu/). Tandem mass spectrometry experiments were carried out on a Q-TOF hybrid mass spectrometer (Micromass, Manchester, United Kingdom) coupled to a nanoflow liquid chromatography instrument essentially as described previously (33). The Mascot algorithm (Matrix Science, London, United Kingdom) was used to interpret mass spectrometry data.
Sequence analysis and molecular modeling.
Searches for sequence similarities were performed with PSI-BLAST (1) from the National Center for Biotechnology Information. The sequence analysis and comparison were further refined by using bidimensional hydrophobic cluster analysis (HCA) (8, 22). This method is particularly efficient in detecting relationships between remote sequences (below the 20 to 25% identity level). The model of the C25/C17 complex was constructed with Modeller4 (57) and Tito (35) on the CBS Meta-server (17) on the basis of the experimental coordinates of the archaeal RpoE/RpoF dimer (67) (PDB identifier, 1go3). Some restraints relative to secondary structures, attributed with P-SEA (34) from the experimental structure coordinates, were added to the modeling procedure to improve the quality of the model.
RESULTS
CGRP-RCP, a new subunit of human RNA Pol III.
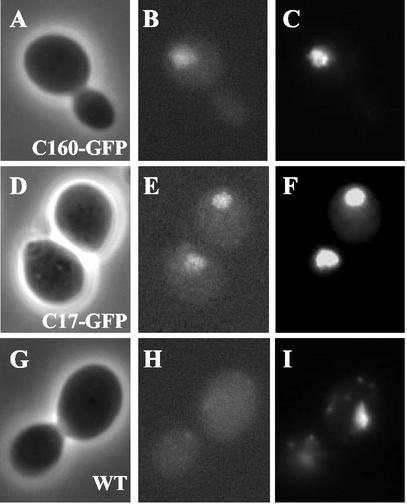
C17 is strongly conserved from yeasts to humans (20). However, the putative mammalian homologue of C17 named CGRP-RCP (for calcitonin gene-related peptide-receptor component protein) appears to be unrelated to transcription (4, 19, 41, 47-49, 51). Human CGRP-RCP was described as an intracellular peripheral membrane protein, expressed in all cell lines examined, that facilitates signal transduction at G protein-coupled receptors, especially CGRP receptors (19). The puzzling sequence similarity between an essential subunit of yeast Pol III and proteins involved in signal transduction prompted us to examine the cellular localization of C17 in yeast cells by using a strain expressing C17 fused at its C terminus to GFP. The in vivo visualization of the GFP-tagged version of C17 showed that most of the C17-GFP fluorescence was localized in the nucleus of yeast cells and visualized by 4′,6′-diamidino-2-phenylindole (DAPI) staining (Fig. 1). The low level of cytoplasmic fluorescence observed for the C17-GFP or C160-GFP fusion was similar (Fig. 1), showing that, in contrast to the cytoplasmic localization reported for CGRP-RCP in mammalian cells (19), the cellular localization of yeast C17 correlated well with its nuclear function in transcription.
FIG. 1.
In vivo visualization of C17 cellular localization. Phase contrast (A), DAPI staining (B), and green fluorescence (C) images of the same C160-GFP-expressing cells. Phase contrast (D), DAPI staining (E), and green fluorescence (F) images of the same C17-GFP-expressing cells. Phase contrast (G), DAPI staining (H), and green fluorescence (I) images of the same control (YPH500) cells.
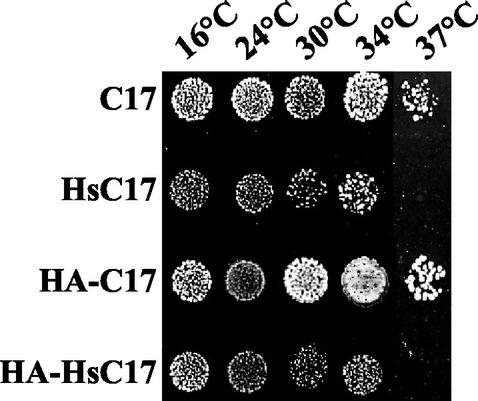
To investigate the possible implication of the human protein in transcription, the cDNA encoding human CGRP-RCP was expressed under the control of a strong constitutive promoter in a multicopy yeast vector and tested for its ability to complement the Δrpc17 null allele. As shown in Fig. 2, the heterocomplemented strain was viable, with a partial growth defect at 30°C (with a generation time of 300 min instead of 120 min for the control strain). Moreover, it was strictly thermosensitive at 37°C. Similar results were obtained when a HA-tagged version of human CGRP-RCP was assayed for complementation (Fig. 2), with a slightly more pronounced growth defect. Therefore, human CGRP-RCP can functionally replace C17 in yeast, indicating that CGRP-RCP (hereafter also named HsC17) is the human orthologue of C17.
FIG. 2.
Heterocomplementation of RPC17 by its H. sapiens orthologue. Haploid strains containing the Δrpc17 null allele and multicopy plasmids expressing C17 (strain YMS1), HsC17 (YMS2), HA-C17 (YMS3), or HA-HsC17 (YMS4) were obtained by plasmid shuffling. Cell growth was tested by spotting 5 μl of liquid cell cultures on YPD plates, and growth was observed after incubation of the plates at 16, 24, 30, 34, or 37°C for 7, 5, 3, 3, or 4 days, respectively.
