Abstract
Exposure of Saccharomyces cerevisiae to increases in extracellular osmolarity activates the stress-activated Hog1 mitogen-activated protein kinase (MAPK), which is essential for cell survival upon osmotic stress. Yeast cells respond to osmotic stress by inducing the expression of a very large number of genes, and the Hog1 MAPK plays a critical role in gene transcription upon stress. To understand how Hog1 controls gene expression, we designed a genetic screen to isolate new transcription factors under the control of the MAPK and identified the MEF2-like transcription factor, Smp1, as a target for Hog1. Overexpression of SMP1 induced Hog1-dependent expression of osmoresponsive genes such as STL1, whereas smp1Δ cells were defective in their expression. Consistently, smp1Δ cells displayed reduced viability upon osmotic shock. In vivo coprecipitation and phosphorylation studies showed that Smp1 and Hog1 interact and that Smp1 is phosphorylated upon osmotic stress in a Hog1-dependent manner. Hog1 phosphorylated Smp1 in vitro at the C-terminal region. Phosphorylation of Smp1 by the MAPK is essential for its function, since a mutant allele unable to be phosphorylated by the MAPK displays impaired stress responses. Thus, our data indicate that Smp1 acts downstream of Hog1, controlling a subset of the responses induced by the MAPK. Moreover, Smp1 concentrates in the nucleus during the stationary phase, and the lack of SMP1 results in cells that lose viability in the stationary phase. Localization of Smp1 depends on HOG1, and consistently, hog1Δ cells also lose viability during this growth phase. These data suggest that Smp1 could be mediating a role for the Hog1 MAPK during the stationary phase.
Mitogen-activated protein kinase (MAPK) cascades are common signaling modules found in both higher and lower eukaryotic cells. Activation of a MAPK results in modification of a set of target proteins, often transcription factors, that allow the generation of appropriate cellular responses to an external stimulus. Stress-activated protein kinases (SAPKs) are a subset of MAPKs activated by environmental and genotoxic stresses (reviewed in references 9 and 20). A prototype of the SAPK family is the yeast p38-related MAPK, Hog1, which specifically responds to increased extracellular osmolarity and is required for cell survival under these conditions.
Activation of the yeast Hog1 MAPK induces diverse osmo-adaptive responses, such as regulation of gene expression. Genome-wide transcriptional analyses showed that a great number of genes are regulated by osmotic stress in a HOG1-dependent manner. Among the genes under the control of Hog1 are genes that encode proteins implicated in carbohydrate metabolism, general stress protection, protein production, and signal transduction (reviewed in reference 8). One mechanism by which SAPKs, and MAPKs in general, modulate gene expression is by direct modification of transcription activators. In Saccharomyces cerevisiae, only four transcription factors, Sko1, Hot1, and the redundant Msn2 and Msn4, have been proposed to be controlled by the Hog1 MAPK. These factors are unrelated, and the mechanisms by which Hog1 regulates their function may differ from one to another. Hot1, Msn2, and Msn4 activate transcription, whereas Sko1 represses and activates different subsets of osmotic-inducible and Hog1-regulated genes (15, 17, 18). Sko1 is an ATF/CREB factor that represses genes under nonstress conditions by the recruitment of the general corepressor complex Cyc8-Tup1. In response to osmotic stress, Sko1 is phosphorylated by Hog1, thereby relieving repression (15, 16). Msn2 and Msn4 are generic stress factors controlled by PKA and Hog1 by an unknown mechanism. Hot1 physically interacts with Hog1, and its binding to DNA and subsequent transactivation activity are regulated by Hog1 kinase activity (1, 19). Global gene expression analyses carried out to dissect the specific roles for each transcription factor have revealed that each of these factors can account for a limited effect on global gene expression by regulation of a small subset of the osmostress-inducible genes (3, 17). Evaluation of the subset of osmostress-responsive genes controlled by each factor revealed that the activators reported to be under the control of Hog1 were not sufficient to explain the impact of Hog1 on gene expression.
The mechanism by which Hog1 regulates gene expression is not completely understood. In particular, there remained the possibility that additional transcription factors were required for gene expression upon stress. Therefore, we undertook a genetic screen to identify new activators under the control of the MAPK. Thus, we found that Smp1, a member of the MEF2C family of transcription factors, is involved in Hog1 signaling. Interestingly, it was reported previously that in metazoan cells, the MEF2C family of transcription factors can be targeted by the mammalian p38 MAPK (11). We show here that Hog1 and Smp1 interact in vivo, and we have identified the relevant Hog1 phosphorylation sites in Smp1 and analyzed their effect on stress-regulated gene expression in vivo. Furthermore, we report that Smp1 may play an important role not only in osmostress responses, but also in a new function for the Hog1 MAPK required for cell survival in the stationary phase.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ), TM141 (MATa ura3 leu2 trp1 his3), TM233 (MAT α ura3 leu2 trp1 his3 lys2 hog1::TRP1), YEN44 (MATa ura3 leu2 trp1 his3 smp1::HIS3), YEN46 (MAT α ura3 leu2 trp1 his3 lys2 hog1::TRP1 smp1::HIS3), and YEN114 (MATa ura3 leu2 trp1 HOG1-HA×6-HIS3). Integration of the STL1-LacZ reporter construct (PEN05) in wild-type, hog1Δ, and smp1Δ strains yielded YEN2 (MATa leu2 trp1 his3 STL1-LacZ URA3), YEN7 (MAT α leu2 trp1 his3 lys2 hog1::TRP1 STL1-LacZ URA3), and YEN48 (MATa leu2 trp1 his3 smp1::HIS3 STL1-LacZ URA3). Genomic disruptions were made by long flanking homology PCR-based gene disruption.
Buffers and media.
