Abstract
Zyxin is an evolutionarily conserved protein that is concentrated at sites of cell adhesion, where it associates with members of the Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) family of cytoskeletal regulators and is postulated to play a role in cytoskeletal dynamics and signaling. Zyxin transcripts are detected throughout murine embryonic development, and the protein is widely expressed in adults. Here we used a reverse genetic approach to examine the consequences of loss of zyxin function in the mouse. Mice that lack zyxin function are viable and fertile and display no obvious histological abnormalities in any of the organs examined. Because zyxin contributes to the localization of Ena/VASP family members at certain subcellular locations, we carefully examined the zyxin−/− mice for evidence of defects that have been observed when Ena/VASP proteins are compromised in the mouse. Specifically, we evaluated blood platelet function, nervous system development, and skin architecture but did not detect any defects in these systems. Zyxin is the founding member of a family of proteins that also includes the lipoma preferred partner (LPP) and thyroid receptor-interacting protein 6 (TRIP6). These zyxin family members display patterns of expression that significantly overlap that of zyxin. Western blot analysis indicates that there is no detectable upregulation of either LPP or TRIP6 expression in tissues derived from zyxin-null mice. Because zyxin family members may have overlapping functions, a comprehensive understanding of the role of these proteins in the mouse will require the generation of compound mutations in which multiple zyxin family members are simultaneously compromised.
Zyxin is a LIM domain protein that is present at sites of cell-substratum and cell-cell adhesion, where it is proposed to dock proteins involved in cytoskeletal organization and dynamics (7, 8, 11, 16). Biochemical studies have identified α-actinin, members of the Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) family, Vav, members of the cysteine-rich protein family, the LATS1 tumor suppressor, and members of the p130cas family as binding partners for zyxin (12, 19, 23, 24, 42, 43, 47, 56). The molecular architecture of zyxin is compatible with the protein's postulated role as a scaffold for the assembly of multimeric protein complexes. Zyxin displays three tandemly arrayed LIM domains, double zinc finger structures that support specific protein interactions (37, 47). In addition, zyxin exhibits at least one nuclear export signal that regulates its excursions between the nucleus and the cytoplasm (35, 36).
Zyxin also displays four proline-rich ActA repeats that can interact directly with members of the Ena/VASP family (14, 19, 34). Members of the Ena/VASP family are concentrated at focal adhesions, the leading edge of lamellipodia, and filopodial tips (19, 42). Ena/VASP proteins are involved in modulation of actin assembly and organization (4, 6, 30, 41, 48) and are required for normal fibroblast and neuron migration (5, 20). Recent work has illustrated that the proper subcellular targeting of Ena/VASP family members is essential for them to fulfill their normal roles in vivo (1, 5, 6).
The Ena/VASP binding capacity of zyxin has been studied in detail, and several lines of evidence suggest that zyxin plays an important role in localizing members of this family to sites of cell-substratum adhesion. For example, microinjection of peptides that cause displacement of zyxin from focal adhesions also results in loss of Ena/VASP family members from these sites (15). In complementary experiments, targeting of zyxin to membranes or mitochondria leads to recruitment of Ena/VASP proteins (14, 18). Moreover, microinjection of peptide inhibitors that interfere with the binding of Ena/VASP proteins to partners with ActA repeats, such as zyxin, disturbs the subcellular distribution of Ena/VASP proteins and affects cell spreading and cell-cell adhesion (14, 52). Collectively, these studies suggest that failure of Ena/VASP proteins to localize properly results in defects in actin-based cell functions and implicate zyxin as an important contributor to the proper localization of Ena/VASP proteins.
The physiological consequences of disturbing Ena/VASP function have been characterized in vertebrates by analysis of mice in which Mena (mammalian Ena) or VASP functions have been compromised by gene disruption or expression of mutant constructs. Although mice that lack either Mena or VASP are viable and fertile, compromising the functions of murine Ena/VASP family members results in disturbed platelet function, nervous system development, and epithelial sheet formation (3, 21, 29, 52). Here we pursued a reverse genetic strategy to probe the role of zyxin in the mouse. We generated mice that lack zyxin function and analyzed the importance of zyxin for embryonic development and adult functions, with particular attention to processes that are known to depend on members of the Ena/VASP family. Mice that lack zyxin are both viable and fertile and do not display defects in processes that are compromised when either VASP or Mena functions are eliminated. The existence of multiple zyxin family members may obscure our ability to detect the contribution of zyxin to these processes.