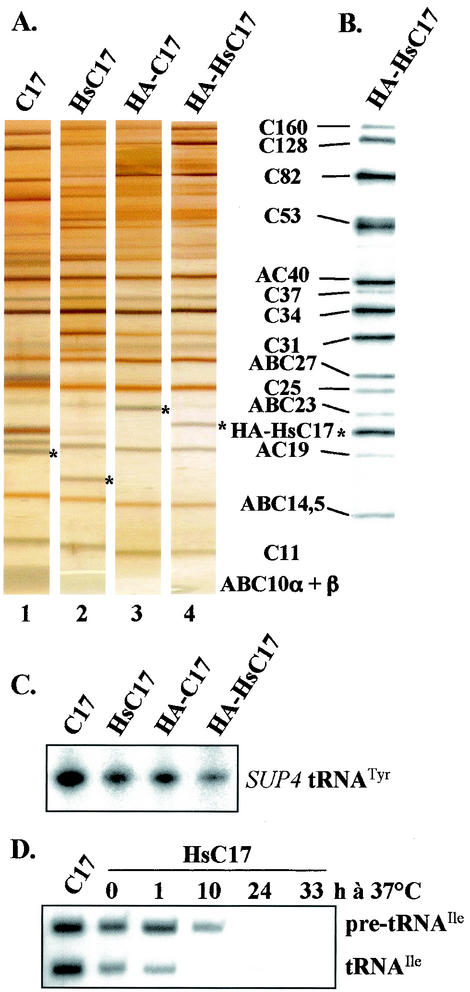
To confirm these results, RNA Pol III was highly purified from four strains containing untagged or HA-tagged versions of C17 or HsC17 by using a micropurification procedure (25). One-third as much enzyme was obtained from yeast cells growing with HsC17 than from wild-type cells. Similar amounts of RNA Pol III were analyzed by SDS-PAGE, and the subunit composition of the four purified enzymes was examined by silver staining. As shown in Fig. 3A, all of the subunits of Pol III were present in the purified fraction from the control strains, including C17 (Fig. 3A, lane 1) or HA-C17 that migrated more slowly (Fig. 3A, lane 3). Pol III from the YMS2 strain, growing with HsC17, lacked the yeast C17 polypeptide as expected but contained instead a new polypeptide migrating faster than C17 (Fig. 3A, lane 2). This new polypeptide was replaced by a more-slowly migrating polypeptide in Pol III harboring the HA-tagged version of HsC17 (Fig. 3A, lane 4). The differential migration of the polypeptides corresponding to the untagged or tagged versions of C17 and HsC17 was in keeping with the calculated molecular masses of C17 (17 kDa), HsC17 (16 kDa), HA-C17 (20 kDa), and HA-HsC17 (19 kDa), respectively. Western blot analysis of purified Pol III prepared from the strain expressing HA-HsC17 by using a mixture of anti-HA and anti-Pol III antibodies confirmed the presence of all of the enzyme subunits and the incorporation of HA-HsC17 protein into the enzyme (Fig. 3B). In vitro transcription experiments performed on nonspecific or specific DNA templates with similar amounts of enzyme showed that the polymerase-containing HsC17 or HA-HsC17 was competent to transcribe the SUP4 tRNATyr gene and was about half as active as the wild-type enzyme (Fig. 3C and data not shown). Primer extension experiments indicated that the mutant enzyme (with HsC17) initiated accurately on a tRNAIle gene in vivo, even after a shift to a nonpermissive temperature (Fig. 3D). However, tRNA synthesis strongly decreased after 10 h at 37°C (Fig. 3D) while the cells remained viable for a longer period of time.
FIG. 3.
Subunit composition and transcriptional activity of mutant RNA Pol III. (A) Subunit composition. RNA Pol III fractions (1 μg), purified from strains YMS1, YMS2, YMS3, and YMS4 containing C17, HsC17, HA-C17, or HA-HsC17 as indicated, were analyzed by SDS-PAGE on a 4 to 15% polyacrylamide gel and revealed by silver staining. The positions of Pol III subunits are indicated on the right. Asterisks point to the bands corresponding to the untagged or HA-tagged versions of C17 or HsC17. (B) Western analysis. An RNA Pol III fraction (1 μg), purified from strain YMS4 expressing HA-HsC17, was analyzed by SDS-PAGE on a 4 to 13% polyacrylamide gel, and subunits were revealed by Western blotting with a mixture of anti-HA and anti-Pol III antibodies. Fourteen subunits, indicated on the left, were revealed in this experiment. The asterisk points to the band revealed by anti-HA antibodies corresponding to HA-HsC17. (C) Specific transcription of the SUP4 tRNATyr gene. The SUP4 tRNATyr gene was transcribed in vitro for 45 min at 25°C in the presence of TFIIIC, TFIIIB, and 50 ng of RNA Pol III purified from strains YMS1 toYMS4 containing C17, HsC17, HA-C17, or HA-HsC17, as indicated. Transcripts were separated in urea gel and revealed by autoradiography. (D) Comparison of in vivo start site selection pattern. YMS1 or YMS2 cells expressing C17 or HsC17 were grown at 37°C for 1, 10, 24, or 33 h, as indicated. RNAs were extracted, and primer extension analysis on the tRNAIle gene was performed as described in Materials and Methods with an oligonucleotide hybridizing within the tRNAIle gene intron. The lower band corresponds to the 5′-end-processed transcript, and the upper band (6 pb longer) corresponds to the primary transcript.
To directly show that CGRP-RCP was a subunit of human RNA Pol III, a highly purified human Pol III fraction (68) was subjected to SDS-PAGE, protein bands migrating between 6 and 20 kDa were isolated, and tryptic peptides were generated and analyzed by mass spectrometry as described in Materials and Methods. Two peptides (HQSPEIVR and HSSGQQNLNTITYETLK) contained in CGRP-RCP were generated from a protein migrating with an apparent mass between 15 to 20 kDa. The presence of CGRP-RCP among human Pol III subunits was also revealed by using an immunological approach (Z. Wang and R. G. Roeder, personal communication). Together, our genetic and biochemical results established CGRP-RCP/HsC17 as a bona fide Pol III subunit in humans and the orthologue of C17.
C17 and C25 interact in genetic suppression assays.
