Abstract
There are several classes of ATP-dependent chromatin remodeling complexes, which modulate the structure of chromatin to regulate a variety of cellular processes. The budding yeast, Saccharomyces cerevisiae, encodes two ATPases of the ISWI class, Isw1p and Isw2p. Previously Isw1p was shown to copurify with three other proteins. Here we identify these associated proteins and show that Isw1p forms two separable complexes in vivo (designated Isw1a and Isw1b). Biochemical assays revealed that while both have equivalent nucleosome-stimulated ATPase activities, Isw1a and Isw1b differ in their abilities to bind to DNA and nucleosomal substrates, which possibly accounts for differences in specific activities in nucleosomal spacing and sliding. In vivo, the two Isw1 complexes have overlapping functions in transcriptional regulation of some genes yet distinct functions at others. In addition, these complexes show different contributions to cell growth at elevated temperatures.
As eukaryotes have evolved larger and more complex genomes, they have required the coevolution of enzymatic and structural mechanisms to physically organize large amounts of genetic material. For example, nearly every human cell packages approximately 2 m of DNA into a nucleus with a diameter 5 to 6 orders of magnitude smaller. This extraordinary level of compaction must, however, be flexible and organized for such processes as replication, repair, mitotic and meiotic chromosome segregation, and transcription to occur throughout the genome. The nucleosome, the primary unit of this chromatin organization, was described over 30 years ago (34). However, the higher-order structure of chromatin and its regulation are still largely undefined (32, 51).
Two groups of factors remodel chromatin to regulate a variety of cellular processes. One such group, the histone-modifying enzymes, covalently modifies histone proteins. In contrast, the ATP-dependent nucleosome-remodeling factors utilize the energy of ATP hydrolysis to reposition or alter the structure of nucleosomes along a DNA template. The first, and best characterized, of this group is the SWI/SNF complex from Saccharomyces cerevisiae, a large (∼2 MDa) complex consisting of at least 10 subunits, including the ATPase Swi2/Snf2p. Homologs of the SWI/SNF complex have been found in nearly all eukaryotes studied, with several members often present in the same organism (16, 20, 40, 45). Three other classes of ATP-dependent chromatin remodeling complexes have also been identified and are classified by the sequences of their Swi2/Snf2p-like ATPase subunits: the imitation-switch (ISWI) class, the chromodomain helicase DNA-binding (CHD)/Mi-2 class, and the most recently identified INO80 class. Like members of the SWI/SNF class, ISWI, CHD/Mi-2, and INO80 homologs have also been found in a wide variety of eukaryotes from yeasts to humans, suggesting that each class has important cellular functions that may have been evolutionarily conserved (for a review, see reference 18).
The founding member of the ISWI class, Drosophila Iswi, was originally identified by sequence homology to brahma, a SWI2/SNF2 homolog (16). It was subsequently found that this ISWI ATPase is shared among three complexes, NURF, CHRAC, and ACF, each with different enzymatic activities (for reviews, see references 7 and 50). How the activities of these complexes are utilized in vivo is not clear, yet ISWI is required for cell viability, showing that it performs critical functions. Polytene staining of ISWI and RNA polymerase II reveals mutually exclusive localization of these proteins in general, consistent with a role for ISWI in transcriptional repression. The structure of the male X chromosome is also grossly aberrant in Iswi mutant cells, suggesting that one or more ISWI-containing complexes play a role in establishment and/or maintenance of higher-order chromatin structures (14).
Both the ACF and CHRAC complexes are composed of ISWI and ACF1, a protein with several sequence motifs commonly found in transcription factors and chromatin-associated proteins. It contains two PHD fingers, a bromodomain, a WAC (WSTF, ACF1, cbp146) motif, and a WACZ (WSTF, ACF1, cbp146, ZK783.4) motif (30). In addition, the CHRAC complex contains two novel histone-fold proteins, CHRAC-14 and CHRAC-16 (10, 15). The largest subunit of the NURF complex, NURF301, has a remarkably similar arrangement of these same domains, and it also contains an N-terminal HMGA domain, which is necessary for biochemical activity of NURF (52). Proteins bearing similar motifs to those of ACF1 are associated with ISWI homologs in human and Xenopus cells as well as one of the S. cerevisiae ISWI homologs, Isw2p (4, 19, 25, 37, 41, 47). The high degree of evolutionary conservation of ISWI complex components suggests important roles for these subunits in ISWI-mediated activity. The Drosophila ISWI monomer possesses many of the activities associated with ISWI-containing complexes, although its specific activity is significantly lower than that of the full complexes (11, 30). In addition, the ISWI monomer alone does not show the directed sliding observed with intact NURF complex (52), again suggesting that the ISWI-associated proteins are required for modulating the function of the ISWI ATPase.
Another level of complexity is introduced when one considers that like Drosophila, all of the organisms studied appear to have several ISWI-containing complexes which must share and/or compete for a common ISWI ATPase. One of the human ISWI homologs, hSNF2h, is found in four separable complexes (RSF, NoRC, hACF/WCRF/hCHRAC, and WICH [4, 6, 37, 38, 41, 47]), while the Xenopus homolog also appears to form four complexes (xACF, xWICH, and xISWI-A, and xISWI-D [6, 25]). Each of the two mouse ISWI homologs, mSnf2h and mSnf2l, has distinct and separate expression patterns, implying that each homolog has unique roles in vivo (36). How these distinct ISWI complexes work within a single organism and how the other subunits modulate ISWI activity remain unknown.
The budding yeast, S. cerevisiae, expresses two homologs of ISWI, Isw1p and Isw2p. Isw2p associates with Itc1p (which contains a WAC motif), to form a heterodimer (19). The Isw2 complex is recruited to the promoters of several early meiotic genes by the transcription factor Ume6p, working in a parallel pathway with the Sin3-Rpd3 histone deacetylase complex to repress these genes during vegetative growth (21). Isw2 complex-dependent transcriptional changes at these meiotic promoters, as well as some nonmeiotic gene promoters, are associated with the formation of a nuclease-inaccessible chromatin structure (17, 31).
In contrast, the functions of Isw1 complexes in vivo are largely unknown. Isw1p copurifies with three other proteins of 110-, 105-, and 74-kDa sizes (49) and has been shown to be required for local remodeling of chromatin (31). Here we identify these associated proteins and show that Isw1p forms two distinct complexes in vivo. These two Isw1 complexes both exhibit equivalent nucleosome-stimulated ATPase activities, suggesting that each complex is purified in its active form. However, the two complexes differ in their abilities to interact with DNA and nucleosomes and have different specific activities to alter chromatin structure in vitro. DNA microarray analysis and phenotypic characterization revealed that the two Isw1 complexes have overlapping, but distinct, functions in vivo.
MATERIALS AND METHODS
Media and strains.
Yeast strains were grown and manipulated according to standard procedures (1). All strains used in this study were derived from W303, in which a weak rad5 mutation was repaired (48, 54), and are listed in Table 1.
TABLE 1.
Strains used
| Strain | Description |
|---|---|
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| W1588-4C | MATa; same as W303-1A but RAD5 |
| YTT227a | MATaisw1::ADE2 isw2::LEU2 chd1::TRP1 |
| YTT396 | MATα pep4::HIS3 |
| YTT441 | MATaisw1::kanMX |
| YTT442 | Same as YTT441 |
| YTT443 | MATaisw1::kanMX isw2::HISG |
| YTT449 | MATα ISW1-3′ 2xFLAG (ISW1-2FL) pep4::HIS3 |
| YTT512 | MATaISW1-2FL ioc2::kanMX pep4::HIS3 |
| YTT522 | MATaioc2::kanMX isw2::LEU2 chd1::TRP1 |
| YTT548 | MATaISW1-2FL ioc4::kanMX pep4::HIS3 |
| YTT610 | MATα ISW1-2FL ioc3::HIS3 pep4::HIS3 |
| YTT642 | MATaISW1-2FL IOC2-3MYC IOC3-3HA |
| YTT823 | MATaioc2::kanMX |
| YTT825 | MATaioc3::natMX |
| YTT829 | MATaioc2::kanMX ioc3::natMX |
| YTT853 | MATaioc3::natMX isw2::LEU2 chd1::TRP1 |
| YTT855 | MATaioc4::hphMX isw2::LEU2 chd1::TRP1 |
| YTT861 | MATaioc2::kanMX ioc3::natMX isw2::LEU2 chd1::TRP1 |
| YTT1167 | MATα IOC2-TAP:TRP1 pep4::HIS3 |
| YTT1168 | MATα IOC3-TAP:TRP1 pep4::HIS3 |
All YTT strains are derived from W1588-4C.
ISW1 was tagged with two copies of the FLAG epitope sequence at its 3′ end (49) and integrated at the ISW1 genomic locus by using the URA3 pop-in/5-FOA pop-out method (5). Integration of the 3′ triple-MYC or 3′ triple-hemagglutinin (HA) epitope sequence for Iswi one complex 2 (IOC2) and IOC3 was done using a PCR-based strategy that utilized pMPY-3xMYC and pMPY-3xHA, respectively (46).
