Abstract
We have isolated a novel Drosophila (d) gene coding for two distinct proteins via alternative splicing: a homologue of the yeast adaptor protein ADA2, dADA2a, and a subunit of RNA polymerase II (Pol II), dRPB4. Moreover, we have identified another gene in the Drosophila genome encoding a second ADA2 homologue (dADA2b). The two dADA2 homologues, as well as many putative ADA2 homologues from different species, all contain, in addition to the ZZ and SANT domains, several evolutionarily conserved domains. The dada2a/rpb4 and dada2b genes are differentially expressed at various stages of Drosophila development. Both dADA2a and dADA2b interacted with the GCN5 histone acetyltransferase (HAT) in a yeast two-hybrid assay, and dADA2b, but not dADA2a, also interacted with Drosophila ADA3. Both dADA2s further potentiate transcriptional activation in insect and mammalian cells. Antibodies raised either against dADA2a or dADA2b both immunoprecipitated GCN5 as well as several Drosophila TATA binding protein-associated factors (TAFs). Moreover, following glycerol gradient sedimentation or chromatographic purification combined with gel filtration of Drosophila nuclear extracts, dADA2a and dGCN5 were detected in fractions with an apparent molecular mass of about 0.8 MDa whereas dADA2b was found in fractions corresponding to masses of at least 2 MDa, together with GCN5 and several Drosophila TAFs. Furthermore, in vivo the two dADA2 proteins showed different localizations on polytene X chromosomes. These results, taken together, suggest that the two Drosophila ADA2 homologues are present in distinct GCN5-containing HAT complexes.
Transcription in eukaryotes is a tightly regulated, multistep process. General transcription factors, gene specific transcriptional activators, and several different cofactors are necessary to access specific loci in the context of eukaryotic chromatin to allow precise initiation of RNA polymerase II (Pol II) transcription. One of the most appealing questions in eukaryotic transcription is how activators transmit their signals to the general transcription machinery to stimulate transcription.
Posttranslational modifications of nucleosomal histones have been correlated with the function of chromatin in transcription activation or repression (18, 34). One of the most extensively studied modifications is the acetylation of the highly conserved amino-terminal histone tails. The steady-state level of acetylation of histone proteins is accomplished by the action of histone acetyltransferases (HATs) and histone deacetylases (9, 37). Acetylation affects higher-order folding of chromatin fibers and histone-nonhistone protein interactions (31, 32). Thus, it can increase the affinity of transcription factors for nucleosomal DNA (40, 61).
A large number of recent studies have provided a direct molecular link between histone acetylation and transcriptional activation (reviewed in references 9 and 30). In these reports, it has been shown that several previously identified coactivators and adaptors of transcription possess intrinsic HAT activity. Among these co-activators are yeast Gcn5 (10), human GCN5 (12), TATA box-binding protein (TBP)-associated factor TAF1 (formerly TAFII250 [58]) (43), p300/CBP (46), ACTR (13), and steroid receptor coactivator 1 (SRC-1) (54). Many of these chromatin-modifying activities have been found within large multiprotein complexes that also contain several components with homology or identity to known transcriptional regulators.
In Saccharomyces cerevisiae the coactivator-adaptor protein Gcn5 is part of large multisubunit complexes, the largest of which is the 1.8- to 2-MDa SAGA complex (27). Yeast SAGA comprises products of at least four distinct classes of genes: (i) the Ada proteins (yAda1, yAda2, yAda3, yGcn5 [yAda4], and yAda5 [ySpt20]), which have been isolated in a genetic screen as proteins interacting functionally with the yeast activator Gcn4 and the herpes simplex virus activation domain VP16 (6); (ii) the TBP-related set of Spt proteins (ySpt3, ySpt7, ySpt8, and ySpt 20), initially identified as suppressors of transcription initiation defects caused by promoter insertions of the Ty transposable element (65); (iii) a subset of TBP-associated factors (TAFs) including scTAF5 (formerly TAFII90), scTAF6 (formerly TAFII60), scTAF9 (formerly TAFII17), scTAF10 (formerly TAFII25), and scTAF12 (formerly TAFII68/61) (28); and (iv) the product of the essential gene Tra1, which has been shown to be a component of SAGA (29, 49).
Another type of GCN5-containing HAT complex identified in yeast is the 0.8-MDa ADA complex (for “alteration/deficiency in activation”) (27). The ADA complex differs from SAGA in many aspects. In contrast to the 1.8 to 2-MDa ySAGA complex, the only components of the 0.8-MDa yADA complex are the three adaptor proteins (Ada2, Ada3, and Gcn5) and Ahc1 (19). The ADA complex does not contain yAda1, yAda5, or the other ySpt proteins found in SAGA. Furthermore, the structural integrity of the yADA complex, but not that of ySAGA, was dependent on the presence of the AHC1 gene product. The SAGA complex physically interacts with the acidic activators yGcn4 and VP16, whereas ADA fails to do so (17, 60). Moreover, ADA and SAGA HAT complexes generate overlapping yet distinct patterns of lysine acetylation on histone H3. These results taken together, strongly suggest that in yeast two distinct ADA-Gcn5 HAT complexes exist.
A number of similar multiprotein complexes have been characterised in mammalian systems as well, such as the human TBP-free TAF-containing complex (TFTC) (7, 64), the PCAF-GCN5 complex (45), and the SPT3-TAF9(TAFII31)-GCN5 acetyltransferase complex (STAGA) (42), which all contain the GCN5 HAT, ADA proteins, SPTs, TAFs, and the human homologue of yTRA1, TRRAP.
TFTC is able to direct preinitiation complex assembly on both TATA-containing and TATA-less promoters in vitro. Similarly to other TBP-free TAFII complexes, TFTC contains the hGCN5 HAT and is able to acetylate histone H3 in both free and nucleosomal contexts (7). The fact that histone acetylation has been linked to the activation of transcription (37) suggests that TFTC is recruited to chromatin templates by activators to acetylate histones and potentiate transcription initiation (68). Additional recently identified TFTC subunits common to other human TAF-HAT complexes include TAF9, hADA3, hSPT3, hPAF65α, hPAF65β, and TRRAP (7). Moreover, it has recently been shown that the Drosophila melanogaster dTAFII24 coimmunoprecipitates with dGCN5, suggesting the existence of a TFTC-like HAT complex in Drosophila (25).
RPB4 is the fourth largest of the 12 subunits of yeast Pol II. Its unusual feature is that in optimally growing cells it is present only in a small fraction of Pol II complexes (14, 36); however, it is required for efficient transcription during temperature extremes and certain other stress conditions like starvation. RPB4 and another subunit, RPB7, are thought to form a subcomplex (20, 33) which is incorporated into the Pol II enzyme under suboptimal growth conditions to play a stress-protective role by inducing a closed Pol II conformation (14, 48). Interestingly, unlike the other subunits of Pol II, the level of RPB4 is posttranscriptionally regulated. In yeast, RPB4 is nonessential under normal growth conditions. Its function and specific role in higher eukaryotes are still unclear. Nevertheless, the recent finding that its homologue is already present in archaea supports the assumption that it plays an important role (63).
In this study we report the identification of two novel Drosophila homologues of the yeast Ada2 protein (dADA2a and dADA2b). Interestingly, two genes that encode ADA2 homologues were also found in the Arabidobsis thaliana and human genomes but not in the fully sequenced Caenorhabditis elegans genome. The analysis of the gene encoding the Drosophila ADA2a protein revealed that in addition to dADA2a, this gene encodes the Pol II subunit dRPB4 by alternative splicing. The N-terminal end of the two proteins is encoded by the same exon. Evolutionarily conserved protein-protein interactions were found among dADA2a and dADA2b proteins and their predicted partners of interaction. We present several lines of evidence which show that the two novel dADA2 homologues are present in different GCN5 HAT-containing complexes.
MATERIALS AND METHODS
Cloning of the Drosophila dAda2a/Rpb4 gene and the determination of its exon-intron boundary.
Overlapping genomic fragments from the 94F cytological region of the Drosophila chromosome were isolated from a lambda genomic library. The transcript map of one 7.4-kb fragment recovered in several independent clones indicated that it gives rise to at least four mRNAs. cDNA clones corresponding to all four transcripts were isolated from a Drosophila embryonic cDNA library. Complete nucleotide sequence of the isolated 7.4-kb genomic DNA (pFF1) and representative cDNA clones were determined and compared to each other to reveal the exon-intron boundaries (data not shown) and show that two of the cDNA populations are related (see Results and Fig. 1). For the isolation of full-length dADA2a and dRPB4 mRNAs, the 5′ rapid amplification of cDNA ends technique was used with gene-specific primers R1 and R2 (see Table 1 and Fig. 1).
FIG. 1.