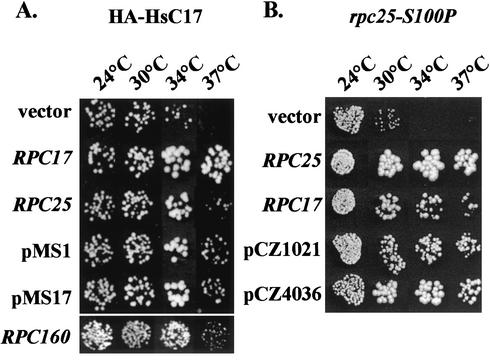
The temperature-sensitive growth defect associated with the tagged HsC17 subunit was used to isolate physiological partners of C17 by dosage-dependent suppression of the thermosensitive strain YMS4. Twenty-four dosage-dependent suppressors were isolated at 37°C. Among them, 20 contained the entire RPC17 ORF and 2 harbored a truncated version of RPC17 with the five C-terminal residues (ISAYA) replaced by one leucine residue. Two suppressors (pMS1 and pMS17) (Fig. 4) harbored overlapping regions of chromosome XI comprising the RPT1, RPC25, and LTV1 genes. RPC25 encodes another Pol III-specific subunit (54). As shown in Fig. 4A, a high-copy-number plasmid carrying only RPC25 (pFL44L-RPC25) suppressed the HsC17-associated growth defect at 37°C as efficiently as pMS1 and pMS17. The finding of 22 different versions of RPC17 among the suppressor collection showed that the suppressor screen was exhaustive, although it generated only one extragenic suppressor candidate (other than RPC17). As two-hybrid screens with C17 as a bait revealed interactions with C11, C31, Brf1, and C160 (see below), we transformed strain YMS4 with high-copy-number plasmids carrying these genes. Only the overexpression of C160 suppressed the thermosensitivity of the HA-HsC17-containing strain (RPC160) (Fig. 4A). To explore the genetic interaction between RPC17 and RPC25, we also used a thermosensitive mutant strain (DS3-6b) harboring the rpc25-S100P mutation in the RPC25 ORF to screen for high-copy-number suppressors. This strain displayed a slow-growth phenotype at 30°C and was strictly thermosensitive even at 34°C (Fig. 4B). Twenty dosage-dependent suppressors were isolated at 37°C. Eighteen of them contained RPC25, and two suppressors (pCZ1021 and pCZ4036) (Fig. 4B) harbored overlapping regions of chromosome VII comprising the entire RPC17 ORF and two tRNA genes [tK(CUU)J and tW(CCA)J]. As shown in Fig. 4B, a high-copy-number plasmid carrying only RPC17 (pFL44L-RPC17) restored the ability of the rpc25-S100P mutant to grow at 34 and 37°C. These results strongly suggested a specific interaction between RPC17 and RPC25 and prompted us to look for a physical interaction between these two Pol III subunits.
FIG. 4.
Dosage-dependent suppression of rpc17 or rpc25 mutants. The growth of the YMS4 (expressing HA-HsC17) or DS3-6b (rpc25-S100P) mutant strain transformed by various multicopy plasmids was tested by spotting 5 μl of liquid cell cultures on YPD plates. Plates were incubated for 3 days at 24, 30, 34, or 37°C as indicated. (A) High-copy-number suppression of the YMS4 mutant by RPC25 or RPC160. The growth of strain YMS4 transformed by the pFL44L void vector was compared to the growth of the same strain transformed by two plasmids isolated from a yeast genomic library (pMS1 and pMS17) or pFL44L harboring RPC17, RPC25, or RPC160, as indicated. (B) High-copy-number suppression of the rpc25-S100P mutation by RPC17. The growth of strain DS3-6b transformed by the pFL44L void vector was compared to the growth of the same strain transformed by two plasmids (pCZ1021 and pCZ4036) isolated from a genomic library or by pFL44L harboring RPC25 or RPC17, as indicated.
C25 and C17 physically interact.
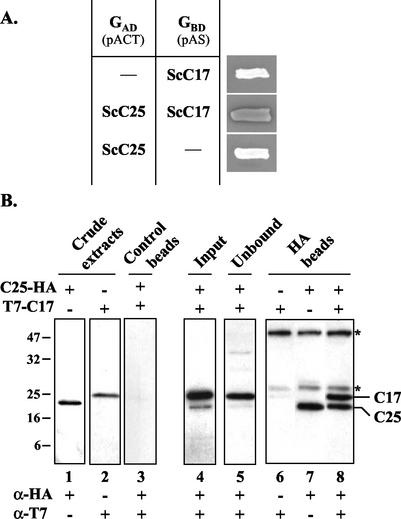
A global two-hybrid screen of a yeast genomic library (21) with C17 as a bait identified two independent clones (residues 1349 to 1460 and 1394 to 1460) corresponding to the C terminus of C160, the counterpart of Rpb1 in Pol II. The overlap of these two clones corresponded to the last 67 residues of C160. A systematic two-hybrid approach was used to uncover interactions of C17 with the other components of the yeast Pol III system (20). An interaction of C17 with TFIIIB70, C31, and C11 was detected in this way (20). However, the interaction of C17 with C25 had not been assayed. As shown in Fig. 5A, when a combination of C17 and C25 fusions was tested, a significant blue coloration was observed, whereas none of the fusion proteins alone activated lacZ transcription. The reciprocal combination of protein fusions gave similar results (data not shown). The physical interaction of C17 with C25 was confirmed by a coimmunopurification experiment. T7-tagged C17 and HA-tagged C25 were expressed separately or coexpressed in E. coli BL21(DE3). Bacterial crude extracts were prepared, and mixed with magnetic beads precoated with anti-HA monoclonal antibodies, and the bound proteins were eluted by SDS. The input and unbound fractions as well as the eluted proteins were subsequently analyzed by SDS-PAGE and immunoblotting with anti-T7 or anti-HA antibodies. The electrophoretic migration of the tagged C17 and C25 proteins is shown in Fig. 5B (lanes 1 and 2). Note that recombinant C25-HA migrated faster than T7-C17, in spite of its larger theoretical molecular mass (24 versus 21 kDa). None of the two recombinant proteins was bound to the control beads (Fig. 5B, lane 3). In contrast, a large proportion of the HA-tagged C25 present in the crude extract was retained on the anti-HA beads (Fig. 5B, compare lanes 4 and 5). When T7-C17 was coexpressed with C25-HA, we observed the coimmunopurification of both proteins (Fig. 5B, lane 8), whereas T7-C17 alone was not retained on the anti-HA beads (Fig. 5B, lane 6). These results confirmed the interaction between C17 and C25 observed with the two-hybrid system (Fig. 5A) and shows that this interaction is based on a direct physical contact between these two proteins. While the coimmunopurification of T7-C17 with C25-HA was reproducibly observed when assayed on crude extracts prepared from bacterial cells coexpressing both proteins, the complex could not be reconstituted by mixing two crude extracts, each containing one of the two proteins.
FIG. 5.
Physical interaction between C17 and C25. (A) Two-hybrid interaction between C25 and C17. The RPC25 ORF was fused in frame with the activating domain (GAD), and the RPC17 ORF was fused in frame with the DNA-binding domain (GBD) of GAL4. Transcriptional activation of the lacZ reporter gene was assayed by growing yeast cells transformed by a combination of plasmids as indicated on selective medium and overlaying the cells with X-Gal agar. White or blue coloration of cell patches on X-Gal plates is shown. (B) Coimmunopurification of C17 with C25. Bacterial crude extracts (240 μg) prepared from cells expressing T7-C17 or C25-HA or coexpressing both recombinant proteins were incubated with magnetic beads coated with anti-HA antibodies (HA beads) or uncoated (Control beads), as indicated. The beads were washed, and bound proteins were eluted by boiling the beads in loading buffer. The input, unbound, and eluted fractions were analyzed by SDS-PAGE and Western blotting with anti-T7 or anti-HA antibodies, as indicated. Asterisks indicate the positions of immunoglobulin heavy or light chains. The positions and molecular masses (in kilodaltons) of marker bands are indicated on the left. The positions of C25-HA and T7-C17 are indicated on the right.
