Abstract
Cells normally restrict the nuclear export and expression of intron-containing mRNA. In many cell lines, this restriction can be overcome by inclusion of cis-acting elements, such as the Mason-Pfizer monkey virus constitutive transport element (CTE), in the RNA. In contrast, we observed that CTE-mediated expression from human immunodeficiency virus Gag-Pol reporters was very inefficient in 293 and 293T cells. However, addition of Sam68 led to a dramatic increase in the amount of Gag-Pol proteins produced in these cells. Enhancement of CTE function was not seen when a Sam68 point mutant (G178E) that is defective for RNA binding was used. Additionally, the effect of Sam68 was inhibited in a dose-dependent manner by coexpression of an activated form of the nuclear kinase Sik/BRK that hyperphosphorylated Sam68. RNA analysis showed that cytoplasmic Gag-Pol-CTE RNA levels were only slightly enhanced by the addition of Sam68, compared to a 60- to 70-fold increase in the levels of Gag-Pol protein expression. Thus, in this system, Sam68 functioned to enhance the cytoplasmic utilization of RNA containing the CTE. These results suggest that Sam68 may interact with specific RNAs in the nucleus to provide a “mark” that affects their cytoplasmic fate. They also provide further evidence of links between signal transduction and RNA utilization.
Sam68 (Src associated in mitosis 68) was originally identified as one of the major targets for the Src tyrosine kinase in mitosis (16, 63, 64). It is a member of the STAR (signal transduction and activation of RNA) family of proteins (65). The STAR proteins are RNA binding proteins that contain an hnRNP K homology domain flanked by regions that are required for high-affinity RNA binding (65). In addition to Sam68 and the closely related proteins SLM-1 and SLM-2 (10), this family includes the quaking proteins (11), Caenorhabditis elegans GLD-1 (15, 32), and the drosophila held out wings protein (68). Some of the STAR proteins appear to be translational regulators (30, 31, 55), whereas others have been proposed to affect alternative splicing (62).
Although the functions of Sam68 are still largely unknown, it has been shown to be a nuclear protein that localizes with splicing-associated factors in nuclear bodies (5, 26, 47). In addition, Sam68 has been proposed to play a role in cell cycle regulation (54) and a recent study indicates that Sam68 is a tumor suppressor (41). Sam68 has also been reported to have effects on human immunodeficiency virus (HIV) gene expression and replication (52, 59). Specifically, it was shown that overexpression of a truncated form of Sam68 inhibited HIV replication and that the wild-type Sam68 protein could substitute efficiently for the HIV Rev protein in assays using chloramphenicol acetyltransferase (CAT)-based reporter constructs that contain the HIV rev response element (RRE). On the basis of these findings, it has been suggested that Sam68 plays a role in nucleocytoplasmic RNA export and may function as a cellular Rev homologue (52).
Retroviral replication requires that intron-containing unspliced or incompletely spliced RNAs are efficiently exported to the cytoplasm of infected cells (21, 24). This occurs despite the restriction on export of RNA containing introns, which appears to normally prevent RNAs from exiting the nucleus until they are completely spliced (37). Complex retroviruses overcome this restriction through the use of cis-acting elements in the viral RNA (RRE in the case of HIV) in combination with virus-encoded trans-acting proteins (Rev in the case of HIV) (49). An interaction between the cis-acting element and the viral protein in the nucleus allows the RNA to be exported by specialized export pathways. In contrast, some of the simpler retroviruses have been shown to achieve export of unspliced RNA through the use of cis-acting RNA elements, termed constitutive transport elements (CTEs), that interact directly with cellular export proteins (24). The best characterized of the CTEs is present in the genome of Mason-Pfizer monkey virus (MPMV) (4, 12, 13). This CTE is 154 nucleotides (nt) in length and forms a distinct secondary stem-loop structure that interacts with the cellular protein Tap/NXF1 (19). This protein has also been proposed to play a role in general cellular mRNA export (29, 34). However, it is likely that additional cellular proteins are involved in the CTE-mediated export pathway. Sam68 has previously been suggested to have effects on CTE function (51). However, this has remained controversial since the initial studies reporting effects of Sam68 on Rev/RRE function concluded that Sam68 had no effects on CTE-mediated expression (52).
In this paper, we demonstrate that expression of protein from mRNA containing the CTE can be significantly increased in certain cell lines by coexpression of Sam68 and that the Sam68-mediated enhancement of expression is abrogated by coexpression of a constitutively active mutant form of the nuclear Src family Sik/BRK kinase that induces hyperphosphorylation of Sam68 (9). Furthermore, a direct analysis of RNA containing the CTE shows that Sam68 does not affect mRNA export directly but functions to alter the fate of RNA in the cytoplasm.
MATERIALS AND METHODS
Plasmid constructs and nomenclature.
To facilitate identification, all of the plasmids used in this study have been given index numbers in the form pHRXXXX. Several have been previously described. These are the subgenomic HIV type 1 reporter constructs pCMVGagPol-RRE (pHR354) and pCMVGagPol-CTE (pHR1361) (60); pCMVRev (pHR30) (58), a plasmid that expresses the HIV Rev protein; and pCMVSEAP (pHR1831) (12), a plasmid that expresses secreted alkaline phosphatase (SEAP) (1); pCMVTap (pHR2128) (20), a plasmid that expresses the Tap protein.
The plasmids expressing wild-type and mutant forms of mouse Sam68, pcDFSam68 (pHR2208), pcDHSam68ΔKH (pHR2212), and pcDHSam68(G178E) (pHR2210), have also been previously described (40). The plasmids expressing wild-type and mutant forms of the Sik/BRK kinase, pcDNA3-Sikwt (pHR2531), pcDNA3-Sik(Y447F) (pHR2533), and pcDNA3-Sik(K219M) (pHR2533), were kind gifts from Angela Tyner (9). The plasmids expressing c-Src proteins (pcDNA3-K-Src [pHR2228] and pcDNA3-SrcY527F [pHR2229]) (2) were kind gifts from Sally Parsons. The plasmids expressing HuR (14), hnRNPH (7), and hnRNPD (35) were kind gifts from Joan Steitz, Douglas Black, and Gary Brewer.
Cell lines and transient transfections.
293, 293T/17, and CMT3-COS cells were maintained in Iscove's minimal essential medium supplemented with 10% bovine calf serum. Qcl3 cells (8) were a kind gift from Bryan Cullen. They were maintained in M199 medium (Life Technologies) supplemented with 4% fetal bovine serum (HyClone Laboratories), 1% chicken serum (Gibco BRL), 10% tryptone phosphate (Life Technologies), and 1% dimethyl sulfoxide (Sigma Chemicals). Transient transfections of CMT3-COS and Qcl3 cells were performed by using modifications of the DEAE-dextran method as previously described (23). 293T/17 cells were transfected by using a calcium phosphate transfection protocol (17). All transfections were performed with the amounts of plasmids described in the figure legends.
p24 ELISA and SEAP quantitation.
Supernatants from transfected cells were collected at 65 to 72 h posttransfection, centrifuged in a microcentrifuge to remove residual cells and debris, and stored at −20°C until assayed. p24 expression levels were determined with either a commercial enzyme-linked immunosorbent assay (ELISA) kit (NEN) or an ELISA protocol involving a p24 monoclonal antibody and pooled human anti HIV immunoglobulin G (67). The p24 antibody was obtained from the AIDS Research and Reference Reagent Program and was contributed by Bruce Chesebro (National Institute of Allergy and Infectious Diseases-Rocky Mountain Laboratories). SEAP activity in the supernatants was measured with the Phospha-Light Chemiluminescent Reporter Kit (Tropix).
Western blot analysis.