Disruption of genes with the dominant drug resistance cassette kanMX, natMX4, or hphMX4 was accomplished using a PCR-based strategy based upon the plasmid pUG6, pAG25, or pAG32, respectively (22, 24).
The ISW1 genomic locus was fused to the tandem affinity purification (TAP) tag by a PCR-based strategy utilizing the plasmid pBS1479 as a template, generously provided by Bertrand Séraphin (43).
Purification of Isw1 complexes for in vitro assays.
All whole-cell yeast extracts described in this paper were created by growing cultures to saturation, unless otherwise noted. Purification of Isw1 complexes was done as previously described (49). Purification of Isw1p monomer was done concurrently to Isw1 complex purification from either strain YTT449 or from congenic strains (strain YTT512, YTT548, or YTT610) bearing deletions in single IOC genes. All such strains yielded identical Isw1p monomers in addition to Isw1a and/or Isw1b complexes. Following anti-FLAG immunoprecipitation (IP) and elution, the purified proteins were applied to a BioRex 70 cation exchange column in buffer H (25 mM HEPES-KOH [pH 7.6], 0.5 mM EGTA, 0.1 mM EDTA, 2 mM MgCl2, 20% glycerol, 0.02% NP-40, 1 mM dithiothreitol, 0.5 mM Na2S2O5, 1 mM phenylmethylsulfonyl fluoride, 2 μM pepstatin A, 0.6 μM leupeptine, 2 μg of chymostatin/ml, 2 mM benzamidine) with a salt concentration of 0.2 M KCl. Isw1p monomer does not bind to the resin under this condition, whereas both complexes are retained. The bound Isw1 complexes were eluted by a linear salt gradient from 0.2 to 0.6 M KCl as previously described (49).
Size exclusion chromatography.
Whole-cell extract of YTT642 grown to saturation was prepared in buffer H-0.3 M KCl. A total of 1 ml of this extract was applied to a Superose 6 10/30-size exclusion column (Amersham Pharmacia Biotech, Piscataway, N.J.) preequilibrated in buffer H-0.3 M KCl, and 0.5-ml fractions were collected. Following complete elution, gel filtration size standards (Bio-Rad Laboratories, Hercules, Calif.) were run under identical conditions.
Fractions were loaded onto nine identical sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-8% PAGE) gels and transferred to nitrocellulose membranes. Detection of Isw1p, Ioc2p, or Ioc3p was done in triplicate using α-FLAG M2, α-MYC 9E10, or α-HA 16B12 monoclonal antibodies, respectively, and quantified using an Odyssey infrared imaging system and fluorescently labeled α-mouse antibody (Li-Cor Biosciences, Lincoln, Nebr.). The integrated intensities of the bands in each gel were normalized to their mean intensity. To account for quantitative differences due to loading variability, these values were averaged with the corresponding lanes for each triplicate blot to determine the elution profiles for each protein, although the elution profiles for any individual blot were similar to those of the averaged values.
IPs.
FLAG, MYC, and HA IPs were done using whole-cell extract from YTT642 grown to saturation. Protein G-Sepharose (Amersham Pharmacia Biotech) was precoupled to either α-HA 16B12 monoclonal antibody or α-MYC 9E10 monoclonal antibody (Covance, Inc., Princeton, N.J.). Each precoupled resin preparation, α-FLAG M2 agarose resin, or protein G resin alone was added to 200 μl of YTT642 extract and incubated at 4°C for 4 h. Each immunoprecipitate was washed once in buffer H-0.3 M KCl and twice in buffer H-0.1 M KCl before being eluted in SDS loading buffer at 95°C for 5 min. Samples of starting material, supernatants, and immunoprecipitates of each IP were loaded on SDS-8% PAGE gels, transferred to nitrocellulose, probed with either α-MYC 9E10, α-HA polyclonal, or α-FLAG M2 and appropriate horseradish peroxidase-conjugated secondary antibodies, and visualized with SuperSignal West Femto chemiluminescent substrate (Pierce, Rockford, Ill.).
TAP precipitations were done using whole-cell extract from 12-liter cultures grown to an optical density at 660 nm of 0.7. For each extract, 0.8 ml of Pharmacia immunoglobulin G (IgG) Sepharose 6 Fast Flow suspension and 0.8 ml of Stratagene calmodulin affinity resin were used. Purification was done as previously described (43), with the following changes: (i) the IgG precipitation was done in extract prepared in buffer H-0.3 M KCl, with NP-40 added to 0.1%, without dialysis; (ii) both precipitations were done by the batch method in 50-ml or 15-ml tubes rather than in a column, and one 5-min wash step was added following both incubations before transference of the mixture to a column for subsequent washing; (iii) the KCl concentration of the buffer used for washing the IgG precipitation was raised to 0.3 M; (iv) NP-40 was omitted from the calmodulin binding and elution buffers; and (v) additional final elution fractions were collected to increase yields.
ATPase assays.
ATPase assays were performed as described previously, using 0.3 pmol of nucleosomes assembled from purified Drosophila histones (49), except that 30 fmol of Isw1 complexes was used in each 5-μl reaction mixture.
Nucleosomal spacing assays.
Nucleosomal spacing assays were done essentially as previously described (49), except 0.22 μg (2 pmol) of recombinant yeast octamer (19) was used with 30 fmol of Isw1 complexes and 0.75 μg of recombinant yeast Nap1p. This mixture was incubated for 30 min on ice, prior to the addition of DNA and ATP.
Nucleosomal sliding assays.
The DNA used for reconstitutions was a 214-bp EcoRI-DdeI fragment of DNA derived from the Xenopus borealis somatic 5S rRNA gene.
Mononucleosomes were assembled at 37°C by salt dilution with 10 μg (92 pmol) of recombinant octamers, 100 fmol of labeled 5S ribosomal DNA (rDNA), 10 μg of salmon sperm DNA, and 1.8 M NaCl in a starting volume of 10 μl. Salmon sperm DNA was prepared as described by Sambrook et al. (44), the DNA being sheared to a 100- to 700-bp length by sonication. Stepwise dilution of NaCl was carried out in three stages (1 M, 714 mM, and 270 mM) by the addition of 6.8 μl, 8.4 μl, and 42 μl of buffer D (25 mM Tris-HCl [pH 8.0], 1 mM β-mercaptoethanol), respectively, at 10-min intervals. Nucleosome assemblies were analyzed on a 4% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 38.9:1.1) in 0.5× Tris-borate-EDTA buffer at 4°C.
In a standard 12.5-μl reaction mixture, Isw1a and Isw1b were incubated in the molar ratios shown with 12 ng of nucleosome assemblies and 400 μM ATP, at 30°C for 30 min, under the following reaction conditions: 30 mM HEPES-KOH (pH 7.6), 5 mM MgCl2, 37 mM KCl, 8.8 mM NaCl, 0.1 mM EGTA, 0.02 mM EDTA, 10% glycerol, and 0.1 μg of bovine serum albumin/μl. Of the 12.5-μl reaction mixture, 4 μl was analyzed by electrophoretic mobility shift assay on a 5% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 60:1) in 0.2× Tris-borate-EDTA with buffer recirculation at 4°C.
Immobilized template interaction assays.
Streptavidin-coated magnetic particles (Dynal Biotech, Lake Success, N.Y.) were bound to linearized pBluescript SK(−) DNA linearized with ClaI/EcoRI, which was end filled with dCTP, dGTP, dTTP, and biotinylated dATP. Recombinant histones were loaded with rNAP1p, followed by a wash in buffer containing 600 mM NaCl to remove Nap1p and nonnucleosomal histones as previously described (19). A total of 12.5 ng (or 75 ng for silver detection) of DNA equivalent of beads with nucleosomal or naked DNA were incubated with 1 ng (7.6 fmol) of Isw1 equivalent (or 6 ng for silver detection) of Isw1 monomer, Isw1a, or Isw1b complex with or without 1.7 mM ATP in 10 mM Tris (pH 7.6)-50 mM NaCl-5 mM MgCl2-1 mM EDTA-0.05% NP-40-0.1 mg of bovine serum albumin/ml for 30 min at 30°C in an Eppendorf Thermomixer at 1,200 rpm. Bound fractions were washed for 1 min in buffer at 30°C before boiling in sample buffer. Proteins were visualized by using a silver-stained SDS-PAGE gel, or Isw1p was detected by Western blotting using α-FLAG antibody.
Electrophoretic mobility shift assays.
Mononucleosomes were assembled onto clone 601, a 255-bp DNA fragment with a high-affinity nucleosome positioning sequence (39). Binding assays were done under conditions similar to those of the sliding assays (see “Nucleosomal sliding assays”) but with no ATP. Nucleosomes (41 fmol) were incubated with 41 fmol, 123 fmol, or 369 fmol of either the Isw1a or the Isw1b complex. These were analyzed by an electrophoretic mobility shift assay on a 4% native polyacrylamide gel in 2× Tris-EDTA buffer (10 mM Tris [pH 8.0], 1 mM EDTA) with buffer recirculation at 4°C. For antibody supershift, 369 fmol of Isw1a complex was first prebound to 41 fmol of mononucleosomes as described above. For a second incubation reaction, the prebound complex was incubated with 46 pmol of α-FLAG antibody for an additional 15 min. The reaction products were analyzed as described above.
