Abstract
The yeast Sir2p protein has an essential role in maintaining telomeric and mating type genes in their transcriptionally inactive state. Mammalian cells have a very large proportion of their genome inactive and also contain seven genes that have regions of homology with the yeast sir2 gene. One of these mammalian genes, sir2α, is the presumptive mammalian homologue of the yeast sir2 gene. We set out to determine if sir2α plays a role in mammalian gene silencing by creating a strain of mice carrying a null allele of sir2α. Animals carrying two null alleles of sir2α were smaller than normal at birth, and most died during the early postnatal period. In an outbred background, the sir2α null animals often survived to adulthood, but both sexes were sterile. We found no evidence for failure of gene silencing in sir2α null animals, suggesting that either SIR2α has a different role in mammals than it does in Saccharomyces cerevisiae or that its role in gene silencing in confined to a small subset of mammalian genes. The phenotype of the sir2α null animals suggests that the SIR2α protein is essential for normal embryogenesis and for normal reproduction in both sexes.
The irreversible inactivation of genes occurs in the mammalian genome in the context of X chromosome inactivation, genomic imprinting, and allelic exclusion. Sporadic silencing of certain tumor suppressor genes also occurs during the development of some cancers (reviewed in reference 31). The events responsible for initiating gene silencing and maintaining silent genes in their inactive form are widely believed to rely on DNA methylation and nucleosome modifications (reviewed in references 9, 19, 22, and 39). The molecular events comprising histone modifications and their relationship to gene inactivation have been intensively investigated in the budding yeast Saccharomyces cerevisiae, in which telomere-dependent gene silencing and maintenance of the silent mating type loci have indicated that the Sir2p protein plays a central role in maintaining genes in their inactive configuration (reviewed in reference 17).
Sir2p has NAD+-dependent histone deacetylase activity (18, 24, 43, 45), and the catalytic domain of Sir2p is present in four other genes in S. cerevisiae (10). This domain is also present in genes encoded by the genomes of the most primitive organisms (7) as well as in mammals (13). Seven mammalian genes carry the SIR2 catalytic domain, and this domain can substitute for the same domain in yeast Sir2p (42).
Yeast Sir2p plays an important role not only in gene silencing but also in a variety of other biological processes. These include regulation of the cell cycle, DNA repair, DNA recombination, and aging (reviewed in references 14, 17, and 33).
Given the extraordinary conservation of the SIR2 catalytic domain and the multitude of biological functions served by the yeast Sir2p, it seems likely that the related proteins in mammalian cells also have important functions. We were particularly interested in determining whether the mammalian sir2 homologues play roles in gene silencing. We therefore selected the murine gene most closely related to the yeast SIR2 and created a null allele of this sir2α gene in embryonic stem (ES) cells. This null allele was introduced into the germ line of mice, and sir2α null animals were created. The characteristics of these mice suggest that the mammalian SIR2α protein has no role in gene silencing but plays a role in the growth and maturation of the embryo and in gametogenesis in both sexes.
MATERIALS AND METHODS
ES cell culture.
ES cells of the R1 line (36) were cultured on irradiated STO feeder cells in the presence of leukemia inhibitory factor. The STO feeder cells had been selected previously for hygromycin resistance following transfection with an expression plasmid (2) encoding the hygromycin resistance (hyg) gene. The R1 cells were electroporated with the linearized plasmid DNA shown in Fig. 1, and colonies were selected for growth in 200 μg of hygromycin per ml. Individual colonies were picked and expanded, and DNA was prepared from each as described previously (23).
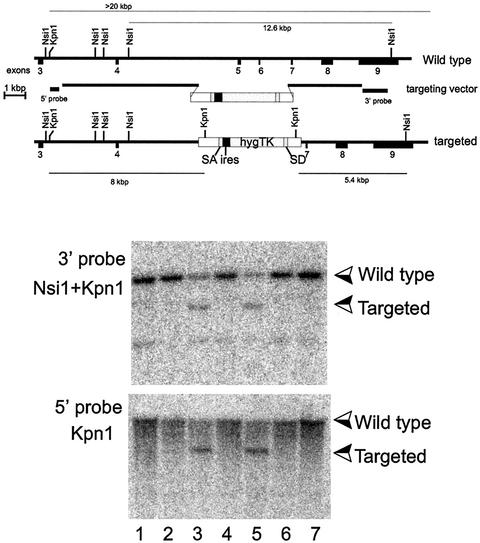
FIG. 1.
Homologous recombination into the sir2α locus. The sequence for sir2α mRNA was inferred from the expressed sequence tag database, and reverse transcription-PCR was used to amplify and clone a cDNA corresponding to exons 3 to 9. This cDNA sequence was confirmed and used to isolate two overlapping genomic clones derived from strain 129/Sv mice. DNA sequencing was used to locate the exons, as shown in the upper panel. The targeting vector was constructed as shown, where the selectable sequence replaces exons 5 and 6, which encode highly conserved regions of the catalytic domain of the SIR2α protein. The selectable sequence consists of splice acceptor (SA) and splice donor (SD) sequences derived from exons 3 and 5 of the mouse Pgk-1 gene (3) surrounding the poliovirus internal ribosome entry site (ires) (35) and the coding region for a gene fused from the hygromycin and herpes simplex virus thymidine kinase (TK) genes (30). The targeting vector was linearized and electroporated into the R1 line of ES cells (36), which were subsequently selected for hygromycin resistance. Individual clones were isolated and expanded, and their DNA was analyzed by Southern blotting following digestion with the enzymes shown in the two lower panels. The two clones shown in lanes 3 and 5 had patterns of hybridizing bands that are consistent with homologous recombination having occurred at both the 5′ and 3′ ends of the targeting vector.
Animal experimentation.
Groups of 5 to 10 ES cells carrying the integrated vector were aggregated in culture along with zona-free 2.5-day embryos of the CD1 outbred mouse strain (36). These aggregates were cultured for 48 h, and the expanded blastocysts that developed were transferred to the uteri of 3.5-day-pseudopregnant foster mothers of the CD1 strain. Male animals born from these manipulated embryos were assessed for coat color chimerism, and these animals were mated with CD1 females to determine which were able to sire black/agouti offspring. The male chimeric animals whose germ lines were derived from the ES cells were subsequently mated with 129/Sv females, and the offspring from these matings were genotyped from DNA obtained by a tail clip at weaning.
Vaginal washes from female animals were acquired daily and assessed essentially as described previously (47). The females were maintained in cages that had previously been occupied by males.
Blot hybridization.
DNA was isolated from cells or tissues by standard procedures (23), digested with restriction enzymes under the conditions recommended by the manufacturer, and separated by electrophoresis on 1% agarose gels (41). The DNA was transferred to membranes, radioactive probes were created by oligonucleotide priming from isolated DNA fragments, and hybridization was carried out as described previously (41). RNA isolation, gel electrophoresis, and blot hybridization were done as described previously (41).
Immunofluorescence.
Immunofluorescence was performed on whole preimplantation embryos or frozen sections from adult or embryonic tissues previously fixed for 1 to 2 h in 4% paraformaldehyde. Rabbit antiserum directed against the SIR2α protein (M.W. McBurney, X. Yang, K. Jardine, M. Bieman, J. Th’ng, and M. Lemieux, unpublished data) was immunopurified by adsorption and elution from recombinant His-tagged SIR2α protein made in bacteria. After incubation with the primary and secondary antibodies, preparations were stained with Hoechst 33258 before mounting and viewing in a Zeiss Axiophot microscope equipped with a deconvolution system.
RESULTS
sir2α gene structure and targeted disruption.
Although there are seven genes in the human (16) and mouse genomes that share a region homologous to the catalytic domain of the yeast SIR2 gene, one of these homologues, sir2α, encodes a protein with a higher degree of sequence identity (40%) to the yeast Sir2p than the other six. The full-length cDNA sequence of sir2α has been published (18). We amplified a partial sir2α cDNA encoding the C terminus of the SIR2α protein from reverse-transcribed mRNA isolated from the R1 line of ES cells (36). This sir2α cDNA was used to screen a bacteriophage genomic library derived from the DNA from strain 129/Sv mice (a gift from Douglas Gray). Two overlapping clones were isolated, subcloned into plasmid vectors, restriction mapped, and partially sequenced. The cloned genomic region consisted of the last seven exons of the sir2α gene (Fig. 1), a gene structure that is the same as that of its human homologue (accession number AL133551).