The Western blot analysis was performed essentially as previously described (22). Briefly, proteins were transferred to an Immobilon-P membrane (Millipore) and the membrane was blocked in 5% milk. For detection of HIV Gag proteins, blots were probed with a mouse anti-p24 monoclonal antibody (183-H12-5C) from the National Institutes of Health AIDS Research and Reference Reagent Program. After washing, the membranes were incubated with rabbit anti-mouse immunoglobulin G and 10 μCi of I125-labeled protein A per ml. For detection of Sam68, blots were probed with an anti-Sam68 monoclonal antibody (SC-333; Santa Cruz Biotechnology, Inc.). These blots were probed with an alkaline phosphatase-conjugated anti-mouse secondary antibody and developed by using 0.33 mg of Nitro Blue Tetrazolium per ml and 0.0825 mg of 5-bromo-4-chloro-3-indolylphosphate per ml.
35S labeling and immunoprecipitations.
293T or CMT3/COS cells were preincubated for 1 h in Met/Cys-free Dulbecco’s modified Eagle medium (Gibco/Invitrogen) containing 10% predialyzed bovine calf serum and then incubated at 37°C in labeling medium containing 200 μCi of [35S]Met/Cys Tran35S-label (ICN) for 3 or 24 h. Cells were lysed in 150 mM NaCl-10 mM Tris HCl-1% NP-40-1% deoxycholate. Lysates were cleared by centrifugation, and protein concentration was determined by colorimetric analysis (Bio-Rad). Sam68 was immunoprecipitated from 200 μg of total cell lysates with 1 μg of mouse anti-Sam68 monoclonal antibody (SC-333; Santa Cruz) and 30 μl of 50% protein G-Sepharose beads (Amersham Biosciences) after incubation at 4°C for 3 h. Samples were washed two times with 500 μl of lysis buffer and one time with 500 μl of 10 mM Tris HCl-100 mM NaCl-1 mM EDTA (pH 8.0) (TNE) with centrifugation at 500 × g for 3 min, resuspended in 1.5× sodium dodecyl sulfate (SDS) sample buffer, boiled for 5 min, centrifuged briefly, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 12% gel.
32P labeling and tryptic peptide mapping.
32P labeling and tryptic peptide mapping were performed as previously described (3), with minor modifications. Thirty-six hours after transfection with a plasmid expressing a FLAG-tagged version of Sam68 (pcDFSam68 [pHR2208]), 293T cells were washed two times in labeling medium (10% predialyzed bovine calf serum in Dulbecco's medium lacking sodium pyruvate and sodium phosphate). Cells were then radiolabeled for 8 h at 37°C in 3 ml of labeling medium containing 2.5 to 3.0 mCi of 32P (ICN), washed three times in phosphate-buffered saline (PBS) at 4°C, and resuspended in 500 μl of 1× RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 1.5 mM MgCl2) containing 1× protein phosphatase inhibitor cocktails I and II and protease inhibitor cocktail as suggested by the supplier (Sigma). A 500-μl volume of 2× NET buffer (40 mM Tris HCl [pH 7.4], 150 mM NaCl, 1% NP-40) was then added to lyse the cells. Protein concentrations were determined by colorimetric analysis (Bio-Rad). Sam68 was immunoprecipitated by a 3-h incubation at 4°C with 100 μl of a 50% agarose bead-conjugated anti-FLAG antibody slurry (Sigma). The beads were then washed two times with 1 ml of TBS (50 mM Tris HCl [pH 7.4], 150 mM NaCl) at 4°C. The pellets were then resuspended in 100 μl of 1.5× SDS sample buffer and boiled for 5 min. The beads were then spun down, and the supernatant was analyzed on an SDS-12% polyacrylamide gel. After electrophoresis, proteins in the gel were transferred to an Immobilon-P (Millipore, Bedford, Mass.) membrane by electroblotting at 15 V for 4 h. The portions of the membrane containing the Sam68 protein were then excised and treated with 5 ml of blocking solution (0.5% polyvinylpyrrolidone 360, 100 mM acetic acid) at 37°C for 30 min. The membrane pieces were treated with trypsin, and then the resulting peptides were collected and subjected to two-dimensional tryptic peptide mapping on thin-layer chromatography plates. The first dimension consisted of electrophoresis at pH 1.9. It was followed by chromatography in the second dimension. The procedures used to prepare and analyze the peptides have been previously described (3).
RNA fractionation and Northern blot analysis.
The methods used for nuclear and cytoplasmic RNA extraction, poly(A) mRNA selection, and Northern blot analysis were previously described (22, 25). Cells were harvested 65 h posttransfection. [32P]CTP-labeled DNA probes were generated by using the T7 Quickprime Kit (Pharmacia). The gagpol-specific probe was generated by using the SacI-BglII (nt 682 to 2093) fragment of the BH10 HIV type 1 proviral clone. The SEAP-specific probe was generated by using the BamHI fragment of the human SEAP-encoding gene (nt 213 to 1698). Visualization and quantitation of Northern blots were performed with a Molecular Dynamics PhosphorImager and ImageQuant analysis software.
RNA in situ hybridization.
Hybridization was performed with a digoxigenin (DIG)-labeled oligonucleotide probe using 3 × 106 293T cells that were cotransfected with 15 μg of a GagPol RRE or CTE reporter plasmid with or without 3 μg of pCMVRev(pHR30) or pcDFSam68(pHR2208). Twenty-four hours posttransfection, cells were seeded onto 0.1% gelatin-coated coverslips and incubated for another 48 h. Cells were then fixed with 4% paraformaldehyde-PBS and stored in 70% ethanol at 4°C until use. Before in situ hybridization, the fixed cells were dehydrated with a graded ethanol series (70, 90, and 100%) and subsequently treated with 100, 90, and 70% ethanol. Cells were then washed in PBS, incubated in 0.5% Triton X-100-PBS for 10 min, and treated with 0.2 N HCl for 20 min at room temperature. The cells were again washed with PBS for 10 min before use.
Prehybridization was performed at 37°C for 1 h in 100 μl of hybridization buffer (50% deionized formamide, 10% dextran sulfate, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10 μg of tRNA per ml, 0.1 mg of bovine serum albumin per ml, 0.1 mg of herring sperm DNA per ml).
The hybridization probe was prepared by labeling a gag-specific oligonucleotide (corresponding to the HIV gag sequence spanning positions 1606 to 1629 5′-GCTGGTAGGGCTATACATTCTTAC-3′ in the antisense orientation) by using 3′-end DIG-dUTP (Roche) in accordance with the manufacturer's protocol. The labeled oligonucleotide was resuspended at 1 to 4 ng/μl in fresh hybridization buffer, heated to 80°C for 10 min, and cooled on ice for 5 min. The coverslips containing fixed cells were immersed in 30 μl of fresh hybridization buffer, denatured at 80°C for 5 min, and incubated overnight at 42°C. Cells were washed four times with 50% deionized formamide-2× SSC for 15 min each time at 37°C. Hybrids were detected by using anti-DIG antibodies conjugated to fluorescein isothiocyanate (FITC). For detection, antibodies were incubated with cell samples for 1 h at 37°C in 1% blocking reagent containing anti-DIG-FITC (1/100 dilution; Roche). Cells were then incubated four times more with 0.15 M NaCl-0.1 M Tris-HCl (pH 7.4) for 15 min each time at room temperature to remove unbound antibodies. Coverslips were mounted in antibleach medium (Vector Lab) and stored in the dark at 4°C. The hybridization signals were visualized by a LSM 5 Pascal Confocal Microscope (Zeiss).
RESULTS
Transfection of a plasmid expressing Sam68 greatly increases p24 expression in 293 and 293T cells from HIV Gag-Pol constructs that utilize the CTE.
In several previous studies, we utilized HIV Gag-Pol reporter constructs to study the requirements for expression and nucleocytoplasmic export of intron-containing RNA in mammalian cells (4, 20, 57, 58, 60, 61). These reporters contain the complete coding region for HIV Gag and Pol, and the expression of these proteins leads to the production of HIV virus-like particles that are efficiently secreted into the medium of transfected cells.