MATERIALS AND METHODS
Northern blot analysis.
zyxin transcript levels were analyzed by probing mouse embryo and adult multiple tissue Northern blots containing 2 μg of polyadenylated RNA per lane (Clontech Laboratories, Palo Alto, Calif.). Both a zyxin LIM domain DNA probe and a zyxin 3′ untranslated region DNA were labeled by nick translation with [32P]dCTP and used to probe the Northern blots by standard methods (2). Glyceraldehyde-3-phosphate dehydrogenase transcript levels were monitored as a control.
zxyin targeting construct and generation of zyxin-null mice.
All mice were maintained in a specific-pathogen-free barrier facility and cared for, handled, and euthanized following guidelines approved by the Institutional Animal Care and Use Committee of the University of Utah. The neomycin resistance selection cassette pKT3lox(A) and the pTK1-pTK2-C targeting vector were provided by Kirk Thomas (University of Utah Transgenic Core Facility). The 19-kb mouse genomic DNA construct was isolated by screening a LambdaFix mouse 129 phage library with a murine zyxin cDNA probe (31).
To generate the targeting construct, a 4.5-kb genomic region that encompasses the translation initiation codon was replaced with a neomycin resistance cassette (neo) for positive selection. Deletion of the 4.5-kb DNA would remove a 5′ noncoding exon and the first four coding exons for zyxin. Two thymidine kinase-negative selection cassettes flanked the targeting construct. To assist in cloning and screening, the SpeI site was deleted from the 5′ end of the neomycin resistance coding sequence, and a new SpeI site was added to the 3′ end of the neomycin resistance cassette in the targeting construct. The linearized zyxin gene targeting construct was electroporated into the 129SVJ GS stem cell line (Genome Systems Inc., St. Louis, Mo.), and recombinant clones were selected.
Positive embryonic stem (ES) cell lines were screened for correct homologous recombination by Southern DNA analysis. Cells from two independently derived ES cell lines (86 and 185) were introduced separately into recipient morulae to generate chimeric mice. The resulting chimeric mice were mated with C57BL/6 mice (Jackson Laboratory, Bar Harbor, Maine). Tail DNAs from agouti mice in the resulting litters were analyzed by Southern blot to identify mice in which the targeted allele was transmitted via the germ line. These heterozygous mice served as the founders for two independent zyxin−/− mouse lines (Zyxin-86 and Zyxin-185). The mouse genome database provided information on the intron-exon boundaries and locations within the zyxin gene and the chromosome locations for zyxin gene family members (ncbi.nlm.nih.gov/genome/guide/mouse).
Characterization of ES cell lines and mice.
Genomic DNA was prepared from mouse tails by 55°C incubation with 10 mg of proteinase K per ml in 0.5% sodium dodecyl sulfate (SDS), 50 mM Tris (pH 8), and 100 mM EDTA (25). For Southern DNA analysis, the DNA was digested with SpeI and then electrophoresed through a 0.8% agarose gel, depurinated (0.25 M HCl for 10 min), followed by capillary transfer (0.4 M NaOH) to nylon filters for 18 h. The DNA was neutralized (20 min in 0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl), washed in 2× SSPE (20 min), dried, and UV cross-linked. The 3′ EcoRI-HindIII DNA fragment was labeled with [32P]dCTP (Ready To Go DNA labeling beads; Amersham Pharmacia, Piscataway, N.J.), and used to probe the DNA filters (1 h at 65°C). After washing at 42°C (0.1% SDS with sequential steps of 2× SSPE, 1× SSPE, and 0.1× SSPE), the blot was developed by autoradiography (wild-type zyxin, 11.8 kb; null mutant, 6.2 kb).