Buffer A consists of 50 mM Tris-HCl (pH 7.5), 15 mM EDTA, 15 mM EGTA, 2 mM dithiothreitol (DTT), 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, 5 μg of pepstatine per ml, and 5 μg of leupeptin per ml. Alkaline phosphatase buffer consists of 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 10 mM MgCl2. Buffer B consists of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 1% Triton X-100, 1 mM PMSF, 1 mM benzamidine, and 5 μg of leupeptin per ml. Kinase buffer consists of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT. Phosphatase inhibitor mixture contains 10 mM NaF, 1 mM sodium pyrophosphate, and 10 mM β-glycerophosphate. Sodium dodecyl sulfate (SDS) loading buffer consists of 50 mM Tris-HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol. Yeast extract-peptone-dextrose (YPD) medium contains, per liter, 10 g of yeast extract, 20 g of peptone, and 20 g of dextrose. Selective medium contains 1.7 g of of yeast nitrogen base (Difco) per liter, 5 g of (NH4)2SO4 per liter, 20 g of dextrose per liter, and supplements (100 mg each of the amino acid[s] uracil or adenine, as appropriate, except where indicated). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solid medium contains selective medium buffered with MES [2-(N-Morpholino)ethanesulfonic acid] at pH 7 plus 0.1 mg of X-Gal per ml. All yeast growth was at 30°C.
Plasmids.
The STL1::LacZ reporter construct PEN05 was generated by cloning the STL1 promoter (base pairs −824 to +4) by PCR into YIp358R (CEN URA3) (12). Plasmid PEN45 carries a 3.4-kbp EcoRI/SalI fragment containing SMP1 in a multicopy vector YEplac181. p426GAG1 (PGAG1-GST URA3+ 2μm) is a gift from M. Takekawa (unpublished). The PGAG1-GST allows the expression of GST-fusion proteins via the yeast PGAL1 promoter. Full-length wild-type and mutant SMP1 genes were cloned into plasmid p426GAG1. Full-length SMP1 and SMP1ΔMADS (which contains a 90-amino-acid terminal deletion of the MADS box domain) were cloned into the SalI site of the pBTM116 plasmid to fuse them to the LexA binding domain. pACTII-HOG1 was obtained by fusion of the full-length HOG1 PCR product with the GAL4 activation domain in pACTII. The bacterial expression plasmid pGEX-4T (Pharmacia) allows the expression of GST-tagged proteins in Escherichia coli. Full-length wild-type and several truncated SMP1 alleles were cloned into the EcoRI site of the pGEX-4T plasmid by PCR. Construction of SMP1 site-directed mutants was carried out by PCR-directed mutagenesis. Each mutation was verified by DNA sequencing. The SMP1-m4 was sequenced to verify that it contained only the desired mutations. The m1 mutant contains a double amino acid substitution, namely, Ser365 and Ser376 to Ala, while m2 contains the Ser348 and the Ser357 amino acids mutated to Ala. The m4 contains a quadruple amino acid substitution, namely, Ser348, Ser357, Thr365 and Ser376 to Ala. Those mutant alleles were cloned into pGEX-4T for bacterial expression and into p426GAG1 (PGAG1-GST URA3+ 2μm) for yeast expression. The SMP1-GFP plasmid contains the SMP1 gene fused to the green fluorescent protein gene (GFP) in a multicopy plasmid pRS426.
Isolation of STL1::LacZ reporter activators.
Wild-type yeast strain TM141 carrying an integrated STL1::LacZ reporter (PEN05) was transformed with a yeast genomic library in the multicopy plasmid YEp13. Colonies were grown in synthetic media containing X-Gal for 5 to 6 days. From approximately 30,000 colonies, 20 positive clones were selected by their ability to induce STL1 and thus to produce beta-galactosidase. Plasmids were isolated, and their HOG1 dependence was evaluated in a hog1Δ strain containing the same reporter construct. Plasmids from HOG1-dependent positives were isolated and partially sequenced. Candidate genes were subcloned and assayed as before.
Two-hybrid analysis.
The two-hybrid analysis was carried out essentially as described previously (5) by using pACTII and pBTM116 as the activation domain plasmid and LexA as the DNA-binding domain plasmid. The plasmids LexA-SMP1 and LexA-ΔMADSSMP1 were cotransformed with pACT-HOG1 using the L40 reporter strain. Positive clones were selected and further tested for β-galactosidase activity as follows. Cells (∼5 × 106) were spotted onto YPD plates, incubated for 5 h at 30°C, and replicated onto nitrocellulose membranes. β-Galactosidase activity was visualized in situ by using X-Gal as described elsewhere (5).
In vivo coprecipitation assays.
In vivo interaction of GST-SMP1 with Hog1 was determined by GST pull-down experiments. Mid-log-phase cells were grown in the presence of 2% galactose for 4 h and subjected or not subjected to a brief osmotic shock (0.4 M NaCl, 10 min). Yeast extract (750 μg, in a mixture of buffer A plus 150 mM NaCl plus phosphatase inhibitor) was incubated with 50 μl of glutathione-Sepharose beads overnight at 4°C. The beads were washed extensively with buffer A plus 150 mM NaCl, resuspended in loading buffer, and separated by SDS-polyacrylamide gel electrophoresis (PAGE). Immunoblotting was done by using anti-HA monoclonal antibody 12CA5 (Roche) and anti-GST monoclonal antibody (Pharmacia) together with ECL reagent (Pharmacia).
In vivo SMP1 phosphorylation assays.
Wild-type and mutant GST-SMP1 proteins were purified as described above but in the presence of buffer A without EGTA and EDTA. Bead-bound proteins were incubated with 40 U of calf-intestinal alkaline phosphatase (Roche) in 50 μl of alkaline phosphatase buffer for 60 min at 30°C. After alkaline phosphatase treatment, the beads were washed with several column volumes and Smp1 was detected by immunoblotting by using an anti-GST monoclonal antibody.
Purification of GST proteins in E. coli and in vitro kinase assays.
GST fusion proteins encoding Pbs2(EE), Hog1, and full-length or truncated Smp1 were constructed by using pGEX-4T (Pharmacia), expressed in E. coli DH5, and purified by using glutathione-Sepharose beads (Pharmacia) in buffer B as described previously (14). Phosphorylation of Smp1 by Hog1 was monitored by the following in vitro kinase assay. One microgram of recombinant GST-HOG1 from E. coli was activated by phosphorylation by using 0.5 μg of GST-PBS2(EE) in the presence of kinase buffer and ATP. After 15 min at 30°C, 5 μg of wild-type or mutant versions of Smp1, purified from E. coli, was added to the previous mixture together with [γ-32P]ATP (0.2 μCi/μl). The mixture was then incubated for 5 min at 30°C, and the reactions were terminated by the addition of 2× SDS loading buffer. The labeled proteins were resolved by SDS-PAGE and detected by autoradiography.
β-Galactosidase assays.