C17 is homologous to Rpb4 and to the archaeal RpoF subunit.
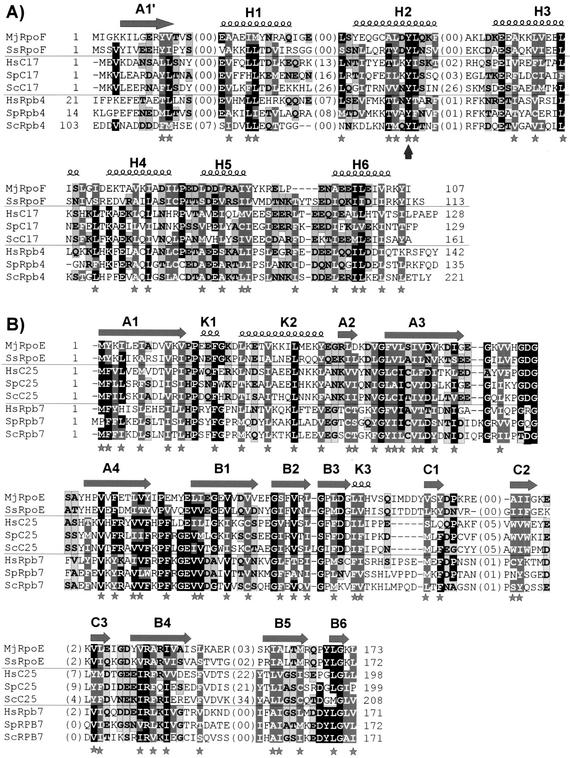
C25 has already been described as the Pol III homologue of Rpb7 and of the archaeal RpoE subunit (36, 54). Previous attempts to find a homologue of Rpb4, the well-established interacting partner of Rpb7 (16, 18, 28, 37) among Pol III subunits have failed. However, our results showing that C17 interacts with C25 suggested that C17 could be the Pol III homologue of Rpb4 and prompted us to further analyze the protein sequence of C17. Searches for similarities that the C17 sequence could share with other proteins with PSI-BLAST first revealed, as already described (20), its orthologues in different species. Similarities were also observed just below the threshold value with the human Rpb4 sequence (E-value, 0.03; 16% sequence identity over 127 amino acids). PSI-BLAST investigations with the C17/Rpb4 family further highlighted the RpoF subunit of the archaeal RNA polymerase whose crystal structure was recently determined (67). Sequence similarities between the Rpb4 and RpoF subunits were previously reported (69). We further examined the proposed sequence alignments by using HCA, a method that focuses on the fold invariants and allows the processing of loops of variable lengths (22). This analysis strengthened the C17/Rpb4 sequence similarity in the C-terminal part of the protein sequences (from residue 96 to 161 in the C17 sequence) and highlighted conserved sequence blocks in the N-terminal part (corresponding to the secondary structures A1′, H1, and especially H2 [see Fig. 7], as inferred from the experimental structure of archaeal subunit RpoF [67]) (see below) that were not detectable by BLAST due to the presence of large insertions in the yeast sequence (Fig. 6A). Indeed, yeast C17 contains two large loops, made exclusively of nonhydrophobic amino acids, between the secondary structure elements H1-H2 and H2-H3, as inferred from the crystal structure of the archaeal subunit RpoF (67). The H2-H3 loop is very specific to the yeast sequence. Interestingly, after completion of our analysis, the C17/Rpb4 relationship was reported in the Smart database (39) (Smart 00657), thereby strengthening our observations. However, the Smart alignments do not extend the C17/Rpb4 family to the archaeal RpoF subunit and do not cover all of the C17/Rpb4/RpoF common structures. Indeed, the alignment reported in this domain database is limited to the C-terminal region (residues 101 to 160 in yeast C17 versus residues 56 to 115 in human Rpb4) and does not project beyond the large insertions existing in the yeast C17 sequence. It is worth mentioning that most of the hydrophobic amino acids that make up the RpoF core structures are conserved in the C17 and Rpb4 sequences (Fig. 6A), further supporting the existence of a significant structural relationship between these proteins.
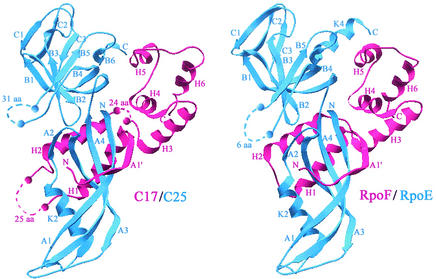
FIG. 7.
Molecular modeling of the C17/C25 complex. Ribbon representation of the model of the three-dimensional structure of the C17/C25 complex (left panel) that was constructed on the basis of the experimental structure of the RpoE/RpoF complex (right panel) (67). The C17/C25 complex was modeled on the basis of the alignments shown in Fig. 6. The large insertions in the C17 (pink) and C25 (blue) sequences, which cannot be modeled accurately, are indicated by broken lines (with the number of residues in the loop). Secondary structures are labeled according to the results of Todone et al. (67). Major differences between the left and right panels are located in the loops linking H1 to H2 and H2 to H3 (C17) as well as in the loop linking B4 to B5 (C25). Two of them (H1-H2 and B4-B5) are located in close proximity, suggesting that these two loops could interact with each other.
FIG. 6.