We constructed two knockout vectors designed to delete exons 5 and 6 from the sir2α gene. These two exons encode a region of the SIR2α protein that comprises a large portion of the catalytic domain, so we predicted that the result of homologous recombination would be a null sir2α allele. Because the sir2α gene is expressed in ES cells (McBurney et al., unpublished data), we created a knockout vector in which the selectable gene encoding hygromycin resistance would be driven by the sir2α promoter following homologous integration into the sir2α locus (35) (see Fig. 1). This vector was linearized and electroporated into R1 cells. Treated cells were selected in hygromycin, and drug-resistant colonies were expanded and screened by Southern blot hybridization with probes that identify both the 5′ and 3′ sides of the homologous recombination event.
A second knockout vector was created with the selectable gene encoding G418 resistance (35) inserted into the same sir2α targeting arms, and this linearized vector was electroporated into one of the R1 clones carrying the correctly targeted hygromycin resistance vector. Cells resistant to G418 were selected and expanded for analysis. Among the G418-resistant clones were those in which both wild-type alleles of sir2α were replaced with the two knockout vectors (Fig. 2). These sir2α null cells grew well in culture and have been maintained for more than a year, indicating that the SIR2α protein is not essential for cell growth, viability, or immortality.
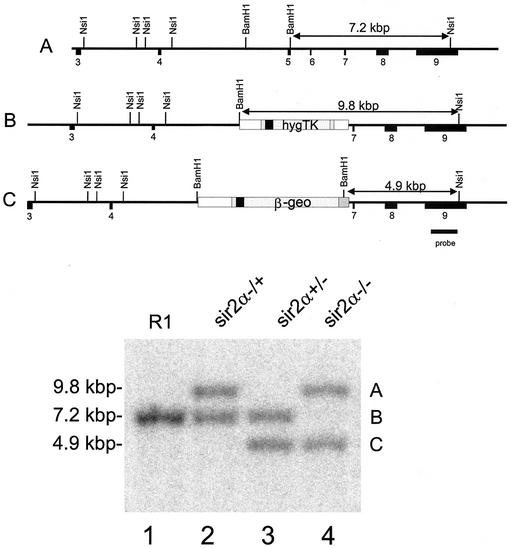
FIG. 2.
Isolation of ES clones deleted for both sir2α genes. R1 ES cells (lane 1) that had homologously integrated the hygromycin resistance vector (lane 2) were electroporated with a second knockout construct carrying the lacZ-neomycin resistance (β-geo) gene (35), and the cells were selected for resistance to G418. Among the clones isolated, some, like that shown in lane 3, had the β-geo sequence in place of the hygromycin resistance-thymidine kinase sequence, while others, like that shown in lane 4, had both the β-geo and the hygromycin resistance-thymidine kinase sequences homologously integrated into the two sir2α genes. The Southern blot shown in the lower panel was derived from DNA digested with NsiI plus BamHI and probed with the sequence shown, derived from DNA outside the region used for the knockout vectors.
We expected that the knockout alleles would be unable to produce SIR2α protein. To confirm this, we created a polyclonal rabbit antibody that recognizes the murine SIR2α protein (McBurney et al., unpublished data) and used this antibody to probe protein blots from the ES knockout cells (Fig. 3). ES cells carrying one knockout allele had approximately half the SIR2α protein present in the diploid wild-type cells, and the double knockout had no detectable SIR2α protein.
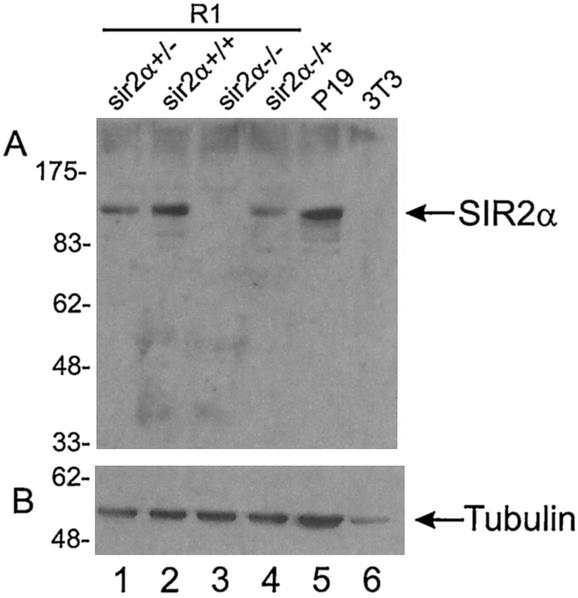
FIG. 3.
SIR2α protein is absent from sir2α−/− ES cells. Protein was isolated from the cells indicated and run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were electrophoretically transferred to membranes, where they were probed with antibody to SIR2α (panel A) and to tubulin (panel B).
The cells from the hygromycin-resistant clone isolated from the first electroporation experiment were aggregated with 2.5-day embryos derived from the CD1 mouse strain, and the mosaic blastocysts were transferred to the uteri of pseudopregnant foster mothers. Several of the male chimeras born from these manipulated embryos had their germ lines derived exclusively from the ES cells, and these were used to introduce the sir2α null allele into both the outbred CD1 line and the inbred 129/Sv strain. Animals carrying the sir2α null allele were identified by Southern blot hybridization, and these heterozygous animals were fertile and apparently normal.
sir2α null genotype is postnatal lethal.
Males and females heterozygous for the sir2α null allele were mated, and offspring were genotyped at weaning by Southern blotting (Fig. 4). Animals carrying two sir2α null alleles were present among the viable offspring, but their proportion was about half what was expected, suggesting that there was prenatal or early postnatal loss of approximately half of the sir2α null offspring.
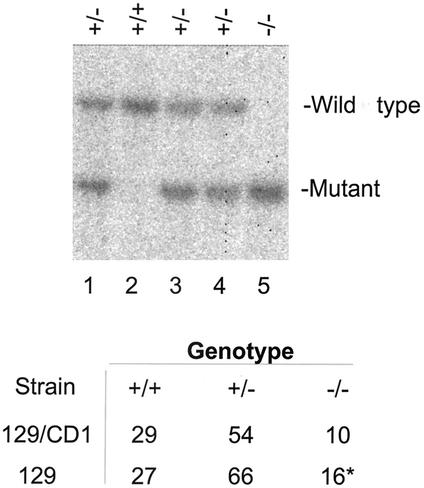
FIG. 4.
Viable sir2α null offspring arise from crosses between heterozygous animals. Males and females that were genotyped as heterozygous for the mutated sir2α alleles were mated, and the DNA from their offspring was used for Southern blots to allow genotyping of individual animals. The DNA was digested with NsiI and KpnI, and the blots were probed with the 3′ probe shown in Fig. 1. In the litter shown in the upper panel, the three expected genotypes were present; lane 2 has two wild-type sir2α alleles, lane 5 has two mutant alleles, and lanes 1, 3, and 4 are heterozygous. The table shows the numbers of offspring of each genotype from these crosses of heterozygous parents. The strain labeled 129/CD1 is outbred and has a mixed genotype, while the strain labeled 129 includes animals in which the targeted sir2α allele was maintained on the 129/Sv background.
To investigate the possible loss of sir2α null embryos during gestation, we mated heterozygous animals and harvested mothers at various times after the appearance of the copulation plug. Each embryo was genotyped from DNA isolated from the fetal membranes, and each embryo was fixed for examination. sir2α null embryos were present at early and late stages of gestation in roughly the expected proportions, but some of these sir2α null embryos were abnormal (Fig. 5) and would not have survived following birth. Thus, the reduced number of sir2 null animals at birth probably reflects the immediate postnatal loss of abnormal fetuses.
FIG. 5.