The transformed yeast strains were grown selectively until mid-log phase in the appropriate selective liquid media and then diluted in YPD for 3 h. Logarithmically growing cells (optical density at 660 nm, 0.5 to 0.8) were treated or not treated with 0.4 M of NaCl for 35 min and permeabilized by ethanol-toluene treatment, and β-galactosidase activity was determined as described previously (6).
Cell viability assays.
Cell viability was scored essentially as described previously (7). Yeast cells were grown in YPD or minimal medium to an optical density of 0.8 and subjected or not subjected to an osmotic shock (1 M NaCl for 60 min). Viability was determined by adding propidium iodide (PI) at 0.004 mg/ml and measuring the number of PI-positive cells by using a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.). Cell viability was also determined by adding Phloxin B at 0.5 mg/ml to the untreated or treated cells as before and counting the stained cells by visual microscopy.
GFP fluorescence microscopy.
GFP was visualized without fixation by using a Nikon E-600 with an ORCA II CCD camera (Hamamatsu). Images were taken at 100× magnification and converted to Photoshop format version 4.0 (Adobe Systems).
RESULTS
The MEF2C-related Smp1 transcription factor regulates HOG-dependent gene expression.
Osmostress gene expression is strongly defective in hog1Δ cells (see above); however, deletion of the transcription factors described as being under the control of the MAPK has only a limited effect on osmostress gene expression. Thus, it was predicted that other transcription factors must exist under the control of the Hog1 MAPK. To identify new Hog1-regulated transcription factors, we conducted a screening by which we isolated genes whose overexpression was able to induce STL1 expression. The STL1 gene encodes a putative sugar transporter, and it is one of the genes that is most strongly induced in response to osmostress and is completely under the control of Hog1 (13).
Yeast cells containing an integrated STL1-LacZ reporter construct were transformed with a multicopy genomic library, and positive clones were selected by their ability to induce STL1 and, therefore, to produce β-galactosidase on X-Gal-containing plates (described in Material and Methods). Thus, 20 positive clones were identified from approximately 30,000 colonies. Plasmids were isolated from positive clones and tested again for their ability to induce STL1 in both wild-type and hog1Δ strains containing the STL1-LacZ reporter. Only clones that induced STL1 in a HOG1-dependent manner were partially sequenced. From the genomic clones, candidate genes were subcloned into a multicopy plasmid and tested as above.
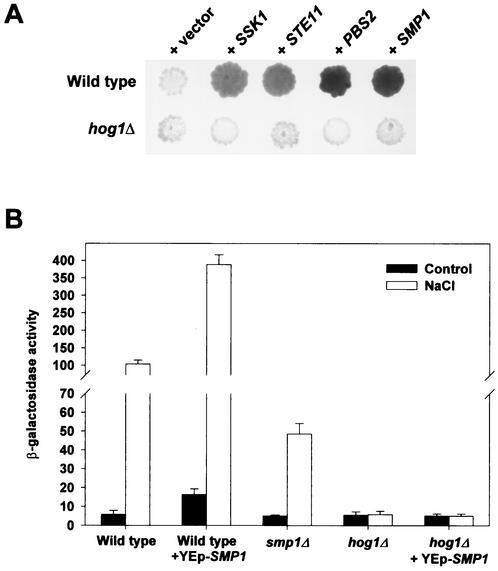
As expected, some of the clones capable of inducing our reporter system encoded upstream components of the HOG pathway, namely STE11, PBS2, and SSK1, that are known to be able to induce activation of Hog1 by overexpression (Fig. 1A). In addition to those upstream components of the HOG pathway, we identified elements that could be acting at the level of transcription regulation. We thus identified some genes encoding components of the basic transcription machinery, such as SRB9 and SRB4 (data not shown) and a gene encoding the Smp1 protein, a member of the MEF2 transcription factor family (Fig. 1A).
FIG. 1.
Smp1 transcription factor modulates HOG1-mediated STL1 gene expression. (A) Yeast multicopy plasmids capable of inducing STL1-LacZ expression in wild-type cells were isolated, and their dependence on HOG1 was assayed in wild-type cells (YEN2) and hog1Δ cells (YEN7). A representative filter β-galactosidase assay demonstrating induction of STL1-LacZ by several positive clones from the screening is shown. (B) Smp1 modulates expression of STL1-LacZ. Wild-type, hog1Δ, and smp1Δ strains containing the STL1-LacZ reporter system (strains YEN2, YEN7, and YEN48) were transformed with a control plasmid or a multicopy plasmid expressing SMP1. β-Galactosidase activity was assayed in cells that were grown to mid-log phase and that were subjected (open bars) or not subjected (control; filled bars) to hyperosmotic stress (0.4 M NaCl for 35 min). β-Galactosidase activity is given in nanomoles per minute per milligram and is the result of the measurement in quadruplicate of results for two independent transformants.
To determine the importance of SMP1 in regulation of gene expression, we quantified STL1 expression in liquid β-galactosidase assays. As shown in Fig. 1B, cells overexpressing SMP1 induced expression of the STL1 reporter gene to approximately fourfold-higher levels than did wild-type cells under both nonstress and stress conditions. Overexpression of SMP1 in hog1Δ cells did not have any effect on STL1 expression. Furthermore, deletion of SMP1 resulted in reduced STL1 expression (Fig. 1B). Similar results were obtained when STL1 expression was analyzed by Northern blotting (data not shown). Deletion of the gene that encodes for the Hot1 transcription factor had an even more extensive impact on STL1 expression than did deletion of the SMP1 gene, indicating that Hot1 plays a major role in the regulation of STL1 (data not shown).
The role of Smp1 on gene expression was also assayed by analyzing the expression of other stress-responsive genes known to be controlled by the Hog1 MAPK (13, 17). We created reporter genes containing the CWP1, ALD3, and HXT1 gene promoters fused to the LacZ reporter system. Similar to the observations with STL1, expression of CWP1 was induced by overexpression of SMP1, whereas deletion of SMP1 partially reduced its expression upon stress. No SMP1-dependent differences on gene expression were observed when the HXT1 and ALD3 gene reporters were used, although these reporters were induced under osmotic stress in a HOG1-dependent manner (data not shown). Thus, the genetic screen yielded a clone, SMP1, encoding a transcription factor of the MEF2 family, capable of regulating a subset of stress-responsive genes under the control of the Hog1 MAPK.
Smp1 is phosphorylated after osmotic stress in a Hog1-dependent manner.