Since the mRNA that expresses the Gag-Pol proteins contains a complete intron, it is retained in the nucleus in the absence of signals that overcome the cellular restriction to the export of such RNAs. We initially showed that the presence of an HIV RRE in cis in the Gag-Pol mRNA, in conjunction with supplying the Rev protein in trans, led to efficient export of the Gag-Pol RNA to the cytoplasm and subsequent expression of large amounts of HIV p24 in the medium of transfected cells (57). Subsequently, we showed that the insertion of the MPMV CTE could efficiently substitute for the requirement for RRE/Rev in these reporter plasmids in both the HeLa and COS cell lines (4). In contrast, we repeatedly observed that the Gag-Pol reporter plasmids containing the CTE gave very low levels of p24 activity in both 293 and 293T cells.
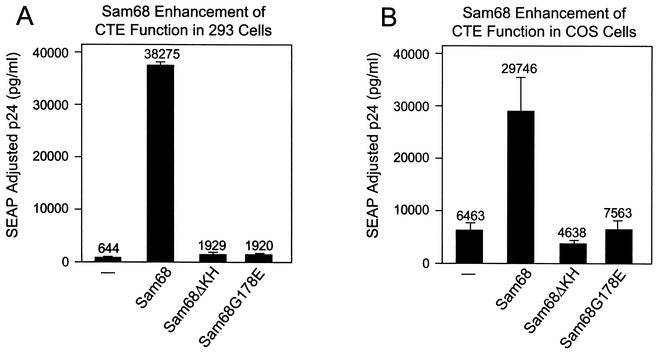
In an effort to try to complement the apparent defect in CTE function in the 293 cell lines, we cotransfected these cells with the Gag-Pol-CTE reporter and several different constructs expressing known RNA binding proteins with proposed roles in RNA metabolism. In all of these transfections, we included a plasmid that expressed SEAP (pCMVSEAP) as an internal control. Most of the cotransfected plasmids were unable to specifically increase p24 expression. Among the proteins tested were HuR, hnRNPD, and hnRNPH (data not shown). However, as shown in Fig. 1A, expression of mouse Sam68 led to a 60-fold increase in the amount of p24 expressed. In contrast, very little enhancement was observed in transfections with two different Sam68 mutants (Sam68ΔKH and Sam68G178E). Sam68ΔKH has a large deletion in the RNA binding KH domain, whereas Sam68G178E corresponds to a mutation in the KH domain of the C. elegans GLD-1 protein that results in a loss of function and development of an oncogenic phenotype (germ line tumor). These results indicate the importance of the RNA binding domain of Sam68 for the observed effects and demonstrate that a single point mutation corresponding to a mutation with a known phenotype in GLD-1 is sufficient to abrogate the enhancement. Similar results were obtained with both 293 and 293T cells.
FIG. 1.
Sam68 enhances CTE function in 293 and COS cells. Five micrograms of the pCMVGag-Pol-CTE reporter plasmid (pHR1361) and 0.25 μg of pCMVSEAP (pHR1831) were transfected into 100-mm-diameter dishes of either 293 cells (A) or COS cells (B) together with 1 μg each of pcDFSam68 (pHR2208), pcDHSam68ΔKH (pHR2212), pcDHSam68(G178E) (pHR2210), or pcDNA3 with no insert (pHR2214). Supernatants were collected at 72 h posttransfection, and p24 and SEAP levels in each sample were determined. p24 values were then adjusted for variations in SEAP levels. The data represent the average of two independent transfections.
To analyze the effect of additional Sam68 expression in COS cells, we performed a cotransfection experiment with Sam68 similar to the one described above. Figure 1B shows that an effect of Sam68 on CTE function was also observed in COS cells. However, basal levels of p24 expression were significantly higher than that observed in 293 or 293T cells and the enhancement seen in this cell line was much more modest (about fivefold). Again, the mutant forms of Sam68 failed to significantly enhance p24 expression. Mutated versions of the CTE that worked less efficiently showed the same differential enhancement by Sam68 in the two cell lines, indicating that the lower enhancement seen in COS cells was not simply the result of the system being saturated for p24 production (data not shown).
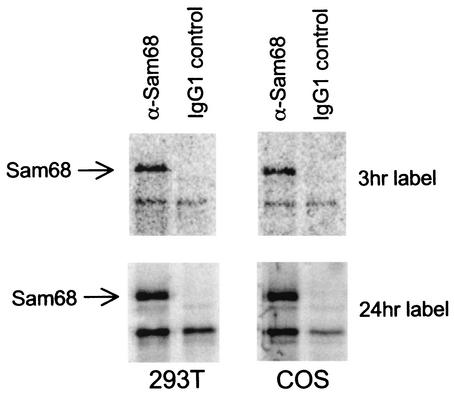
To test whether there was a difference in endogenous expression of Sam68 between 293 or 293T cells and COS cells that could explain these results, nonconfluent cultures of CMT3-COS cells and 293T cells were metabolically labeled with [35S]Met/Cys Tran35S-label for either 3 or 24 h. Sam68 protein was then immunoprecipitated from extracts of these cells by using an anti-Sam68 monoclonal antibody. After electrophoresis of the immunoprecipitates on an SDS-containing polyacrylamide gel, Sam68 was detected by PhosphorImager analysis. Figure 2 shows that similar amounts of Sam68 were detected in 293T and COS cells after labeling of either for 3 or 24 h. These results indicate that there were no major differences in either the rate of synthesis or the levels of Sam68 in these cells that could explain the differences in CTE function.
FIG. 2.
Comparison of endogenous Sam68 produced in 293T and COS cells. Sam68 was immunoprecipitated with anti-Sam68 monoclonal antibodies from lysates of 293T and COS cells metabolically labeled with [35S]Met/Cys Tran35S-label for either 3 or 24 h. Protein was resolved by SDS-PAGE and visualized by PhosphorImager analysis. IgG1, immunoglobulin G1.
Sam68 enhances expression from an integrated Gag-Pol-CTE reporter in 293T cells and synergizes with Tap in quail cells.
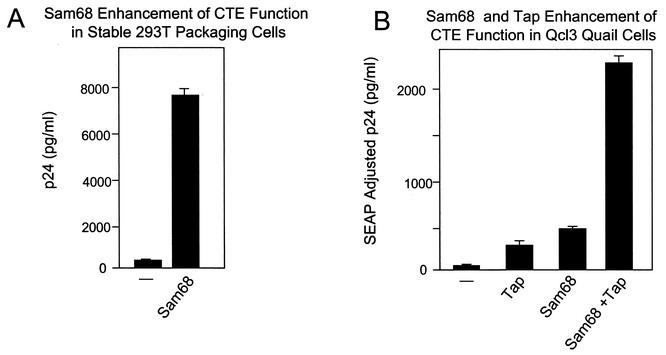
We also analyzed the effects of Sam68 on CTE expression in a 293T-based cell line (B2.10) where the CTE-containing Gag-Pol reporter was stably integrated. Figure 3A shows that a very low basal level of p24 is secreted into the supernatant of this cell line. Transfection of 1 μg of the plasmid expressing the Sam68 wild-type protein produced a large increase in p24 production, demonstrating that Sam68 is also able to dramatically increase CTE function in cells where the CTE reporter is integrated into the genome.
FIG. 3.
Sam68 enhances CTE function in a stable 293T packaging cell line and Qcl3 quail cells. (A) Dishes (100-mm diameter) of B2.10 cells were transfected with 1 μg of pcDFSam68 (pHR2208) or pcDNA empty vector (pHR2214). B2.10 is a cell line derived from 293T cells by stable transfection of pCMVGag-Pol-CTE (pHR1361) and pCMVEnv-CTE (pHR1374). p24 levels in the supernatant were determined at 72 h posttransfection. The data represent the average of two independent transfections. (B) Dishes (100-mm diameter) of Qcl3 quail fibroblast cells were transfected with 5 μg of the pCMVGag-Pol-CTE reporter (pHR1361) and 0.25 μg of pCMVSEAP (pHR1831) together with 2 μg of pCMVTap (pHR2128), 1 μg of pcDFSam68 (pHR2208), or both. At 72 h posttransfection, supernatants were collected and analyzed for p24 and SEAP expression. The data represent the average of two independent transfections.