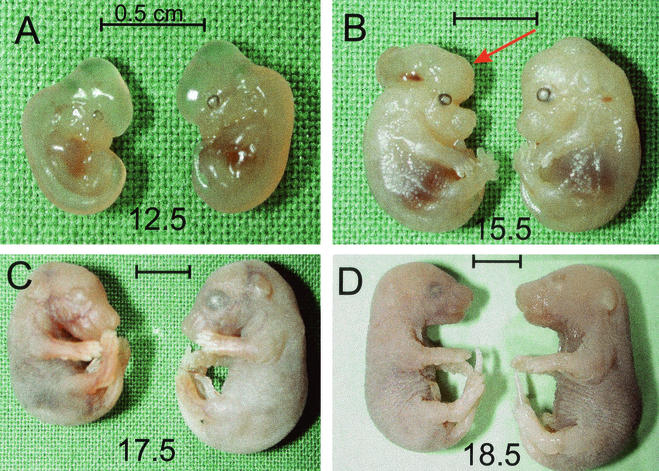
sir2α null animals are not lost during fetal development but are smaller and occasionally have exencephaly. Animals heterozygous for the mutant sir2α allele were mated, and fetuses were harvested at daily intervals after embryonic day 9.5. The fetal membranes were used to isolate DNA for genotyping, and the corresponding embryo was fixed. The genotypes were present in the expected ratios, and there was no evident downward trend in the proportion of the sir2α null genotype. The sir2α null embryos were usually smaller than their littermates. This size difference was evident as early as embryonic day 12.5. Two of the sir2α null embryos had exencephaly, as indicated with the arrow in panel B. Bars, 0.5 cm.
Most of the sir2α null embryos looked normal but were smaller than their wild-type or heterozygous littermates (Fig. 5). Examination of some of the sir2α null fetuses indicated that they were retarded in certain developmental processes. For example, the mineralization of the digits in the sir2 null animals was delayed relative to their wild-type littermates (K. Reuhl et al., unpublished observations).
Following birth, the sir2α null mice were easily identified, as they were smaller than their littermates and their subsequent development was slower than that of their littermates. The most obvious developmental delay was eyelid opening. In the sir2α null animals, eyelids remained closed forever or for several months after birth.
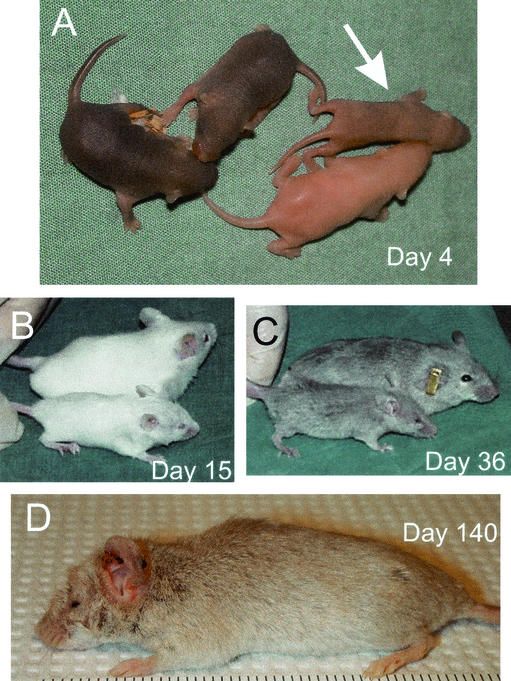
The postnatal development of the sir2α null animals was dependent on their genetic background. On the inbred 129/Sv genetic background, the sir2α null animals invariably died before reaching 1 month of age. On the outbred genetic background from a mix of the CD1 and 129/Sv strains, the sir2α null mice were much more likely to thrive, although their stature was smaller than that of their littermates (Fig. 6) and they all had the characteristic eyelid defect. Many of these sir2α null animals had a short or deviated snout as well but otherwise appeared normal. During the early postnatal period, these sir2α null animals were inevitably smaller than their littermates, but most thrived and eventually approached the size of the wild-type animals (Fig. 7).
FIG. 6.
Postnatal sir2α null animals are developmentally abnormal. sir2α null animals on the outbred background often survived past weaning but were characterized by a number of common developmental defects. Null animals were smaller than their littermates (both wild type and heterozygotes), and all null animals failed to open one or both eyelids. The snout of sir2α null animals was often shorter than normal. The photographs show sir2α null animals of various ages along with their normal littermates.
FIG. 7.
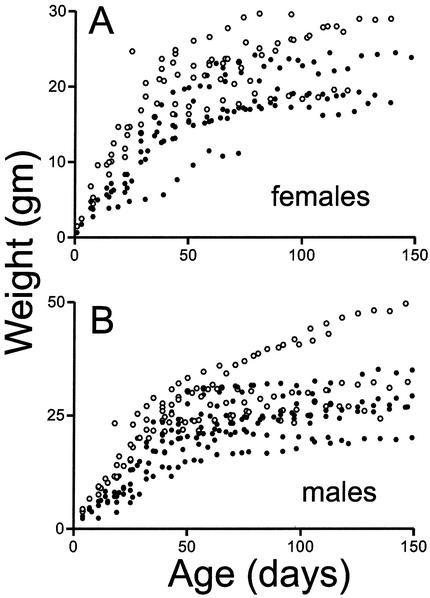
Some sir2α null animals thrive on the outbred genetic background. sir2α null animals (•) and their wild-type littermates (○) were weighed at weekly intervals. During the early postnatal development period, sir2α null animals were uniformly smaller than their littermates, but many of the null animals gained weight and eventually reached sizes commensurate with those of their littermates.
Examination of the internal tissues of the sir2 null animals indicated that many organs were smaller than those of wild-type animals, but the histology of most appeared normal. In some animals, the eyes, although normal in size, had abnormalities of the cornea, lens, and retina, but these may have been a secondary consequence of the fact that the eyelids of sir2α null mice do not open in a timely manner. Two organs consistently affected in the mutants were the lung and pancreas. Neutrophil infiltration of the lungs suggested chronic pulmonary infection that led to pneumonitis, pulmonary edema, and right ventricular hypertrophy. The pancreas showed patchy atrophy of the exocrine epithelium. These lung infections and pancreatic defects are probably responsible for the early postnatal lethality of the sir2α null animals. An examination of the lymphoid system in sir2α null animals indicated that the thymus contained normal numbers and proportions of T cells and that the spleen complement of B cells was normal. The only consistent abnormality appeared to be a reduced proportion of CD8-positive T cells in the spleens of sir2α null animals.
sir2α null animals do not reexpress a silenced transgene or prematurely erode their telomeres.
The yeast Sir2p is essential for maintaining the transcriptional inactivity of genes in this organism. The fact that the sir2α null mice develop relatively normally and that a very large proportion of the genes in a mammalian tissue remain silent suggest that the mammalian SIR2α may not be required to maintain gene inactivity in the mammalian genome. We probed Northern blots of RNA isolated from sir2α null and wild-type day-12.5 embryos and adult tissues. These blots yielded no evidence for ectopic expression of any of the endogenous genes that we assayed (IGF-II, IGF-IIR, endogenous retrovirus, gtl-2, SNRPN, Peg-3, H19, Lo-1, Zfp127, necdin, UBE3, and β2-microglobulin). Microarray analysis of RNA isolated from day-12.5 embryos also indicated that the vast majority of genes are normally expressed in the sir2α null animals.
Nevertheless, we set out to directly test whether the inactivation of a silent transgene could be modulated in sir2α null animals. For this experiment, we used a previously described transgenic strain of mice that carry eight copies of a transgene consisting of the regulatory sequences of the mouse Pgk-1 gene driving the Escherichia coli lacZ reporter sequence (32). This Pgk-1,2-lacZ transgene is expressed in all tissues investigated and encodes a fusion protein with β-galactosidase activity.
The Pgk-1,2-lacZ transgene remains active provided it is inherited through the male germ line; however, if this transgene is inherited through the female germ line, its expression is irreversibly abolished (26). We interbred female mice carrying the sir2α null allele with male animals carrying the active Pgk-1,2-lacZ transgene. Animals heterozygous for the sir2α null allele were mated with each other so that the Pgk-1,2-lacZ gene was inherited from either the male or the female parent. Embryos were harvested at 9.5 days of gestation, each embryo was fixed and stained for β-galactosidase expression, and the fetal membranes of each embryo were used to isolate DNA, from which the genotype of each embryo was determined.