For PCR analysis, the genomic DNA was incubated with three primers, WTFzyx (5′-TAC AAG GGC GAA GTC AGG GCG AGT G-3′), WTRzyx (5′-TGG ACG AAG TTT CCG TGT GTT G-3′), and NEOFzyx (5′-GAC CGC TTC CTC GTG CTT TAC-3′) and a 30-cycle PCR regimen of 95°C for 60 s, 58°C for 30 s, and 72°C for 50 s, followed by 95°C for 1 min and 72°C for 3 min. The PCR products (wild-type zyxin, 327 bp; null mutant, 473 bp) were electrophoresed on a 1.8% agarose gel and visualized with ethidium bromide.
For analysis of proteins, tissues were dissected from mice (euthanized by Halothane inhalation followed by cervical dislocation), weighed, homogenized in 5 parts H2O with protease and phosphatase inhibitors (100 mM phenylmethylsulfonyl fluoride, 100 mM benzamidine HCl, 1 mg of pepstatin A per ml, 1 mg of phenanthroline per ml, 0.1 M NaF, 0.2 mM sodium orthovanadate), and diluted with an equal amount of Laemmli sample buffer (28). The tissue homogenates were boiled for 5 min, and the DNA was sheared with a 26-gauge needle. Protein extracts (10 μl) were electrophoresed through denaturing 10% polyacrylamide gels and either Coomassie stained or transferred to nitrocellulose by standard methods (51). Western immunoblot analysis was performed by enhanced chemiluminescence (Amersham Pharmacia, Piscataway, N.J.) for detection.
Antibodies.
Polyclonal rabbit antizyxin serum B71 was generated by immunizing a rabbit with keyhole limpet hemocyanin coupled to peptide CSPGAPGPLTLKEVEELEQLT (human zyxin amino acids 344 to 363, encompassing the nuclear export sequence). Polyclonal rabbit antizyxin serum B72 was generated by immunizing a rabbit with keyhole limpet hemocyanin coupled to peptide CDFPLPPPPLAGDGDDAEGAL (human zyxin amino acids 70 to 89, encompassing the first ActA repeat). Rabbit immunizations and serum collection (serum samples B71 and B72) were performed at Harlan Bioproducts for Science (Madison, Wis.). Polyclonal rabbit serum B38 (31) is now known to recognize both zyxin and lipoma preferred partner (LPP) and is therefore referred to as a panzyxin antibody. Zyxin antisera were diluted 1:10,000 for Western blot analysis.
Mouse monoclonal antizyxin antibody 1D1 was generated by immunizing mice with bacterially expressed glutathione S-transferase (GST)-human zyxin; the epitope maps to VVAPKPKVNPFRPGDSEPPP (human zyxin amino acids 28 to 48). Mouse monoclonal antibody 12A12 recognizes LIM3 of both zyxin and TRIP6. Mouse monoclonal antibody 164D4 against human zyxin was supplied by Matthias Krause and Jürgen Wehland (45). Polyclonal rabbit serum MP2 against LPP was supplied by Marleen Petit and Wim Van de Ven (38). Polyclonal rabbit serum B65 (supplied by Susanne Kloeker) is directed against mouse TRIP6 peptide KQPEPSRLPQGRSLPR and was affinity purified for these studies. Antibodies directed against laminin (Sigma-Aldrich, St. Louis, Mo.), keratin 1, keratin 5, loricrin (BabCo, Berkeley, Calif.), and fillagrin (BabCo, Berkeley, Calif.) were used as described previously (40).
Platelets.
Platelet-rich plasma was prepared from blood collected from wild-type and zyxin−/− mice by established procedures (3). Platelets were exposed to collagen, and the timing of the resulting shape change and aggregation response was measured with a lumiaggregometer (Aggrecorder II PA-3220; Kyoto Kaiichi Kagaku, Kyoto, Japan) as described previously (3, 57). Both collagen and ADP were used as agonists.