Sequence alignments of the C17/Rpb4/RpoF and C25/Rpb7/RpoE families. Protein sequences (the first and last residues shown in the alignments are indicated) of C17, Rpb4, and RpoF (A) or C25, Rpb7, and RpoE (B) from several species were aligned with PSI-BLAST and HCA (22). The lengths of the loops not included in the alignments are indicated within brackets. Identical amino acid residues (at least five at the same position) are highlighted with a black background. Conserved residues are highlighted with a gray background. White letters indicate hydrophobic amino acids or amino acids that can substitute for them in some circumstances (A, C, T, and S). The secondary structures experimentally determined by Todone et al. (67) for the MjRpoF/MjRpoE complex are indicated above the sequences. The highly conserved hydrophobic amino acids mainly involved in the maintenance of the fold are marked with stars. The vertical arrow shows the strictly conserved tyrosine of the C17/Rpb4/RpoF family, which makes contact with the other subunit. GenBank identifiers are as follows: MjRpoF, 17943305; MjRpoE, 17943304; SsRpoF, 15897651; SsRpoE, 13813566; HsC17, 7656977; HsC25, 5304853; HsRpb4, 4758574; HsRpb7, 929921; SpC17, 13624765; SpC25, 7490488; SpRpb4, 9297039; SpRpb7, 2529241; ScC17, 6322449; ScC25, 516561; ScRpb4, 6322321; ScRpb7, 464672. Mj, Methanococcus jannaschii; Ss, Sulfolobus solfataricus; Hs, H. sapiens; Sp, S. pombe; Sc, S. cerevisiae
Similar PSI-BLAST searches were performed with the yeast C25 sequence as the query, and significantly, they detected conserved sequences with Rpb7, with the A43 subunit of yeast RNA Pol I, and with the RpoE subunit of archaeal RNA polymerase (54, 62, 67). The resulting alignments (Fig. 6B), which were refined by using HCA, particularly for the most divergent C-terminal sequences, were used for the molecular modeling of the C17/C25 complex.
Molecular modeling of the C17/C25 complex.
A model of the C25/C17 complex was constructed on the basis of the alignments defined above by using the experimental coordinates of the RpoE/RpoF dimer (67). As a consequence of the observed sequence similarities, the C25/C17 complex should have a structure similar to that of the RpoE/RpoF complex, with the C25 subunit having an elongated two-domain structure and C17 wrapping on one side of C25, at the interface between the two domains (Fig. 7). The N-terminal amino acids of C17 should also, like those of RpoF, contribute an extra strand A1′ to the N-terminal β-sheet A of C25. One of the two subdomains of C25, including sheets B and C, corresponds to an OB fold, and the second one resembles a truncated RNP fold, suggesting that this domain could be involved in RNA binding (67).
The major differences between the C25/C17 and RpoE/RpoF heterodimers are concentrated in loops of considerable length (Fig. 7, left panel), two of them being located in close proximity. In addition to amino acid positions where hydrophobicity is conserved and which are mostly involved in the maintenance of the fold, the majority of the other highly conserved amino acids are located at the dimer interface, making bonds between C25 and C17 (interchain) or within C25 (intrachain). For example, like in the RpoF/RpoE complex, the hydroxyl group of the conserved Tyr65 of C17 (Fig. 6A) should be hydrogen bonded to the Phe80 (main chain CO) of C25, whereas the conserved Glu83 of C25 should make a hydrogen bond through its oxygen (Oɛ1) with the Thr86 (main chain NH) of C25 (data not shown).
DISCUSSION
Pursuing the analysis of yeast RNA polymerase subunits, we propose here that two small subunits of Pol III, C25 and C17, are the counterparts of the Rpb7/Rpb4 heterodimer in Pol II. These polypeptides appear to have evolved within the different forms of RNA polymerases to allow specific interactions with cognate auxiliary factors. Our observation that the human protein CGRP-RCP reported to be involved in signal transduction can replace C17 in yeast Pol III and copurifies with human Pol III raises the question of the functional evolution of CGRP-RCP/HsC17 in mammalian cells.
The proposal that the Pol II subunit heterodimer Rpb7/Rpb4 has been conserved in Pol III simplifies our view of the molecular evolution of eukaryotic RNA polymerases (Table 1). This conservation is supported by biochemical and genetic evidence and sequence analysis. (i) C25 and C17 interact physically by coimmunopurification and by two-hybrid analysis. (ii) Extensive dosage-dependent suppressor screens selectively generated RPC17 as a suppressor of the rpc25-S100P mutation and, conversely, RPC25 as a suppressor of the HsC17-associated growth defect. (iii) Sequence analysis and molecular modeling showed that the C25/C17 complex can adopt a structure similar to that of the archaeal counterpart of the Rpb7/Rpb4 complex. Therefore, the C25/C17 pair of subunits appears to match the Rpb7/Rpb4 complex. A related heterodimer is also likely conserved in Pol I (subunits A43 and A14) (Table 1), as documented elsewhere (50a). As shown in Table 1, a 12-subunit core RNA polymerase has been conserved from the archaea (except for ABC14.5) to eukaryotes and yeast Pol I and Pol III containing two and five additional specific subunits, respectively.
TABLE 1.
Structural relationship of multisubunit RNA polymerasesa
| S. cerevisiae Pol II subunit | Conserved subunit in:
|
|||
|---|---|---|---|---|
| E. coli | Methanococcus jannaschiib |
S. cerevisiae
|
||
| Pol Ic | Pol IIId | |||
| Rpb1 | β′ | RpoA′/" | A190 | C160 |
| Rpb2 | β | RpoB′/" | A135 | C128 |
| Rpb3 | α | RpoD | AC40 | AC40 |
| Rpb4 | RpoF | A14 | C17 | |
| Rpb5 | RpoH | ABC27 | ABC27 | |
| Rpb6 | ω | RpoK | ABC23 | ABC23 |
| Rpb7 | RpoE | A43 | C25 | |
| Rpb8 | ABC14.5 | ABC14.5 | ||
| Rpb9 | RpoX | A12.2 | C11 | |
| Rpb10 | RpoN | ABC10β | ABC10β | |
| Rpb11 | α | RpoL | AC19 | AC19 |
| Rpb12 | RpoP | ABC10α | ABC10α | |
The 12 subunits of yeast RNA Pol II (Rpb1 to Rpb12) and the conserved (or shared) subunits in E. coli, M. jannaschii, and yeast Pol I and III are listed. The 12-subunit Pol II structure is fully conserved in Pol III and probably also in Pol I, as the Rpb4/Rpb7 heterodimer is likely conserved in Pol I. Although the A43 sequence can be significantly related to the C25/Rpb7 sequences, the A14 sequence seems to be far more divergent from the C17/Rpb4 sequences (50a).