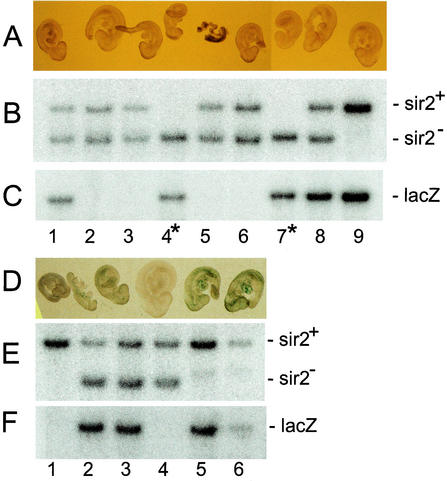
Embryos who inherited their Pgk-1,2-lacZ transgene from the male parent expressed β-galactosidase as determined by the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of these embryos (Fig. 8D). Embryos that inherited their Pgk-1,2-lacZ transgene from the female parent are expected to inherit this transgene in an inactive form. It is evident that the embryos shown in Fig. 8A, numbers 1, 8, and 9, have inherited the Pgk-1,2-lacZ transgene along with at least one wild-type copy of sir2α. As expected, these embryos failed to express the transgene, as indicated by the absence of X-Gal staining. The embryos labeled 4 and 7 also inherited the Pgk-1,2-lacZ transgene, but these two embryos were also sir2α null. Because neither embryo 4 nor 7 was stained with X-Gal, we conclude that the absence of SIR2α did not result in compromised repression of the silent Pgk-1,2-lacZ transgene.
FIG. 8.
sir2α null embryos do not reactivate expression of silent transgenes. Female mice carrying the sir2α null mutation were intercrossed with male mice carrying the Pgk-1,2-lacZ transgene, and animals heterozygous for the sir2α null mutation were mated so that the transgene was inherited from either the male parent, where it remains active, or the female parent, where it becomes inactivated (26). Embryos were harvested at day 9.5 of gestation, the membranes were used to isolate DNA for genotyping, and the fetus was fixed and stained with X-Gal. The upper three panels show a litter in which the Pgk-1,2-lacZ transgene was inherited through the female germ line, during which it is inactivated. Embryos 1, 8, and 9 inherited the transgene along with at least one wild-type sir2α allele. As expected, none was stained with X-Gal, indicating that the transgene had been inactivated. Embryos 4 and 7 inherited the transgene along with two sir2α null alleles. Neither embryo was stained with X-Gal, indicating that the absence of SIR2α in these embryos did not reactivate expression from the silenced transgene. The embryos in the lower three panels were derived from a cross in which the Pgk-1,2-lacZ transgene was inherited from the male parent. In this case, the transgene remained active, and the fetuses that inherited the transgene (lanes 2, 3, 4, and 6) were stained with X-Gal.
Some embryos from these matings were allowed to develop to term, and the pups were allowed to mature. The internal tissues from these animals were analyzed for β-galactosidase activity. The results were similar to those seen in the embryos; sir2α null animals that had inherited the Pgk-1,2-lacZ transgene from their mothers did not express it, while the sir2α null animals that inherited the Pgk-1,2-lacZ transgene from their fathers did show staining with X-Gal in their internal tissues. Thus, the sir2α null animals did not express the silenced transgenes or modulate the expression of active transgenes.
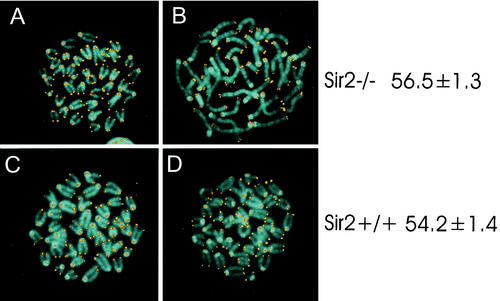
The yeast Sir2p also plays a role in determining the rate at which yeast mother cells undergo senescence (20). Work with Caenorhabditis elegans also suggests that the sir2 homologue in worms may also play a similar role in regulating aging (46). Because many of the sir2α null animals died before weaning, one possibility is that they are aging prematurely. To investigate this possibility, we compared the lengths of the telomeres in both B and T lymphocytes from wild-type and sir2α null littermates with animals of 4 months of age on the outbred CD1 background. The mean telomere lengths in these cells were identical (Fig. 9). In addition, the chromosomes from the cells appeared normal, consistent with the absence of telomere erosion that might be predicted to occur if the sir2α null animals underwent premature aging. Our oldest sir2α null animals are now 17 months old.
FIG. 9.
Telomeres do not erode prematurely in sir2α null mice. Splenocytes from sir2α null (panels A and B) and wild-type (panels C and D) mice were stimulated with concanavalin A (panels A and C) or lipopolysaccharide (panels B and D) to activate T and B lymphocytes, respectively. Chromosome spreads were stained for telomeres by fluorescence in situ hybridization with a peptide nucleic acid telomere probe (53). DNA was counterstained with 4′,6′-diamidino-2-phenylindole. Quantitation of telomere content per cell was done by flow cytometry. The values on the right represent total telomere lengths in kilobase pairs per cell along with the standard error.
sir2α expression is widespread, but the protein is associated with euchromatin.
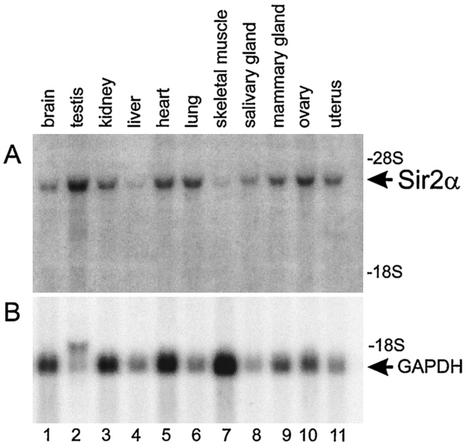
To attempt to understand the phenotype of the sir2α null mice, we investigated the normal expression pattern of the sir2α gene by probing Northern blots of RNA isolated from various tissues. The sir2α transcript was readily detected in all tissues investigated (Fig. 10) and was particularly evident in the testis and ovary.
FIG. 10.
SIR2α expression is widespread throughout adult tissues. RNA was isolated from a variety of tissues from wild-type mice and subjected to electrophoresis, blotting, and hybridization to cDNA probes specific for sir2α (upper panel) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, lower panel). The sir2α transcript was detected in all tissues examined and was particularly abundant in the testis and ovary.
The polyclonal rabbit antibody that recognizes the murine SIR2α protein (McBurney et al., unpublished data) was used for immunoblots of protein isolated from various murine tissues and to carry out immunofluorescence studies on these tissues. Despite the prevalence of the sir2α transcript in adult mouse tissues, we were able to detect protein only in the testis and ovary (see below).