Platelet-rich plasma was prepared for Western blot analysis as follows. After anesthetization and cardiac puncture of three wild-type and three zyxin−/− mice, blood was collected into 1.5-ml microcentrifuge tubes containing 20 μl of anticoagulant citrate dextrose (110 mM sodium citrate [pH 7.4], 2.45% dextrose) plus 250 μl of blood per tube. Phosphate-buffered saline containing 4.4 mM EDTA and 0.5% bovine serum albumin was added to the blood-anticoagulant citrate dextrose mixture (400 μl per tube). Centrifugation at 1,000 rpm (Eppendorf microcentrifuge) for 10 min sedimented the red and white blood cells. The supernatant was recentrifuged at 2,700 rpm for 4 min and aspirated. Next, 300 μl of ammonium chloride solution (150 mM ammonium chloride, 1 mM potassium bicarbonate, 0.1 mM EDTA, pH 7.4) was added to each pellet and mixed, followed by the addition of 400 μl of phosphate-buffered saline-EDTA-bovine serum albumin and centrifugation at 2,700 rpm for 4 min. After the platelet pellets were washed twice in phosphate-buffered saline-EDTA-bovine serum albumin and evaluated by microscopic inspection, they were resuspended in SDS sample buffer, electrophoresed (approximately 400,000 platelets per lane), transferred, and probed as described above.
Histology and immunocytochemistry.
For general histological analysis to compare wild-type and zyxin−/− mice, tissues were dissected, fixed, embedded, and stained as described previously (9). For examination of brain morphology, silver staining was performed on five adult brains sectioned along the horizontal plane to evaluate possible commissure defects (29). Immunocytochemical analysis of tail skin from newborn mice was performed as described previously (40, 52).
Flow cytometry.
Blood samples (0.5 ml) from mice of various zyxin genotypes were incubated with labeled antibodies specific for mouse blood cell markers. The labeled blood cells were then sorted into populations of B cells, T cells (CD4+ and CD8+), granulocytes, and monocytes by flow cytometry (University of Utah Flow Cytometry Core Facility). The blood cell populations were analyzed for standard error means and paired t tests with Prism software (Graph Pad, San Diego, Calif.).
RESULTS
Knowledge of zyxin's binding partner repertoire as well as its molecular architecture suggests that the protein serves as a scaffold for the docking of multiple protein partners (Fig. 1A) (12, 19, 23, 24, 42, 43, 46, 47, 56). One of zyxin's notable features is the presence of four proline-rich motifs called ActA repeats. These repeats were first identified in the ActA protein of Listeria monocytogenes, where they make a contribution to the intracellular motility of this pathogenic bacterium (30, 34). ActA repeats serve as docking sites for members of the Ena/VASP family (34, 44). Ena/VASP proteins contribute to the control of actin assembly and organization (4, 44) and have been shown to play important roles in platelets, neurons, and keratinocytes in the mouse (3, 29, 42, 52). Because zyxin plays a central role in localizing Ena/VASP proteins to certain subcellular locations (15), we postulated that elimination of zyxin function may perturb processes that depend on Ena/VASP proteins.
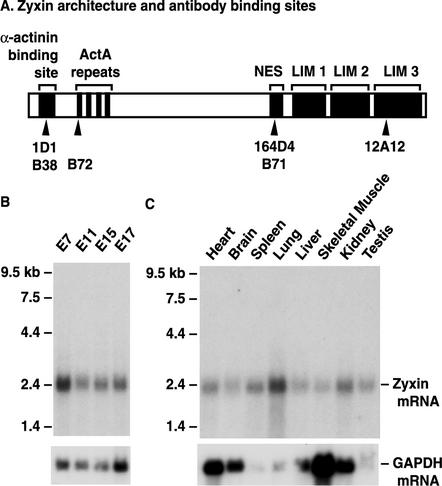
FIG. 1.