Expression analyses.
RNA was harvested from cultures grown to an optical density at 660 nm of 0.7, and Northern blots were run as described previously (21) except that 30 μg of RNA was loaded into each lane. Blots were hybridized with [α-32P]dCTP-labeled probes for the genes indicated and quantified with a PhosphorImager.
DNA microarray analyses were done using 30 μg of total RNA from each strain, as previously described (17). We utilized a number of different previously described methods to normalize our expression data. Briefly, a Bayesian background correction method was applied to reduce the variance of spots of low intensity (17, 33). These corrected data from each microarray slide were then normalized to account for bias due to spot intensity (intensity-dependent normalization using a Lowess smoother to account for nonlinearity) and the position of each cDNA on the array grid (within-print-tip-group normalization). In addition, conversely labeled slide pairs were normalized to each other to account for dye-specific differences in labeling efficiency and/or dye stability (paired-slides normalization) and separate slides were normalized to each other to reduce absolute expression differences that introduce bias when making slide-to-slide comparisons (multiple slide normalization) (53).
The expression changes for each gene were then determined by calculating the median value from three (isw1 mutants) or four (ioc2, ioc3, and ioc2ioc3 mutants) independent microarray hybridizations. Clustering analysis was done utilizing a GeneSpring version 4.0.4 program (Silicon Genetics, Redwood City, Calif.) and data that had undergone only Bayesian background correction to allow direct comparison with previously published microarray data for isw2, rpd3, isw2rpd3, and sin3 strains (17, 33). Clustering was weighted more heavily for the Isw1 complex mutants to prevent bias due to stronger phenotypes of Sin3/Rpd3 mutations.
RESULTS
Isw1p copurifies with three previously uncharacterized proteins.
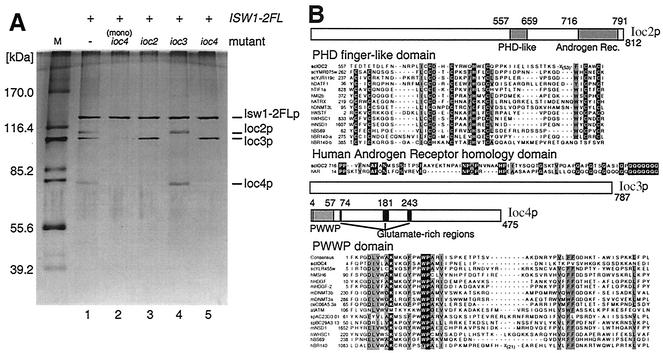
Isw1p was previously shown to copurify with three proteins, p110, p105, and p74 (49). To identify these three proteins, Isw1p was purified as described previously (Fig. 1A, lane 1) and each subunit was subjected to mass spectrometric analysis. The sequences of the 110-kDa, 105-kDa, and 74-kDa proteins were encoded by previously uncharacterized open reading frames identified as YLR095c, YFR013w, and YMR044w, respectively (yeast proteome database) (12, 13). We have designated them as IOC2, IOC3, and IOC4, respectively (Fig. 1B). Ioc2p has a putative PHD finger motif between amino acids 557 to 659, a domain found in many chromatin-associated proteins. PHD finger domains typically have seven universally conserved cysteine residues and one histidine residue (C4HC3), which are thought to coordinate two zinc ions (9). While Ioc2p has a high degree of conservation around the central four conserved residues, the homology to other PHD fingers is lost in the N-terminal region and separated by 70 residues (12 to 46 residues is normal) from the C-terminal region (see alignment in Fig. 1B). It is unclear whether this represents a recent degeneration of an ancestral PHD finger structure or whether this region of Ioc2p is an atypical PHD-like domain. A truncated PHD finger is also found in human Peregrin (hBR140-b [Fig. 1B]), though this protein also contains a separate intact PHD finger (hBR140-a [Fig. 1B]). Ioc2p also has homology to a human androgen receptor near its C-terminal end, though this region has a polyglutamine tract that accounts for much of this homology. The function of this region of human androgen receptor is unclear, but mutations within it are associated with prostate cancer (23). Ioc3p has significant homology (27%) to a protein involved in silencing, Esc8p (yeast proteome database [12, 13]), yet has no identifiable conserved motifs. Ioc4p is a glutamic acid-rich protein with a PWWP motif between amino acids 4 and 76. The PWWP motif is a putative DNA-binding domain and is found in a number of proteins involved in transcriptional and chromatin regulation, such as the putative de novo DNA methyltransferase 3 (Dnmt3); the nuclear corepressor, adenovirus E1A binding protein (BS69); and the TAF250 homolog, Peregrin (BR140) (Fig. 1B). The PWWP domain of murine Dnmt3b can bind DNA in vitro, and mutations of this domain within Dnmt3a prevent its heterochromatin association (42). In addition to PWWP homology, Ioc4p also has overall homology to PWWP-containing S. cerevisiae and S. pombe proteins Ylr455p and SPBC29A3.13 (24% identity for both), respectively (yeast proteome database [12, 13]). In addition to Isw1p associated with Ioc proteins, a significant amount of Isw1p was purified as an apparent monomer (Fig. 1A, lane 2) from both wild-type and ioc mutants using FLAG affinity and Bio-Rex cation exchange chromatography (see Materials and Methods).
FIG. 1.
Identification of the proteins associated with Isw1p: Ioc2p, Ioc3p, and Ioc4p. (A) Isw1-2xFLAGp (Isw1-2FLp) was purified from strain YTT449 (lane 1) or congenic strains bearing ioc2 (YTT512; lane 3), ioc3 (YTT610; lane 4), or ioc4 (YTT548; lane 5) mutations. Isw1-2FLp monomer (mono) was also purified from all of these strains (Isw1-2FLp from ioc4 deletion strain YTT548 is shown; lane 2). (B) Protein motifs for Ioc2p, Ioc3p, and Ioc4p were determined by database searching. Residue positions for each motif are shown. Ioc2p has a PHD finger-like motif and limited homology to human androgen receptor in its C-terminal region. Alignment to other PHD finger-containing proteins is shown (the core PHD cysteine and histidine residues are shown in white type on black, while other conserved residues are shown in black type on grey), as is alignment to the human androgen receptor homology region. Ioc3p has no recognized sequence and/or structural motifs. Ioc4p is a glutamate-rich protein with a PWWP domain near its N terminus. Alignment to other PWWP domains is shown. Residues conserved in >80% of aligned sequences are shown in white type on black when identical or black type on grey when similar (National Center for Biotechnology Information conserved domain database [2] and Blocks database [27]). Accession numbers and descriptions for the aligned proteins are shown in Table 2.
Isw1p forms two distinct complexes in vivo.
To reveal interactions among Isw1 complex subunits, Isw1p was immunoprecipitated from strains from each of which the IOC2, IOC3, or IOC4 gene was deleted (Fig. 1A, lanes 3, 4, and 5). When Isw1p was immunoprecipitated from each deletion strain, the corresponding protein was absent from the complex, confirming the identities of the subunits. However, in an ioc4 strain we also found that Ioc2p failed to coimmunoprecipitate with Isw1p. Conversely, Ioc4p did not coimmunoprecipitate with Isw1p in an ioc2 strain. This suggests that Ioc2p and Ioc4p are mutually dependent for their stable association with Isw1p.
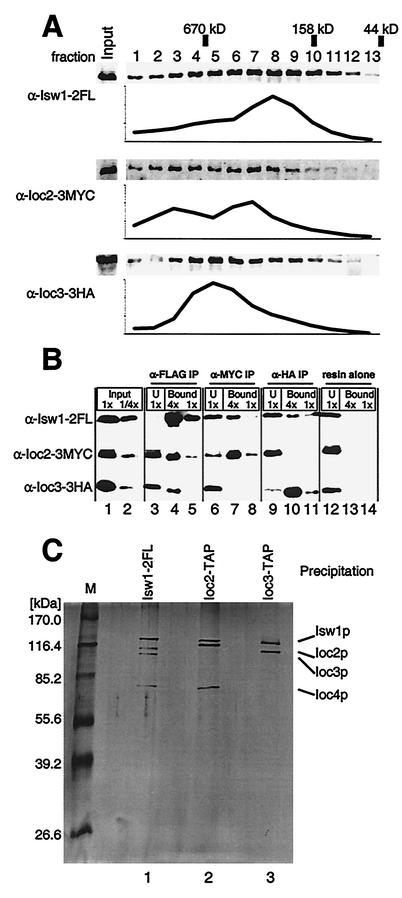
We hypothesized that the complexes purified from ioc3 strains (composed of Isw1p, Ioc2p, and Ioc4p) and those purified from either ioc2 or ioc4 strains (composed of Isw1p and Ioc3p) might represent two distinct and separable complexes which normally coexist in wild-type strains. We constructed a strain (YTT642) that bears unique epitope tags on Isw1p, Ioc2p, and Ioc3p to directly test this hypothesis. Whole-cell extract was made from this strain and subjected to size exclusion chromatography. The presence of Isw1p, Ioc2p, and Ioc3p was detected by Western blotting, and the relative level of each protein across the collected fractions was determined. As shown in Fig. 2A, the elution profiles of Isw1p, Ioc2p, and Ioc3p overlap significantly. Ioc3p elutes in a single peak in which Isw1p is also detected (fraction 5); however, Ioc2p reproducibly forms two apparently distinct peaks (fractions 3 and 7) that are out of phase with the Ioc3p peak. It is probable that one such population represents its associated Isw1 complex while the other may represent Ioc2p in either monomeric form or in another as yet unidentified complex. Isw1p was detected in all of the fractions in which Ioc2p and Ioc3p are detected; however, the peak of Isw1p (fraction 8) is smaller in molecular mass than any of the Ioc2p and Ioc3p peaks (fractions 3 and 7 and fraction 5, respectively). This suggests the presence of an Isw1p population that is not associated with Ioc2p or Ioc3p; therefore, this likely represents a monomer form of Isw1p.