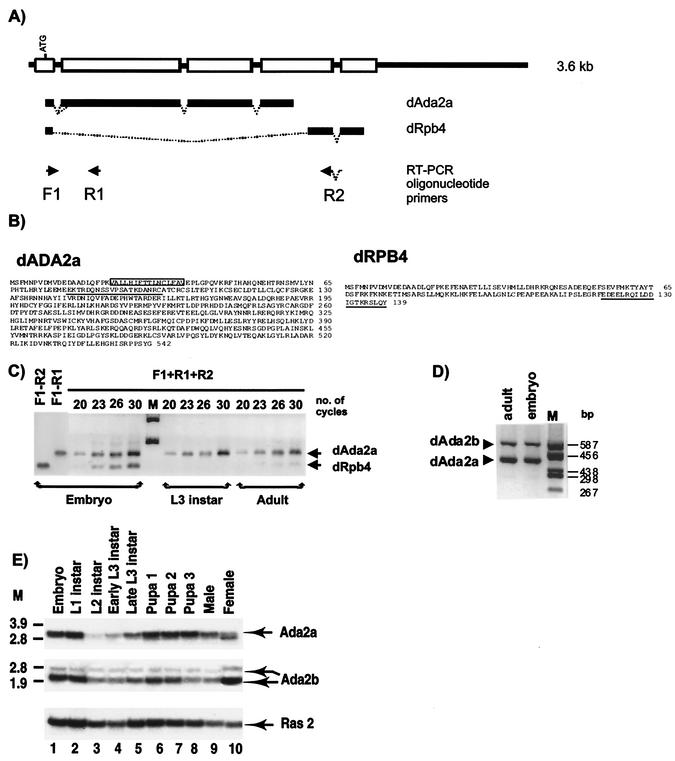
The dAda2a/dRpb4 gene encodes two putative transcription factors, dADA2a and dRPB4. (A) Schematic representation of the dAda2a/dRpb4 gene structure. Exons are shown as boxes on the thick line representing the genomic sequence. The common ATG located in the first exon is indicated. The dAda2 and the dRpb4 transcripts produced by alternative splicing are indicated as bold lines under the genomic structure. F1, R1, and R2 show the locations of oligonucleotide primers used for RT-PCR analysis of the dAda2 or dRpb4 mRNA. (B) Deduced amino acid sequences of the Drosophila ADA2a and RPB4 proteins. The 15-amino-acid stretch that is generated by alternative 3′ splice site selection in certain dADA2a isoforms is boxed. The oligopeptides used for immunization to produce specific antibodies against dADA2a and dRPB4 are underlined. (C) RT-PCR analysis of total RNA prepared from Drosophila at different developmental stages, as indicated. The arrows show the dAda2a- and dRpb4-specific PCR products. Aliquots from each PCR amplification reaction were taken at 20, 23, 26, and 30 cycles, resolved on a 1.2% agarose gel, and visualized by ethidium bromide staining. M, molecular weight marker. (D) RT-PCR analysis using total RNA prepared from developmental stages, as indicated above the lanes. The dAda2a and dAda2b PCR products with different sizes are indicated. Aliquots from each PCR amplification reaction were resolved on a 2% agarose gel and visualized by ethidium bromide staining. M, molecular weight marker. (E) Northern blot analysis of dAda2a and dAda2b mRNAs during Drosophila development. cDNA fragments from either dADA2a or dADA2b cDNAs were labeled and hybridized with mRNA samples prepared from the indicated stages of the Oregon R strain. The same blot was also hybridized with the Ras2 probe as a control (53). M, molecular weight marker.
TABLE 1.
Oligonucleotide primers used in PCR amplifications for plasmid construction
| Name | Sequencea | Directionb |
|---|---|---|
| F1 | GAACCCCGTGGATATGGTGG | fw |
| R1 | CATGTGGCACACCGATTGGC | rv |
| R2 | CTGCATCAGCAAGCTTCGAG | rv |
| 2bF | CGCGGAGTGCGAAAACTT | fw |
| 2bR | GGGCCAGCTTAAGCATCA | rv |
| dGCN | CATGCGAATTCTCTGCCGATCTTGGA | fw |
| dGCN | CTGTCGGATCCTTCGGCCTTATGCAG | rv |
| dADA2b-RI | GCATGAATTCATGACCACAATCGCGGATTT | fw |
| dADA2b-BHI | CGATGGATCCCCGACAGCTATCCAA | rv |
| dADA3F | GATCGAATTCATGAGTGCGAACCTGAAGAA | fw |
| dADA3R | TGACGAATTCTCCTGTCCTACATCTTTGGC | rv |
| dRPB4F | GCTAGGATCCCCGTGGATATGGTGGAT | fw |
| dRPB4R | CGATGTCGACTTAGTATTGTAAGCTGCGTTTAGT | rv |
| dRPB7F | GCTAGGATCCGAATATCGCTGGAGCAAGAGATT | fw |
| dRPB7R | CGATGTCGACTTAGTTGGACACCAATCCCAA | rv |
The gene-specific sequences are underlined.
fw, forward; rv, reverse.
Construction of recombinant plasmids.
Drosophila ada2b, ada3, and rpb7 cDNAs were generated by PCR using oligonucleotide primers based on expressed sequence tag (EST) sequences (Table 1). Similarly, plasmid constructs for yeast two-hybrid assays were generated using gene-specific primers with the appropriate restriction sites incorporated (Table 1). Detailed descriptions of the LexA DNA-binding domain fusion of dADA2a, dADA2b, dADA3, dGCN5, and dRPB4 in the pBTM116 vector (39) and the Gal4 activation domain fusion of dADA2b, dADA3, dGCN5, and dRPB7 in the pGAD424 vector (Clontech) are available on request. pBTM-yADA2, pBTM-yADA3, pASV-yGCN5, and pBTM-hADA2a are described in reference 62. hGCN5-S and dGCN5 cDNAs were kind gifts from S. Berger and from E. Smith and D. Allis, respectively, and the pASV3-hGCN5-S vector was a gift from E. vom Baur and R. Losson.
The reporter plasmids for transient-transfection assays in insect (S2) and human HeLa cells were pIND-Luc and 17M-Glob-Luc (2), respectively. pIND-Luc carries the luciferase reporter gene under the control of an ecdysone-inducible promoter which contains five EcREs (Invitrogen).
For the expression of dADA2a and dADA2b in S2 cells, the corresponding cDNA fragments were cloned into pMT/V5 (Invitrogen). For HeLa cell expression, the corresponding cDNAs were cloned into pXJ41 (67). Detailed descriptions of plasmid constructs are available on request.
RNA isolation, RT-PCR, and Northern analysis.
Total RNA was isolated from Drosophila at various stages of development as described previously (15, 41). For reverse transcriptase PCR (RT-PCR), 3 μg of RNA was reverse transcribed with 50 ng of random primers (GIBCO) in a final volume of 50 μl. The reaction was carried out for 1 h with 20 U of Moloney murine leukemia virus RT (Fermentas) at 37°C as specified by the manufacturer. For PCR amplification of dada2a and drpb4 mRNAs, gene-specific primers were designed which allowed us to distinguish between the two mRNAs (see Fig. 1). The forward primer F1 (Table 1) lies in the common first exon present in both mRNAs, while the reverse primer R1 lies in the second exon of the dAda2a coding sequence. The dRpb4-specific mRNA was amplified using the F1 primer as forward primer and the reverse primer R2, which lies at the junction of the last two exons of dRpb4 cDNA. For RT-PCR detection of the dAda2b mRNA, the 2bF and 2bR oligonucleotide primers were used (Table 1). Northern blot analysis was carried out as described earlier (53).
Antibody production.
To generate specific polyclonal antibodies (PAbs), specific peptides for each protein were synthesized: dADA2a (EKTRDQNSSVPSATKDANRC) (underlined in Fig. 1B), dRPB4 (EDEELRQILDDIGTKRSLQY(C)) (underlined in Fig. 1B), dADA2b (PAQSQRPRLIDHTGDDDA(C)) (underlined in Fig. 2A), and dGCN5 (accession number AAC39102) from amino acids 778 to 795 (SNCRFYNSPDTEYYRCAN) (52). The peptides were coupled to an ovalbumin carrier protein and used for immunization of rabbits. Collected sera were purified on SulfoLink columns (Pierce) to which the synthesized peptides had previously been conjugated through their C-terminal cysteines. Affinity columns were prepared as specified by the manufacturer. Each rabbit serum was affinity purified except for the dGCN5 serum. Antibodies against dTBP and dTAFs were previously described (25, 35).
FIG. 2.
The novel dADA2s and dRPB4 are evolutionarily well conserved. ADA2 (A) and RPB4 (B) amino acid sequences from various species were extracted from the GenBank, EMBL, DDBJ, and Genome Sequencing Center databases by iterative searches using the PSI-BLAST program and hADA2a or yRPB4 as query. The one-letter amino acid code is used. The different ADA2 sequences are as follows: A. thaliana (At) (accession no. q9sfd5 and o23486), Z. mays (Zm) (accession no. aw055700), P. falciparum (Pf) (accession no. q9u611), S. pombe (Sp) (accession no. q9p7j7), S. cerevisiae (Sc) (accession no. q02336), H. sapiens (Hs) ADA2a (accession no. o75478), D. melanogaster (Dm) ADA2a (accession no. AF544017), D. melanogaster (Dm) ADA2b (accession no. q9vhv0), C. elegans (Ce) (accession no. q19953), D. rerio (Dr) (accession no. aw421870), H. sapiens (Hs) ADA2b (accession no. ac048379), M. musculus (Mm) (accession no. be532918). Residues with the same physicochemical properties conserved among the different ADA2 homologues at between 100 and 91% have a black background, those conserved at between 90 and 76% have a dark gray background, and those conserved at between 75 and 60% have a light gray background. The highly conserved ZZ and SANT domains are boxed. The amino acid positions in the different proteins are labeled on the left in the sequences represents the deletion of a nonconserved region in the corresponding protein. In panel A, the nonconserved N-terminal end of certain ADA2 factors is not represented. The conserved secondary structures predicted (Sec. Struct. Pred.) for the ADA2 proteins are labeled such that the positions of the α-helices (h) and the β-strands (b) are shown under the sequences. The amino acid sequence in dmADA2b that was used to generate the anti-dADA2b PAbs is underlined.