It has previously been demonstrated that Qcl3 quail cells do not support CTE function, and this has been attributed to a failure of the quail Tap protein to function in conjunction with the CTE element (33). To analyze whether Sam68 could enhance expression from the CTE in these cells, we transfected Qcl3 cells with the Gag-Pol-CTE reporter either alone or in cotransfections with plasmids expressing Sam68 and/or human Tap. The results of this experiment are shown in Fig. 3B. Very low basal levels of p24 were observed with the reporter plasmid alone, confirming the previous studies that showed inefficient CTE function in these cells. Surprisingly, coexpression of Sam68 alone stimulated p24 expression to the same extent as coexpression of Tap and a synergistic effect was obtained when both Sam68 and Tap were coexpressed together with the Gag-Pol-CTE reporter. Thus, the addition of human Tap to Qcl3 cells is not absolutely essential for CTE function.
Expression of constitutively active Sik/BRK abrogates Sam68 enhancement of CTE function.
Sam68 has been shown to be a major target for Src and related kinases. Thus, we decided to analyze the effects of Src on Sam68 enhancement of CTE function. To do this, we transfected 293 cells with plasmids expressing c-Src proteins that were either kinase dead or constitutively activated, respectively (pcDNA3-K-Src and pcDNA3-SrcY527F). We did not observe any effects of either of these plasmids on CTE function in 293 cells, either in the presence or in the absence of a plasmid expressing Sam68 (data not shown).
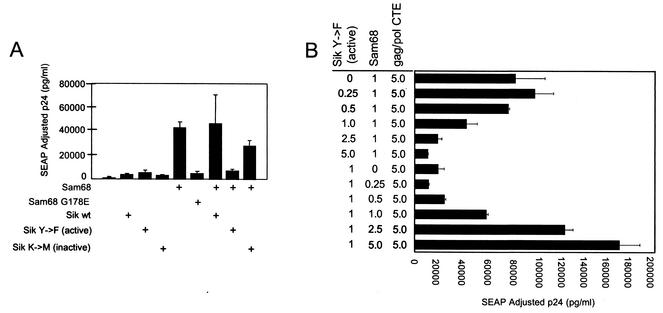
Recent studies have indicated that Sam68 is also a substrate for the nuclear Src family kinase known as Sik/BRK (9). To investigate whether expression of Sik/BRK would affect Sam68-mediated enhancement of CTE function in 293T cells, cells were transfected with the Gag-Pol-CTE vector with or without Sam68 in the presence of plasmids expressing three different Sik/BRK proteins [wild-type, constitutively active mutant Sik/BRK(Y447F), or kinase-inactive mutant Sik/BRK(K219M)]. These plasmids were a kind gift from Angela Tyner. Neither of the Sik/BRK plasmids had any effect on p24 expression from the Gag-Pol-CTE plasmid in the absence of cotransfected Sam68 (Fig. 4A). However, in the Sam68 cotransfections, a significant decrease in the level of p24 expression was observed in the presence of constitutively active Sik/BRK(Y447F). In contrast, neither the wild-type nor the K219M mutated form of the protein had any significant effect on Sam68 enhancement. These results support the notion that Sam68 enhancement of CTE function can be regulated by the Sik/BRK kinase.
FIG. 4.
Coexpression of the constitutively active Sik(Y447F) mutant protein with Sam68 inhibits Sam68-mediated enhancement of CTE function. (A) Five micrograms of pCMVGag-Pol-CTE reporter (pHR1361) and 0.25 μg of pCMVSEAP (pHR1831) were transfected into 293T cells with 1 μg of a plasmid that expresses either the wild-type (pcDNA3-Sikwt, pHR2531), the kinase-inactive [pcDNA3-Sik(K219M), pHR2533], or the constitutively active [pcDNA3Sik(Y447F), pHR2533] form of Sik. These plasmids were expressed either alone or together with 1 μg of pcDSam68 (pHR2208). At 72 h posttransfection, supernatants were collected and analyzed for p24 and SEAP expression. The data represent the average of two independent transfections. (B) Increasing amounts of pcDFSam68 or pcDNA3-Sik(Y447F) were cotransfected with 5 μg of pCMVGag-Pol-CTE reporter (pHR1361) and 0.25 μg of pCMVSEAP (pHR1831) plasmid into 293T cells. At 72 h posttransfection, supernatants were collected and analyzed for p24 and SEAP expression. The data represent the average of two independent transfections.
To further investigate the effect of Sik(Y447F) on Sam68-mediated enhancement of CTE function, we performed a dose-response experiment in which the ratio of the plasmid expressing Sik(Y447F) to the plasmid expressing Sam68 was varied. The result of this experiment is shown in Fig. 4B. The top part of the figure shows that increasing amounts of Sik(Y447F) reduced p24 expression in a dose-responsive fashion for a given amount of Sam68 (1 μg of transfected plasmid). The lower part of the figure illustrates that the inhibitory effect seen for a given amount of Sik(Y447F) (1 μg of transfected plasmid) could readily be titrated out by increasing the levels of the Sam68 plasmid. These results demonstrate that the enhancement function of Sam68 can be modulated by Sik(Y447F) in a dose-dependent manner and thus lend further support to the hypothesis that Sik serves to functionally regulate the effects of Sam68 in this system.
Sam68 is hyperphosphorylated in the presence of Sik(Y447F) in 293T cells.
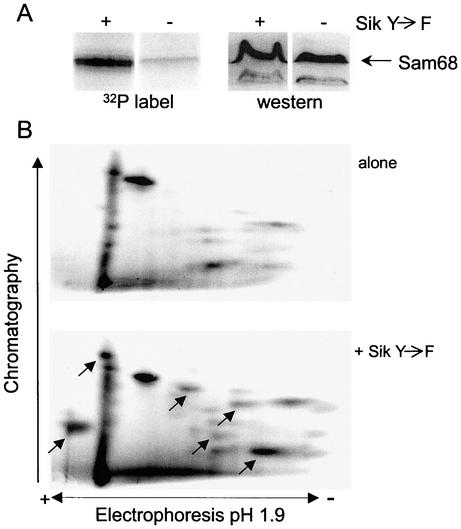
It has previously been shown that the function and RNA binding activity of Sam68 are regulated through phosphorylation by Src (6, 66). In addition, a recent report showed that Sam68 is a substrate for Sik/BRK in vivo and that this phosphorylation also inhibits its RNA binding activity (9). To directly analyze if Sik(Y447F) affected Sam68 phosphorylation in our system, we transfected 293T cells with pcDFSam68 in the absence or presence of the plasmid expressing Sik(Y447F) and labeled these cells with [32P]orthophosphate. Sam68 was immunoprecipitated from cellular extracts by using anti-FLAG agarose beads, and proteins eluted from these beads were separated by SDS-PAGE (Fig. 5A). Similar amounts of Sam68 were detected in the absence or presence of Sik(Y447F) by Western blot analysis (Fig. 5A, right side). In contrast, the amount of 32P label detected in this protein was dramatically increased in the presence of Sik(Y447F) (Fig. 5A, left side). To further analyze how Sik affected Sam68 phosphorylation, we performed a two-dimensional phosphopeptide analysis of trypsin-digested Sam68 (Fig. 5B). Several novel phosphopeptides were detected in the presence of Sik(Y447F), indicating that Sam68 is phosphorylated at multiple novel sites in response to the expression of this kinase.
FIG. 5.