FIG. 2.
Isw1p is found in two distinct complexes in vivo. (A) Whole-cell extract from strain YTT642 was separated by size exclusion chromatography. Isw1p, Ioc2p, and Ioc3p were detected by Western blotting using α-FLAG, α-MYC, and α-HA antibodies, respectively. The relative amounts of protein in each fraction were quantified, and the results were averaged across three identically loaded Western gels for each antibody to generate the elution profiles shown. The positions of size standards run in identical conditions are indicated. Note that the protein levels shown are in arbitrary units and are relative to the other fractions for each antibody-protein pair only; therefore, direct comparison of the relative levels of different proteins is not possible. (B) IP of YTT642 whole-cell extract was done usingα-FLAG (lanes 3 to 5), α-MYC (lanes 6 to 8), or α-HA (lanes 9 to 11) resin or resin alone as a control (lanes 12 to 14). The presence of Isw1p, Ioc2p, or Ioc3p in each experiment was determined by analysis of separate Western blots of identically loaded SDS-8% PAGE gels. Starting material was loaded at either 1× (undiluted) or at 1/4× dilutions (lanes 1 and 2, respectively), and undiluted unbound (U) samples from each IP were loaded for comparison (lanes 3, 6, 9, and 12). The immunoprecipitate was eluted in SDS-PAGE sample buffer, and appropriate volumes were loaded to correspond to 4× and 1× concentrations of the starting material (lanes 4 and 5, lanes 7 and 8, lanes 10 and 11, and lanes 13 and 14, respectively) as indicated. (C) Isw1-2FLp was purified from strain YTT449 as described before and loaded in lane 1 of an SDS-8% PAGE gel. Ioc2p and Ioc3p were purified from strains YTT1167 and YTT1168, respectively, by using the TAP method. The most concentrated eluates from each purification were loaded into lanes 2 and 3, respectively. The proteins were visualized by silver staining. Note that the 2× FLAG epitope on Isw1p that adds ∼2 kDa to this protein is in lane 1 only. In addition, the calmodulin binding peptide that remains after TAP purification adds ∼5.1 kDa to Ioc2p (lane 2) and to Ioc3p (lane 3).
The distinct elution profiles of Ioc2p and Ioc3p are consistent with their presence in two separable complexes; however, the extensive overlap of the profiles does not rule out a single four-subunit Isw1 complex. To more rigorously separate these complexes, we utilized the distinct epitope tags in this extract to immunoprecipitate each protein individually. As shown in Fig. 2B (lanes 3 to 5), the IP of Isw1p coprecipitated Ioc2p and Ioc3p, as observed previously (Fig. 1A) (49). Although Isw1p was nearly immunodepleted from the whole-cell extract, a significant portion of Ioc2p and Ioc3p remained in the supernatant. This suggests that some of these proteins may exist either as monomers or in another complex in vivo; however, it is also possible that their associations with Isw1p became unstable during IP. Anti-Myc antibodies were used to immunoprecipitate Ioc2p. As expected, Isw1p was coprecipitated; however, no detectable Ioc3p was precipitated (Fig. 2B, lanes 6 to 8). Conversely, the IP of Ioc3p with anti-HA antibodies pulled down Isw1p but not Ioc2p (Fig. 2B, lanes 9 to 11). This is consistent with our hypothesis that Isw1p forms two distinct complexes in vivo.
To independently verify these results, as well as to identify what other proteins, if any, Ioc2p and Ioc3p associate with in vivo, we used the TAP method to purify each protein separately. TAP-tagged strains were created for each protein, as previously described (43). TAP purification from whole-cell extracts of mid-log-phase cells resulted in highly purified complexes containing the tagged protein. Purification from either mid-log- phase or saturated cultures yielded identical complexes (data not shown). Ioc2p copurified two other proteins in approximately stoichiometric amounts (Fig. 2C). These proteins were subsequently identified by mass spectrometry as Isw1p and Ioc4p (data not shown). Notably, there was no band corresponding to Ioc3p. Ioc3p copurified one other protein in near-stoichiometric amounts, which was subsequently identified by mass spectrometry as Isw1p (data not shown). Ioc2p and Ioc4p were notably absent from this purified complex, as were any other proteins with which Ioc3p might associate. A control purification from an untagged strain (YTT396) had no detectable proteins by silver staining (data not shown).
We saw no evidence by the approaches described above that Isw1p forms a single complex with all three Ioc proteins. As a result, we conclude that there are two stable complexes containing Isw1p that coexist within yeast organisms. We have designated the Isw1p-Ioc3p and Isw1p-Ioc2p-Ioc4p complexes as Isw1a and Isw1b, respectively. This nomenclature is used throughout the remainder of this article.
Both Isw1a and Isw1b complexes show nucleosome-dependent ATPase activity.
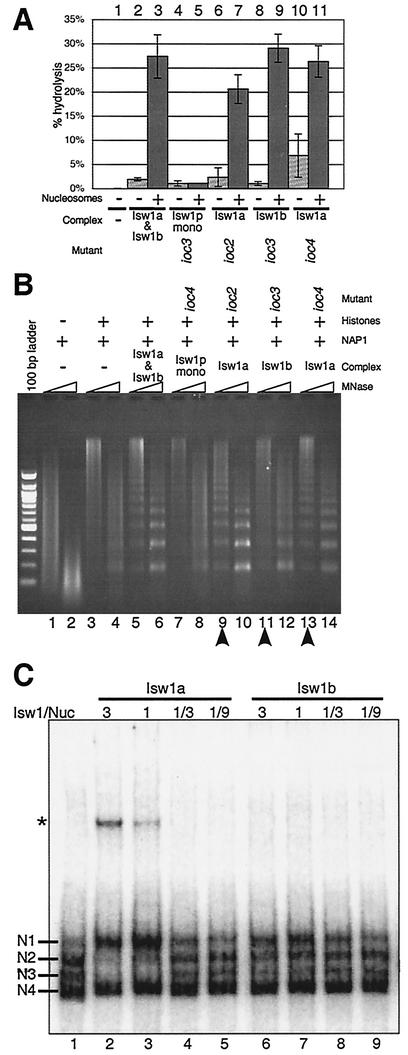
We sought to compare the biochemical properties of the two Isw1p-containing complexes. It had previously been shown that a mixture of the two Isw1 complexes purified from a wild-type strain exhibited an ATPase activity that was stimulated by nucleosomes but not by histones or DNA alone (49). Approximately equimolar amounts of Isw1p monomer and Isw1a or Isw1b complex (as determined by normalizing the Isw1p subunit in silver-stained SDS-PAGE gels) were assayed for ATPase activity in the presence or absence of nucleosomes. Both Isw1 complexes had similar nucleosome-stimulated ATPase activities, while the ATPase activity of Isw1 monomer was not stimulated by nucleosomes (Fig. 3A). Using various amounts of Isw1 complexes, we confirmed that these ATPase activities were within linear ranges (data not shown). This suggests that Ioc3p as well as Ioc2p and/or Ioc4p play essential roles in stimulating the ATPase activity of Isw1 complexes in a nucleosome-dependent manner. As complexes purified from either ioc2 or ioc4 strain backgrounds had identical subunit compositions (consisting of Isw1p and Ioc3p only) (Fig. 1A), we expected that these complexes would have similar activities (Fig. 3A, lanes 6 and 7 and lanes 10 and 11). The slight differences we observed were likely due to variability in quantitation of the complexes and/or experimental procedures.
FIG. 3.