Immunolocalization of dADA2a and dADA2b proteins on polytene chromosomes.
Immunostaining of polytene chromosomes was performed as described previously (5, 53), and the anti-dADA2a and anti-dADA2b antibodies were diluted 100- and 500-fold, respectively.
Transient-transfection and reporter gene assay.
Drosophila Schneider S2 and HeLa cells were transfected using the calcium-phosphate transfection protocol. Routinely, 1 or 2 μg of reporter constructs was used with the indicated amounts of ADA2 expression plasmids and 50 ng of the GAL-VP16 expression plasmid (59) (in HeLa cells). For the overproduction of dADA2a and dADA2b in transfected S2 cells, the metallothionein promoter of pMT/V5-dAda2a and pMT/V5-dADA2b was induced with copper sulfate. Transient expression of the luciferase reporter gene was detected in total-cell extracts 2 days posttransfection by using a luciferase detection kit (Promega) as recommended by the manufacturer.
Extract preparations, immunoprecipitation, chromatographic purifications, and Western blot analysis.
Preparation of Drosophila nuclear extracts (TRAX) from 0- to 12-h embryos and immunoprecipitations (IP) were previously described (25, 50). Briefly, 100- to 500-μl volumes (approximately 500 to 800 μg) of the indicated protein fractions were immunoprecipitated with 50 to 80 μl of protein A-Sepharose (Pharmacia) and approximately 5 to 10 μg of specific antibody. Protein A-Sepharose antibody-bound complexes were extensively washed with an IP buffer containing 500 mM KCl and eluted from the beads by boiling 5 to 10 μl of beads in 30 μl of sodium dodecyl sulfate (SDS) sample buffer. Following electrophoresis, samples were subjected to Western blot analysis. Protein separation by heparin Ultrogel chromatography was performed as described previously (8), except that a 500-μl column was used. The gel filtration on a Superose 6 column using the Smart System (Pharmacia) was carried out under standard conditions (16). For Western blot analysis, protein extracts were prepared by boiling in SDS sample buffer. The samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were electroblotted to a nitrocellulose membrane and incubated with primary antibodies as indicated. Peroxidase-conjugated goat anti-rabbit immunoglobulin (heavy plus light chain)-specific antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies. Detection was performed with an ECL kit (Amersham).
Sedimentation in glycerol gradients of dADA2-containing complexes.
High-molecular weight (HMW) markers (Pharmacia) and Drosophila nuclear extract (200 μl, approximately 2 mg of protein) dialyzed overnight at 4°C against buffer D (50 mM Tris-HCl [pH7.9], 50mM KCl, 0.5 mM dithiothreitol, 0.1 mM EDTA, 5% glycerol) were layered on 3.8 ml of a 10 to 30% linear glycerol gradient and centrifuged for 2.5 h at 55 krpm, using an SW60 Ti rotor (Beckman). Gradients were fractionated by collecting 200-μl aliquots, and 20 μl from every second fraction of the separated HMW markers was separated by SDS-PAGE (10% polyacrylamide) and stained with Coomassie brilliant blue. This allowed the calculation of the apparent molecular weights corresponding to particular fractions of the gradient. Unfractioned extract (2 μl) and 20-μl volumes from the fractions of the separated extract were analyzed by Western blotting using specific antibodies, as indicated in the figure legends. The glycerol concentration in each fraction was determined by refractometry to ensure that the gradients were linear.
Nucleotide sequence accession numbers.
The accession numbers of the dAda2a and dRpb4 cDNAs and corresponding proteins are AF544017 and AF544019, respectively.
RESULTS
The same gene encodes a Drosophila ADA2 homologue (dADA2a) and the Pol II subunit dRPB4.
In an independent screen for genes encoding proteins involved in transcription regulation, we recently isolated a genomic fragment of a novel Drosophila gene hereafter called dAda2a/dRpb4. Surprisingly, a number of clones representing two distinct types of cDNAs which corresponded to the novel dAda2a/dRpb4 gene were isolated when Drosophila embryonic cDNA libraries were screened with genomic probes (Fig. 1A and B). Nucleotide sequence alignments revealed that the two types of cDNAs isolated have a common exon encoding 21 amino acids with a potential translation start site at their 5′ ends (Fig. 1A and B). The rest of the two cDNAs hybridized with different parts of the genomic sequence, indicating that the two types of cDNAs represent two distinct alternatively spliced mRNAs from the same gene (Fig. 1A). By using 5′ rapid amplification of cDNA ends with primers from the second exons of the two cDNAs (primers R1 and R2 in Fig. 1 and Table 1), we recovered several independent cDNA clones corresponding to the two mRNAs. Each of these cDNAs had exactly the same 5′ end, suggesting that the two types of mRNAs are transcribed from the same initiation site. The two types of mRNAs have a common 148-nucleotide long first exon and contain an open reading frame coding for either 542 or 139 amino acids, respectively.
Blast searches in databases revealed that the 542-amino-acid putative protein sequence encoded by the longer mRNA is a Drosophila homologue of the yeast transcriptional adaptor ADA2 (6) (hereafter called dADA2a [Fig. 1B]). Moreover, we noticed a small variation among the recovered cDNA clones coding for dADA2a. Specifically, as a result of two alternatively used 3′ splice acceptor sites (Fig. 1A), some of the dADA2a cDNAs encode an isoform which is 15 amino acids shorter than the full-length dADA2a (the 15-amino-acid difference is boxed in Fig. 1B).
The shorter cDNA encodes another protein involved in transcription, the Drosophila Pol II subunit dRPB4 (33, 66) (Fig. 1A and B). Thus, the dAda2a/dRpb4 gene encodes two putative transcription factors, dADA2a and dRPB4, both of which are supposedly involved in Pol II transcription. Interestingly, due to their common first exon, the dADA2a and the dRPB4 proteins may have a 21-amino-acid common N-terminal end.
Drosophila contains two distinct genes which encode ADA2 homologues.
The position of the dAda2a/dRpb4 gene is 90F4 on the Drosophila cytological map. Sequence alignments of the isolated genomic clone with nucleotide sequence data of the Berkeley Drosophila Genome Project and independent genetic mapping (data not shown) confirmed this position. Surprisingly, when we used the published human ADA2 amino acid sequence (12) as a query sequence in PSI-BLAST searches of GenBank, EMBL, DDBJ, and Genome Sequencing Center databases, we found another gene coding for a second Drosophila ADA2 homologue which is located at position 84F6 in the Drosophila genome. Thus, in Drosophila there are two distinct genes encoding two ADA2 homologues (Fig. 2A). We named the second Drosophila ADA2 homologue dADA2b. In the same search, in addition to the previously described ADA2s, we found a number of factors from different species with a significantly high similarity to ADA2s (E values were lower than 10−17). The result of this analysis not only confirmed that the two novel dADA2s are indeed homologous to each other and to the Saccharomyces cerevisiae ADA2, as originally hypothesized, but also revealed the occurrence of several homologues from different species, including A. thaliana, Zea mays, Plasmodium falciparum, C. elegans, Schizoaccharomyces pombe, Danio rerio, and Mus musculus. Interestingly, in this search we found that in addition to the Drosophila genome, the Arabidobsis and human genomes contain two distinct genes which encode proteins with a significant homology to the ADA2 family members. The second putative human ADA2 homologue was called hADA2b to distinguish it from the one identified earlier (12) (hereafter called hADA2a). Pairwise alignments showed that dADA2a and dADA2b are 55 and 50% similar to human ADA2a, respectively; 51 and 60% similar to the novel human ADA2b, respectively; and 51 and 53% similar to yeast ADA2, respectively (Fig. 2A). Thus, dADA2a is more similar to hADA2a while dADA2b is more similar to hADA2b. Note that both putative ADA2s from Arabidobsis were more similar to ADA2a (human or Drosophila) than to ADA2b and that the other vertebrate ADA2s identified from zebrafish (Dr) or mouse (Mm) were more homologues to hADA2b.
All identified ADA2 homologues contain conserved domains which include the previously identified putative zinc finger domain, called the ZZ domain (47), and the so-called SANT domain, which is often found in transcriptional cofactors (1). (Fig. 2A). The notion that both the ZZ and SANT domains show similarity to the DNA-binding domain of Myb-related proteins (44) and that the minimal DNA-binding domain of Myb is composed of two repeats might suggest that the ADA2 proteins bind to DNA (see also Discussion). In addition to the ZZ and SANT domains, we identified three conserved domains among all the ADA2 homologues, which we called ADA boxes (Fig. 2A). All ADA boxes contain several well-conserved α-helical secondary-structure motifs separated by shorter or longer loops, as predicted by a program that takes into account conservation among all the family members.
RPB4 of Drosophila shows a striking similarity to its human counterpart. The 139-amino-acid Drosophila and the 2-amino-acid-longer human proteins are 75.4% identical. In addition, a comparison of the known genomic and cDNA sequences indicates that the exon-intron structure of the Drosophila and human RPB4 genes are similar (33). Note that the first exons in both the Drosophila and human genes end at exactly the same position. The similarity between the yeast and Drosophila RPB4 proteins is weaker (60.1% similarity, 36% identity). As a result of N-terminal regions not present in the Drosophila protein, the yeast (Sc) RPB4 is about 80 amino acids longer (Fig. 2B).