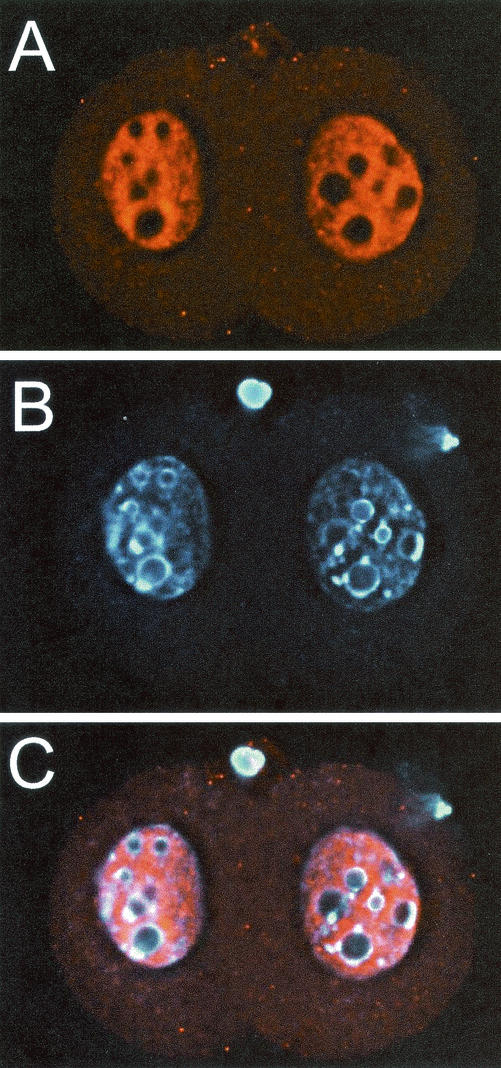
We had noted previously that the SIR2α protein is expressed in abundance in embryonal carcinoma and embryonic stem cells (McBurney et al., unpublished data), so we carried out immunofluorescence experiments in early embryonic cells. The SIR2α protein was present and easily detected in zygotes, two-cell embryos (Fig. 11), and cells of the blastocyst. At all stages, the distribution of the SIR2α protein was nuclear, but it was not concentrated in those regions of the nucleus thought to contain heterochromatin and identified by intense staining with DNA intercalating dyes such as Hoechst 33258. SIR2α also appeared to be excluded from the nucleolus. There was no staining with this antibody in cells from embryos that were sir2α null (data not shown), consistent with the prediction that no stable protein could be made from the knockout allele.
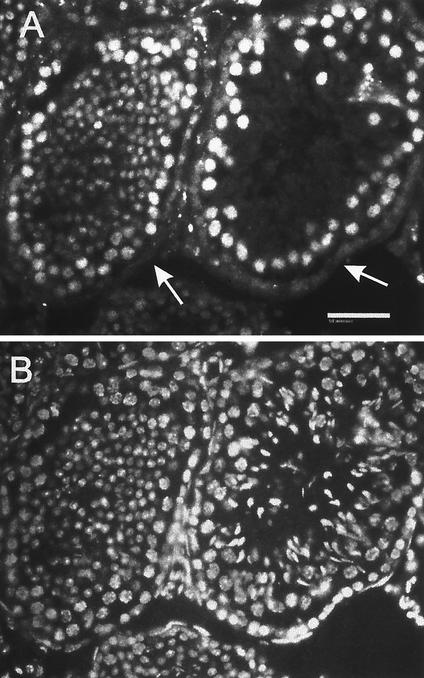
FIG. 11.
SIR2α protein is expressed at high level in two-cell embryos. Wild-type two-cell embryos were recovered from the oviducts 24 h following mating. The zona pellucida was removed, and the embryos were fixed and stained with antibody to SIR2α (panel A) and with Hoechst 33258 (panel B). Vertical stacks of images were captured and subsequently deconvolved. Panel C shows the merged image. Note that the SIR2α protein is exclusively nuclear and that it appears to be excluded from regions of heterchromatin, as identified by intense DNA stain. SIR2α appears to have been excluded from the two polar bodies.
In the ovary, the SIR2α protein was evident in large oocytes as well as in proliferating granulosa cells in larger follicles (Fig. 12). Within both cell types, the SIR2 protein was nuclear and the intranuclear distribution was similar to that seen in early embryonic cells; the protein was nuclear but appeared to be associated with the euchromatin rather than the heterochromatin.
FIG. 12.
SIR2α is present at high level in proliferating granulosa cells. Ovaries from wild-type mice were sectioned and stained with antibody to SIR2α (panel A) and with Hoechst 33258 (panel B). The merged image is shown in panel C. The box in panel C was used to deconvolve the image, and the result is shown in panels D to F. Note that the SIR2α protein is nuclear but appears to be excluded from the nucleolus and those regions of the nucleus that stain intensely with Hoechst dye.
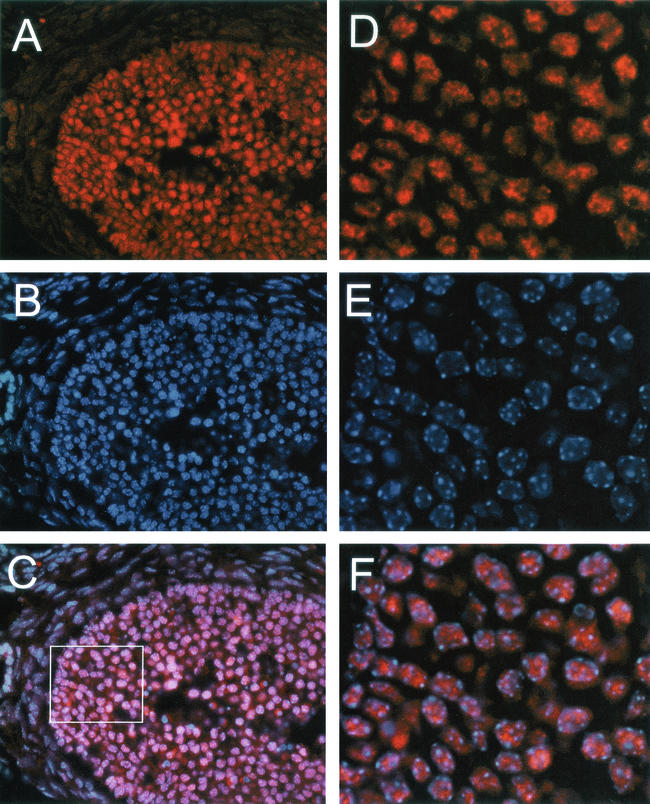
In the testis, the SIR2α protein was present in the nuclei of spermatogenic cells only (Fig. 13). In the developmental sequence of germ cells, a weak, finely speckled nuclear staining was observed in type A spermatogonia, while type B and intermediate spermatogonia displayed a slightly more intense nuclear staining with coarser positive spots. Spermatocytes and round spermatids were intensely positive with diffusely speckled nuclear staining. Soon after the spermatids began to elongate (step 9), the nuclear staining decreased to only a few spots and by step 11 was lost altogether. This staining pattern identifies the nuclear localization of the SIR2 protein in late spermatogonia, spermatocytes, and round spermatids, with the main expression in pachytene spermatocytes. There was no staining of Sertoli cells or of peritubular cells. Due to inherent autofluorescence, the staining of Leydig cells could not be assessed.
FIG. 13.
SIR2α is expressed at high level during spermatogenesis. Frozen sections of testis from wild-type mice were stained with polyclonal antibody to SIR2α (panel A) and with Hoechst 33258 to stain DNA (panel B). In the two seminiferous tubules shown, it is evident that the somatic cells of the tubules do not stain for SIR2α (arrows in panel A), while the immature spermatogenic cells stain to various extents before losing SIR2α expression in the elongate spermatocyte stage (panel A, right tubule). Bar, 50 μm.
sir2α knockout mice are sterile.
Although many sir2α null mice died before reaching maturity, some of those on the outbred background did survive, and these mature animals were caged with wild-type animals of the opposite sex. Both male and female sir2α null animals were sterile. Eleven sir2α null male animals were caged with wild-type females for up to 6 months (mean, 4 months), and none has sired a single litter despite evidence of copulation plugs in a number of the female mates. Seven sir2α null female animals were caged with wild-type males for up to 7 months (mean, 3.5 months). Only one of these females was fertile. She produced three litters but failed to suckle her offspring. However, some of these pups were fostered to another mother, with whom they were able to suckle and thrive and develop into fertile males and females.
The female reproductive tract in sir2α null animals was characterized by small ovaries in which the corpora lutea were conspicuously absent and the uterus was thin walled. Microscopic examination indicated that early, intermediate, and late stages of follicle development were present, but the absence of the corpus luteum indicated that ovulation did not occur. We examined the vaginal washes from two sir2α null females and two wild-type females maintained for 18 days in cages previously occupied by males. The wild-type females cycled through estrus every 4 to 6 days, but the sir2α null animals appeared to be arrested in diestrus. To determine if the absence of an estrous cycle was the cause of the infertility, we injected two sir2α null and two wild-type animals with hormones (5 IU of pregnant mare serum gonadotropin followed 46 h later with 5 IU of human choriogonadotropin) to induce ovulation. The next day, ovulated eggs were present in the oviducts of both sir2α null animals (16 and 19 eggs) as well as in both wild-type animals (29 and 35 eggs). This is consistent with the idea that female sterility in the sir2α null animals is due to hormonal inadequacy.