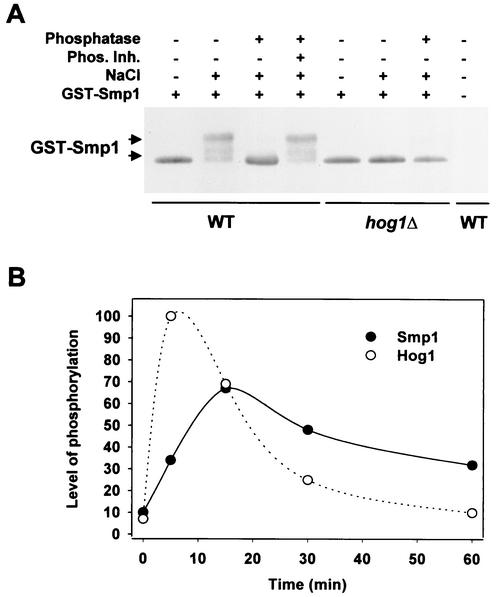
When yeast cells are exposed to osmotic stress, the Hog1 MAPK is rapidly phosphorylated and activated (10) (Fig. 2B). To determine whether Smp1 is regulated in response to osmotic stress, we tested whether Smp1 was also phosphorylated upon osmotic stress. Wild-type and hog1Δ strains were subjected to a brief osmotic shock, and GST-tagged Smp1 protein was monitored by Western blotting by using anti-GST antibodies. The GST-Smp1 fusion protein was fully functional as tested for its ability to induce STL1-LacZ expression (data not shown). As shown in Fig. 2A, when subjected to osmotic stress, the mobility pattern of Smp1 was altered. The mobility change of Smp1 was induced by phosphorylation, because when extracts from osmotic-stressed cells were treated with alkaline phosphatase, the mobility pattern could be reversed (Fig. 2A). Interestingly, Smp1 phosphorylation was rapidly induced upon stress but the kinetics lagged behind those observed for the phosphorylation of Hog1. This kinetic relationship is consistent with Smp1 being dependent on Hog1 (Fig. 2B). Moreover, phosphorylation of Smp1 in response to osmotic stress depends on Hog1, since Smp1 from hog1Δ cells did not undergo the mobility shift observed in wild-type cells. Thus, these results showed that Smp1 is rapidly phosphorylated upon hyperosmotic shock in vivo and that this modification depends on the Hog1 MAPK.
FIG. 2.
In vivo phosphorylation of Smp1 upon osmotic stress depends on Hog1. (A) GST-SMP1 was expressed under the PGAL1 promoter in wild-type (TM141) and hog1Δ (TM233) cells. Yeast cells were grown in the presence of galactose for 4 h and subjected (+) or not subjected (−) to a brief osmotic shock (0.4 M NaCl for 10 min), and the extracts were treated (+) or not treated (−) with 10 U of alkaline phosphatase or phosphatase inhibitors (Phos. Inh). Extracts were separated by SDS-PAGE, and GST-tagged Smp1 was detected by the use of polyclonal GST-specific antibodies. The control strain (without GST-SMP1) was the wild-type strain (TM141). (B) Time course of Hog1 and Smp1 phosphorylation upon osmotic stress. Cells expressing GST-SMP1 in a wild-type strain were subjected to osmotic stress (0.4 M NaCl), and cells were collected at different times. Phosphorylation of Smp1 and Hog1 was monitored by Western blotting by using anti-GST antibodies and anti-phospho-p38 MAPK (Thr180/Tyr182) antibodies (New England BioLabs), respectively. The level of phosphorylation was measured from scanned films by using Quantity One software (Bio-Rad).
Hog1 interacts with the putative transcription activator domain of Smp1.
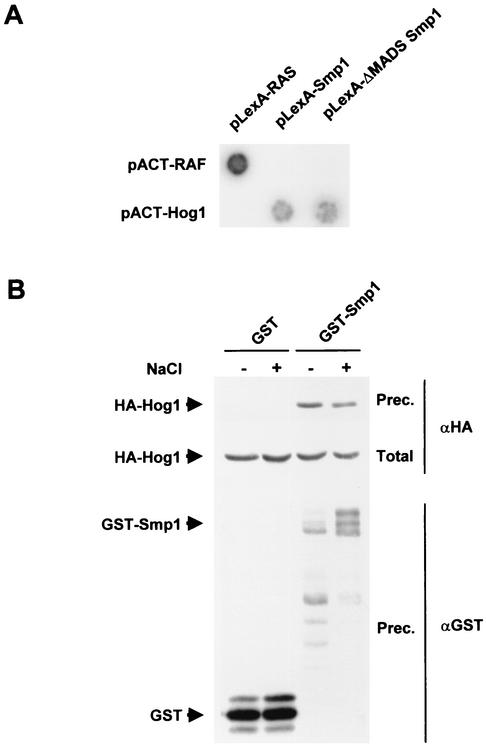
To obtain direct evidence for the interaction of Smp1 and Hog1, we tested by two-hybrid analysis whether these proteins interacted. Smp1 or a mutant containing a deletion on the MADS box and MEF2 domains (amino acids 1 to 90) were fused to the LexA-DNA binding domain, and their interaction with the full-length Hog1 fused to the GAL4 activator domain was tested. Figure 3A shows a typical result and indicates that Smp1 interacts with Hog1 through its C-terminal domain and not through the MADS and MEF2 domains.
FIG. 3.
In vivo binding of Hog1 to Smp1. (A) Interaction as shown by two-hybrid analysis of Hog1 and Smp1. The wild-type version and a mutant that lacks the MADS and MEF2 domains (ΔMADS Smp1) fused to the LexA DB were expressed with the full-length HOG1 fused to the GAL4 activator domain. A representative filter β-galactosidase assay demonstrating interactions between Hog1 and Smp1 is shown. Proteins encoded by the control plasmids pLexA-RASV12 and pACT-RAF, which are known to interact with each other, are shown for comparison. (B) Smp1 coprecipitates with Hog1. Strain YEN114 (which expresses HA-tagged Hog1 from the wild-type locus) was transformed with a plasmid expressing GST or GST-SMP1 under the PGAL1 promoter. Cells were grown in the presence of galactose, and samples were taken before (−) or 10 min after (+) the addition of NaCl to a final concentration of 0.4 M. GST proteins were affinity purified through a glutathione-Sepharose matrix; the presence of HA-Hog1 in the precipitates was probed by immunoblotting with anti-HA (indicated by αHA at right), and GST-containing proteins (indicated by αGST at right) were detected by using antibodies against GST. Total, <10% of the input protein; Prec., total amount of Hog1 or GST precipitated.