Both dADA2 homologues and the dRPB4 are expressed at the mRNA and protein levels.
The unexpected observations that a transcription adaptor and a Pol II subunit are encoded by the same gene and that the Drosophila genome harbors two Ada2-related genes prompted us to test the expression of the two dADA2 homologues as well as dRPB4 in Drosophila at the mRNA and protein levels. First, we used RT-PCR analysis to detect and distinguish the accumulation of dAda2a and dRpb4 transcripts, which are apparently transcribed from the same gene. Total RNA samples isolated from embryos and adults were investigated for the presence of specific mRNAs by using primers designed to distinguish between dAda2a and dRpb4 mRNA (Fig. 1A). Fragments of of dAda2a and dRpb4 mRNAs of the expected sizes were detected in similar amount by RT-PCR amplification of embryonic samples, whereas in adult samples similar amounts of dAda2a product were detected to those in embryos; however, the amount of dRpb4-specific fragment was hardly detectable (Fig. 1C). Thus, while the identical 5′ ends of the dAda2a and dRpb4 mRNAs suggest that they are transcribed from the same promoter, their mRNA levels seem to be differentially regulated during development. Similar RT-PCR analysis was carried out for the detection of dAda2b transcripts in embryonic and adult samples. As shown in Fig. 1D, no variation was observed in the expression of the dAda2b transcript compared to the amount of dAda2a transcript in embryonic and adult samples.
A more detailed analysis of the developmental stage-specific distribution of the dAda2a and dAda2b mRNAs was carried out by using Northern blot analysis. Hybridization of dAda2a-and dAda2b-specific probes to poly(A)+ RNA revealed differences in the amounts of the two mRNAs at different stages of Drosophila development (Fig. 1E). Although present at different levels, both dAda2b and dAda2a mRNAs were detected at all analyzed stages of development. dAda2b transcripts seem to be more evenly expressed, while for the dAda2a mRNA an apparent peak of expression was detected at the pupal stages of development. In contrast, only very low levels of Ada2a mRNA could be detected at the midlarval stages (Fig. 1E).
In conclusion, the differences observed in the expression of the three mRNAs by RT-PCR and Northern analysis suggest unique stage-specific regulation of the expression of the dAda2a/dRpb4 and dAda2b genes. Moreover, the fact that RT-PCR demonstrated differences in the levels of the two transcripts originating from the dAda2a/dRpb4 gene suggests that posttranscriptional control plays a role in the production of dAda2a and dRpb4 mRNAs from the same gene.
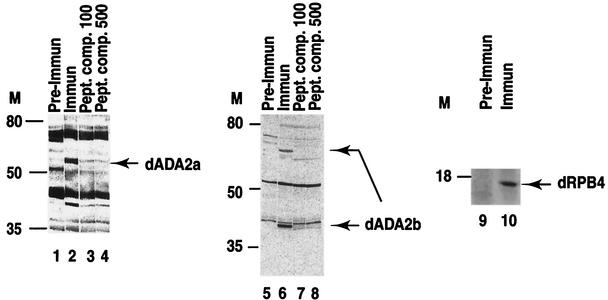
To test whether both of the dADA2 homologues and the dRPB4 proteins are expressed in Drosophila, we developed rabbit PAbs which recognize these proteins specifically (see Materials and Methods). Nuclear extracts prepared from 0- to 12-h-old Drosophila embryos were analyzed by Western blotting using dADA2a, dADA2b, or dRPB4-specific immune sera (Fig. 3). To verify the specificity of the PAbs, we used preimmune sera taken from rabbits before immunization as negative controls. For the ADA2a-and ADA2b-specific PAbs, we also included the specific peptides, used for immunization, as competitors during the Western blot analysis. The migration of the protein product recognized specifically by the anti-dADA2a PAb on SDS-PAGE corresponds to that of a protein of 58 to 60 kDa (Fig. 3, lanes 1 to 4), which is in good agreement with the calculated molecular mass of 59.6 kDa. In certain Drosophila protein extract preparations, we also observed a second protein recognized specifically by the dADA2a PAb, migrating around 75 kDa (data not shown). The anti-dADA2b PAb recognized two protein species specifically in nuclear extracts: a more abundant polypeptide with an apparent molecular mass of about 42 to 44 kDa and another polypeptide migrating around 70 kDa (Fig. 3, lanes 5 to 8). The estimated size of the faster-migrating protein is in good agreement with the calculated molecular mass of dADA2b (44.8 kDa), whereas the slower-migrating form could be either a posttranslationally modified form or a splice variant of ADA2b. Note that in the Northern blot analysis we also detected two mRNAs for dAda2b (Fig. 1E). In good agreement with the calculated size of dRPB4 (15.3 kDa), a 16-kDa protein product was identified on Western blots obtained with the anti-dRPB4 PAb (Fig. 3, lanes 9 and 10). Thus, from these experiments, we concluded that both of the dADA2 homologues and the dRPB4 protein are expressed in Drosophila nuclei.
FIG. 3.
The two dADA2 homologues and dRPB4 are expressed in Drosophila. Drosophila embryo extracts were resolved by SDS-PAGE on 10% (ADA2a and ADA2b) or 15% (RPB4) polyacrylamide gels and blotted with purified anti-dADA2a, anti-dADA2b, or anti-dRPB4 PAbs (Immun) or with preimmune (Pre-Immun) sera taken from the corresponding rabbits before immunization. Molecular mass markers (M) are indicated in kilodaltons. To test the specificity of the sera raised against dADA2a and dADA2b, competition experiments with peptides used for the immunization (see Fig. 1B and 2) were done. Polyclonal sera raised against either ADA2a (500× dilution) or ADA2b (1,000× dilution) were preincubated with 100 μg (Pept. comp. 100; lanes 3 and 7) or 500 μg (Pept. comp. 500; lanes 4 and 8) of the corresponding peptides, respectively, prior to incubation with the Western blots.
Both dADA2a and dADA2b interact with GCN5, but only dADA2b interacts with dADA3.
To study whether the two Drosophila ADA2 homologues interact with GCN5 and ADA3, as their yeast and human homologues do, we carried out a yeast two-hybrid analysis. In our assays, we first compared the interactions of dADA2a or dADA2b with full-length dGCN5, human hGCN5-S(hort), or yeast GCN5. hGCN5-S-pASV3, a VP16 activation domain fusion plasmid, was used together with either LexA-dADA2a or LexA-dADA2b DNA-binding domain fusion plasmids. Both LexA-dADA2a and LexA-dADA2b interacted strongly with VP16-hGCN5-S in this assay (Table 2). In contrast, neither the VP16-containing plasmid nor the LexA-dADA2s alone resulted in any β-galactosidase activity (Table 2). The interactions between dADA2a and hGCN5-S or dADA2b and hGCN5-S were comparable to that observed between hADA2a and hGCN5 or yADA2 and yGCN5 (Table 2). In good agreement, both GAL4AD-dADA2 fusion proteins (pGAL424-dADA2a, and pGAL424-dADA2b) interacted with the LexA fusion of the full-length Drosophila GCN5 (pLexA-dGCN5) (Table 2), however this interaction was somewhat weaker, possibly due to the long N-terminal domain of dGCN5 that was absent from the human GCN5-S fusion tested. In good agreement with the lack of interaction between human ADA2a and yGCN5 (12), neither of the Drosophila ADA2s interacted with yeast GCN5 (Table 2). Next, the Drosophila ADA3 protein was tested for its pairwise interaction with the two Drosophila ADA2 homologues. For this purpose, dADA3 cDNA was inserted into the pGAD424 expression vector to produce a fusion protein with the GAL4 activation domain. Surprisingly, a two-hybrid interaction was observed between dADA2b and dADA3 but not between dADA2a and dADA3 (Table 2). Thus, these in vivo two-hybrid interactions demonstrated a functional and evolutionarily conserved homology in the interaction pattern between the two novel Drosophila ADA2 proteins and their putative Drosophila interaction partners (i.e., dGCN5 and dADA3) or their human counterparts. Furthermore, this two-hybrid test also revealed differences in the partners of dADA2 proteins. In conclusion, dADA2a interacts with dGCN5 but not with dADA3 whereas dADA2b interacts with both dGCN5 and dADA3.
TABLE 2.
Results of two-hybrid interaction assays with ADA2 and RPB4 fusion proteins
| Proteina | β-Galactosidase activityb | |
|---|---|---|
| DBD | AAD | |
| dAda2a | None | − |
| dAda2b | None | − |
| dRpb4 | None | − |
| yAda2 | None | − |
| hAda2a | None | − |
| dGcn5 | None | |
| None | dAda3 | − |
| None | dRpb4 | − |
| None | dRpb7 | − |
| None | yGcn5 | − |
| None | hGcn5 | − |
| hAda2a | hGcn5 | +++ |
| yAda2 | yGcn5 | +++ |
| dAda2a | hGcn5 | +++ |
| dAda2b | hGcn5 | +++ |
| dGcn5 | dAda2a | ++ |
| dGcn5 | dAda2b | ++ |
| dAda2a | yGcn5 | − |
| dAda2b | yGcn5 | − |
| dAda2a | dAda3 | − |
| dAda2b | dAda3 | +++ |
| dAda2a | dRpb4 | − |
| dAda2b | dRpb4 | − |
| dRpb4 | dRpb7 | ++ |
DBD and AAD indicate DNA-binding and activation domain fusions, respectively.