Zyxin structure and expression. (A) Zyxin architectural features including the α-actinin binding site, four proline-rich ActA repeats for binding to Mena and VASP, the leucine-rich nuclear export sequence (NES), and three double zinc finger LIM domains. Epitopes recognized by several antizyxin antibodies used in our analysis are marked with arrowheads; mouse monoclonal antibodies (1D1, 164D4, and 12A12) are above the rabbit polyclonal antibodies (B38, B72, and B71). (B) Northern blot of 2 μg of polyadenylated RNA isolated from mouse embryos (day 7, day 11, day 15, and day 17 of gestation) reveals that zyxin transcripts are present throughout embryogenesis. (C) Northern blot of 2 μg of polyadenylated RNA isolated from adult mouse tissues illustrates widespread expression of zyxin transcripts in the adult mouse. The Northern blots shown were probed with a zyxin cDNA that encodes the C-terminal LIM domains. To ensure that no transcripts encoding irrelevant LIM proteins were contributing to the signal, the blots were stripped and reprobed with a zyxin-specific 3′ untranslated region probe; identical signals were obtained (data not shown). As an additional control, the blots were stripped and reprobed to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts, which are present at variable levels depending on the tissue type.
In order to test this hypothesis and to learn more about the physiological role of zyxin in vivo, we examined zyxin function in a mouse model system. By Northern analysis, we determined that murine zyxin is encoded by a single transcript of 2.4 kb that is expressed throughout embryonic development (Fig. 1B). Evaluation of zyxin RNA expression in adult mouse tissues indicated that zyxin is ubiquitously expressed (Fig. 1C).
Targeted disruption of zyxin gene.
In order to study the role of zyxin in the mouse, we generated a deletion allele by standard methods for engineering gene disruptions by homologous recombination (Fig. 2A) (10, 32). To abolish zyxin function, approximately 4.5 kb of zyxin gene sequence, including coding sequence for the translational initiation codon, was replaced with a neomycin resistance selection cassette. Inspection of the available mouse genome sequence confirmed that a 5′ noncoding exon plus four exons encoding amino acids 1 to 333 would be deleted by this strategy. Properly targeted ES cells were identified by Southern blot analysis (Fig. 2B) and were incorporated into recipient morulae. The resulting chimeric animals were bred to C57BL/6 mice to generate heterozygous mice, which were subsequently interbred. The progeny of zyxin+/− intercrosses were genotyped by either Southern blot (Fig. 2C) or PCR (Fig. 2D) analysis.
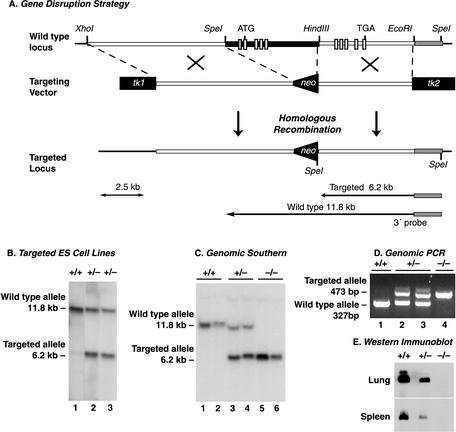
FIG. 2.
Disruption of murine zyxin gene. (A) Partial restriction map of the zyxin genomic region used for generation of the targeting construct. Restriction enzyme sites on the wild-type zyxin gene, the targeting vector, and the correctly targeted DNA following homologous recombination are shown. This approach deleted 4.5 kb of zyxin genomic sequence, which includes a 5′ noncoding exon (small shaded box) and the first four coding exons (small white boxes) for amino acids 1 to 333. The ATG methionine start codon in the first coding exon and the TGA stop codon in the final coding exon are shown. (B) Southern blot of SpeI-digested genomic DNA from two neomycin-resistant embryonic stem (ES) cell lines probed with the 3′ 1.2-kb EcoRI-HindIII DNA identified correctly targeted DNA (wild type, 11.8 kb; targeted mutant, 6.2 kb). (C) Southern blot of SpeI-digested genomic DNA from mouse tail samples distinguishes wild-type (+/+), heterozygous (+/−), and homozygous-null (−/−) genotypes. (D) Genomic PCR with zyxin- and neo-specific primers with mouse tail genomic DNA confirms the wild-type (+/+), heterozygous (+/−), and homozygous-null (−/−) genotypes (wild type, 327 bp; zyxin-null, 473 bp). (E) Western blot analysis of lung and spleen tissue extracts from zyxin wild-type (+/+), heterozygous (+/−), and homozygous-null (−/−) mice with antizyxin antibody 164D4. No zyxin protein was detected in samples derived from the zyxin-null mouse with any of the antibodies specific for epitopes located throughout the zyxin protein.