Analysis of spermatozoa from the cauda epididymis of wild-type and sir2α null mice showed striking differences in number, appearance, and motility. Wild-type mice had sperm counts of 48.8 × 106 ± 12.4 × 106, whereas the sir2α null animals had dramatically reduced numbers of mature sperm, with average counts of 2.6 × 106 ± 3.6 × 106 (data from three animals of each genotype). Virtually none of the sperm from the sir2α null mice were motile. Wild-type spermatozoa cell bodies had a consistent shape with a characteristic hook (Fig. 14a). In contrast, the majority of the sperm of the sir2α null mice were abnormal, with small, rounded, or smudged cell bodies and blunted or absent hooks (Fig. 14b). In addition, many of the sperm from sir2α null mice had retained cytoplasm around the nucleus, and many of the flagella appeared very fine and had no cell body attached.
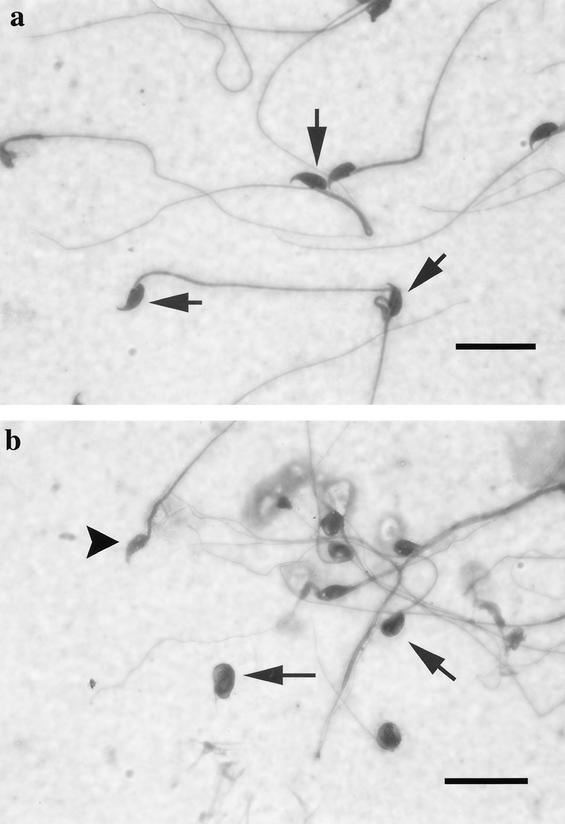
FIG. 14.
sir2α null mice have abnormal sperm morphology. Spermatozoa from the cauda epididymis were spread on a slide and stained with Dif-Quick stain. (a) Wild-type spermatozoa have a consistent nuclear shape with a characteristic hook. In contrast, the spermatozoa of sir2α null mice (b) had small, rounded, or smudged nuclear shapes. Some had a blunted (arrowhead) or absent hook. The flagella were also variable in shape, and many were no longer attached to sperm heads. Bar, 20 μm.
The male reproductive tract of sir2 null animals was characterized by organs significantly smaller than those of the wild-type littermates. All testes from sir2α null mice were subnormal in weight (wild type, 0.087 ± 0.013 g, n = 3; sir2 null, 0.056 ± 0.011 g, n = 3). Flow cytometry of testicular cells stained for DNA content indicated that all stages of spermatogenesis were present (Fig. 15), but a histological inspection indicated that the spermatogenesis occurring in the testes of sir2α null mice was abnormal. In the least affected testis, only subtle evidence of abnormal spermatogenesis was apparent, manifested as an increased incidence of spermatocyte apoptosis, occasional multinucleated germ cells, decreased numbers of mature elongated spermatids, and increased numbers of retained elongated spermatids.
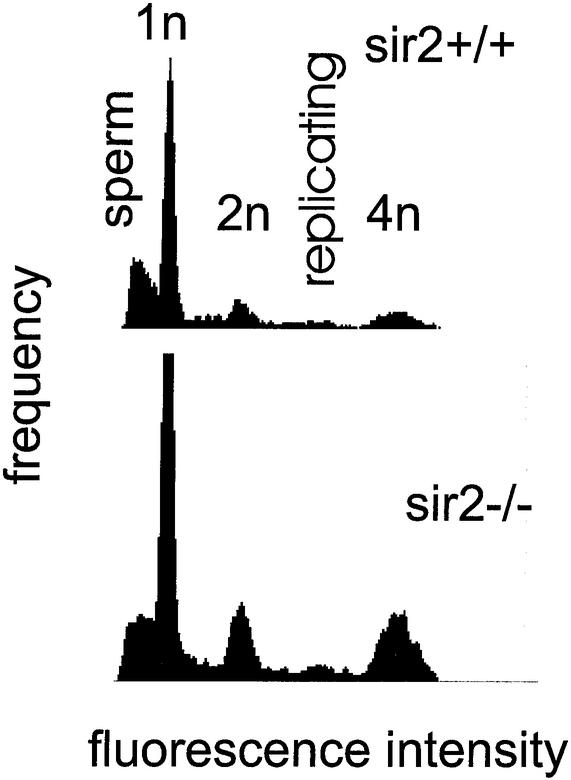
FIG. 15.
DNA content distribution of testes from sir2α null mice indicates that all stages of spermatogenesis are present. Single-cell suspensions were created from the testes of 7-month-old wild-type (upper panel) and sir2α null (lower panel) animals. Cells were fixed and stained with 4′,6′-diamidino-2-phenylindole before analysis by flow cytometry for DNA content per cell. The peaks corresponding to 1n, 2n, and 4n genomes are shown. Mature sperm stained less efficiently than expected and formed the broad peak at the left of the pattern. Replicating cells have a DNA content between 2n and 4n.
In the most affected testis, many seminiferous tubules showed an obvious depletion of the normal complement of spermatocytes, round spermatids, and elongated spermatids, while the spermatogonia and the supportive cells, Sertoli cells, Leydig cells, and peritubular cells, were present in normal numbers (Fig. 16a and b). In addition to the obvious depletion of differentiating germ cells, abnormalities included an increased number and size of vacuoles within the seminiferous epithelium, an increased incidence of apoptotic spermatocytes and spermatids, retention of highly condensed spermatids, and morphologically abnormal round spermatids (Fig. 16b, c, d, and e). The round spermatid abnormalities consisted of the sharing of acrosomes (Fig. 16c) and larger-than-normal cells in the stages right after meiosis (Fig. 16d).
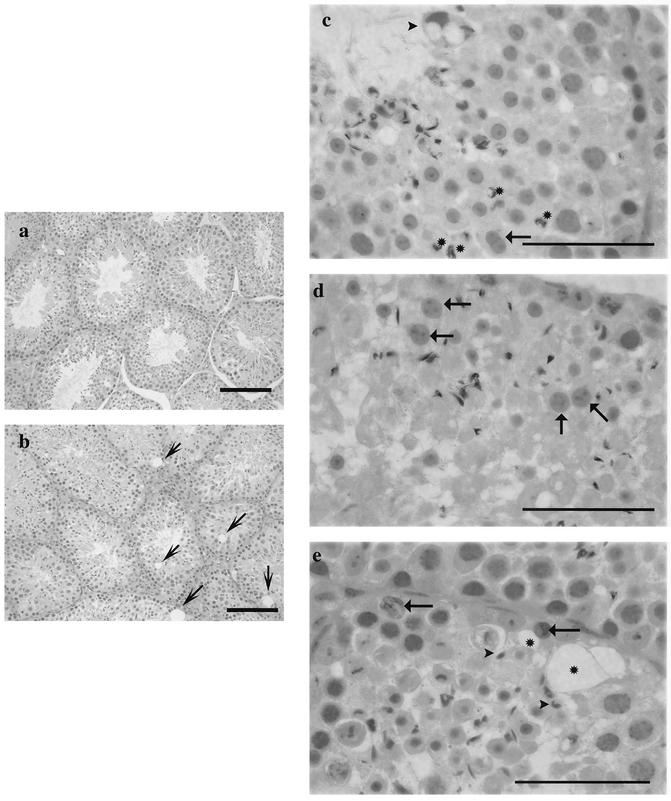
FIG. 16.