M. jannaschii also contains the specific subunits RpoG and RpoI.
S. cerevisiae Pol I also contains the specific subunits A49 and A34.5.
S. cerevisiae Pol III also contains the specific subunits C82, C53, C37, C34, and C31.
A difference electron density map analysis between the complete 12-subunit Pol II and the Rpb7/Rpb4-depleted enzyme showed these two subunits forming part of the DNA-binding cleft at the active center (27). Somewhat at variance with this conclusion, our two-hybrid screen with C17 as a bait suggested an interaction between C17 and the C-terminal domain (67 amino acid residues) of the C160 subunit. This interaction between C17 and C160 was confirmed genetically by suppression assays (Fig. 4A). Based on the atomic structure of Pol II, the C-terminal domain of the C160 subunit is distant from the DNA-binding cleft but rather close to where TFIIB is expected to lie (13). Binding of C17 (and of C25) in this area would explain its interaction with the TFIIB-related factor Brf1 (20) and the interaction of the archaeal RpoE subunit with TFB (42). The location of C17 at the C-terminal end of C160 would also be in keeping with the observation that S. pombe Rpb4 interacts with the carboxy-terminal domain (CTD)-specific phosphatase Fcp1 (32).
The Rpb4/C17 relationship was unexpected because these two subunits are apparently unrelated at the functional level. While C17 was shown to be essential and to interact with the Pol III factor Brf1, Rpb4 is dispensable for cell viability and, for many years, was thought to be essentially involved in heat stress responses (11, 12, 29, 52, 59). As shown recently for S. cerevisiae, Rpb4 is essential for the global activity of Pol II at high temperatures, probably in part by stabilizing Rpb7 within the enzyme (43, 46). Indeed, the deletion of RPB4 caused the loss of Rpb7 in the purified Pol II, and overdosage of RPB7 partially compensated for the absence of Rpb4 at high temperatures (43, 59). The basal function of the Rpb7/Rpb4 complex in promoter-directed transcription initiation is therefore likely fulfilled by Rpb7 in Pol II and, by extension, also by C25 in the Pol III system. Recent studies with S. pombe, however, revealed the important role of SpRpb4 (which is essential for cell growth in fission yeast) for the assembly of the CTD-phosphatase Fcp1 into the Pol II complex, the dephosphorylation of the CTD of Rpb1 in vivo, and thereby, for RNA polymerase recycling (32). C17 and Rpb4, therefore, appear to have gained distinct functions within the two forms of enzyme. There are hints that Rpb7/Rpb4 may also have some regulatory role. Human Rpb7 is expressed in a highly tissue-specific manner, in a pattern that differs from that of Rpb1 (29). In S. cerevisiae, the level of Rpb4 is regulated in a different manner from that of the Pol II core subunits (11, 12, 52). In S. pombe, Rpb4 was found in excess of core subunits and was detected both in the nucleus and in the cytoplasm (31, 55). The possible role of such a pool of free Rpb4 is unclear but is reminiscent of the claim that the homologue of C17 in mammalian cells, CGRP-RCP, has a role in signal transduction.
The case of the mammalian CGRP-RCP is intriguing. This CGRP-RCP was originally found to be required for CGRP receptor activation in a Xenopus oocyte expression system (41). The protein has been detected in all immortalized cell lines tested (19) but only in distinct populations of cells in the cochlea, brain, and eye (41, 49, 51). Mouse tissues contain highly variable levels of CGRP-RCP mRNA, the highest levels being found in testis and spermatozoa (4). Stable cell lines expressing antisense RNA were shown to have greatly diminished levels of CGRP-RCP (19). In addition, CGRP-RCP was found to be a peripheral membrane protein (easily extracted from membrane material by mild detergents or salts) and to coimmunoprecipitate with the CGRP receptor (19). These results do not fit well with a ubiquitous, housekeeping, nuclear function as expected for a Pol III subunit. Therefore, it was striking to find that human CGRP-RCP could functionally replace in vivo the C17 subunit that is essential for growth in S. cerevisiae. This was a particularly significant result since previous attempts to replace other yeast Pol III subunits (C53, C34, and C11) with their mammalian counterparts had failed (10, 26; unpublished data). Furthermore, the human protein was found to be stably incorporated in transcriptionally active purified Pol III in the place of C17. These results convincingly established that CGRP-RCP (or HsC17) is the mammalian functional homologue of yeast C17, which should imply that HsC17 is a subunit of human Pol III. Indeed, a polypeptide of the appropriate size present in highly purified preparations of mammalian Pol III (68) was shown to correspond to CGRP-RCP through the use of mass spectrometry (this work) or with an immunological approach (Wang and Roeder, personal communication). As for the cell- and tissue-specific accumulation of CGRP-RCP mRNA in the brain and testis, one may note that the expression of the TATA-binding protein-related factor TRF1, which directs Pol III transcription in Drosophila melanogaster (65), becomes restricted to the cells that form the nervous system and gonads during embryonic development (23). In adult flies, the TRF1 mRNA level is also the highest in primary spermatocytes (14). Elevated levels of critical Pol III components may be related to a rapid cell growth rate, as during spermatogenesis, or to the need for the selective expression of Pol III transcripts in the nervous system, like identifier RNA (64), BC200 RNA (45), and BC1 RNA (44). Clearly, the functions of CGRP-RCP/HsC17 deserve to be further clarified, as a dual role in Pol III transcription and signal transduction would represent an unusual example of functional divergence.
Acknowledgments
We thank Martin Teichmann from R. G. Roeder's laboratory for his generous gift of human Pol III; Jacques H. Camonis, Anne Chevalier, Sébastien Fribourg, Olivier Gadal, and Michèle Minet for providing a human cDNA library, plasmids, or strains; Claire Boschiero for help with the two-hybrid screen and DNA sequencing; Emmanuel Favry for transcription factors; Jérome Garin for help with protein identification with mass spectrometry; Florence Bordon-Pallier, Christophe Carles, and Michel Riva for helpful discussions; and R. G. Roeder and Z. Wang for communicating unpublished results.