Isw1a and Isw1b complexes exhibit different specific activities in biochemical assays. (A) In vitro nucleosome-stimulatedATPase assays for each complex. Reconstituted nucleosomes (+; lanes 3, 5, 7, 9, and 11) or buffer alone (−; lanes 1, 2, 4, 6, 8, and 10) were incubated with [γ-32P]ATP and either no complex (lane 1) or 30 fmol of Isw1 complexes in the form of copurified Isw1a and Isw1b complexes (lanes 2 and 3), Isw1p monomer (lanes 4 and 5), Isw1a complex from an ioc2 strain (lanes 6 and 7) or an ioc4 strain (lanes 10 and 11), or Isw1b complex from an ioc3 strain (lanes 8 and 9). The percentages of ATP hydrolysis were determined by the 32Pi/[γ-32P]ATP ratios. The results shown are the averages of three typical assays ± 1 standard deviation. (B) Nucleosome spacing assays for each complex. Recombinant Nap1p (lanes 1 to 14) was allowed to assemble yeast histones (lanes 3 to 14) onto λ DNA in the presence of either no complex (lanes 1 to 4) or 30 fmol of Isw1 complexes in the form of copurified Isw1a and Isw1b complex (lanes 5 and 6), Isw1p monomer (lanes 7 and 8), Isw1a complex from an ioc2 strain (lanes 9 and 10) or ioc4 strain (lanes 13 and 14), or Isw1b complex from an ioc3 strain (lanes 11 and 12). After assembly, DNA was digested with micrococcal nuclease (MNase) for 3 min (odd lanes) or 15 min (even lanes), purified, and run on 1.3% agarose and visualized with ethidium bromide. (C) Nucleosome sliding assays for Isw1a and Isw1b complexes. Recombinant octamer was assembled onto 5S rDNA (214 bp). The efficiency of sliding was checked with 48 fmol of labeled nucleosomes and either Isw1a (lanes 2 to 5) or Isw1b (lanes 6 to 9) in the ratios shown and 400 μM ATP. Nucleosome positions prior to mobilization are labeled N1 to N4. The asterisk denotes a band that is specific for the Isw1a complex. It is not observed with DNA alone.
The Isw1a complex exhibits greater specific activity in nucleosome spacing.
To further compare these two complexes, we tested different forms of Isw1 for the ability to facilitate the regular spacing of nucleosomes. We used an in vitro system to assemble recombinant yeast histones onto naked DNA in the presence of a yeast histone chaperone, Nap1p (19). In the absence of any exogenous spacing activity, Nap1p will randomly assemble nucleosomes along a DNA template. This array can then be digested with micrococcal nuclease (MNase), which preferentially cuts between nucleosomes. Digestion of such a randomly spaced array results in a population of differently sized fragments of DNA, which appear as a smear when the purified DNA is run on an agarose gel (see Fig. 3B, lanes 3 and 4). However, the addition of a mixture of the two Isw1 complexes and ATP produces a nucleosome ladder in which distinct bands are observed approximately every 175 bp (49). This is due to an ATP-dependent activity of an Isw1 complex(es) that positions a majority of the assembled nucleosomes in a regularly spaced manner.
To determine whether this spacing activity depends upon one or both Isw1 complexes, we tested each Isw1 complex in such a nucleosome spacing assay. As previously shown, a clear nucleosomal ladder was observed when the mixture of Isw1 complexes was used (Fig. 3B, lanes 5 and 6). However, consistent with its ATPase activity, Isw1p monomer did not affect the MNase digestion pattern (compare Fig. 3B, lanes 3 and 4 and lanes 7 and 8). The Isw1a complex, purified from either ioc2 or ioc4 strains, exhibited nucleosome spacing activity (Fig. 3B, lanes 9 and 10 and lanes 13 and 14). However, the spacing activity of equimolar amounts of the Isw1b complex (Fig. 3B, lanes 11 and 12) was less than that of the Isw1a complex. The qualitative difference between the two complexes was reproducible in several independent assays and was most noticeable after short MNase digestions (Fig. 3B; compare lanes 9 and 13 to lane 11), for which the ladder is visible much higher in the gel. This suggests that of the two complexes, the Isw1a complex has greater specific activity and precision in spacing multiple nucleosomes as these higher bands represent long arrays of regularly spaced nucleosomes.
The Isw1a complex exhibits greater specific activity in nucleosome sliding.
Remodeling of nucleosomal arrays, as in the spacing assay, could occur by a sliding mechanism in which nucleosomes track along the DNA template. Several ATP-dependent chromatin remodeling complexes exhibit such sliding activity, often differing from each other by the direction in which nucleosomes are moved (i.e., whether they slide nucleosomes to the ends of a DNA fragment or to the middle). The directionality of this activity can be modified by the ATPase-associated proteins. For example, Drosophila ISWI monomer slides histones to the DNA ends while ISWI-containing NURF, ACF, and CHRAC complexes slide histones to a central position (15, 26, 35). To investigate which, if any, of the Isw1 complexes could mobilize nucleosomes, we incubated Isw1a or Isw1b complexes with yeast nucleosomes assembled by salt dialysis with recombinant histones and a 214-bp 5S rDNA fragment. In the absence of an Isw1 complex, nucleosomes adopted four separable positions we have designated as N1 to N4 (Fig. 3C, lane 1). Previous work has shown that DNA fragments with nucleosomes positioned at either end (e.g., N4) migrate more quickly than do those with nucleosomes positioned more centrally (e.g., N1) (26, 35). Following incubation with increasing amounts of Isw1a complex, the N4 position remained unchanged while nucleosomes positioned at N2 and N3 were reduced and the N1 population was increased (compare Fig. 3C, lanes 1 to 5). This shows that the Isw1a complex exhibits a nucleosome mobilization activity, in agreement with the observed nucleosome spacing activity (Fig. 3B). Interestingly, using equivalent amounts of Isw1b complex we saw relatively little change in positions (Fig. 3C, lanes 6 to 9), though at higher Isw1b/nucleosome ratios there may have been a slight increase in the N1 population (compare lane 6 to lanes 1 and 9). We estimate that the Isw1b complex has a specific activity at least ninefold lower than that of the Isw1a complex in this assay. These results support those obtained in our nucleosome spacing assay, which may be mechanistically similar. We also noted the presence of a lower-mobility band that possibly represents the Isw1a complex bound to the nucleosomal substrate.
Isw1a complex binds to DNA and nucleosomes in an ATP-independent manner.
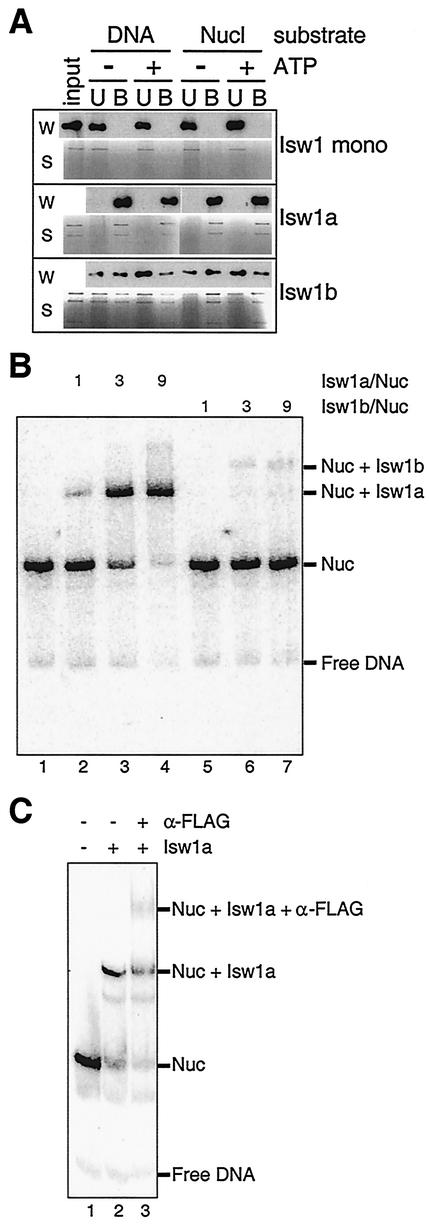
To determine the mechanisms underlying the differences in specific activities that we observed for the two Isw1 complexes as well as to investigate the possible binding of Isw1a complex to its nucleosomal substrates observed in the sliding assay, we tested how these complexes interact with DNA and nucleosomes. To this end, we employed two assays, an immobilized template assay (19) and an electrophoretic mobility shift assay.
In the first assay, nucleosome arrays were assembled using recombinant histones and DNA immobilized onto magnetic beads. After mixing immobilized naked DNA or nucleosomal templates with Isw1 complexes, unbound and bound Isw1 species were detected by α-FLAG Western blotting or by silver-stained gels (Fig. 4A). Consistent with our ATPase and nucleosome spacing assays, Isw1 monomer failed to interact with DNA or nucleosome arrays. However, the Isw1a complex interacted strongly with nucleosomal as well as naked DNA templates. The Isw1b complex interacted with both templates as well, though this interaction was not as robust as that of Isw1a. The Ioc proteins associated with Isw1p are visible by silver staining in all fractions in which Isw1p is detected by Western blotting, confirming that binding requires the intact complexes. All of these interactions were ATP independent, suggesting that the enzymatic activity of Isw1 complexes is utilized at a step following binding, as observed previously for the Isw2 complex (19). None of the Isw1 species interacted with beads alone (data not shown).