The interaction between Hog1 and Smp1, shown by the two-hybrid data, was confirmed by in vivo coprecipitation experiments. Yeast YEN114 cells (which express HA-tagged Hog1 from their own genomic locus) were transformed with a plasmid that expresses a GST-tagged Smp1 from the GAL1 promoter. Cells were subjected to a brief osmotic shock, and Smp1 was precipitated by using glutathione-Sepharose beads. The presence of Hog1 in the precipitates was probed with an anti-HA monoclonal antibody. As shown in Fig. 3B, Smp1 was able to coprecipitate Hog1 irrespective of the environmental conditions. Thus, these in vivo binding assays confirmed the conclusion of the two-hybrid analysis indicating that Hog1 binds to the Smp1 transcription factor.
Phosphorylation of multiples sites at the C terminus of Smp1 by Hog1 affects Smp1 function.
Smp1 is phosphorylated upon osmotic stress in a Hog1-dependent manner and interacts physically with the MAPK. We then tested whether Smp1 phosphorylation was carried out directly by the Hog1 MAPK by using purified proteins in an in vitro kinase assay. For this purpose, Hog1 and a constitutively activated version of Pbs2 [PBS2(EE)] were purified as GST-fusion proteins from E. coli. In the first step of the reaction, Hog1 was activated by phosphorylation in the presence of PBS2(EE) and ATP (2). Then Smp1, purified from E. coli as a GST-tagged protein, and [γ-32P]ATP were added to the reaction. As shown in Fig. 4A, lane 1, full-length Smp1 was phosphorylated by Hog1 (see the asterisk), whereas when a catalytically impaired Hog1 (hog1KN) was used instead of the wild-type Hog1, no phosphorylation of Smp1 was observed (data not shown), thus suggesting that Smp1 is a direct substrate for the MAPK Hog1.
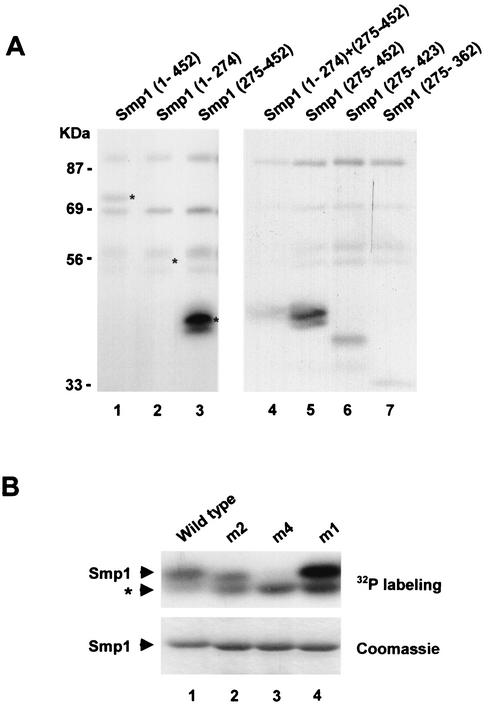
FIG. 4.
Hog1 phosphorylates the C-terminal domain of Smp1. (A) In vitro-activated Hog1 phosphorylates Smp1. Various Smp1 fragments were tested for their ability to be phosphorylated by an in vitro-activated Hog1. The positions of the Smp1 fragments included in the constructs are indicated in parentheses. Recombinant tagged proteins were purified from E. coli and subjected to phosphorylation by activated Hog1 as described in Materials and Methods. Phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. Asterisks indicate the positions of Smp1 proteins detected by Coomassie staining. (B) Mutation of Smp1 Ser348, Ser357, Thr365, and Ser376 to Ala abolishes Hog1 phosphorylation. The wild-type Smp1 fragment (amino acids 275 to 452) and various Smp1 site-directed mutants were tested for Hog1 phosphorylation as described in the legend for panel A. The Smp1 mutants and their corresponding mutations (indicated in parentheses) are as follows: m2 (Ser348 and Ser357 to Ala), m4 (Ser348, Ser357, Thr365, and Ser376 to Ala), and m1 (Thr365 and Ser376 to Ala). After phosphorylation, the proteins were resolved by SDS-PAGE, and phosphorylated proteins were detected by autoradiography (upper panel). GST-tagged Smp1 proteins were detected by Coomassie blue stain (lower panel). The asterisk indicates a contaminant phosphorylation not corresponding to Smp1.
To map the phosphorylation site(s) for Hog1 in Smp1, we created several truncated SMP1 alleles and expressed them as GST-tagged proteins in E. coli. After purification, the same amounts of the pure proteins were subjected to in vitro phosphorylation by activated Hog1 (as described above). A truncated form of Smp1 containing the N-terminal region (amino acids 1 to 274) was unable to be phosphorylated by Hog1, whereas a C-terminal region (amino acids 275 to 452) was strongly phosphorylated by the MAPK (Fig. 4, lanes 2 and 3). Thus, phosphorylation of Smp1 by Hog1 occurs in a region which is coincident with the Hog1 binding domain. It is worth noting that when both N-terminal and C-terminal fragments were present in the same assay, the amount of phosphorylation of the C-terminal fragment was strongly reduced (Fig. 4, lane 4).
To further determine the phosphorylation sites in Smp1, we created several truncated versions of the C-terminal domain (amino acids 275 to 452) and expressed them as GST-tagged proteins in E. coli. Truncated forms were purified and assayed as described previously. A 30-residue C-terminal deletion had little effect on Smp1 phosphorylation, whereas deletion of the last 90 residues had a stronger effect on phosphorylation (Fig. 4, lanes 6 and 7). Four sequences corresponding to the consensus phosphorylation site for MAPKs (Ser-Pro/Thr-Pro) are present in the region comprised of residues 275 to 423. We created point mutant versions to replace Ser and Thr residues with Ala, and we tested them for phosphorylation by Hog1. Single mutations of Ser 348, Ser 357, Thr 365, and Ser 376 did not alter Smp1 phosphorylation (data not shown). We then constructed several combinations of mutant sites and tested them by phosphorylation. Mutation of both Thr 365 and Ser 376 (m1) did not change the level of phosphorylation, whereas mutation on both Ser 348 and Ser 357 (m2) slightly decreased the phosphorylation of Smp1 (Fig. 4B). Smp1 phosphorylation was only abolished in the Smp1-m4 mutant, which contains mutations in all four sites (Fig. 4B).