To evaluate the consequences of loss of zyxin function, zyxin+/− mice were intercrossed. The resulting progeny displayed a normal Mendelian ratio of zyxin+/+, zyxin+/−, and zyxin−/− animals (Table 1), indicating that zyxin is not absolutely essential for development. To demonstrate that zyxin protein is indeed missing from the zyxin-null mice, tissue extracts from wild-type, heterozygous, and zyxin-null mice were analyzed by Western blot. Analysis of two tissues, lung and spleen, that displayed relatively high levels of zyxin mRNA is shown in Fig. 2E. Tissues isolated from zyxin+/− mice displayed reduced zyxin levels, and no zyxin was detected in tissues from zyxin−/− animals. The absence of zyxin protein expression was confirmed with multiple antizyxin antibodies whose epitopes mapped to the N-terminal, central, and C-terminal regions of the protein (Fig. 1A and data not shown). Since the mouse genome sequence suggests that our targeting strategy would leave five coding exons for the C-terminal half of zyxin, it is important that our Western blot analysis confirmed that there is no detectable expression of these partial zyxin products.
TABLE 1.
Genotypes of progeny resulting from heterozygous (zyxin+/−) intercrosses
| Genotype | No. of mice generated (% of total) | No. predicted by Mendelian genetics (% of total) |
|---|---|---|
| zyxin+/+ | 72 (23) | 78 (25) |
| zyxin+/− | 162 (52) | 155 (50) |
| zyxin−/− | 77 (25) | 78 (25) |
zyxin−/− animals can mate and produce offspring, indicating that zyxin expression is not essential for either male or female fertility.
Histological examination of the tissues evaluated by Northern analysis in Fig. 1C revealed that the zyxin-null mice displayed no obvious abnormalities in tissue architecture (data not shown). Since zyxin is highly expressed in blood cells (31), we examined the white blood cell population of four sets of littermates with assorted zyxin genotypes (+/+, +/−, and −/−). Flow cytometric analysis indicated that loss of zyxin did not significantly affect resting populations of B cells, T cells (CD4+ or CD8+), granulocytes, or monocytes (Table 2).
TABLE 2.
Flow cytometry analysis of blood cell populations from wild-type (zyxin+/+) heterozygous (zyxin+/−) and homozygous-null (zyxin−/−) littermatesa
| Genotype | Mean % of total ± SEM
|
||||
|---|---|---|---|---|---|
| B cells | T cells (CD4+) | T cells (CD8+) | Granulocytes | Macrophages | |
| Wild type | 43.4 ± 4.8 | 26.9 ± 3.2 | 14.3 ± 1.1 | 15.5 ± 2.0 | 7.1 ± 1.5 |
| Heterozygous | 49.2 ± 4.8 | 24.2 ± 1.3 | 18.1 ± 2.9 | 20.8 ± 2.7 | 7.4 ± 0.9 |
| Null | 36.7 ± 4.3 | 29.5 ± 4.0 | 15.8 ± 1.2 | 14.0 ± 1.8 | 5.5 ± 0.6 |
Four mice per genotype were tested. The standard error of the mean was calculated with the Graph Pad Prism statistical analysis software.
Expression of zyxin family members is not upregulated in zyxin−/− animals.