β-Gal activities were estimated on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates and are expressed as follows: −, no activity; ++, intermediate activity; +++, strong activity.
Similarly to the two dADA2 proteins, we also tested whether interaction would exist between dRPB4, encoded by the alternatively spliced mRNA, and its putative partner dRPB7 (36). For this purpose, the dRPB7 cDNA was inserted in frame in the GAL4 expression vector. In the two-hybrid assay, these two Pol II subunits interacted as expected (Table 2). These results, together with the sequence similarity (Fig. 1B), further show that the alternatively spliced dRpb4 mRNA transcribed from the dAda2a/dRpb4 gene encodes a functional Drosophila homologue of the Pol II subunit RPB4. Note that dRPB4 did not interact with either dADA2a or dADA2b (Table 2).
dADA2a and dADA2b are present in multiprotein complexes containing TAFs and GCN5 HAT.
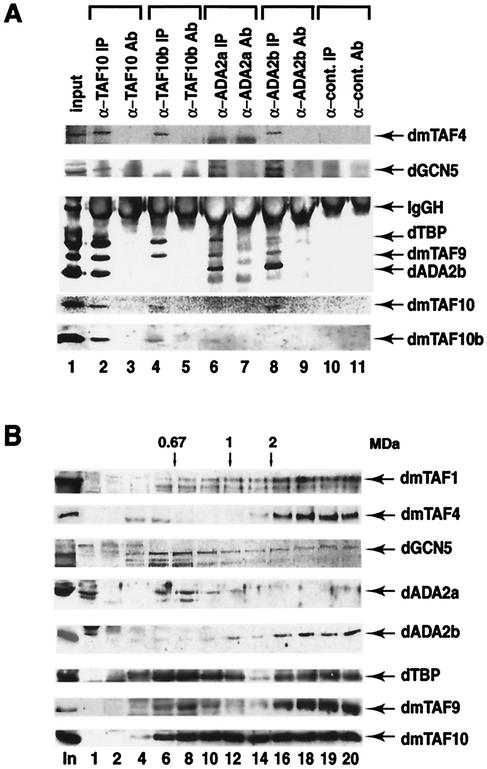
To determine whether the newly identified Drosophila ADA2s are indeed associated with multiprotein complexes containing the GCN5 HAT and TAFs, we first investigated whether dADA2a or dADA2b would be associated with dmTAF10 (formerly dTAFII24), a TAF that was suggested to be a component of both the Drosophila TBP-free GCN5 HAT-containing TAF complex (dTFTC) and the dTFIID (25). To this end, IP experiments were carried out using an anti-dmTAF10 PAb and anti-dmTAF10b (formerly dTAFII16) PAb as control, since dmTAF10b is present only in dTFIID (25). In the anti-dmTAF10 IP, dADA2b could be found associated with dmTAF10 together with GCN5 (Fig. 4A, lane 2), indicating that dADA2b is associated with a dmTAF10-containing multiprotein complex. In contrast, dADA2b did not coimmunoprecipitate with dmTAF10b (lane 4), whereas dmTAF1 (dTAFII230), dmTAF4 (dTAFII110), dmTAF6 (dTAFII80), dmTAF9 (dTAFII40), and dTBP were specifically coimmunoprecipitated with both dmTAF10 and dmTAF10b (lanes 2 and 4 and data not shown). Moreover, in the control IP, none of these proteins could be detected (lane 10). Unfortunately, in these IP experiments we could not test the presence of dADA2a because it migrates at the same position as the heavy chains of the different antibodies, which are always present in our immunopurifications (Fig. 4A and data not shown). To overcome this problem and to further study the different proteins associated with the two novel dADA2s, IP experiments were carried out with the purified antisera raised against either dADA2a or dADA2b. Both antibodies coimmunoprecipitated dADA2b, GCN5, and TAF9 from the nuclear extract (lanes 6 and 8); however, the anti-dADA2b PAb also coimmunoprecipitated dTAF10 and dTAF4 (lane 8). Note that in the control IP using an unrelated PAb bound to protein A-Sepharose (lane 10) or in the “only-antibody” control experiments (lanes 3, 5, 7, 9, and 11), none of these proteins were detected. These results, in good agreement with the two-hybrid results, demonstrate that the two newly identified dADA2s are associated with GCN5 HAT complexes. Moreover, dADA2b seems to be associated with a different complex(es) from dADA2a, since it coimmunoprecipitated with TAF10 and TAF4 in addition to the proteins that were also found in the anti-dADA2a IP (lanes 6 and 8). Thus, our results suggest that a TFTC-type multiprotein complex(es) exists in Drosophila, which would contain dADA2b, GCN5, TAF4, TAF9, TAF10, and possibly other proteins, but not TAF10b. Our results further suggest that in Drosophila there may be a different multiprotein complex in which dADA2a would be associated with dGCN5, dmTAF9, and dADA2b. Surprisingly, from the crude nuclear extract, both the anti-dADA2a and the anti-dADA2b PAbs coimmunoprecipitated some TBP as well, even though the immunopurified complexes had been extensively washed with buffers containing 500 mM KCl and the anti-dTAF10b IP did not coimmunopurify any dADA2b (lanes 4, 6, and 8). This suggests that in Drosophila embryo extracts, some TBP may associate with the ADA2-containing complexes (see Discussion).
FIG. 4.
dADA2a and dADA2b are subunits of different GCN5 HAT-containing multiprotein complexes. dADA2a and dADA2b coimmunoprecipitate with dGCN5 and with several different dTAFs. Proteins from nuclear extracts were immunoprecipitated with affinity-purified anti-dTAF10, anti-dTA10b, anti-dADA2a, and anti-dADA2b PAbs and a nonrelated PAb raised against a human protein, as indicated. The input nuclear fraction (input), the immunoprecipitated protein A-Sepharose antibody-bound proteins (IP), and the protein A-Sepharose bound PAbs alone (Ab) were resolved by SDS-PAGE (10 or 15% polyacrylamide) and transferred. The blots were then probed with antibodies raised against the indicated proteins. (B) Glycerol gradient sedimentation analysis of Drosophila embryo nuclear extract. A 200-μl volume of TRAX was loaded onto a 10 to 30% glycerol gradient and centrifuged for 4 h at 50 krpm, using an SW60 Ti rotor. The sedimentation standards, thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and albumin (67 kDa), were centrifuged in parallel gradients. Fractions of 200 μl were collected. Aliquots (20 μl) from the indicated fractions were resolved in parallel with the unfractioned nuclear extract (In) by SDS-PAGE (10% polyacrylamide), transferred to a filter, and probed with the indicated PAbs. The position of the 0.67-MDa molecular mass marker and some extrapolated molecular masses are indicated at the top. The numbers of the analyzed fractions are indicated at the bottom. Note that only the 42-kDa form of dADA2b is shown; however, the 70-kDa form showed exactly the same migration pattern.
dADA2a and dADA2b exist in different multiprotein complexes.
Since yeast ADA2 is known to be present in two different multiprotein complexes, the 0.8-MDa ADA and the 1.8- to 2-MDa SAGA complex (27), we decided to test whether the two novel dADA2 homologues also participate in different complexes. To this end, high-molecular-weight complexes of Drosophila nuclear extracts were separated either on glycerol gradients or by chromatography followed by gel filtration.
Drosophila nuclear extract was centrifuged through a 10 to 30% glycerol gradient, and to ensure that high-molecular-weight complexes remained intact, fractions of the gradients were analyzed by SDS-PAGE without further manipulation. The sedimentation of dADA2a and dADA2b was compared with that of TFIID-specific components (TBP and TAF1), TAF components which may be present in both TFIID and the Drosophila TFTC complexes (TAF4, TAF9, and TAF10), and a dTFTC-specific component (dGCN5), as well as markers of known molecular mass (Fig. 4B). Immunoblot analysis of the different fractions using a number of PAbs specific for the above-mentioned proteins showed dADA2a in fractions containing complexes with molecular masses between 400 and 800 kDa (Fig. 4B, fractions 6 to 12) which also contained a portion of dGCN5, TAF9, TAF10, and TBP, together with detectable levels of dADA2b (Fig. 4B). However, dADA2b was detected mainly in fractions corresponding to high molecular masses (greater than 2 MDa) (fractions 16 to 20). Note that both specific forms of dADA2b (42 and 70 kDa) showed the same pattern of sedimentation (Fig. 4B and data not shown). In the fractions which correspond to masses greater than 2 MDa (fractions 16 to 20), dADA2b cosedimented with the second peak of dGCN5, TBP, and all the other TAFs tested, suggesting that these fractions contain both dTFTC and dTFIID. Moreover, dADA2b was also weakly detectable in fractions corresponding to molecular masses smaller than 1 MDa (between fractions 6 and 14).