Expression of the Sik(Y447F) mutant increases phosphorylation of Sam68 at multiple residues. 293T cells were transfected with plasmids expressing Flag-Sam68 either alone or with a plasmid expressing Sik(Y447F) and labeled with [32P]orthophosphate. (A, left side) 32P analysis of immunoprecipitated Sam68. Sam68 was immunoprecipitated from transfected cells by using an agarose bead-conjugated anti-FLAG antibody. Proteins were resolved by SDS-PAGE, and 32P incorporation was visualized by PhosphorImager analysis. (A, right side) Western blot analysis of Sam68 in the absence or presence of Sik(Y447F). Proteins from whole-cell lysates were resolved by SDS-PAGE and transferred to Immobilon. The blot was probed with an anti-Sam68 monoclonal antibody, and Sam68 was visualized by using an alkaline phosphatase-conjugated anti-mouse secondary antibody. (B) Two-dimensional phosphopeptide analysis of Sam68 expressed in the presence or absence of Sik(Y447F). Tryptic peptide analysis was carried out on Sam68 immunoprecipitated from 32P-labeled 293T cells by using an agarose bead-conjugated anti-FLAG antibody. The peptides were resolved in two dimensions, first by electrophoresis in pH 1.9 buffer and second by chromatography in phosphochromatography buffer. Labeled peptides were visualized by PhosphorImager analysis. Arrows indicate peptides that appeared de novo or increased in intensity in the presence of Sik(Y447F).
Sam68-mediated enhancement of CTE function is a consequence of increased cytoplasmic RNA utilization, not increased RNA export.
To address the mechanism by which Sam68 enhances CTE function, 293T cells were transfected with the Gag-Pol-CTE reporter plasmid in the presence or absence of the plasmid expressing Sam68. As controls, cells were also transfected with a Gag-Pol plasmid containing the RRE either alone or in cotransfections with a plasmid expressing the Rev protein. We have previously shown that Rev increases the levels of cytoplasmic Gag-Pol-RRE RNA by at least 10-fold in these cells as measured by Northern blot analysis (20).
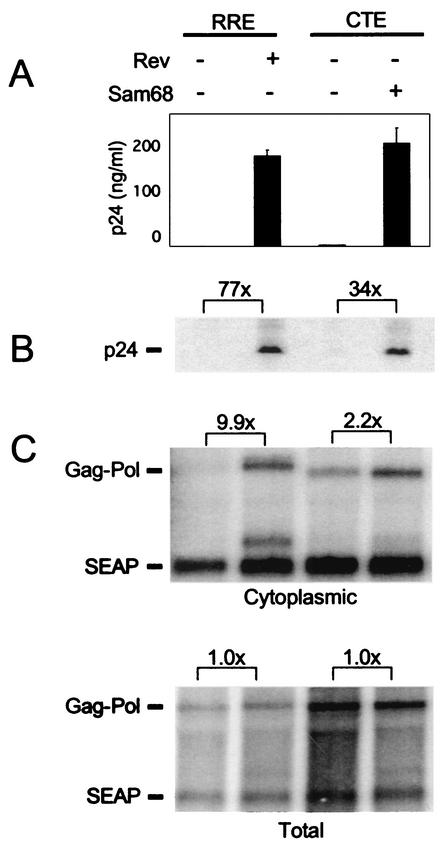
Total and cytoplasmic RNAs were prepared from the transfected cells, and Northern blot analyses were performed. In addition, cells were harvested for determination of intracellular p24 levels and supernatant medium was collected for the measurement of secreted p24 and SEAP. The results of these analyses are shown in Fig. 6. As expected, very low levels of p24 were expressed from the Gag-Pol-RRE plasmid alone, whereas large amounts were expressed in response to Rev both in the medium and intracellularly (panels A and B). With the Gag-Pol-CTE plasmid, very little p24 was produced when it was transfected alone and Sam68 expression again dramatically increased the p24 levels in the medium of transfected cells (120-fold; panel A). A significant increase in intracellular p24 was also observed (34-fold; panel B).
FIG. 6.
Analysis of p24 and Gag-Pol RNA in 293T cells. 293T cells were transfected with the reporter plasmids pCMVGag-Pol-RRE (pHR354) or pCMVGag-Pol-CTE (pHR1361) and pCMVSEAP (pHR1831) in the presence or absence of Rev (pHR30)- and/or Sam68 (pHR2208)-expressing plasmids essentially as described in the legend to Fig. 1. (A) Supernatants were harvested at 72 h posttransfection, and p24 levels were analyzed by ELISA. (B) Cells were harvested at 72 h and analyzed by Western blot assay. The Western blot analysis was performed on whole-cell lysates prepared from transfected cells with an anti-p24 primary monoclonal antibody, a rabbit anti-mouse secondary antibody, and 125I-coupled protein A for detection. Brackets show the fold difference in p24 levels between indicated lanes. (C) Northern blot analysis of cytoplasmic and total poly(A)+ RNA isolated from 293T cells 60 h after transfection. Gag-Pol- and SEAP-specific mRNAs were detected by using specific radiolabeled DNA probes. The blot was analyzed with a Molecular Dynamics PhosphorImager and ImageQuant software. Brackets show the fold differences in the levels of the Gag-Pol RNA bands between the indicated lanes after adjustment for variations in SEAP RNA levels.
The RNA analysis demonstrated that Rev caused a nearly 10-fold specific increase in the levels of cytoplasmic Gag-Pol-RRE RNA (panel C), consistent with our previous results and the well-established role of Rev in nucleocytoplasmic export of RRE-containing RNA (20, 49). In contrast, cytoplasmic Gag-Pol-CTE RNA levels were only slightly increased in response to expression of Sam68 (panel C). In both cases, total Gag-Pol RNA levels were unaffected. Thus, although little or no p24 was expressed from the Gag-Pol-CTE plasmid in the absence of Sam68 (panel A), substantial amounts of Gag-Pol-CTE RNA were detected in the cytoplasm, even in the absence of this protein (panel C). Taken together, the results of the experiments depicted in Fig. 6 show that the main effect of Sam68 is not to increase nuclear export of the Gag-Pol mRNA, indicating that the enhancement of p24 production must be through a different mechanism. The fact that a twofold increase in cytoplasmic RNA levels led to a 34- to 120-fold increase in p24 protein expression suggested that Sam68 functions to increase the utilization of CTE-containing RNA in the cytoplasm.
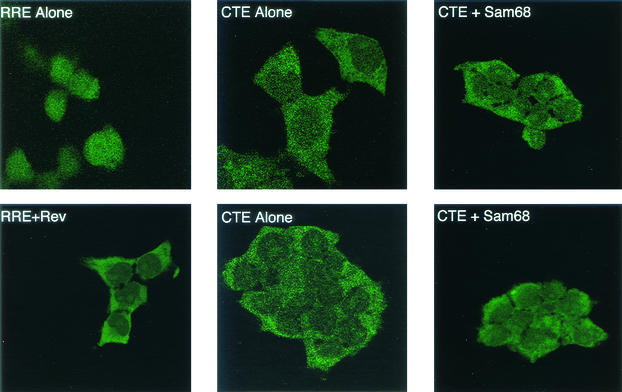
To further analyze the localization of Gag-Pol-CTE RNA in the absence or presence of Sam68, we performed an RNA in situ hybridization experiment with transfected 293T cells. Cells expressing Gag-Pol-RRE RNA with or without Rev coexpression were used as controls in this experiment. The results of this experiment are shown in Fig. 7. As expected, cells transfected with the Gag-Pol-RRE plasmid alone showed only nuclear fluorescence (upper left panel). In contrast, the fluorescence was almost exclusively cytoplasmic in the presence of Rev (lower left panel). This is consistent with previous studies and the Northern blot results presented above and confirms that Rev is essential for the cytoplasmic appearance of Gag-Pol-RRE RNA. In cells transfected with pCMVGag-Pol-CTE alone, a signal was observed mainly in the cytoplasm and the localization did not change visibly in the presence of Sam68 (Fig. 7, middle and right panels). In fact, the localization and intensity of the signal observed in the cells expressing Gag-Pol-CTE RNA both in the absence and in the presence of Sam68 were very similar to those observed with Gag-Pol-RRE in the presence of Rev. These results confirm that Sam68 has no major effect on RNA export in this system and support the hypothesis that Sam68 functions to enable efficient translation of the RNA once it reaches the cytoplasm.