Spermatogenesis is abnormal in sir2α null mice. Testes from both wild-type (a) and sir2 null (b to e) mice were examined for histopathological abnormalities. At low power, the seminiferous tubules in the wild-type mouse appeared almost uniform in size, while those of the sir2α null mice varied in size and contained large vacuoles in the basal epithelium (arrows). At higher power, significant abnormalities were seen in the testes from sir2α null mice: acrosomal sharing in round spermatids (arrow, c); distorted residual bodies (arrowhead, c); larger-than-normal round spermatids (arrows, d); abnormal retained elongated spermatids (c, asterisks); and numerous vacuolated spaces (asterisks, e). Sections were stained with periodic acid Schiff-hematoxylin. Bars: 100 μm (a and b); 50 μm (c to e).
It has been proposed that p53 is a substrate for the mammalian SIR2α deacetylase activity, that p53 activity is dependent on acetylation, and that p53 function is normally modulated by SIR2α deacetylation (25, 29, 50). Apoptosis in the testis is dependent on p53 (4, 8, 52), and therefore the elevated frequency of apoptotic cells might be a consequence of the absence of SIR2α in the testes of sir2α null animals. We also predicted that the testes of the sir2α null mice might be hypersensitive to radiation. Wild-type and sir2α null mice were irradiated with 3 Gy and sacrificed 12 h later, and seminiferous tubules were examined for their incidence of apoptosis. X-irradiation increased the frequency of apoptotic cells in the testes of both wild-type and sir2α null mice (Fig. 17). However, testes from the the sir2α null mice did not appear to be hypersensitive to the radiation.
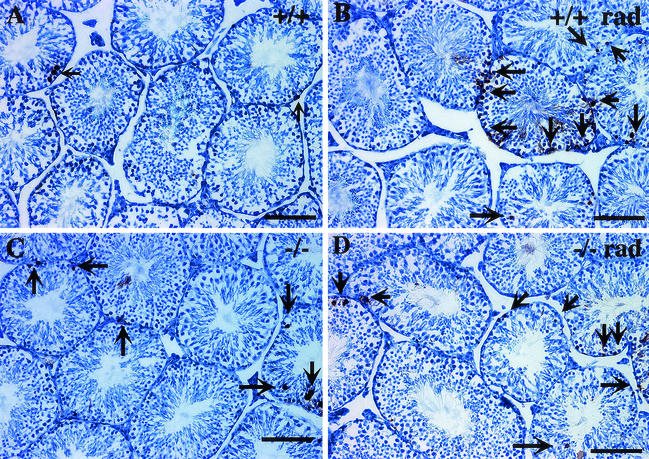
FIG. 17.
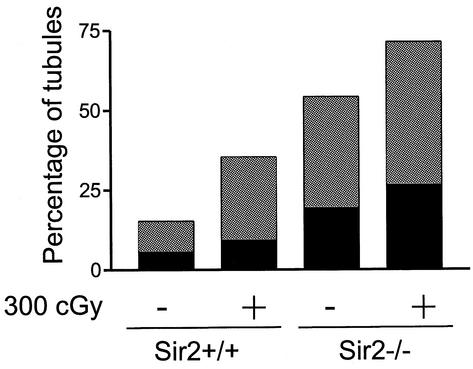
Increased numbers of apoptotic germ cells are present in the seminiferous tubules of sir2α null mice. Testis cross sections from wild-type (a) and sir2α null (b) mice were analyzed by the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling (TUNEL) assay to determine the frequency of apoptotic cells. Following exposure to 3 Gy of X-irradiation, the number of TUNEL-positive cells was increased (b and d) in both wild-type and sir2α null mice. The graph at the bottom shows the percentage of seminiferous tubule cross sections with one to three (grey portion of each bar) and more than three (black portion of bar) TUNEL-positive cells. Significantly more apoptotic cells were present in the sir2α null testes than in the wild-type testes (P < 0.05 by Student's unpaired t test).
DISCUSSION
Mammals maintain large proportions of their genomes in a transcriptionally silent state in each cell. If SIR2α were required for the maintenance of gene silencing, we would expect that the sir2α null animals would die very early during embryogenesis as a consequence of ectopic expression of genes normally maintained in an inactive state. The relatively mild phenotype of these null animals suggests that the SIR2α protein is not involved in the global silencing of genes in the mammalian genome, and this was partly confirmed by the normally regulated expression of a number of individual genes assessed both on Northern blots and in microarray assays. We directly tested the role of SIR2α in maintaining the inactivity of a silent transgene and found that the lack of expression of the transgene was not influenced by the sir2α genotype. Additional studies with sir2α null ES cells also indicated that these cells were not compromised in their ability to establish or maintain genes in their inactive state (McBurney et al., unpublished data).
Immunolocalization of the SIR2α protein to the euchromatin of mammalian cells is also consistent with the idea that this protein plays a role different from that of the yeast Sir2p. The Drosophila homologue of SIR2 is also associated with active genes and euchromatin (48), although it appears to be required for the downregulation of certain genes (40). The yeast Sir2p is not localized only to inactive regions of the genome such as the telomere; it is also found in multiprotein complexes that can be isolated as large stable assemblages (15). The Drosophila SIR2p is not found in a large complex but can be isolated in native conditions in a 200-kDa fraction, perhaps as a dimer (6). The mouse SIR2α protein runs on gel filtration columns at about 160 kDa, a size consistent with its being a dimer and not a member of a large multiprotein complex (McBurney et al., unpublished data).
The yeast Sir2p is part of a complex of proteins that associates with chromatin in regions that are maintained transcriptionally inert. The histone deacetylase activity of Sir2p is essential for the proper function of these protein complexes, and the substrate is thought to be the N termini of core histones. In mammals, a large number of nuclear proteins in addition to histones are acetylated (44), and in the cases of some important nuclear proteins such as retinoblastoma (11) and p53 (38, 51), it is thought that acetylation plays a role in regulating the function of these transcription regulators. In fact, there is evidence from transfection experiments suggesting that p53 is a substrate for SIR2α and that deacetylation of p53 downregulates its transactivation function (25, 29, 50). If SIR2α were part of the network regulating p53 activity, one might predict that the sir2α null mice would have hyperactive p53. Our finding that the testes of sir2α null mice have high spontaneous levels of apoptosis is consistent with this idea.
The reproductive viability of yeast mother cells is regulated by Sir2p (20), and the prolongation of cell viability brought about by caloric restriction is dependent on the presence of Sir2p (27). This effect of Sir2p is thought to be due to its ability to suppress recombination between tandem copies of the ribosomal genes, as aging mother cells show loss of ribosomal DNA, presumed to be due to recombinational excision from the genome (20). In S. cerevisiae, extra copies of the sir2 gene extend the life span of yeast mother cells. In C. elegans, an extra copy of the sir2 gene also resulted in extended life spans (46), but the evidence from this organism suggests that this effect of Sir2p acts via a mechanism unrelated to gene silencing or suppression of recombination. It appears that in C. elegans, Sir2p is part of the insulin-like growth factor signaling pathway, and the hyperactivity of this pathway forestalls normal aging.
The involvement of Sir2p in growth factor signaling in C. elegans might be an important clue to the role of SIR2α in mammalian cells. The phenotype of the sir2α null mice that we described above is strikingly similar to the phenotypes of mice bearing null alleles of IGF-1, IGF-2, and IGF-2R (1, 5, 12, 28, 37). The shared phenotypes include infertility, retarded growth rates, reduced survival to adulthood, reduced adult weight, and slowed rates of bone mineralization. The levels of expression of IGF-2 and IGF-2R are the same in wild-type and sir2α null animals (unpublished results), consistent with the genetic results from C. elegans that indicate that Sir2p is downstream of the growth factor in the pathway. The phenocopy of null alleles does not necessarily imply that SIR2α is in the IGF signaling pathway; for example, mice null for the activin βB gene also have reproductive and eyelid opening defects, similar to the sir2α null mice (49).