Recently, two proteins that show a high degree of structural similarity to zyxin have been identified (Fig. 3A). LPP is the closest zyxin relative, with conserved proline-rich ActA repeats, LIM domains, and almost identical mobility in SDS-PAGE (38). Zyxin and LPP exhibit overlapping tissue expression and a similar subcellular distribution, and they also share some binding partners (31, 38, 39). Thyroid receptor-interacting protein 6 (TRIP6), also called zyxin-related protein (ZRP-1), has significant amino acid identity with human zyxin, particularly in the three C-terminal LIM domains (33, 53, 55). Zyxin, LPP, and TRIP6 have all been shown to localize at sites of cell adhesion (11, 38, 53). The shared architectural features and subcellular localizations of these proteins suggest that they may have common functions in vivo.
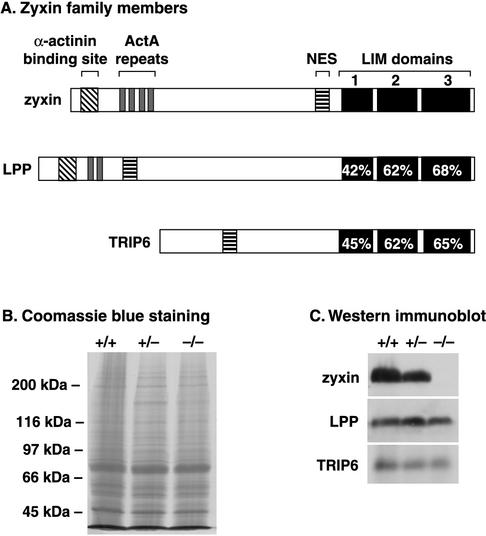
FIG. 3.
Zyxin family member structure and protein expression. (A) Alignment of human zyxin family members. Certain sequence motifs present in zyxin, including an α-actinin binding site (diagonal bars), proline-rich ActA repeats (shaded), nuclear export sequence (NES) (horizontal bars), and three C-terminal LIM domains (solid), are also observed in zyxin's closest relative, LPP. Another zyxin family member, TRIP6, displays a nuclear export signal and three C-terminal LIM domains. Accession numbers are X94991 (human zyxin) (31), NM_005578 (human LPP) (39), and AJ001902 (human TRIP6) (55). The mouse genome sequence database identifies the zyxin gene on mouse chromosome 6, the LPP gene on mouse chromosome 16, and the TRIP6 gene on mouse chromosome 5. (B and C). Expression of zyxin family members in mice. In panel B, a Coomassie blue-stained gel of total lung protein from zyxin+/+, zyxin+/−, and zyxin−/− mice is shown to illustrate comparable protein loading. As can be seen in the Western immunoblots shown in panel C, zyxin (B71 antiserum), LPP (MP2 antiserum), and TRIP6 (affinity-purified B65 antibody) are all expressed in wild-type lung tissue. Zyxin protein is reduced in samples from zyxin+/− animals and is absent in zyxin−/− animals. LPP and TRIP6 levels are unaffected when zyxin expression is eliminated.
We examined whether the expression of either LPP or TRIP6 is elevated in the zyxin-null mice because upregulation of these proteins could theoretically compensate for elimination of zyxin expression. Total protein isolated from the lung, spleen, bladder, kidney, heart, and liver samples derived from zyxin+/+, zyxin+/−, and zyxin−/− animals were probed with antibodies that specifically recognize zyxin, LPP, and TRIP6. Western blot analysis of lung, a tissue that expresses particularly high levels of zyxin (Fig. 1C), is shown in Fig. 3B and C. Zyxin levels were reduced by approximately one half in the heterozygote, and, as expected, no zyxin protein was observed in the sample derived from the zyxin−/− lung. No change in either LPP or TRIP6 protein levels was detected when zyxin function was compromised by mutation (Fig. 3C). Results similar to these shown with lung tissue were also found for bladder, spleen, kidney, heart, and liver (data not shown). These results illustrate that upregulated expression of zyxin family members does not occur in response to mutation of the zyxin gene. Interestingly, we found that all of the wild-type tissues examined displayed expression of at least two zyxin family members (data not shown). Therefore, although no compensatory upregulated expression of zyxin family members was observed, it remains possible that the presence of multiple family members contributes to the survival of the mice.