FIG. 4.
Isw1a and Isw1b bind DNA and nucleosomes in an ATP-independent manner. (A) Immobilized template assays for DNA and nucleosome interactions with Isw1 complexes. Magnetic beads were bound with either naked DNA or nucleosomal DNA. Isw1 monomer, Isw1a, or Isw1b was allowed to interact with beads in the presence or absence of ATP. Following incubation, unbound (U) and bound (B) fractions were detected by using silver-stained SDS-PAGE gels(S) or by Western blotting using α-FLAG antibodies to detect Isw1p (W). A total of 7.6 fmol (1 ng of Isw1p equivalent) or 46 fmol (6 ng of Isw1p equivalent) of each Isw1 complex was used for either Western blotting or silver staining detection, respectively. (B) Electrophoretic mobility shift assay for Isw1a and Isw1b binding to mononucleosomes. Mononucleosomes (41 fmol) were assembled from recombinant histone octamer and a labeled 255-bp DNA fragment and incubated without ATP in the indicated ratios with either Isw1a (lanes 2 to 4), Isw1b (lanes 5 to 7), or no complex (lane 1). Reaction products were analyzed on a 4% native polyacrylamide gel. (C) Antibody supershift assay. Mononucleosomes (41 fmol) were incubated alone (lane 1) or with 369 fmol of Isw1a complex (lanes 2 and 3). To one prebound sample, 46 pmol of α-FLAG antibody was added for an additional 15 min (lane 3). Reaction products were analyzed on a 4% native polyacrylamide gel.
The electrophoretic mobility shift assay utilized mononucleosomes assembled from a 255-bp DNA fragment and recombinant histone octamer. These were incubated with increasing amounts of Isw1a or Isw1b complexes in the absence of ATP and separated on a 4% native polyacrylamide gel. The presence of slower-migrating bands was associated with increasing amounts of either Isw1a or Isw1b, suggesting that each complex was binding to mononucleosomes (Fig. 4B, lanes 2 to 4 and 5 to 7). This is consistent with our previous results, as is the observation that the Isw1a complex appeared to bind more avidly than the Isw1b complex. As this reaction mixture contained no ATP, it is unlikely that these slower-migrating bands were due to Isw1-mediated remodeling of the labeled nucleosomes; it is more likely that the bands rather represented Isw1a or Isw1b bound to nucleosomes. In addition, these bands could be competed away with excess DNA, consistent with our hypothesis that they represent Isw1a or Isw1b bound to nucleosomes (data not shown). To confirm the identity of the putative nucleosome-Isw1a band, α-FLAG antibody was added to this reaction mixture, resulting in the supershift of some of the putative nucleosome-Isw1a species (Fig. 4C). The band caused by Isw1b was too weak to characterize by an antibody supershift assay.
The behavior of each Isw1 complex in the template interaction assays correlated well with its activities in both our nucleosome spacing and sliding assays. This suggests that the differences we observe in specific activity can be attributed to their abilities to interact with their substrates in vitro rather than to inherent differences in their enzymatic activities. These results also highlight the requirement of the Ioc proteins in the interaction of Isw1p with templates. Although either Ioc2p and Ioc4p or Ioc3p alone is sufficient to target Isw1p, there is an apparent lack of amino acid homology among them, suggesting that each may utilize unique substrate interaction domains.
The Isw1a and Isw1b complexes show different genetic interactions with other chromatin remodeling factors.
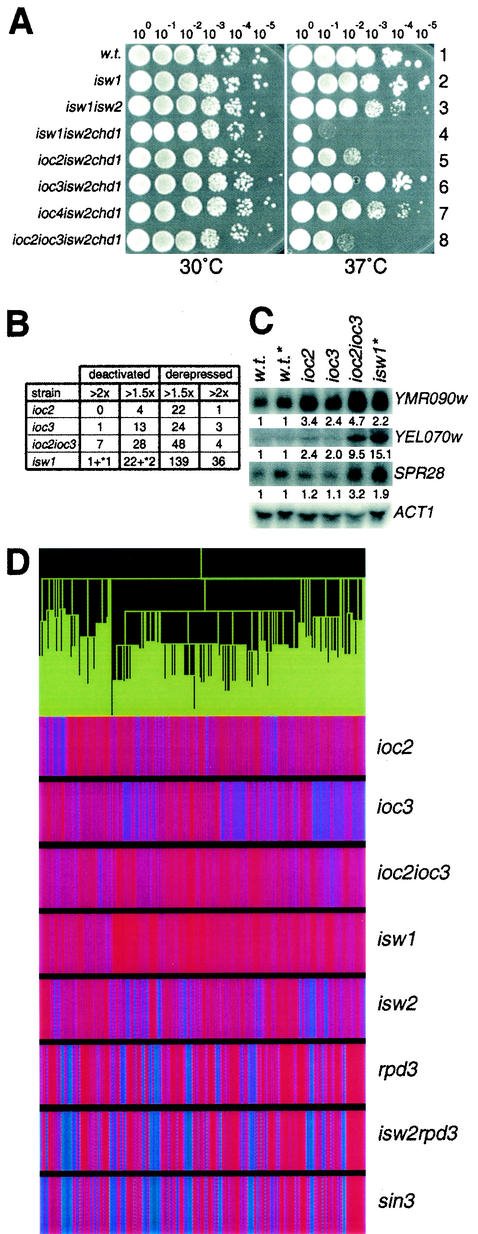
The differences in the biochemical properties of Isw1a and Isw1b complexes prompted us to compare their functions in vivo. Tsukiyama et al. previously found that an isw1isw2chd1 triple mutant fails to form single colonies at elevated temperatures and that any double-mutant combinations are affected to significantly lesser degrees (49). To determine whether defects in one or both Isw1 complexes are required for this phenotype, we created strains YTT522, YTT853, YTT855, and YTT861 that had deletions in specific IOC genes as well as ISW2 or CHD1 (Table 1). Deletion of either IOC2 or IOC4 specifically disrupts the Isw1b complex, and deletion of IOC3 disrupts the Isw1a complex (Fig. 1A, lanes 3 to 5). Substitution of these ioc mutations for the ISW1 deletion, therefore, allows us to separately examine the contribution of each Isw1 complex to this phenotype.
In our serial dilution assay, all strain backgrounds grew as well as the wild-type strain at 30°C. As expected, the isw1isw2chd1 strain exhibited a severe growth defect when grown at 37°C (Fig. 5A, row 4); it was unable to form single colonies and only grew at the most saturated dilution (100). When a deletion of IOC2 was substituted for the ISW1 deletion, the cells still showed a strong growth defect though it was not as severe, growing at the 10−2 dilution (Fig. 5A, row 5). Deletion of IOC3 rather than ISW1 resulted in no detectable growth defects, suggesting that the presence of the Isw1b complex is sufficient for cells to grow at elevated temperature (Fig. 5A, row 6). However, deletion of IOC3 in an ioc2isw2chd1 background made the temperature-sensitive phenotype more severe (Fig. 5A; compare rows 5 and 8), showing that the Isw1a complex has a minor involvement in growth at elevated temperature. We would expect that deletion of IOC4 in this assay would phenocopy an IOC2 deletion. However, IOC4 deletion in an isw2chd1 background caused only slight growth retardation at 37°C (Fig. 5A, row 7), suggesting either that Ioc4p function makes only a minor contribution to this particular Isw1b function or that Ioc2p has functions in vivo that are independent of its role in the Isw1b complex. We also noticed that isw1isw2chd1 cells exhibited a significantly more severe phenotype than ioc2ioc3isw2chd1 cells (or than ioc2ioc3ioc4isw2chd1 cells, which showed no additional growth defects [data not shown]) (compare Fig. 5A, rows 4 and 8). This suggests that a portion of the ISW1 requirement at elevated temperatures is due to functions of Isw1p monomer or another, as yet unidentified, Ioc-independent complex. It is interesting that this temperature sensitivity is completely eliminated by plating on media containing 1 M sorbitol (data not shown). This type of temperature-dependent osmotic sensitivity has also been observed in rsc3 and rsc30 mutants (3) and suggests that this phenotype may result from effects on the integrity of the cell wall.
FIG. 5.