FIG. 7.
In situ hybridization of Gag-Pol-CTE RNA in cells with or without Sam68 coexpression. Cells were transfected with plasmids expressing Gag-Pol-RRE RNA (with or without Rev) or Gag-Pol-CTE RNA (with or without Sam68), as indicated. At 72 h after transfection, cells were fixed and hybridized to an antisense DIG-labeled oligonucleotide probe specific for a sequence within gag. Hybridization was detected with an anti-DIG antibody conjugated to FITC and visualized by LSM5 Pascal confocal microscopy (Zeiss).
Sam68 has no major effect on Rev/RRE-mediated RNA export and expression in 293 cells.
In previous reports, Wong-Staal and coworkers suggested that Sam68 functions in RNA export and that this protein is a cellular Rev homologue (52, 53). Those studies used a reporter construct that contained the RRE in conjunction with the cat gene to measure Rev function, and it was shown that expression of Sam68 alone could achieve expression of cat from the RRE reporter that was 50% of that obtained with Rev. That group has also reported that expression of Sam68 and that of Rev have a synergistic effect on expression from the CAT-RRE plasmid. However, none of these studies directly measured whether Sam68 promoted RNA export of the RRE-containing RNA.
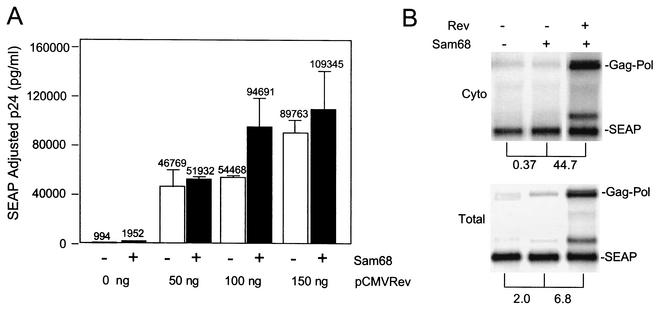
Since the results described above indicate a role for Sam68 in promoting protein translation rather than RNA export, we decided to perform additional experiments to further analyze the role of this protein in Rev/RRE function. To this end, we examined p24 production and RNA expression from a Gag-Pol-RRE reporter construct that required Rev for expression of p24. In this experiment, the Gag-Pol-RRE reporter plasmid was transfected with or without a plasmid expressing Sam68 and increasing amounts of a plasmid expressing Rev. The results are presented in Fig. 8. As expected, very little p24 expression was observed when the RRE-containing plasmid was transfected alone into 293 cells in the absence of Rev (Fig. 8A). Addition of Sam68 alone increased p24 production about twofold. In other experiments, increases in p24 production of up to eightfold were observed (data not shown), indicating that Sam68 is indeed able to enhance expression in constructs containing the RRE. However, in every transfection, the increases were small compared to the 45- to 90-fold increase that was observed with Rev. As shown in Fig. 8A, the combination of Sam68 and Rev resulted in little increase in p24 levels beyond that mediated by Rev alone.
FIG. 8.
Effect of Sam68 on Rev/RRE function. (A) Five micrograms of pCMVGag-Pol-RRE (pHR354) and 0.25 μg of pCMVSEAP (pHR1831) were transfected into 100-mm-diameter dishes of 293 cells with the indicated amounts of pCMVRev (pHR30) with or without 1 μg of pcDFSam68 (pHR2208). p24 analysis was carried out as described in the legend to Fig. 1. (B) Dishes (150-mm diameter) of 293T cells were transfected with 20 μg of pCMVGag-Pol-RRE and 5 μg of pCMVSEAP in the presence or absence of 5 μg of pCMVRev and/or pcDFSam68. Northern blot analysis was performed on cytoplasmic (Cyto) and total poly(A)+ mRNA isolated at 48 h after transfection and detected with a radiolabeled gag/pol-specific probe. Gag-Pol- and SEAP-specific mRNAs were detected by using specific radiolabeled DNA probes. The blot was analyzed with a Molecular Dynamics Phosphorimager and ImageQuant software. Brackets show the fold differences in the levels of the Gag-Pol RNA bands between the indicated lanes after adjustment for variations in SEAP RNA levels.
To directly determine effects of Rev and Sam68 on RNA export in conjunction with the RRE reporter, total and cytoplasmic RNA fractions were prepared as described above from cells transfected with different combinations of the Gag-Pol-RRE plasmid and plasmids expressing Sam68 and Rev. One of these experiments is shown in Fig. 8B. As expected, only a very small amount of Gag-Pol-RRE RNA was detected in the cytoplasmic fraction from cells transfected with the Gag-Pol-RRE reporter alone (Fig. 8B). Cotransfection with Sam68 did not increase these levels (in this particular experiment, Gag-Pol RNA levels actually decreased somewhat). In other experiments, no effect was seen (data not shown). In contrast, addition of Rev increased the cytoplasmic levels of Gag-Pol-RRE RNA more than 40-fold in the experiment shown in Fig. 8B.
When total RNA was examined, addition of Sam68 increased the levels of Gag-Pol-RRE RNA slightly (in other experiments, no effect was seen [data not shown]). In contrast, total RNA levels were increased when Rev was added. It has previously been shown (42, 56) that Rev expression frequently increases total RNA levels. This is presumed to reflect an increased stability of the RNA in the cytoplasm compared to the nucleus, where it is retained in the absence of the Rev protein. However, even when stability differences are taken into account, the specific increase in cytoplasmic accumulation in the presence of Rev was about sevenfold, compared to no increase when only Sam68 was expressed. Thus, these experiments fail to support the notion that Sam68 promotes efficient export of RNA containing the RRE, as Rev does. Rather, these results support the hypothesis that Sam68 also in this case functions to enhance translation of the RNA once it reaches the cytoplasm.
DISCUSSION
The results presented here clearly demonstrate that Sam68 can act in conjunction with the MPMV CTE to mediate efficient expression of proteins expressed from RNA containing a complete intron. However, our results indicate that Sam68 exerts its main effect on CTE function at the cytoplasmic level rather than at the level of export out of the nucleus. Although cytoplasmic levels of Gag-Pol-CTE RNA were only marginally increased by expression of Sam68, protein levels were increased 50-fold or more. The mechanism of this enhancement is not known. However, it has been well established, in several systems, that cytoplasmic mRNAs can be translationally masked and that unmasking of these mRNAs can increase translation up to 100-fold without an increase in the levels of mRNA (45, 50). Several of the Sam68-related STAR proteins (e.g., GLD-1 and Quaking) have been shown to be involved in translational regulation (30, 55). Furthermore, Sam68 is specifically relocalized to the cytoplasm in poliovirus-infected cells, where it interacts with the poliovirus polymerase. For this reason, Sam68 has been proposed to potentially play a role in poliovirus replication which is strictly cytoplasmic (46).
There is also accumulating evidence that the nuclear history of an mRNA can affect its subsequent fate in the cytoplasm, suggesting that nuclear and cytoplasmic processes are coupled (43). Several recent studies have shown that protein “marks” deposited at mRNA exon-exon junctions after splicing facilitate the export of the RNA and also influence its cytoplasmic fate (44). As an example, exon-exon marks downstream of termination codons can elicit nonsense-mediated decay as the RNA reaches the cytoplasm (38, 39). It thus appears that cytoplasmic mechanisms are in place to restrict the utilization of aberrant RNAs. In the Xenopus oocyte system, it has also been shown that the presence and position of introns in mRNAs dramatically affect the ability of these RNAs to be translated (45). Intron-containing RNAs are usually restricted from exiting the nucleus (37). This restriction is likely in place to limit the export of aberrant RNAs that could give rise to defective or transdominant negative proteins. In contrast, intron-containing RNAs are efficiently exported if they contain elements such as the CTE (12). The requirement for Sam68 may signify that these RNAs encounter additional restrictions once they reach the cytoplasm. Multiple restrictions to expression of RNA containing introns could reflect the importance of keeping such RNAs from being translated if they do not have the appropriate nuclear history.