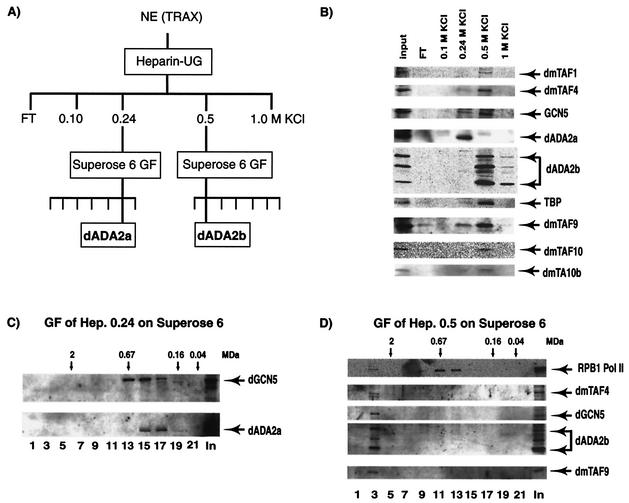
To further characterize the ADA2a- and ADA2b-containing complexes, Drosophila nuclear extract was chromatographed on a heparin-ultrogel column (Fig. 5A). Bound proteins were eluted from the column with stepwise elution using buffers containing 0.1, 0.24, 0.5, and 1 M KCl. The input and the eluted fractions were then tested by Western blot analysis for the presence of ADA2a and ADA2b as well as for GCN5 and the different TAFs (Fig. 5B). Interestingly, the majority of ADA2a was present in the heparin 0.24 M KCl (Hep0.24) elution whereas ADA2b eluted from the column at a higher salt concentration, mainly in the 0.5 M KCl (Hep0.5) fraction. In the ADA2a-containing Hep0.24-derived fraction, we also detected some GCN5, TAF9, and TBP. In contrast, in the ADA2b-containing Hep0.5-derived fraction, we detected more GCN5 together with TAF1, TAF4, TAF9, TAF10, TAF10b, and TBP. To determine the sizes of the separated ADA2a- and ADA2b-containing complexes, the Hep0.24-and Hep0.5-derived fractions were subjected to Superose 6 gel filtration, respectively. ADA2a eluted from the Superose 6 column a between 0.2 and 0.7 MDa, and in fractions 14 to 17 it coeluted with GCN5 (Fig. 5C). When the Hep0.5-derived fraction was separated on the Superose 6 column, ADA2b, together with the tested TAFs and GCN5, eluted from the column as a large multiprotein complex with a molecular mass larger than 2 MDa (Fig. 5D). Note that the Pol II complex with a molecular mass of 0.65 MDa eluted from our gel filtration column at around 0.67 MDa, confirming the calibration of the column. Thus, these results, together with the results of the two-hybrid and IP experiments, suggest that the two distinct Drosophila ADA2 homologues are present in different GCN5 HAT-containing multiprotein complexes. dADA2a-containing complexes have native molecular masses between 0.3 and 0.7 MDa, whereas the dADA2b-containing complexes have native molecular masses of 2 MDa or larger.
FIG. 5.
dADA2a- and dADA2b-containing complexes are chromatographically separable. (A) The chromatography protocol used to separate the dADA2-containing complexes is outlined. All procedures were performed at 4°C. The KCI concentrations used in the heparin-Ultrogel (UG) elution are indicated. NE (TRAX), Drosophila 0- to 12-h embryo nuclear extract; GF, gel filtration. (B) Portions (10-μl) of the input nuclear extract and of the different fractions eluting from the heparin-Ultrogel column were tested by Western blot analysis for the presence of the different factors (as indicated). FT, flowthrough fraction. (C) The heparin 0.24 M KCl-derived fraction was further separated on a Superose 6 gel filtration column. Portions of the input (In) (10 μl) and of every second fraction (as indicated) eluting from the column (20 μl) were tested by Western blot analysis for the presence of the different factors (as indicated). (D) The heparin 0.5 M KCl-derived fraction was also separated on a Superose 6 gel filtration column. Portions of the input (In) (10 μl) and of every second fraction eluting from the column (20 μl) was tested by Western blot analysis for the presence of the different factors (as indicated). Pol II was monitored by using an antibody recognizing the C-terminal repeat of the largest Pol II subunit, RPB1. In panels C and D, the positions of known molecular mass markers are indicated above the panels.
Both ADA2a and ADA2b can mediate transcription activation and do not always target the same set of genes on chromatin.
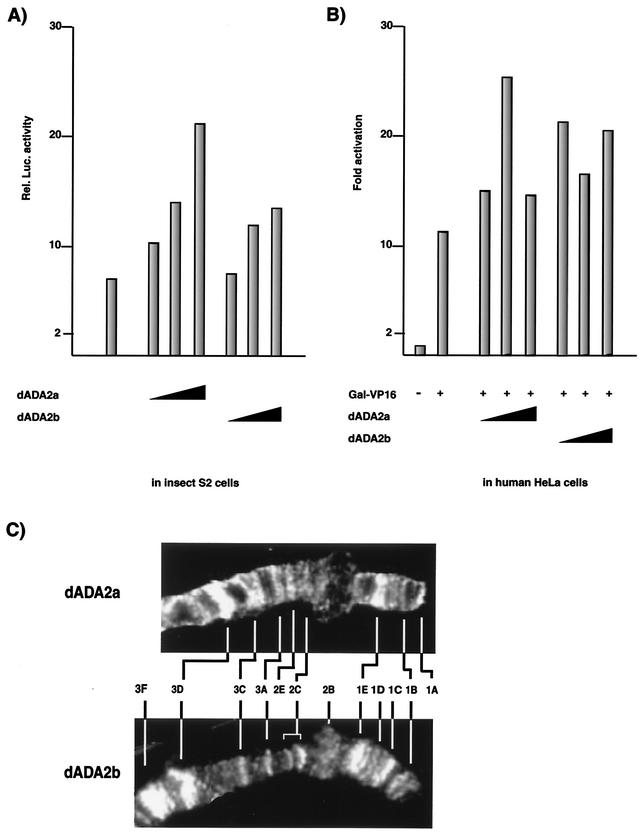
The differences observed between ADA2a and ADA2b in their expression, their ability to interact with dADA3, and their association with multiprotein complexes suggest different roles for these proteins in the nucleus. Since yeast and human ADA proteins are thought to be involved in mediating the transcriptional activity of different activators (i.e., nuclear receptor or acidic activators), we tested whether the ADA2 proteins of Drosophila would also work as coactivators and whether they would show differences in this respect. To this end, a reporter plasmid containing a luciferase reporter under the control of an ecdysone-inducible promoter was cotransfected with increasing amounts of recombinant plasmids expressing either dADA2a or dADA2b into Drosophila S2 cells (Fig. 6A). The luciferase activity of cell extracts was measured 40 h after transfection. These experiments indicated that this ecdysone-inducible promoter which contains 5 ERE (pIND; Invitrogen) had a relatively high basal activity (more than 20-fold higher than that of the promoterless plasmid [data not shown]) in S2 cells (which contain endogenous ecdysone receptors and their heterodimeric interaction partner ultraspiracle [USP]) and that this activity was further increased in the presence of either dADA2a or dADA2b. The cotransfection of the highest concentration of the dADA2a expression vector resulted in a more than twofold increase of the promoter activity and somewhat higher activation than that equal for amounts of dADA2b.
FIG. 6.
dADA2a and dADA2b work as cofactors and localize to different but overlapping loci on the distal region of Drosophila X chromosome. dADA2a and dADA2b stimulate transcriptional activity in Drosophila S2 cells (A) and human HeLa cells (B). (A) Reporter plasmid containing a luciferase reporter under the control of an ecdysone-inducible promoter was cotransfected with increasing amounts (0.5, 1.0, and 2.0 μg) of recombinant plasmids expressing either dADA2a or dADA2b into Drosophila S2 cells. The luciferase activity of the cell extracts was measured 40 h after transfection. (B) HeLa cells were cotransfected with a luciferase reporter gene under the control of the rabbit β-globin promoter harboring one GAL4-binding site, a GAL-VP16 expression plasmid, and increasing amounts (50, 100, and 500 ng) of dADA2a and dADA2b expression plasmids (as indicated). Luciferase activity was measured. (C) Association of dADA2a and dADA2b with specific loci of the Drosophila polytene chromosome. Immunostaining of the polytene X chromosome from wild-type larvae (Oregon R) with antibodies raised against dADA2a (upper panel) and dADA2b (lower panel) and Cy3-conjugated secondary antibodies is shown. The labeled regions of chromosome X are indicated for both panels.
In similar experiments, overexpression of dADA2a or dADA2b was tested on a strong acidic activator, GAL-VP16, activation in human HeLa cells by using a luciferase reporter gene under the control of the rabbit β-globin promoter harboring one GAL4-binding site. Coexpression of the GAL-VP16 expression plasmid with the reporter construct stimulated transcription about 12-fold. When either increasing amounts of the dADA2a or dADA2b expression vector were included in the experiment, they further increased the activation by GAL-VP16 approximately twofold again (Fig. 6B). Note that dADA2a was more efficient at the lower expression vector concentration (100 ng) whereas the tested dADA2b expression vector concentrations (from 50 to 500 ng) resulted in a similar coactivation of transcription.