The reproductive defects in the two sexes are nicely correlated with the fact that gonads from the two sexes are the only adult tissues expressing detectable amounts of the SIR2α protein. However, the defects in the two sexes seem to be caused by different SIR2α-mediated effects. In females, oocytes appear to mature normally but fail to be ovulated. Female animals do not appear to cycle efficiently through estrus, perhaps indicating a hormonal defect. Granulosa cells are important sources of and responders to the hormones responsible for ovulation, and granulosa cells normally express high levels of the SIR2α protein. It is perhaps pertinent that IGF signaling occurs in granulosa cells (21, 34), and the hypothesized role of SIR2α in this pathway may be responsible for the defect in ovulation.
Infertility in male sir2α null animals appears to be due to inefficient spermatogenesis and the failure of sperm to mature properly. SIR2α is expressed in all stages of sperm development and disappears only during the late stages of nuclear condensation. However, it is these late stages that appear to be particularly defective, perhaps indicating that the spermatogenesis defect is in part mediated by a systemic problem. This is consistent with the observation that most of the sex organs are smaller in sir2α null animals than in their wild-type littermates.
The early postnatal lethality of sir2α null animals is difficult to reconcile with the observation that the SIR2α protein seems to be abundantly expressed only during early development and becomes undetectable in somatic tissues from midgestation on. It is possible that the SIR2α protein is expressed in mature somatic tissues at levels below the limit of detection of our antibody or that its expression is tightly regulated at the posttranscriptional level (the sir2α mRNA is readily detected in all somatic tissues tested). Alternatively, the absence of SIR2α during early embryogenesis may set in motion events whose legacy is not felt until much later during development. Were the SIR2α protein involved in the IGF signaling pathway, one might envisage how its absence might contribute to unbalanced tissue growth during early embryogenesis and subsequent failure of those tissues at later developmental times.
Acknowledgments
We are indebted to Douglas Gray for the gift of a phage genomic library from 129/Sv mouse DNA, to Barbara Vanderhyden for help in analysis of the ovary and female reproductive tract, to Manon Dubé for help in creating the ES cell embryos, and to Andras Nagy for the gift of the R1 cells.
This work was supported by the National Cancer Institute of Canada and the Canadian Institutes of Health Research.
REFERENCES
- 1.Accili, D., J. Drago, E. J. Lee, M. D. Johnson, M. H. Cool, P. Salvatore, L. D. Asico, P. A. Jose, S. I. Taylor, and H. Westphal. 1996. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 12:106-109. [DOI] [PubMed] [Google Scholar]
- 2.Adra, C. N., P. H. Boer, and M. W. McBurney. 1987. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60:65-74. [DOI] [PubMed] [Google Scholar]
- 3.Adra, C. N., N. A. Ellis, and M. W. McBurney. 1988. The family of mouse phosphoglycerate kinase genes and pseudogenes. Somat. Cell Mol. Genet. 14:69-81. [DOI] [PubMed] [Google Scholar]
- 4.Allemand, I., A. Anglo, A. Y. Jeantet, I. Cerutti, and E. May. 1999. Testicular wild-type p53 expression in transgenic mice induces spermiogenesis alterations ranging from differentiation defects to apoptosis. Oncogene 18:6521-6530. [DOI] [PubMed] [Google Scholar]
- 5.Baker, J., J. P. Liu, E. J. Robertson, and A. Efstratiadis. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73-82. [PubMed] [Google Scholar]
- 6.Barlow, A. L., C. M. van Drunen, C. A. Johnson, S. Tweedie, A. Bird, and B. M. Turner. 2001. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 265:90-103. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 8.Beumer, T. L., H. L. Roepers-Gajadien, I. S. Gademan, P. P. van Buul, G. Gil-Gomez, D. H. Rutgers, and D. G. de Rooij. 1998. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ. 5:669-677. [DOI] [PubMed] [Google Scholar]
- 9.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 10.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 11.Chan, H. M., M. Krstic-Demonacos, L. Smith, C. Demonacos, and N. B. La Thangue. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 3:667-674. [DOI] [PubMed] [Google Scholar]
- 12.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 13.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793-798. [DOI] [PubMed] [Google Scholar]
- 14.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 17.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 18.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khamsi, F., S. Roberge, Y. Yavas, I. C. Lacanna, X. Zhu, and J. Wong. 2001. Recent discoveries in physiology of insulin-like growth factor-1 and its interaction with gonadotropins in folliculogenesis. Endocrine 16:151-165. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 23.Laird, P. W., A. Zidjerveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau, S., K. Jardine, and M. W. McBurney. 1999. DNA methylation pattern of a tandemly repeated lacZ transgene indicates that most copies are silent. Dev. Dyn. 215:126-138. [DOI] [PubMed] [Google Scholar]
- 27.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J. P., J. Baker, A. S. Perkins, E. J. Robertson, and A. Efstratiadis. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59-72. [PubMed] [Google Scholar]
- 29.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 30.Lupton, S. D., L. L. Brunton, V. A. Kalberg, and R. W. Overell. 1991. Dominant positive and negative selection with a hygromycin phosphotransferase-thymidine kinase fusion gene. Mol. Cell. Biol. 11:3374-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBurney, M. W. 1999. Gene silencing in the development of cancer. Exp. Cell Res. 248:25-29. [DOI] [PubMed] [Google Scholar]
- 32.McBurney, M. W., W. A. Staines, K. Boekelheide, D. Parry, K. Jardine, and L. Pickavance. 1994. The murine Pgk-1 promoter drives widespread but not uniform expression in transgenic mice. Dev. Dyn. 200:278-293. [DOI] [PubMed] [Google Scholar]
- 33.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 34.Monget, P., and C. Bondy. 2000. Importance of the IGF system in early folliculogenesis. Mol. Cell. Endocrinol. 163:89-93. [DOI] [PubMed] [Google Scholar]
- 35.Mountford, P., B. Zevnik, A. Düwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell-Braxton, L., P. Hollingshead, C. Warburton, M. Dowd, S. Pitts-Meek, D. Dalton, N. Gillett, and T. A. Stewart. 1993. IGF-I is required for normal embryonic growth in mice. Genes Dev. 7:2609-2617. [DOI] [PubMed] [Google Scholar]
- 38.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell 107:815-818. [DOI] [PubMed] [Google Scholar]
- 39.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg, M. I., and S. M. Parkhurst. 2002. Drosophila Sir2 is required for heterchromatic silencing and by euchromatic hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109:447-458. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sherman, J. M., E. M. Stone, L. L. Freeman-Cook, C. B. Brachmann, J. D. Boeke, and L. Pillus. 1999. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell 10:3045-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 47.Tyl, R. W. 2002. Guidelines for mating rodents. Curr. Protocols Toxicol. Suppl. 11:162-210. [DOI] [PubMed] [Google Scholar]
- 48.Van Steensel, B., J. Delrow, and S. Henikoff. 2001. Chromatin profiling with targeted DNA adenine methyltransferase. Nat. Genet. 27:304-308. [DOI] [PubMed] [Google Scholar]
- 49.Vassalli, A., M. M. Matzuk, H. A. R. Gardner, K.-F. Lee, and R. Jaenisch. 1994. Activin/inhibin βB subunit gene disruption leads to defects in eyelid development and female reproduction. Genes Dev. 8:414-427. [DOI] [PubMed] [Google Scholar]
- 50.Vaziri, H., S. K. Dessain, E. N. Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 51.Wang, T., T. Kobayashi, R. Takimoto, A. E. Denes, E. L. Snyder, W. S. el Deiry, and R. K. Brachmann. 2001. hADA3 is required for p53 activity. EMBO J. 20:6404-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin, Y., B. C. Stahl, W. C. DeWolf, and A. Morgentaler. 1998. p53-mediated germ cell quality control in spermatogenesis. Dev. Biol. 204:165-171. [DOI] [PubMed] [Google Scholar]
- 53.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large interchromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]