M.S. received a CEA-Aventis grant, and C.Z. and E.L. were supported by fellowships from the Ministère de la Recherche et de l'Enseignement Supérieur.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrau, J.-C., and M. Werner. 2001. B"-associated factor(s) involved in RNA polymerase III preinitiation complex formation and start-site selection. Eur. J. Biochem. 268:5167-5175. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Balkan, W., E. L. Oates, G. A. Howard, and B. A. Roos. 1999. Testes exhibit elevated expression of calcitonin gene-related peptide receptor component protein. Endocrinology 140:1459-1469. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomew, B., D. Durkovich, G. A. Kassavetis, and E. P. Geiduschek. 1993. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 13:942-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609-615. [DOI] [PubMed] [Google Scholar]
- 7.Brun, I., A. Sentenac, and M. Werner. 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16:5730-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callebaut, I., G. Labesse, P. Durand, A. Poupon, L. Canard, J. Chomilier, B. Henrissat, and J. P. Mornon. 1997. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 53:621-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chédin, S., M. L. Ferri, G. Peyroche, J. C. Andrau, S. Jourdain, O. Lefebvre, M. Werner, C. Carles, and A. Sentenac. 1998. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol. 63:381-389. [DOI] [PubMed] [Google Scholar]
- 10.Chédin, S., M. Riva, P. Schultz, A. Sentenac, and C. Carles. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 12:3857-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choder, M. 1993. A growth rate-limiting process in the last growth phase of the yeast life cycle involves RPB4, a subunit of RNA polymerase II. J. Bacteriol. 175:6358-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choder, M., and R. A. Young. 1993. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 14.Crowley, T. E., T. Hoey, J. K. Liu, Y. N. Jan, L. Y. Jan, and R. Tjian. 1993. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature 361:557-561. [DOI] [PubMed] [Google Scholar]
- 15.Darcy, T. J., W. Hausner, D. E. Awery, A. M. Edwards, M. Thomm, and J. N. Reeve. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181:4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dezélée, S., F. Wyers, A. Sentenac, and P. Fromageot. 1976. Two forms of RNA polymerase B in yeast. Eur. J. Biochem. 65:543-552. [DOI] [PubMed] [Google Scholar]
- 17.Douguet, D., and G. Labesse. 2001. Easier threading through web-based comparisons and cross-validations. Bioinformatics 17:752-753. [DOI] [PubMed] [Google Scholar]
- 18.Edwards, A. M., C. M. Kane, R. A. Young, and R. D. Kornberg. 1991. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266:71-75. [PubMed] [Google Scholar]
- 19.Evans, B. N., M. I. Rosenblatt, L. O. Mnayer, K. R. Oliver, and I. M. Dickerson. 2000. CGRP-RCP, a novel protein required for signal transduction at calcitonin gene-related peptide and adrenomedullin receptors. J. Biol. Chem. 275:31438-31443. [DOI] [PubMed] [Google Scholar]
- 20.Ferri, M.-L., G. Peyroche, M. Siaut, O. Lefebvre, C. Carles, C. Conesa, and A. Sentenac. 2000. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 20:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores, A., J.-F. Briand, O. Gadal, J.-C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaboriaud, C., V. Bissery, T. Benchetrit, and J. P. Mornon. 1987. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 224:149-155. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, S. K., S. Takada, R. H. Jacobson, J. T. Lis, and R. Tjian. 1997. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91:71-83. [DOI] [PubMed] [Google Scholar]
- 24.Hermann-Le Denmat, S., M. Werner, A. Sentenac, and P. Thuriaux. 1994. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol. 14:2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huet, J., N. Manaud, G. Dieci, G. Peyroche, C. Conesa, O. Lefebvre, A. Ruet, M. Riva, and A. Sentenac. 1996. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 273:249-267. [DOI] [PubMed] [Google Scholar]
- 26.Ittmann, M., J. Ali, A. Greco, and C. Basilico. 1993. The gene complementing a temperature-sensitive cell cycle mutant of BHK cells is the human homologue of the yeast RPC53 gene, which encodes a subunit of RNA polymerase C (III). Cell Growth Differ. 4:503-511. [PubMed] [Google Scholar]
- 27.Jensen, G. J., G. Meredith, D. A. Bushnell, and R. D. Kornberg. 1998. Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J. 17:2353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khazak, V., J. Estojak, H. Cho, J. Majors, G. Sonoda, J. R. Testa, and E. A. Golemis. 1998. Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol. 18:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khazak, V., P. P. Sadhale, N. A. Woychik, R. Brent, and E. A. Golemis. 1995. Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol. Biol. Cell 6:759-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoo, B., B. Brophy, and S. P. Jackson. 1994. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 8:2879-2890. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M., H. Sakurai, and A. Ishihama. 2001. Intracellular contents and assembly states of all 12 subunits of the RNA polymerase II in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 268:612-619. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, M., H. Suzuki, and A. Ishihama. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA Polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22:1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koller, A., M. P. Washburn, B. M. Lange, N. L. Andon, C. Deciu, P. A. Haynes, L. Hays, D. Schieltz, R. Ulaszek, J. Wei, D. Wolters, and J. R. Yates III. 2002. Proteomic survey of metabolic pathways in rice. Proc. Natl. Acad. Sci. USA 99:11969-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labesse, G., N. Colloc'h, J. Pothier, and J. P. Mornon. 1997. P-SEA: a new efficient assignment of secondary structure from C alpha trace of proteins. Comput. Appl. Biosci. 13:291-295. [DOI] [PubMed] [Google Scholar]
- 35.Labesse, G., and J. P. Mornon. 1998. Incremental threading optimization (TITO) to help alignment and modelling of remote homologues. Bioinformatics 14:206-211. [DOI] [PubMed] [Google Scholar]
- 36.Langer, D., J. Hain, P. Thuriaux, and W. Zillig. 1995. Transcription in archaea: similarity to that in eucarya. Proc. Natl. Acad. Sci. USA 92:5768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin, R. M., and T. J. Guilfoyle. 1998. Two small subunits in Arabidopsis RNA polymerase II are related to yeast RPB4 and RPB7 and interact with one another. J. Biol. Chem. 273:5631-5637. [DOI] [PubMed] [Google Scholar]