If transcriptional selectivity can be achieved at the level of distinct ADA2-containing multiprotein complexes, then the different dADA2 homologues might be expected to be associated with different loci of the genome. Immunostainings of Drosophila salivary gland polytene chromosomes show that both dADA2a and dADA2b are located at a large number of loci (Fig. 6C and data not shown). Antibody stainings revealed that both dADA2a and dADA2b are associated with a unique subset of loci. As exemplified by the localization of the binding sites of the two dADA2s on the distal region of the polytene X chromosome in wild-type strains, there are loci (i) which are stained by both anti-dADA2a and anti-dADA2b antibodies (i.e., puff sites 1B, 1E, 2C, 3A, 3C, and 3D) and (ii) which are recognized only by either the anti-dADA2a (i.e., puff sites 1A and 2E) or anti-dADA2b (i.e., puff sites 1C, 2B, and 3F) antibodies (Fig. 6C and data not shown). Note also that the staining intensities of some loci stained by the two antibodies may be very different. These observations taken together, further suggest that these two novel dADA2s have overlapping, not identical, functions and that functionally different ADA2-containing complexes exist in Drosophila.
DISCUSSION
Here we report the isolation and partial characterization of two novel Drosophila ADA2 homologues, dADA2a and dADA2b. We show that the dAda2a mRNA is encoded by the dAda2a/dRbp4 gene, which also, by alternative splicing, gives rise to another transcript, which encodes a Pol II subunit, dRPB4. dAda2a and dAda2b mRNAs are differentially expressed during different stages of Drosophila development. Similarly to their yeast and human counterparts, both dADA2a and dADA2b interacted with the dGCN5 HAT, whereas dADA2b, but not dADA2a, also interacted with Drosophila ADA3 in a yeast two-hybrid assay. Antibodies raised either against dADA2a or dADA2b both immunoprecipitated the GCN5 HAT and several Drosophila TAFs. Moreover, IP, glycerol gradient sedimentation experiments, and chromatographic separation of Drosophila extracts strongly suggest that the two different Drosophila ADA2 homologues are present in distinct GCN5-containing HAT complexes. dADA2a and dADA2b mediated transient transcriptional activation of transfected reporter genes in insect and mammalian cells. In addition, dADA2 proteins associated with the Drosophila chromosome, showing colocalization at certain sites and different distributions at other loci. These observations taken together, show that the two novel dADA2s are present in distinct GCN5-containing HAT complexes and that they have overlapping but not identical functions, and thus that functionally different ADA2-containing complexes exist in Drosophila.
Unique posttranscriptional regulation of the dAda2a/dRbp4 gene.
We observed an unusual posttranscriptional regulation when studying the dAda2a/dRpb4 gene, which encodes the dAda2a and dRpb4 transcripts (Fig. 1A). Although these mRNAs encode two distinct proteins with separate functions, their pre-mRNA transcripts are transcribed from the same gene, most probably regulated by the same promoter. Thus, the fact that the dAda2a and dRpb4 transcript levels are not always identical during different developmental stages (Fig 1C) suggests that there is a developmentally regulated posttranscriptional regulation for the maturation of the dAda2a/dRpb4 pre-mRNA transcript. It is interesting that all other reported homologues of dADA2a and dRPB4 proteins are products of separate genes. This unusual posttranscriptional coregulation of the two mRNA species coding for dADA2 and dRPB4 may be related to their related functions. Previous studies showed that both yeast ADA2 and RPB4 mediate rapid responses by altering transcription regulation according to changes in the environment and that neither was absolutely necessary for cell viability. yRPB4, together with yRPB7, was reported to play a crucial role in promoter-directed transcription initiation (20) and to play a stress-protective role under suboptimal growth conditions (14, 48, 66). Yeast ADA2 was suggested to have an adaptor-like function that is important during activation of transcription initiation (3). Interestingly, in a recent mutagenesis screen, the yeast Ada2 gene was also identified as one of the few genes required for growth under ethanol stress conditions (57). Thus, the two proteins may have complementary or overlapping functions in eukaryotic gene regulation, which would explain why this unusual posttranscriptional coregulation of the two transcripts evolved in Drosophila. Further comparative functional analysis of these two proteins, especially using Drosophila genetics combined with biochemical approaches, is needed to elucidate their functional relationship.
Several different species have two ADA2 homologues with evolutionarily conserved domains.
By sequence similarity searches in Drosophila databases, we identified two Drosophila homologues of the known yeast and human ADA2s. Surprisingly, two different ADA2 homologues encoded by two distinct genes were found not only in the Drosophila genome but also in the human and the Arabidobsis genomes; they showed significant homology to the ADA2 family members (Fig. 2A) (56). This finding is in agreement with a previous report that two specific hADA2 protein products may exist in human cell extracts (23). Moreover, the partial cDNA sequences identified in mice and zebra fish databases seem to encode the homologue of human ADA2b (Fig. 2A). Therefore, our results suggest that most metazoan organisms and land plants have two ADA2 homologues, similar to Drosophila. Note, however, that in the fully sequenced C. elegans genome, only one gene encoding an ADA2 homologue was found (Fig. 2A).
Nevertheless, the high sequence conservation among the ADA2 proteins from different species ranging from yeast to humans indicates an important evolutionarily conserved role for this protein in eukaryotic gene regulation. In all the ADA2 family members, there are two previously identified conserved domains: the ZZ zinc finger domain and the SANT domain. No precise function has been assigned to either of these conserved domains in ADA2 protein-protein interactions; however, both the ZZ and the SANT domains are found in the minimal domain of ADA2 (amino acids 1 to 114 for yADA2 and 1 to 147 for hADA2a [Fig. 2A]), which was shown to be sufficient for interaction with yGCN5 or the VP16 activation domain (3, 11, 12). Moreover, the SANT domain of yADA2 was shown to be required for normal histone acetylation by SAGA, suggesting a role for ADA2 in nucleosomal substrate recognition (55). The ZZ domain is necessary for the binding of CBP/p300 to CREB or to TFIIB (38); however, yADA2 does not interact with TFIIB (3). This suggests that the ZZ domain of ADA2 plays a different role in directing protein-protein or, possibly together with the SANT/Myb domain, protein-DNA interactions. It is noteworthy that in addition to these known motifs, we have found several other α-helical conserved motifs (labeled as ADA boxes in Fig. 2A) which are conserved in all the ADA2 members, suggesting that they may play important roles in the interactions between ADA2 proteins and other subunits of their respective complexes. Interestingly, several of these ADA boxes contain (α-helix)1-(loop)1-(α-helix)2-(loop)2-(α-helix)3-like motifs that are often signatures of histone fold motifs found not only in histones but also in TAFs and other TFTC/SAGA components (e.g., ADA1, SPT3, and SPT7) (24). The histone fold motifs in the different transcription factors are in general not well conserved evolutionarily and thus are very difficult to predict (4).
The two Drosophila ADA2s are present in functionally different multiprotein complexes.
Drosophila GCN5 was shown to interact with the two different dADA2 proteins both in two-hybrid experiments and in IPs. Several independent lines of evidence suggest that the two dADA2 proteins associate with distinct GCN5 HAT-containing multiprotein complexes and thus may fulfill different functions. First, in two-hybrid experiments, only dADA2b, but not dADA2a interacted with Drosophila ADA3. Second, in IP experiments, antibodies raised against dADA2a coimmunoprecipitated GCN5, TBP, TAF9, and dADA2b whereas PAbs raised against dADA2b coprecipitated GCN5, TAF4, TBP, TAF9, and TAF10, together with dADA2b. Third, the two ADA2 proteins associate with only partially overlapping loci of the genome. Fourth, in glycerol gradient sedimentation experiments, the complexes containing dADA2a sedimented in a mass range of 0.4 to 0.8 MDa while the majority of the dADA2b-containing complexes sedimented at around the 2-MDa range. Finally, in full agreement with the results obtained by glycerol gradient sedimentation, different complexes containing either ADA2a (0.4 to 0.8 MDa) or ADA2b (2 MDa) protein can be separated by chromatography and gel filtration techniques. In agreement with our results, an additional higher-molecular-mass Ada2 protein complex with a migration profile different from the detected 0.8-MDa complex was also detected in a previous study (23). Therefore, we propose that the Drosophila ADA2a protein encoded by the identified dAda2a/dRbp4 gene resides in a multisubunit complex that might be the Drosophila homologue of the yeast 0.8-MDa ADA complex. For the second ADA2 variant, dADA2b, glycerol gradient sedimentation resolved two peaks. The dADA2b protein was considerably more abundant in fractions where the masses of the multiprotein complex should correspond to at least 2 MDa. Thus, it seems that dADA2b is present mostly in the 2-MDa multiprotein complexes, which would have the same size as the human TFTC or yeast SAGA complexes. Furthermore, the glycerol gradient sedimentation experiments, together with the IPs, strongly suggest that the 2-MDa Drosophila TFTC complex would contain, similarly to its human and yeast counterparts (7), dADA2b together with dGCN5, dmTAF4, dmTAF9, and dmTAF10. However, dADA2b was also detected between 0.4 and 0.8 MDa in the glycerol gradient sedimentation experiments using a crude nuclear extract but not after chromatography and gel filtration. Moreover, the anti-dADA2a PAbs coimmunoprecipitated dADA2b from nuclear extract (Fig. 4A). Therefore, dADA2b may transiently associate with the 0.4 to 0.8-MDa dADA complex in crude nuclear extracts but can be dissociated from it after more extensive purification.