Isw1a and Isw1b complexes have partially overlapping yet distinct functions in vivo. (A) Temperature sensitivity assays. Tenfold serial dilutions of overnight cultures were spotted onto yeast extract-peptone-dextrose plates and allowed to grow at 30 or 37°C for 2 or 3days, respectively. The strains used were YTT166 (row 1), YTT441 (row 2), YTT443 (row 3), YTT227 (row 4), YTT522 (row 5), YTT853 (row 6), YTT855 (row 7), and YTT861 (row 8). (B) DNA microarray expression analyses. RNA was prepared and labeled from YTT823 (ioc2), YTT825 (ioc3), YTT829 (ioc2ioc3), or YTT441 or YTT442 (isw1) strains and hybridized to a microarray of yeast open reading frames. Shown are the numbers of yeast genes that were changed either 1.5- or 2.0-fold in each mutant background. Genes whose expression was increased in a mutant were defined as “derepressed,” while those whose expression was decreased in a mutant were defined as “deactivated.” The numbers marked by an asterisk represent two genes directly adjacent to ISW1 which show reduced expression possibly due to the ISW1 deletion rather than to loss of Isw1p. (C) Northern hybridizations. RNA from these mutant strains was probed for the corresponding genes shown and quantified with a PhosphorImager. The ratios of expression relative to wild-type strain levels (normalized to ACT1) are shown below each image. The two strains marked by an asterisk were prepared and normalized separately from the other samples. (D) Expression clustering analysis. Microarray data for ioc2, ioc3, ioc2ioc3, and isw1 mutants from this study were clustered with data from isw2, rpd3, isw2rpd3, and sin3 strains as previously published. Shown is a cluster of 667 genes that were derepressed in the isw1 strains. The green tree at the top of the panel represents smaller clusters of gene subgroups. For each deletion strain, the colored vertical lines correspond to the expression levels of individual genes. Red is indicative of genes which were derepressed in the mutant, while blue is indicative of genes which were deactivated in the mutant. Purple indicates little to no change.
Mutation of Isw1a and Isw1b complex subunits causes distinct expression profiles.
The Isw2 complex is required for transcriptional repression at a number of loci, including those of early meiotic genes. ISW1 has also been shown to affect transcriptional regulation (28), although it is not clear whether each Isw1 complex has unique roles in vivo. To study the relative contributions of each complex, we prepared RNA from ioc2, ioc3, ioc2ioc3, and isw1 strains. These samples were used for Northern blotting and DNA microarray analysis using a PCR-based array of all putative yeast open reading frames. As previously observed for mutants of the Isw2 complex (17, 21, 28), mutations of components of the Isw1 complexes resulted in modest derepression of many genes and they resulted in deactivation of relatively few (Fig. 5B and http://parma.fhcrc.org/JVary). If the Isw1a and Isw1b complexes have parallel functions in transcriptional repression, one would expect that deletion of either single mutant would result in little change and that deletion of both complexes would result in a greater number of genes being derepressed. This did appear to be the case for many genes, as the number of genes derepressed at least 1.5-fold was higher in both ioc2ioc3 and isw1 mutants than in either ioc2 or ioc3 mutants (Fig. 5B). This trend was confirmed by Northern analysis, and the results for three such genes, YEL070w, YMR090w, and SPR28, are shown (Fig. 5C).
We also found evidence that each complex may function independently at other loci. Cluster analysis identified a large group of 667 genes that were derepressed in isw1 and ioc mutants; this cluster includes a significant number of genes involved in carbohydrate transport (P < 0.00002). While the ioc2ioc3 double-mutant strain exhibited a transcriptional profile similar to that of an isw1 single mutant, ioc2 and ioc3 single mutants had distinct transcriptional profiles that, when additively combined, produce a profile similar to that of an ioc2ioc3 strain. This suggests that at many loci within this cluster, Isw1a and Isw1b complexes have independent functions. In addition, the transcriptional profiles of the isw1 and ioc mutants were dramatically different than those of an isw2 mutant or mutants of the Sin3/Rpd3 complex (17) within this cluster as well as the rest of the genome (Fig. 5D and data not shown).
DISCUSSION
We have shown that yeast Isw1p associates with the previously uncharacterized proteins Ioc2p, Ioc3p, and Ioc4p to form two distinct Isw1 complexes in vivo. By utilizing epitope tags on individual Ioc proteins, each complex can be purified to near homogeneity without any evidence of its sibling, showing that these are not subsets of a larger complex. Recently, Ito et al. found that Ioc2p interacts with Ioc3p in a high-throughput two-hybrid screen (29). As we have seen no evidence of this interaction in our work, this may have resulted from endogenous Isw1p acting as a bridge between the overexpressed fusion proteins of Ioc2p and Ioc3p in the screen. If this were the case, it would suggest that Isw1p associates with each protein via unique interaction domains. It is possible, however, that unstable interactions between these complexes may occur or that direct association of Ioc2p and Ioc3p may exist under growth conditions different from those we have tested.
None of the Ioc proteins have similarity to each other nor do they have overall homology to any other proteins for which a function has been described. However, Ioc2p has a putative PHD finger motif and Ioc4p has a PWWP motif; these domains are found in a number of proteins involved in chromatin or transcriptional regulation and have been proposed to be involved in protein-protein interaction. Several proteins in higher eukaryotes such as mNSD1, hBR140, hWHSC1, and hBS69 contain both of these domains, suggesting that both domains might be cooperatively required for a cellular process(es). It is possible that in yeast, this requirement is fulfilled by the proximity of each separate protein within the Isw1b complex. Ioc3p does not have any known conserved motifs, yet it is required for the strong DNA and nucleosome interactions of the Isw1a complex. This suggests that Ioc3p may contain as yet unidentified DNA and/or chromatin association domains.
To characterize each separate complex, we utilized several established assays to study their biochemical activities. Both Isw1a and Isw1b showed equivalent nucleosome-stimulated ATPase activities, suggesting that each can utilize the catalytic activity of Isw1p and that Ioc proteins are required for full specific activity of Isw1p. However, we found that at equimolar levels the Isw1a complex exhibited stronger nucleosome spacing and sliding activities than the Isw1b complex. These in vitro differences are consistent with differences in the interaction of each complex to immobilized DNA and nucleosome arrays. Indeed, our spacing assay shows a dose-dependent effect for the Isw1b complex (data not shown), suggesting that the differences in specific activity we observed are a direct result of their differing abilities to interact with substrates in our system. As the recombinant histones utilized in this study did not bear modifications, it is also possible that the Isw1b complex requires covalently modified histones for full activity.
We found that Isw1a and Isw1b have different roles in vivo. By exploiting a synthetic temperature-sensitive phenotype to study the contributions of each complex, we found that deletion of components of the Isw1a and Isw1b complexes resulted in different phenotypes. While the Isw1a complex appeared to play a very minor role for growth at elevated temperature, deletion of components of the Isw1b complex resulted in strains that grew more slowly at 37°C. The Sth1p ATPase of the yeast RSC chromatin remodeling complexes also forms complexes with distinct functions in vivo. At least three forms of the RSC complex have been purified, two of which are distinguished only by their associations with either Rsc1p or Rsc2p (3, 8). Mutants of RSC1 and RSC2 differed not only in their sensitivities to elevated temperature and hydroxyurea but further showed different genetic interactions with components of the SAGA histone acetyltransferase complex, suggesting unique roles in vivo for each complex (8).
In both in vivo assays, we also found evidence that Isw1p may have functions within the cell that are independent of the Ioc subunits. The isw1isw2chd1 mutant was significantly more temperature sensitive than any similar combination with ioc mutants. Also, our expression data consistently showed that deletion of ISW1 resulted in transcription changes in a greater number of genes than those observed in an ioc2ioc3 mutant. Given that a significant portion of Isw1p is purified as a monomer in wild-type as well as ioc mutant strains, it is possible that these differences are due to functions of Isw1p monomer. Although this could result from disruption of intact Isw1 complex during IP, we believe that Isw1p monomer exists in vivo. When whole-cell extract is applied at 0.2 M KCl to a Bio-Rex 70 cation exchange column prior to α-FLAG IP, Isw1p monomer is found in the flowthrough fraction and intact Isw1 complexes are retained in the column matrix (data not shown). In addition, our size exclusion chromatography data (Fig. 2A) suggest that Isw1p monomer may be the most abundant form of Isw1p within the cell. Other groups have shown that Drosophila ISWI monomer exhibits enzymatic activity in vitro, albeit at lower specific activities than the complete complexes (11, 15).
An Isw1 complex-independent role was also suggested for Ioc2p. Deletion of either IOC2 or IOC4 results in disruption of the Isw1b complex, so we would expect these phenotypes to be identical if their activities were limited to the Isw1b complex. However, the ioc2isw2chd1 mutant was more temperature sensitive than the ioc4isw2chd1 mutant. (Addition of an ioc4 mutation to the ioc2isw2chd1 background did not result in additional growth defects [data not shown].) Size exclusion chromatography revealed that Ioc2p elutes in two apparently distinct populations of similar abundance, consistent with the presence of either Ioc2 monomer or a separate Ioc2-containing complex. TAP purification of Ioc2p failed to yield any non-Isw1b complex proteins at near-stoichiometric levels, so we conclude that Ioc2p also has roles either as a monomer or in another complex that is unstable during TAP purification.