Although recent studies suggest that Sam68 plays an important role in cell cycle regulation (54) and acts as a tumor suppressor (41), no specific function has been attributed to this protein. However, Sam68 can be found in nuclear bodies that also contain splicing factors (5, 26). These bodies also contain SLM-1 and SLM-2 (10), and these proteins have been proposed to play a role in alternative splicing (62). Sam68 has been observed in splicing complexes that form on regulatory splicing elements in certain RNAs (7, 18). It is thus possible that Sam68 enters RNA complexes during the transcription process either through direct RNA binding or through protein-protein interactions. Sam68 could then influence the handling of this RNA by continued association with the RNP complex throughout its entire pathway through the splicing machinery and the nuclear pore, ultimately affecting the fate of the RNA in the cytoplasm. Alternatively, Sam68 could help to recruit other factors to the RNA to serve as a specific protein mark that then allow these RNAs to be efficiently translated once they reach the cytoplasmic compartment. In either case, the fact that the G178E mutant, which has previously been shown to be defective for RNA binding (40), failed to enhance CTE function suggests that RNA binding is essential for the observed phenotype.
Sam68 has previously been reported to substitute for, and synergize with, the effects of the HIV Rev protein (52, 59). Since Rev has been clearly shown to function to promote the export of intron-containing HIV RNAs, these results were taken to imply that Sam68 functions as an RNA export protein. However, in the present study, we failed to observe a significant effect of Sam68 on RNA export per se. Also, in contrast to previous studies, we saw only a minor increase in protein expression from RRE-containing reporter constructs when Sam68 was added. This might be explained by the fact that previous studies analyzing the effects of Sam68 on Rev/RRE function have utilized CAT-based reporter constructs. Such constructs have been shown to be very “leaky” at the level of RNA export (i.e., significant amounts of RNA can be observed in the cytoplasmic RNA fraction even in the absence of Rev) (27, 28, 48). It is thus possible that the observed effects of Sam68 in the absence of Rev when these reporters were used reflect an ability of Sam68 to enhance the utilization of this RNA. With the Gag-Pol-RRE plasmids, a similar enhancement would not be expected, since very little RNA reaches the cytoplasm in the absence of Rev (Fig. 6 and 8). It was also previously reported that Sam68 binds directly to the HIV RRE (52). We have, to date, failed to show a direct interaction between Sam68 and the RRE or the CTE. However, it remains possible that Sam68 interacts with other elements in the intron-containing reporter RNAs.
The results presented here indicate that there is a clear difference in the requirement for Sam68 overexpression, depending on whether the Gag-Pol reporters utilize a CTE or Rev/RRE. Whereas basal expression was very low in 293 and 293T cells in the case of the CTE, high levels of expression were observed when the Rev/RRE was utilized. This may reflect the fact that the Rev/RRE and CTE pathways have been shown to be largely nonoverlapping. Thus, RNA that is exported through Rev in a Crm1-dependent fashion may not require Sam68. However, it is also possible that Rev is able to overcome a cytoplasmic restriction through interaction with as yet unidentified cellular proteins.
It is not clear why the endogenous Sam68 protein present in 293 and 293T cells fails to efficiently promote translation of the Gag-Pol-CTE RNA. Our results indicate that similar levels of endogenous Sam68 are expressed in the 293T cells and COS cells used in the present experiments. However, it remains possible that the Sam68 protein is inactivated in 293 and 293T cells as a consequence of posttranslational modifications or interaction with other cellular proteins. Sam68 has been shown to interact with the related proteins SLM-1 and SLM-2 and also forms heterodimers with other cellular proteins (10). Interestingly, it is has recently been observed that SLM-2 RNA and protein levels are reduced in many transformed cell lines (36). The results of this study indicate that SLM-2 is specifically down-regulated as a consequence of the crisis that such cells frequently go through before they become truly immortal. The crisis is supposed to reflect changes in cellular gene expression necessary for long-term survival of the transformed cell and may thus implicate SLM-2 in the regulation of “death genes.” We do not know if 293 or 293T cells express SLM-2, but 293 cells are likely to have undergone crisis. Also, our preliminary results indicate that expression of SLM-2 enhances expression from the Gag-Pol-CTE reporter in a fashion similar to that of Sam68. SLM-2 has also previously been reported to enhance expression from RRE-containing CAT reporter constructs in these cells (59).
Our results show that the activity of Sam68 on the CTE is regulated by the nuclear Src family kinase Sik/BRK. Sam68 was shown to be hyperphosphorylated on several residues in 293T cells in the presence of constitutively active Sik/BRK, confirming the previous study showing that Sam68 is a target for this kinase. In contrast, we failed to observe effects of activated Src kinase on Sam68 function. It has previously been demonstrated that Sam68 is a major target for Src only in mitosis. This has been suggested to be an effect of nuclear envelope breakdown. Since Sam68 is a nuclear protein and attempts to show that this protein shuttles between the nucleus and cytoplasm have been unsuccessful to date, regulation of Sam68 in interphase cells may require interaction with nuclear kinases such as Sik/BRK. If Sam68 is functionally down regulated as a direct consequence of tyrosine phosphorylation, we would have expected to need only a certain catalytic amount of active Sik/BRK to get maximal down regulation. In contrast, we observed that increasing amounts of the plasmid expressing Sik/BRK gave rise to increased inhibition of the Sam68 effect and that inhibition could be readily overcome by increasing the amount of the Sam68 plasmid. In view of these results, it seems possible that Sik/BRK affects Sam68 function by direct binding to this protein or a cellular Sam68 partner.
Quail cells have previously been reported to fail to support CTE function because the Tap protein in these cells is incapable of CTE binding (33). It was thus surprising that transfection Sam68 alone was able to give rise to as much p24 expression in quail cells as expression of the human Tap protein. This indicates that Tap binding to the CTE is not absolutely essential for CTE function. Furthermore, the synergistic effect seen in these cells with a combination of Tap and Sam68 suggests that these proteins may act at different steps in the CTE pathway.
The results presented here provide yet another example of the links that exist between signal transduction and RNA regulation at the posttranscriptional level. It will be of interest to analyze how activation of the Sik/BRK kinase is regulated in the cell and the exact mechanism by which this kinase regulates Sam68 function. We believe that the CTE reporter systems will provide a useful tool in these studies that might ultimately shed light on the role of these proteins in cell cycle regulation and differentiation.
Acknowledgments
The first two authors contributed equally to this work.
We thank Angela Tyner for the gift of plasmids expressing wild-type and mutant forms of Sik/BRK and Sally Parsons, Joan Steitz, Doug Black, Gary Brewer, and Bryan Cullen for other materials used in this study. Susan Prasad and Joy Morgenegg provided expert technical assistance.
This work was supported by National Institutes of Health grants AI34721 to M.-L.H. and AI47008 to D.R. J.C. is the recipient of NSRA fellowship AI10630. Salary support for M.-L.H. and D.R. was provided by the Charles H. Ross Jr. and Myles H. Thaler Endowments at the University of Virginia.