Although Drosophila seems to contain two GCN5-containing ADA complexes, similar to yeast, we observed some interesting differences when comparing the compositions of the dADA or the dTFTC complexes to those of their yeast or human counterparts. One of these differences is that the 0.4 to 0.8-MDa dADA complex, when isolated from crude extracts, seems to copurify with dmTAF9; however, its yeast homologue, scTAF9, was not identified in the yADA complex. It is noteworthy that a dmTAF9-containing complex has recently been partially characterized and seemed to be smaller than TFIID (26). The fact that dmTAF9 was also present in our glycerol gradient fractions with an apparent molecular mass between 0.4 and 0.8 MDa (fractions 6 to 12), different from the TFIID-containing fractions (fractions 16 to 20), indicates that dmTAF9 may exist in smaller complexes together with dADA2a (Fig. 4B). Further experiments using highly purified 0.4- to 0.8-MDa dADA complexes are needed before we can decide whether TAF9 is a bone fide subunit of the dADA2a complex or, together with dADA2a, is a subunit of another, unknown complex. Another difference is that both the anti-dADA2a and the anti-dADA2b PAbs coimmunoprecipitated some (although substoichiometric amounts of) TBP, while in the control experiments no TBP was found (Fig. 4A). However, this finding may not be surprising since in similar IP experiments using yeast antibodies raised against yADA2 or yGCN5, substoichiometric amounts of yTBP coimmunopurified with yADA2 or yGCN5 from yeast extracts (51). Moreover, the strong genetic interactions demonstrated in yeast between two SAGA-specific subunits, ySPT3 and ySPT8 (21, 22), also suggest an interaction between the 2-MDa ySAGA complex and TBP.
Thus, this study demonstrates the existence of two different GCN5-ADA2-containing HAT complexes in Drosophila embryo extracts and suggests that their subunit compositions and molecular masses are very similar to those of their yeast and human counterparts. Moreover, the observation that dADA2a and dADA2b cannot always be found associated with the same transcriptionally active loci on polytene chromosomes indicates that the different dADA2a- and dADA2b-containing complexes have distinct functions in gene regulation. The discovery of a well-conserved Pol II regulatory subunit, dRPB4, which has an unusual posttranscriptional coregulation with dADA2a, uncovers novel levels of regulatory pathways in Pol II transcription. The fact that the dADA2a and dRPB4 transcription factors, being either subunits of HAT complexes or Pol II, are evolutionarily highly conserved suggests that these factors have important well-conserved regulatory functions in eukaryotic gene regulation. The discovery of these proteins in Drosophila opens the possibility of further analysis of the functions of these large multiprotein complexes in intact metazoan organisms by using the powerful tools of Drosophila genetics.
Acknowledgments
We are grateful to D. Allis, E. Smith, S. Berger, Y. Nakatani, A. Imhof, P. Becker, E. vom Baur, C. Gaudon, and R. Losson for reagents and advice; to O. Poch for help in the sequence alignments; and to Katalin Ökrösné for technical help. We also thank P. Eberling for peptide synthesis and G. Duval for polyclonal antibody production.
S.M. was supported by an EMBO short-term fellowship (ASTF 9604). This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the CNRS, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the FRM, European Community RTN grants (RTN1-1999-00401 and RTN-1999-00171), and the Human Frontier Science Program (RG 196/98) (to L.T.) and by Hungarian Ministry of Education OTKA grants (T29939 and T29207 to I.B. and to A.U.) and FKFP grant (0060/2000 to I.B.).
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Balaguer, P., A. M. Boussioux, E. Demirpence, and J. C. Nicolas. 2001. Reporter cell lines are useful tools for monitoring biological activity of nuclear receptor ligands. Luminescence 16:153-158. [DOI] [PubMed] [Google Scholar]
- 3.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 4.Baxevanis, A. D., and D. Landsman. 1998. Histone Sequence Database: new histone fold family members. Nucleic Acids Res. 26:372-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belenkaya, T., A. Soldatov, E. Nabirochkina, I. Birjukova, S. Georgieva, and P. Georgiey. 1998. P-element insertion at the polyhomeotic gene leads to formation of a novel chimeric protein that negatively regulates yellow gene expression in P-element-induced alleles of Drosophila melanogaster. Genetics 150:687-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 7.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 8.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. E., I. Lechner, I. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 10.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gen5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 11.Candau, R., and S. L. Berger. 1996. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J. Biol. Chem. 271:5237-5245. [DOI] [PubMed] [Google Scholar]
- 12.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 14.Choder, M., and R. A. Young. 1993. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate- phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 16.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 17.Drysdale, C. M., B. M. Jackson, R. McVeigh, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, P. A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gen5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin: second in review series on chromatin dynamics. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards, A. M., C. M. Kane, R. A. Young, and R. D. Kornberg. 1991. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266:71-75. [PubMed] [Google Scholar]
- 21.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 22.Eisenmann, D. M., C. Chapon, S. M. Roberts, C. Dollard, and F. Winston. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsberg, E. C., L. T. Lam, X. J. Yang, Y. Nakatani, and E. H. Bresnick. 1997. Human histone acetyltransferase GCN5 exists in a stable macromolecular complex lacking the adapter ADA2. Biochemistry 36:15918-15924. [DOI] [PubMed] [Google Scholar]
- 24.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 25.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgieva, S., E. Nabirochkina, F. J. Dilworth, H. Eickhoff, P. Becker, L. Tora, P. Georgiev, and A. Soldatov. 2001. The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol. Cell. Biol. 21:5223-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gen5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 28.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 29.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 30.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 31.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637-17641. [DOI] [PubMed] [Google Scholar]
- 32.Hayes, J. J., and J. C. Hansen. 2001. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 11:124-129. [DOI] [PubMed] [Google Scholar]
- 33.Khazak, V., J. Estojak, H. Cho, J. Majors, G. Sonoda, J. R. Testa, and E. A. Golemis. 1998. Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol. 18:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kingston, R. E., C. A. Bunker, and A. N. Imbalzano. 1996. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 10:905-920. [DOI] [PubMed] [Google Scholar]
- 35.Kokubo, T., D.-W. Gong, J. C. Wootton, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Molecular cloning of Drosophila TFIID subunits. Nature 367:484-487. [DOI] [PubMed] [Google Scholar]
- 36.Kolodziej, P. A., N. Woychik, S. M. Liao, and R. A. Young. 1990. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell. Biol. 10:1915-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 38.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 39.Le Douarin, B., D. M. Heery, C. Gaudon, E. vom Baur, and R. Losson. 2001. Yeast two-hybrid screening for proteins that interact with nuclear hormone receptors. Methods Mol. Biol. 176:227-248. [DOI] [PubMed] [Google Scholar]
- 40.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 41.Maes, M., and E. Messens. 1992. Phenol as grinding material in RNA preparations. Nucleic Acids Res. 20:4374.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage- binding-factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 44.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639-648. [DOI] [PubMed] [Google Scholar]
- 45.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 47.Ponting, C. P., D. J. Blake, K. E. Davies, J. Kendrick-Jones, and S. J. Winder. 1996. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21:11-13. [PubMed] [Google Scholar]
- 48.Rosenheck, S., and M. Choder. 1998. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol. 180:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saleh, A., D. Schieltz, N. Ting, S. B. McMahon, D. W. Litchfield, J. R. Yates, S. P. Lees-Miller, M. D. Cole, and C. J. Brandl. 1998. Tra1p is a component of the yeast Ada. Spt transcriptional regulatory complexes. J. Biol Chem. 273:26559-26565. [DOI] [PubMed] [Google Scholar]
- 50.Sandaltzopoulos, R., and P. B. Becker. 1995. Analysis of protein-DNA interaction by solid-phase footprinting. Methods Mol. Cell. Biol. 5:176-181. [Google Scholar]
- 51.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and A. P. Weil. 2002. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of the general transcription factor TFIID. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soldatov, A., E. Nabirochkina, S. Georgieva, T. Belenkaja, and P. Georgiev. 1999. TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mtations. Mol. Cell. Biol. 19:3769-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 55.Sterner, D. E., X. Wang, M. H. Bloom, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 56.Stockinger, E. J., Y. Mao, M. K. Regier, S. J. Triezenberg, and M. F. Thomashow. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidobsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi, T., H. Shimoi, and K. Ito. 2001. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:1112-1119. [DOI] [PubMed] [Google Scholar]
- 58.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 59.Tora, L., J. White, C. Brou, D. Tasset, N. Webster, E. Scheer, and P. Chambon. 1989. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell 59:477-487. [DOI] [PubMed] [Google Scholar]
- 60.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 61.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 62.vom Baur, E., M. Harbers, S. J. Um, A. Benecke, P. Chambon, and R. Losson. 1998. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 12:1278-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Werner, F., J. J. Eloranta, and R. O. Weinzierl. 2000. Archaeal RNA polymerase subunits F and P are bona fide homologs of eukaryotic RPB4 and RPB12. Nucleic Acids Res. 28:4299-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 65.Winston, F., and P. Sudarsanam. 1998. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:553-561. [DOI] [PubMed] [Google Scholar]
- 66.Woychik, N. A., and R. A. Young. 1989. RNA polymerase II subunit RPB4 is essential for high- and low- temperature yeast cell growth. Mol. Cell. Biol. 9:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao, J. H., I. Davidson, H. Matthes, J. M. Garnier, and P. Chambon. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65:551-568. [DOI] [PubMed] [Google Scholar]
- 68.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol Cell. 9:553-562. [DOI] [PubMed] [Google Scholar]