Analyses of transcription profiles in isw1 and ioc mutants revealed that the two Isw1 complexes function in parallel pathways to affect transcription of a number of genes, while they appeared to act separately at many genes. The effects on gene expression that resulted from deletion of Isw1 complex components were subtle, rarely exhibiting more than twofold changes compared to those for wild-type cells. There are at least two models to explain these results. First, it is possible that Isw1 complexes function in parallel with an unidentified factor(s) in transcriptional regulation. If this were the case, these factors would mask the effects of mutations in the Isw1 components. Indeed, parallel functions of the Isw2 and Sin3-Rpd3 complexes make the phenotypes of isw2 mutants subtle due to this very reason (17, 21, 28). We have investigated rpd3 mutations in an isw1 background and have not found evidence that they operate in parallel to repress transcription (J. C. Vary, Jr., and T. Tsukiyama, unpublished data). However, S. cerevisiae has at least nine other putative histone deacetylases that could serve such a function. It is also possible that the Isw1 complexes regulate nuclear processes other than transcription. Kent et al. recently reported that the deletion of ISW1 results in changes in chromatin structure at several genes in vivo (31). However, these changes are not associated with changes in transcription levels (reference 31 and http://parma.fhcrc.org/JVary), suggesting that there may be many Isw1-dependent chromatin changes that do not affect transcription in vivo.
The differences we observed between two Isw1p-containing yeast chromatin remodeling complexes may parallel the situation found in higher eukaryotes. Drosophila, Xenopus, and humans all express several forms of ISWI complexes, which share a common ATPase subunit yet have distinct biochemical functions. Our data also demonstrate that lower specific activities in biochemical assays do not necessarily correlate with lesser in vivo activity, as the Isw1b complex has more significant roles in growth at elevated temperatures and transcription in vivo than the more biochemically active Isw1a complex.
TABLE 2.
Sequences used for alignments in Fig. 1
| Proteina | GI accession no. | Description |
|---|---|---|
| atATM | 7529272 | Ataxia-telangiectasia mutated protein |
| ceC06A5.3a | 14573766 | |
| hAR | 113830 | Androgen receptor |
| hARTX | 6274548 | α-Thalassemia/mental retardation X-linked syndrome |
| hBR140 | 4757865 | Peregrin/BRPF1 |
| hBS69 | 10719919 | Adenovirus 5 E1A-binding protein (BS69 protein) |
| hDATF1 | 2224607 | Death-associated transcription factor |
| hDNMT3L | 7019367 | DNA methyltransferase 3-like protein |
| hHDGF | 1708157 | Hepatoma-derived growth factor |
| hMI2b | 5921744 | CHD4 chromodomain helicase DNA-binding protein 4 |
| hMSH6 | 4504190 | MutS homolog 6 |
| hTIF1a | 12746552 | Transcription intermediary factor 1a |
| hWHSC1 | 4378019 | Wolf-Hirschhorn syndrome critical region protein |
| hWSTF | 4049922 | Williams syndrome transcription factor |
| mDNMT3a | 6681209 | DNA methyltransferase 3A |
| mDNMT3b | 6753662 | DNA methyltransferase 3B |
| mHDGF-2 | 6680201 | Hepatoma-derived growth factor, related protein 2 |
| mNSD1 | 6679138 | Nuclear receptor-binding SET-domain protein 1 |
| scYJR119c | 1352920 | YJ89 |
| scYLR455w | 6323488 | |
| scYMR075w | 2497139 | YMW5 |
| spAC23D3.01 | 2130333 | |
| spBC29A3.13 | 3006149 |
Abbreviations: at, Arabidopsis thaliana; ce, Caenorhabditis elegans; h, Homo sapiens; m, Mus musculus; sp, Schizosaccharomyces pombe; sc, Saccharomyces cerevisiae
Acknowledgments
We thank Tom Fazzio, Marnie Gelbart, Cedar McKay, and Sue Biggins for helpful discussions. We also thank Bertrand Séraphin for providing us with the plasmid for TAP epitope labeling.
This work was supported by a Pew Charitable Trust Biomedical Scholars fellowship and National Institutes of Health grants GM58465 to T.T., 5T32 HD07183 to J.C.V., CA74841 to C.K., and GM48413 to B.B. In addition, B.B. was supported by the American Cancer Society grant RPG-99-199-01-GMC.
REFERENCES
- 1.Adams, A., D. Gottschling, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A rsc3/rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler rsc in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 4.Bochar, D. A., J. Savard, W. Wang, D. W. Lafleur, P. Moore, J. Cote, and R. Shiekhattar. 2000. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Bozhenok, L., P. A. Wade, and P. Varga-Weisz. 2002. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, B. R. 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23:20-25. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 9.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corona, D. F., A. Eberharter, A. Budde, R. Deuring, S. Ferrari, P. Varga-Weisz, M. Wilm, J. Tamkun, and P. B. Becker. 2000. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J. 19:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corona, D. F., G. Langst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo, M. C., M. E. Crawford, J. E. Hirschman, J. E. Kranz, P. Olsen, L. S. Robertson, M. S. Skrzypek, B. R. Braun, K. L. Hopkins, P. Kondu, C. Lengieza, J. E. Lew-Smith, M. Tillberg, and J. I. Garrels. 2001. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29:75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo, M. C., J. D. Hogan, M. E. Cusick, B. P. Davis, A. M. Fancher, P. E. Hodges, P. Kondu, C. Lengieza, J. E. Lew-Smith, C. Lingner, K. J. Roberg-Perez, M. Tillberg, J. E. Brooks, and J. I. Garrels. 2000. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 28:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas, M. Verardo, S. Moseley, M. Berloco, T. Tsukiyama, C. Wu, S. Pimpinelli, and J. W. Tamkun. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5:355-365. [DOI] [PubMed] [Google Scholar]
- 15.Eberharter, A., S. Ferrari, G. Langst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 20:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elfring, L. K., R. Deuring, C. M. McCallum, C. L. Peterson, and J. W. Tamkun. 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 14:2225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 19.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelius, B., P. Wade, A. Wolffe, O. Wrange, and A. K. Ostlund Farrants. 1999. Characterization of a chromatin remodelling activity in Xenopus oocytes. Eur. J. Biochem. 262:426-434. [DOI] [PubMed] [Google Scholar]
- 21.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb, B., H. Lehvaslaiho, L. K. Beitel, R. Lumbroso, L. Pinsky, and M. Trifiro. 1998. The androgen receptor gene mutations database. Nucleic Acids Res. 26:234-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guschin, D., T. M. Geiman, N. Kikyo, D. J. Tremethick, A. P. Wolffe, and P. A. Wade. 2000. Multiple ISWI ATPase complexes from Xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J. Biol. Chem. 275:35248-35255. [DOI] [PubMed] [Google Scholar]
- 26.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff, S., and J. G. Henikoff. 1994. Protein family classification based on searching a database of blocks. Genomics 19:97-107. [DOI] [PubMed] [Google Scholar]
- 28.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 29.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 33.Kooperberg, C., T. G. Fazzio, J. J. Delrow, and T. T. Tsukiyama. 2002. Improved background correction for spotted DNA microarrays. J. Comput. Biol. 9:57-68. [DOI] [PubMed] [Google Scholar]
- 34.Kornberg, R. D. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184:868-871. [DOI] [PubMed] [Google Scholar]
- 35.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 36.Lazzaro, M. A., and D. J. Picketts. 2001. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 77:1145-1156. [DOI] [PubMed] [Google Scholar]
- 37.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 38.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 39.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 40.Okabe, I., L. C. Bailey, O. Attree, S. Srinivasan, J. M. Perkel, B. C. Laurent, M. Carlson, D. L. Nelson, and R. L. Nussbaum. 1992. Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S. cerevisiae. Nucleic Acids Res. 20:4649-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poot, R. A., G. Dellaire, B. B. Hulsmann, M. A. Grimaldi, D. F. Corona, P. B. Becker, W. A. Bickmore, and P. D. Varga-Weisz. 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19:3377-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu, C., K. Sawada, X. Zhang, and X. Cheng. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 9:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sawa, H., H. Kouike, and H. Okano. 2000. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol. Cell 6:617-624. [DOI] [PubMed] [Google Scholar]
- 46.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 47.Strohner, R., A. Nemeth, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Langst, and I. Grummt. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varga-Weisz, P. D., and P. B. Becker. 1998. Chromatin-remodeling factors: machines that regulate? Curr. Opin. Cell Biol. 10:346-353. [DOI] [PubMed] [Google Scholar]
- 51.Wolffe, A. P., and J. C. Hansen. 2001. Nuclear visions: functional flexibility from structural instability. Cell 104:631-634. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, H., R. Sandaltzopoulos, H. M. Wang, A. Hamiche, R. Ranallo, K. M. Lee, D. Fu, and C. Wu. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8:531-543. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Y. H., S. Dudoit, P. Luu, and T. P. Speed. 2001. Normalization for cDNA microarray data, p. 141-152. In M. L. Bittner, Y. Chen, A. N. Dorsel, and E. R. Dougherty (ed.), Microarrays: optical technologies and informatics. Proceedings of SPIE, vol. 4266. International Society for Optical Engineering, San Diego, Calif.
- 54.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]