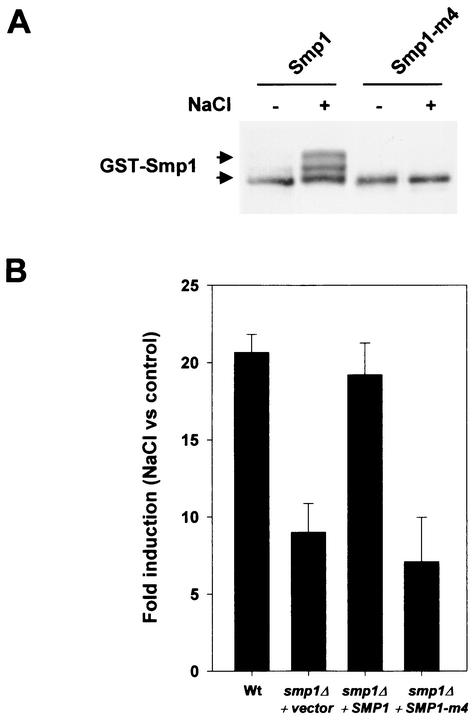
In vivo phosphorylation assays carried out as described previously in wild-type cells transformed with a wild-type GST-tagged Smp1 or the mutant Smp1-m4 protein demonstrated that mutation of the four sites in the Smp1-m4 protein eliminated most of the mobility shift due to phosphorylation of Smp1 in response to osmotic stress (Fig. 5A). Taken together, these results indicate that there is a specific cluster of four phosphorylation sites for Hog1 that is located within 28 amino acids in the C terminus of Smp1.
FIG. 5.
Smp1 phosphorylations are required for Smp1 transcriptional activity. (A) Mutation of Ser348, Ser357, Thr365, and Ser376 to Ala abolishes in vivo Hog1 phosphorylation. Full-length Smp1 and the quadruple-mutant protein Smp1-m4 (mutations: Ser348, Ser357, Thr365, and Ser376 to Ala) were tested for Hog1 phosphorylation as described in the legend for Fig. 2A. After phosphorylation, the proteins were resolved by SDS-PAGE and detected by immunoblotting by using anti-GST polyclonal antibodies. (B) Hog1 phosphorylation modulates Smp1 function. A wild-type strain and the smp1Δ mutant strain (YEN44) containing the reporter STL1::lacZ were transformed with a centromeric plasmid expressing wild-type SMP1 or the quadruple-mutant SMP1-m4 (unphosphorylatable by Hog1). The cells were subjected to a brief osmotic shock, and β-galactosidase activity was assayed as described in the legend for Fig. 1B. β-Galactosidase activity is given as fold induction of control versus that of NaCl-treated cells and is the result of the measurement in quadruplicate of results for two independent transformants.
To assess the role of Hog1 phosphorylation in Smp1, we studied the effect of the mutation of the phosphorylation sites in gene expression. Wild-type and smp1Δ cells were transformed with a vector containing full-length SMP1 or the SMP1-m4 mutant allele (unphosphorylatable by Hog1) under the wild-type promoter, and the effect on transcription was measured by expression of the STL1::LacZ reporter assay as before. As shown previously, deletion of SMP1 resulted in a decrease of STL1 expression upon osmotic stress. Expression of wild-type SMP1 restored the levels of STL1 expression to wild-type levels upon stress, whereas expression of the SMP1-m4, which is no longer phosphorylatable by Hog1, was completely unable to restore STL1 expression in smp1Δ cells (Fig. 5B). Smp1 alleles carrying simultaneous mutations of Ser 348 and Ser 357 (m2) or Thr 365 and Ser 376 altered expression of STL1 only to a minor extent (STL1 expression was 76 and 90% of that of the wild-type allele, respectively). These results indicated that the Hog1 phosphorylation plays an important role in the regulation of Smp1 transcriptional activity.
Smp1 controls part of the Hog1-mediated osmostress responses.
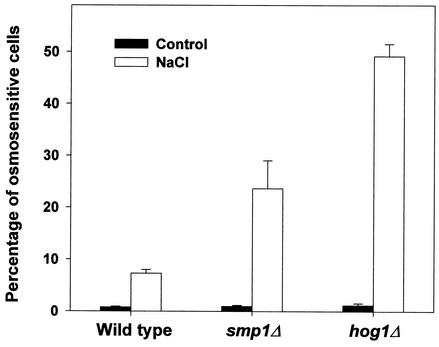
Smp1 regulates the expression of several stress-responsive genes. To understand the role of Smp1 in osmostress adaptation, we aimed to determine the importance of the presence of Smp1 to the generation of osmostress responses. Deletion of SMP1 did not cause an obvious osmosensitivity phenotype on plates containing sorbitol or NaCl (data not shown). This was similar to the results observed with cells containing deletions of any of the known transcription factors under the control of the MAPK. However, it was previously reported that hot1-deficient cells were more sensitive than wild-type cells to a severe osmotic shock (19). Thus, we tested whether smp1Δ cells were viable after an osmotic shock. Exponentially growing wild-type, smp1Δ, or hog1Δ cells were subjected to an osmotic shock (1 M NaCl for 60 min), and viability was determined as described in Materials and Methods. As depicted in Fig. 6, hog1Δ cells were highly sensitive to an osmotic shock compared to the wild-type cells. Interestingly, smp1Δ cells were also sensitive to an osmotic shock, although less sensitive than hog1Δ cells. Similarly to the results observed for the STL1-LacZ expression, a wild-type allele of SMP1 was able to suppress smp1Δ osmosensitivity, whereas the SMP1-m4 mutant allele was unable to restore cell viability. Double-mutant hog1Δ smp1Δ cells were as sensitive as hog1Δ cells (data not shown). Thus, these data indicated that Smp1 plays a role in the generation of osmostress responses, most likely by the regulation of a subset of the osmostress-responsive genes controlled by the Hog1 MAPK.
FIG. 6.