REFERENCES
- 1.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Biscardi, J. S., M. C. Maa, D. A. Tice, M. E. Cox, T. H. Leu, and S. J. Parsons. 1999. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 274:8335-8343. [DOI] [PubMed] [Google Scholar]
- 3.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 4.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M.-L. Hammarskjöld. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 91:1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, T., F. Boisvert, D. Bazett-Jones, and S. Richard. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cells. Mol. Biol. Cell 10:3015-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T., B. B. Damaj, C. Herrera, P. Lasko, and S. Richard. 1997. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol. Cell. Biol. 17:5707-5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, M.-Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R., A. M. Skalka, and G. Ju. 1983. Endogenous avian retroviruses contain deficient promoter and leader sequences. Proc. Natl. Acad. Sci. USA 80:2946-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derry, J. J., S. Richard, H. Valderrama Carvajal, X. Ye, V. Vasioukhin, A. W. Cochrane, T. Chen, and A. L. Tyner. 2000. Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol. Cell. Biol. 20:6114-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Fruscio, M., T. Chen, and S. Richard. 1999. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc. Natl. Acad. Sci. USA 96:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebersole, T. A., Q. Chen, M. J. Justice, and K. Artzt. 1996. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 12:260-265. [DOI] [PubMed] [Google Scholar]
- 12.Ernst, R., M. Bray, D. Rekosh, and M.-L. Hammarskjöld. 1997. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 17:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst, R. K., M. Bray, D. Rekosh, and M.-L. Hammarskjöld. 1997. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA 3:210-222. [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis, R., M. K. Barton, J. Kimble, and T. Schedl. 1995. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139:579-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fumagalli, S., N. F. Totty, J. J. Hsuan, and S. A. Courtneidge. 1994. A target for Src in mitosis. Nature 368:871-874. [DOI] [PubMed] [Google Scholar]
- 17.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 18.Grossman, J. S., M. I. Meyer, Y. C. Wang, G. J. Mulligan, R. Kobayashi, and D. M. Helfman. 1998. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA 4:613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 20.Guzik, B. W., L. Levesque, S. Prasad, Y. C. Bor, B. E. Black, B. M. Paschal, D. Rekosh, and M.-L. Hammarskjöld. 2001. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammarskjöld, M.-L. 1997. Regulation of retroviral RNA export. Semin. Cell Dev. Biol. 8:83-90. [DOI] [PubMed] [Google Scholar]
- 22.Hammarskjöld, M.-L., J. Heimer, B. Hammarskjöld, I. Sangwan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammarskjöld, M.-L., S.-C. Wang, and G. Klein. 1986. High-level expression of the Epstein-Barr virus EBNA1 protein in CV1 cells and human lymphoid cells using a SV40 late replacement vector. Gene 43:41-50. [DOI] [PubMed] [Google Scholar]
- 24.Hammarskjöld, M.-L. 2001. Constitutive transport element-mediated nuclear export. Curr. Top. Microbiol. Immunol. 259:77-93. [DOI] [PubMed] [Google Scholar]
- 25.Hammarskjöld, M.-L., H. Li, D. Rekosh, and S. Prasad. 1994. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J. Virol. 68:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann, A. M., O. Nayler, F. W. Schwaiger, A. Obermeier, and S. Stamm. 1999. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 10:3909-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, Y., and G. C. Carmichael. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18:1642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaurralde, E. 2002. Nuclear export of messenger RNA. Results Probl. Cell Differ. 35:133-150. [DOI] [PubMed] [Google Scholar]
- 30.Jan, E., C. K. Motzny, L. E. Graves, and E. B. Goodwin. 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, A. R., R. Francis, and T. Schedl. 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180:165-183. [DOI] [PubMed] [Google Scholar]
- 32.Jones, A. R., and T. Schedl. 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9:1491-1504. [DOI] [PubMed] [Google Scholar]
- 33.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiledjian, M., C. T. DeMaria, G. Brewer, and K. Novick. 1997. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 17:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kool, J., W. van Zaane, A. J. van der Eb, and C. Terleth. 2001. Down-regulation of T-STAR, a growth inhibitory protein, after SV40-mediated immortalization. Cell Growth Differ. 12:535-541. [PubMed] [Google Scholar]
- 37.Legrain, P., and M. Rosbash. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57:573-583. [DOI] [PubMed] [Google Scholar]
- 38.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, Q., S. J. Taylor, and D. Shalloway. 1997. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J. Biol. Chem. 272:27274-27280. [DOI] [PubMed] [Google Scholar]
- 41.Liu, K., L. Li, P. E. Nisson, C. Gruber, J. Jessee, and S. N. Cohen. 2000. Neoplastic transformation and tumorigenesis associated with sam68 protein deficiency in cultured murine fibroblasts. J. Biol. Chem. 275:40195-40201. [DOI] [PubMed] [Google Scholar]
- 42.Malim, M. H., and B. R. Cullen. 1993. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol. Cell. Biol. 13:6180-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 44.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104:173-176. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto, K., K. M. Wassarman, and A. P. Wolffe. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17:2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride, A. E., S. J. Taylor, D. Shalloway, and K. Kirkegaard. 1998. KH domain integrity is required for wild-type localization of Sam68. Exp. Cell Res. 241:84-95. [DOI] [PubMed] [Google Scholar]
- 48.Otero, G. C., M. E. Harris, J. E. Donello, and T. J. Hope. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 72:7593-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 50.Ranjan, M., S. R. Tafuri, and A. P. Wolffe. 1993. Masking mRNA from translation in somatic cells. Genes Dev. 7:1725-1736. [DOI] [PubMed] [Google Scholar]
- 51.Reddy, T. R., H. Tang, W. Xu, and F. Wong-Staal. 2000. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene 19:3570-3575. [DOI] [PubMed] [Google Scholar]
- 52.Reddy, T. R., W. Xu, J. K. Mau, C. D. Goodwin, M. Suhasini, H. Tang, K. Frimpong, D. W. Rose, and F. Wong-Staal. 1999. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 5:635-642. [DOI] [PubMed] [Google Scholar]
- 53.Reddy, T. R., W. D. Xu, and F. Wong-Staal. 2000. General effect of Sam68 on Rev/Rex regulated expression of complex retroviruses. Oncogene 19:4071-4074. [DOI] [PubMed] [Google Scholar]
- 54.Resnick, R. J., S. J. Taylor, Q. Lin, and D. Shalloway. 1997. Phosphorylation of the Src substrate Sam68 by Cdc2 during mitosis. Oncogene 15:1247-1253. [DOI] [PubMed] [Google Scholar]
- 55.Saccomanno, L., C. Loushin, E. Jan, E. Punkay, K. Artzt, and E. B. Goodwin. 1999. The STAR protein QKI-6 is a translational repressor. Proc. Natl. Acad. Sci. USA 96:12605-12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 66:150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, A. J., M.-I. Cho, M.-L. Hammarskjöld, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, A. J., N. Srinivasakumar, M.-L. Hammarskjöld, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soros, V. B., H. V. Carvajal, S. Richard, and A. W. Cochrane. 2001. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J. Virol. 75:8203-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srinivasakumar, N., N. Chazal, M. C. Helga, S. Prasad, M.-L. Hammarskjöld, and D. Rekosh. 1997. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 71:5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srinivasakumar, N., M.-L. Hammarskjöld, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoss, O., M. Olbrich, A. M. Hartmann, H. Konig, J. Memmott, A. Andreadis, and S. Stamm. 2001. The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J. Biol. Chem. 276:8665-8673. [DOI] [PubMed] [Google Scholar]
- 63.Taylor, S. J., M. Anafi, T. Pawson, and D. Shalloway. 1995. Functional interaction between c-Src and its mitotic target, Sam 68. J. Biol. Chem. 270:10120-10124. [DOI] [PubMed] [Google Scholar]
- 64.Taylor, S. J., and D. Shalloway. 1994. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368:867-871. [DOI] [PubMed] [Google Scholar]
- 65.Vernet, C., and K. Artzt. 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13:479-484. [DOI] [PubMed] [Google Scholar]
- 66.Wang, L. L., S. Richard, and A. S. Shaw. 1995. P62 association with RNA is regulated by tyrosine phosphorylation. J. Biol. Chem. 270:2010-2013. [DOI] [PubMed] [Google Scholar]
- 67.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 68.Zaffran, S., M. Astier, D. Gratecos, and M. Semeriva. 1997. The held out wings (how) Drosophila gene encodes a putative RNA binding protein involved in the control of muscular and cardiac activity. Development 124:2087-2098. [DOI] [PubMed] [Google Scholar]