- 38.Le Masson, Y., C. Saveanu, A. Chevalier, A. Namane, R. Gobin, M. Fromont-Racine, A. Jacquier, and C. Mann. 2002. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol. 43:1431-1443. [DOI] [PubMed] [Google Scholar]
- 39.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 41.Luebke, A. E., G. P. Dahl, B. A. Roos, and I. M. Dickerson. 1996. Identification of a protein that confers calcitonin gene-related peptide responsiveness to oocytes by using a cystic fibrosis transmembrane conductance regulatory assay. Proc. Natl. Acad. Sci. USA 93:3455-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magill, C. P., S. P. Jackson, and S. D. Bell. 2001. Identification of a conserved archaeal RNA polymerase subunit contacted by the basal transcription factor TFB. J. Biol. Chem. 276:46693-46696. [DOI] [PubMed] [Google Scholar]
- 43.Maillet, I., J. M. Buhler, A. Sentenac, and J. Labarre. 1999. Rpb4p is necessary for RNA polymerase II activity at high temperature. J. Biol. Chem. 274:22586-22590. [DOI] [PubMed] [Google Scholar]
- 44.Martignetti, J. A., and J. Brosius. 1995. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol. Cell. Biol. 15:1642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martignetti, J. A., and J. Brosius. 1993. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc. Natl. Acad. Sci. USA 90:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyao, T., J. D. Barnett, and N. A. Woychik. 2001. Deletion of the RNA polymerase subunit RPB4 acts as a global, not stress-specific, shut-off switch for RNA polymerase II transcription at high temperatures. J. Biol. Chem. 276:46408-46413. [DOI] [PubMed] [Google Scholar]
- 47.Naghashpour, M., and G. Dahl. 2000. Sensitivity of myometrium to CGRP varies during mouse estrous cycle and in response to progesterone. Am. J. Physiol. Cell Physiol. 278:C561-C569. [DOI] [PubMed] [Google Scholar]
- 48.Naghashpour, M., M. I. Rosenblatt, I. M. Dickerson, and G. P. Dahl. 1997. Inhibitory effect of calcitonin gene-related peptide on myometrial contractility is diminished at parturition. Endocrinology 138:4207-4214. [DOI] [PubMed] [Google Scholar]
- 49.Oliver, K. R., A. Wainwright, A. M. Kinsey, R. P. Heavens, D. J. Sirinathsinghji, and R. G. Hill. 1999. Regional and cellular localization of calcitonin gene-related peptide-receptor component protein mRNA in the guinea-pig central nervous system. Brain Res. Mol. Brain Res. 66:205-210. [DOI] [PubMed] [Google Scholar]
- 50.Orlicky, S. M., P. T. Tran, M. H. Sayre, and A. M. Edwards. 2001. Dissociable Rpb4-Rpb7 subassembly of RNA polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 276:10097-10102. [DOI] [PubMed] [Google Scholar]
- 50a.Peyroche, G., E. Levillain, M. Siaut, I. Callebaut, P. Schultz, A. Sentenac, M. Riva, and C. Carles. 2002. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl. Acad. Sci. USA 99:14670-14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenblatt, M. I., G. P. Dahl, and I. M. Dickerson. 2000. Characterization and localization of the rabbit ocular calcitonin gene-related peptide (CGRP)-receptor component protein (RCP). Investig. Ophthalmol. Vis. Sci. 41:1159-1167. [PubMed] [Google Scholar]
- 52.Rosenheck, S., and M. Choder. 1998. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol. 180:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruet, A., A. Sentenac, P. Fromageot, B. Winsor, and F. Lacroute. 1980. A mutation of the B220 subunit gene affects the structural and functional properties of yeast RNA polymerase B in vitro. J. Biol. Chem. 255:6450-6455. [PubMed] [Google Scholar]
- 54.Sadhale, P. P., and N. A. Woychik. 1994. C25, an essential RNA polymerase III subunit related to the RNA polymerase II subunit RPB7. Mol. Cell. Biol. 14:6164-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakurai, H., and A. Ishihama. 2001. Transcription organization and mRNA levels of the genes for all 12 subunits of the fission yeast RNA polymerase II. Genes Cells 6:25-36. [DOI] [PubMed] [Google Scholar]
- 56.Sakurai, H., H. Mitsuzawa, M. Kimura, and A. Ishihama. 1999. The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol. 19:7511-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sali, A., L. Potterton, F. Yuan, H. van Vlijmen, and M. Karplus. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23:318-326. [DOI] [PubMed] [Google Scholar]
- 58.Sentenac, A. 1985. Eukaryotic RNA polymerases. Crit. Rev. Biochem. 18:31-90. [DOI] [PubMed] [Google Scholar]
- 59.Sheffer, A., M. Varon, and M. Choder. 1999. Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4. Mol. Cell. Biol. 19:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 61.Shpakovski, G. V., J. Acker, M. Wintzerith, J.-F. Lacroix, P. Thuriaux, and M. Vigneron. 1995. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shpakovski, G. V., and E. K. Shematorova. 1999. Characteristics of the cDNA of the Schizosaccharomyces pombe rpa43+ gene: structural similarity of the Rpa43 subunit of RNA-polymerase I with the Rpc25 subunit of RNA-polymerase III. Bioorg. Khim. 25:791-796. [PubMed] [Google Scholar]
- 63.Stettler, S., N. Chiannilkulchai, S. Hermann-Le Denmat, D. Lalo, F. Lacroute, A. Sentenac, and P. Thuriaux. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239:169-176. [DOI] [PubMed] [Google Scholar]
- 64.Sutcliffe, J. G., R. J. Milner, J. M. Gottesfeld, and R. A. Lerner. 1984. Identifier sequences are transcribed specifically in brain. Nature 308:237-241. [DOI] [PubMed] [Google Scholar]
- 65.Takada, S., J. T. Lis, S. Zhou, and R. Tjian. 2000. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101:459-469. [DOI] [PubMed] [Google Scholar]
- 66.Thuillier, V., S. Stettler, A. Sentenac, P. Thuriaux, and M. Werner. 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 14:351-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Todone, F., P. Brick, F. Werner, R. O. Weinzierl, and S. Onesti. 2001. Structure of an archaeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex. Mol. Cell 8:1137-1143. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Z., and R. G. Roeder. 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11:1315-1326. [DOI] [PubMed] [Google Scholar]
- 69.Werner, F., J. J. Eloranta, and R. O. Weinzierl. 2000. Archaeal RNA polymerase subunits F and P are bona fide homologs of eukaryotic RPB4 and RPB12. Nucleic Acids Res. 28:4299-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Werner, M., N. Chaussivert, I. M. Willis, and A. Sentenac. 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268:20721-20724. [PubMed] [Google Scholar]
- 71.Werner, M., S. Hermann-Le Denmat, I. Treich, A. Sentenac, and P. Thuriaux. 1992. Effect of mutations in a zinc binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell. Biol. 12:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zawel, L., and D. Reinberg. 1995. Common themes in assembly and function of eukaryotic transcription complexes. Annu. Rev. Biochem. 64:533-561. [DOI] [PubMed] [Google Scholar]