Cell survival upon osmotic stress is reduced in hog1Δ and smp1Δ strains. To analyze the role of Smp1 in osmotic stress adaptation, we measured the incorporation of PI, as described in Material and Methods, as a measure of cell viability in wild-type, hog1Δ mutant, and smp1Δ mutant strains. Yeast cells were grown in YPD and subjected (open bars) or not subjected (filled bars) to an osmotic shock (1 M NaCl for 60 min). Viability was determined by the addition of PI and measurement of the percentage of PI-positive cells by using a flow cytometer. The results are the measurements in duplicate from six independent experiments. The data were confirmed by Phloxin B staining and visual microscopy (see Materials and Methods).
Both Smp1 and Hog1 are relevant for cell viability in stationary phase.
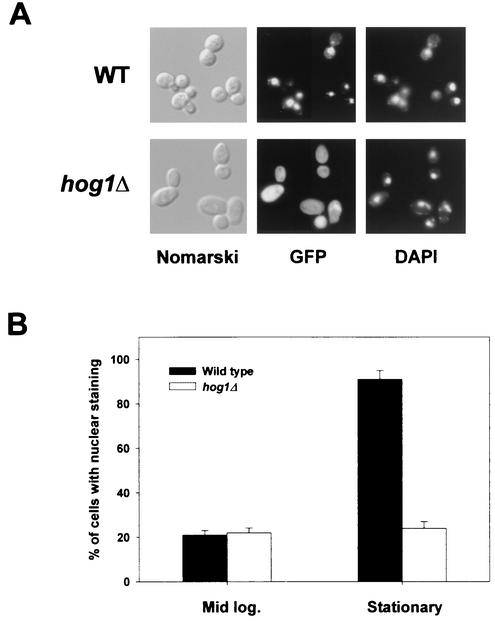
To study the mechanism of Smp1 action, we analyzed the cellular distribution of Smp1. For this purpose, we fused SMP1 to the N terminus of the GFP and expressed the fusion in yeast under its own promoter. The Smp1-GFP fusion protein was fully functional as tested for STL1-LacZ expression (data not shown). Microscopic examination of cells expressing Smp1-GFP revealed that Smp1 localizes throughout the cytoplasm and nuclei of unstressed cells. When cells carrying Smp1-GFP were exposed to a brief osmotic shock (0.4 M NaCl for 5 min), Smp1 did not change its distribution significantly. However, strong nuclear localization was observed in most of the wild-type cells when they were grown in stationary phase (Fig. 7). Nuclear localization of Smp1-GFP was abolished by elimination of Hog1 activity, either by gene deletion or by impairing its catalytic activity (Fig. 7). However, it is worth noting that distribution of the nonphosphorylatable Smp1-m4 protein fused to GFP was similar to that observed in the wild type (data not shown). These results suggested that Hog1 could be playing a role during stationary phase by indirectly controlling Smp1 localization.
FIG. 7.
Smp1 nuclear localization depends on Hog1. The wild-type and the hog1Δ mutant strains were transformed with a plasmid carrying a GFP-tagged Smp1 expressed under its own promoter. The cells were grown and Smp1-GFP was detected by fluorescence microscopy as described in Materials and Methods. (A) Smp1 localizes into the nuclei of wild-type cells in the stationary phase. Cells of the indicated types carrying Smp1-GFP were grown to the stationary phase, and images were taken. Representative images showing the localization of Smp1-GFP are presented. The positions of the nuclei were determined by DAPI staining. (B) Quantitative data were obtained from wild-type or hog1Δ cells growing at the mid-logarithmic (Mid log.) or stationary phase of growth. The data represent the results of three independent experiments which included counting 300 cells each.
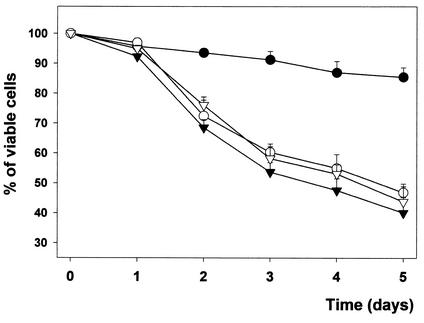
It was reported that deletion of the Hog1 homologue Spc1/StyI MAPK from Schizosaccharomyces pombe resulted in cells with less viability during the stationary phase (22). To study whether Hog1 and Smp1 were playing a role during this phase of growth, we monitored the cell viabilities of wild-type, smp1Δ, and hog1Δ cells during 5 days of culture in stationary phase as before. As shown in Fig. 8, wild-type cells maintained cell viability throughout the culture period, whereas smp1Δ and hog1Δ cells lost viability during growth in the stationary phase. Similar results were obtained with the W303 strain background (data not shown). Furthermore, smp1Δhog1Δ double-mutant cells did not show differences compared to hog1Δ cells. It is worth noting that whereas a plasmid containing wild-type SMP1 was able to restore the cell viability of a smp1Δ strain, a plasmid containing the SMP1-m4 allele (encoding the Smp1 version unphosphorylatable by Hog1) was not able to complement smp1Δ mutation (data not shown). Thus, these data suggest that apart from controlling osmostress responses, Smp1 plays an important role for cell survival during growth in the stationary phase and that this role could be mediated by the MAPK through regulation of Smp1 localization.
FIG. 8.
Hog1 and Smp1 are required for cell viability in stationary phase. Wild-type (•), hog1Δ (▿), smp1Δ (○), or hog1Δ smp1Δ (▾) cells were grown in minimal medium to stationary phase and allowed to grow for the indicated period of time. Viability was determined by adding PI and measuring the percentage of PI-positive cells by using a flow cytometer. The results are the measurements in duplicate from four independent experiments. The data were confirmed by Phloxin B staining and visual microscopy (see Materials and Methods).
DISCUSSION
Yeast cells respond to increases in osmolarity in the extracellular environment by activating the stress-activated MAPK Hog1. A major outcome of the activation of the Hog1 MAPK is the regulation of gene expression. One of the most common mechanisms by which SAPKs regulate gene expression is by modification of specific transcription factors (9), and several transcription regulators have been proposed to be controlled by the Hog1 MAPK. However, due to their DNA binding specificities and the profile of gene induction upon stress shown by DNA microarrays, they cannot account for the regulation of all of the genes under the control of Hog1. In addition, a mutant strain carrying deletions in hot1, msn1, msn2, and msn4 is not osmosensitive (19).
From these considerations, we suspected that additional transcription factors were required for the osmostress-induced regulation of gene expression by the MAPK Hog1. In this report, we describe a genetic screen in which we identified Smp1, a MEF2-like protein, as one such factor. The results of this study demonstrate that both overexpression and deletion of SMP1 result in altered expression of osmoresponsive genes such as STL1 and CWP1 but not of others, such as ALD3 or HXT1. It was reported that Smp1 has MEF2-related DNA-binding specificities (4), and a conserved sequence similar to the predicted DNA binding site for Smp1 was consistently present on the promoters of the osmoresponsive STL1 and CWP1 but not HXT1 and ALD3 genes (Fig. 1 and data not shown).
Several lines of evidence suggest that Smp1 is actually a direct substrate for the MAPK. In vivo studies suggested that Hog1 was able to interact with Smp1 and that Smp1 was phosphorylated upon stress in a HOG1-dependent manner. The relationship of Smp1 and Hog1 in vivo are further supported by the in vitro evidence that Hog1 phosphorylates Smp1 directly. Four independent phosphorylaton sites were mapped within 28 amino acids. Similarly, phosphorylation of Sko1 by Hog1 was restricted to three phosphorylation sites clustered within 19 amino acids (15). The introduction of several phosphates in a small pocket of amino acids might result in conformational changes required for switching the function of those transcriptional regulators. In vitro phosphorylation studies also showed that full-length Smp1 is phosphorylated less efficiently by Hog1 than by the C-terminal domain alone (Fig. 3, lanes 1 and 3). This observation raised the possibility that a region of Smp1 could be limiting the access of the MAPK to the phosphorylation sites. Consistent with this observation was the fact that simultaneous incubation of the C-terminal domain with an N-terminal domain (which is not phosphorylated by Hog1) resulted in a dramatic decrease of C-terminal phosphorylation (Fig. 4, lane 4). This might suggest that Smp1 must be in a preactivated state (i.e., bound to DNA or interacting with other factors) to be accessible for phosphorylation and activation by the MAPK.
Phosphorylation of Smp1 by Hog1 is important for Smp1 function. Upon osmotic stress, Smp1 is strongly phosphorylated, and mutation of the phosphorylation sites to Ala results in an Smp1 that is unable to regulate gene expression (Fig. 5). In yeast, there exists a second MEF2-related protein, Rlm1, that is under the control of the Slt2/Mpk1 MAPK. Interestingly, Mpk1 phosphorylates Rlm1 in a region similar to that found for Smp1, and this phosphorylation results in an increase of its transcriptional activity (4, 23). Thus, two independent MAPK signaling pathways could be controlling the two MEF2-related factors by a similar mechanism. In mammals, regulation of MEF2A and MEF2C factors has been shown to be under the control of the p38 MAPKs, among other kinases (reviewed in references 9 and 11). Phosphorylation of the transcription activator domain of these factors by p38 stimulates MEF2 activity, which is analogous to the mechanism proposed for the yeast Smp1.
Apart from the role of Smp1 in the regulation of a subset of osmoresponsive genes under the control of the Hog1 MAPK (and thus a role in osmoadaptation), we found that smp1Δ cells lose viability in the stationary phase, as occurs with hog1Δ cells (Fig. 8). This phenotype is reminiscent of that observed upon deletion of the Spc1/StyI MAPK in Schizosaccharomyces pombe (21, 24), and although the molecular mechanism(s) of this deficiency remains uncharacterized, the formal possibility exists that this could be caused by the lack of some specific transcript required for this phase of growth. As shown in Fig. 7, Smp1 concentrates into the nucleus when cells enter into the stationary phase, and this could be of relevance for the induction of specific genes. In hog1Δ cells, Smp1 nuclear accumulation is clearly diminished, and thus this could impede the normal function of Smp1. Because smp1Δ and smp1Δ hog1Δ cells display similar viability, it is likely that the deficiency observed in hog1Δ cells could be caused by the improper function of Smp1 rather than other mechanisms.
Taken together, our results show that Smp1 is a direct target for the Hog1 MAPK and that two different levels of regulation can be controlling the activity of this transcription factor. Under stress, Hog1 phosphorylates the putative transcription activation domain stimulating Smp1 activity, and, when entering into the stationary phase, Hog1 is required for proper localization of the transcription factor.
Acknowledgments
We thank Mercè Carmona for her technical assistance.
This work was supported by grants PM99-0028 from the Dirección General de Investigación Científica y Técnica (Ministry of Science and Technology, Spain), Distinció de la Generalitat de Catalunya per a la Promoció de la Recerca Universitària, Joves Investigadors DURSI (Generalitat de Catalunya), and the EMBO Young Investigator Program to F.P. L.C. is the recipient of an F.P.U. predoctoral fellowship from the MEC, Spain.
The first two authors contributed equally to this work.
REFERENCES
- 1.Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell 7:767-777. [DOI] [PubMed] [Google Scholar]
- 2.Bilsland-Marchesan, E., J. Arino, H. Saito, P. Sunnerhagen, and F. Posas. 2000. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 20:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 6.Gaxiola, R., I. F. de Larrinoa, J. M. Villalba, and R. Serrano. 1992. A novel and conserved salt-induced protein is an important determinant of salt tolerance in yeast. EMBO J. 11:3157-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groot, P. W. J., et al. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in S. cerevisiae. Comp. Funct. Genomics 2:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 11.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 12.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 13.Posas, F., J. R. Chambers, J. A. Heyman, J. P. Hoeffler, E. de Nadal, and J. Arino. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275:17249-17255. [DOI] [PubMed] [Google Scholar]
- 14.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component”osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 15.Proft, M., A. Pascual-Ahuir, E. de Nadal, J. Arino, R. Serrano, and F. Posas. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proft, M., and K. Struhl. 2002. Hog1 kinase converts Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1-20. [DOI] [PubMed] [Google Scholar]
- 17.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290-8300. [DOI] [PubMed] [Google Scholar]
- 18.Rep, M., M. Proft, F. Remize, M. Tamas, R. Serrano, J. M. Thevelein, and S. Hohmann. 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40:1067-1083. [DOI] [PubMed] [Google Scholar]
- 19.Rep, M., V. Reiser, U. Gartner, J. M. Thevelein, S. Hohmann, G. Ammerer, and H. Ruis. 1999. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 19:5474-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 21.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 22.Toone, W. M., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]