As discussed above, one of the best-characterized biochemical activities of zyxin is its ability to bind members of the Ena/VASP family and contribute to the proper subcellular localization of these proteins to sites of cell adhesion. Like zyxin-null animals, mice that lack either VASP or Mena are viable and fertile. However, higher-resolution analysis revealed contributions of Ena/VASP proteins to platelet function (21), axon guidance (29), and epithelial sheet formation (52). Therefore, we examined the consequences of eliminating zyxin function for these three systems in which zyxin is postulated to contribute via its capacity to dock Ena/VASP proteins.
Analysis of zyxin-deficient platelet function.
Two groups have generated mice that lack VASP function (3, 21). These mice are viable and fertile but display an altered platelet activation response. Resting platelets are discoid, nonadhesive cells. When exposed to an agonist such as collagen, thrombin, or ADP, they become activated; they rapidly change their shape and begin to aggregate with each other. The precise timing and extent of these two responses to agonist can be monitored by the use of an aggregometer, which detects these two stages as changes in light transmission (57). Although the patterns of expression of Mena and VASP often overlap, platelets provide an example of a cell that expresses primarily VASP (3), enhancing the likelihood of being able to detect a VASP loss-of-function phenotype. Indeed, as can be seen in Fig. 4A, platelets that lacked VASP function underwent shape change precociously but ultimately aggregated to the same extent as their wild-type counterparts (3). In contrast, the activation profiles of wild-type and zyxin−/− platelets were completely coincident, and the platelets achieved comparable aggregation maxima (Fig. 4B), illustrating that zyxin is not required for normal platelet response.
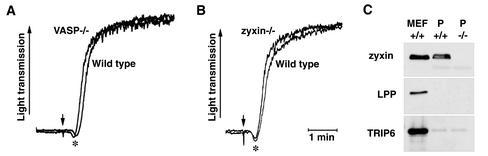
FIG. 4.
Platelet activation response is not altered by elimination of zyxin function. Platelets isolated from wild-type, VASP−/−, and zyxin−/− animals were stimulated with collagen (arrow) in the presence of calcium, and the platelet activation and aggregation responses were monitored by measuring light transmission. (A) As has been reported previously (3), VASP−/− platelets display a precocious shape change response (∗) but then proceed to aggregate to the same extent as wild-type platelets. (B) In contrast, the timing of the platelet shape change of zyxin−/− platelets is similar to what is observed for wild-type platelets. The slight leftward shift of the aggregation trace shown for the zyxin−/− platelets relative to wild-type platelets was not a consistent finding. (C) Expression of zyxin family members in platelets. Approximately 10 μg of wild-type mouse embryo fibroblast (MEF) protein and comparable amounts of protein from wild-type platelets (P +/+) and zyxin-null platelets (P −/−) were electrophoresed through SDS-10% polyacrylamide gels, transferred to nitrocellulose, and probed for zyxin family member expression. Zyxin protein (B71 antiserum) is detectable in mouse embryo fibroblasts and wild-type platelets but not in zyxin-null platelets. LPP (MP2 antiserum) is detectable in mouse embryo fibroblasts but is not detectable under these conditions in wild-type or zyxin-null platelets. TRIP6 (affinity-purified B65 antibody) is detectable in mouse embryo fibroblasts and is seen at low levels in both wild-type and zyxin-null platelets. A faint immunoreactive band that migrates at 66 kDa is detected with antizyxin (B71) antibody in both zyxin+/+ and zyxin−/− platelets. This band was not observed in any other murine tissue that we analyzed and may reflect a platelet-expressed protein that has some similarity to zyxin. As expected for platelets, these platelet extracts were positive for VASP expression and negative for Mena expression by Western blot analysis (data not shown).
To determine if multiple zyxin family members are expressed in platelets and to determine if their levels are altered in the absence of zyxin, a Western blot analysis was performed (Fig. 4C). Zyxin was readily detected in wild-type platelets but not in zyxin−/− platelets. In equivalently loaded samples, small amounts of TRIP6 were observed but no LPP was detected. Thus, as in other tissues, multiple zyxin family members are expressed in platelets.
Analysis of nervous system development.
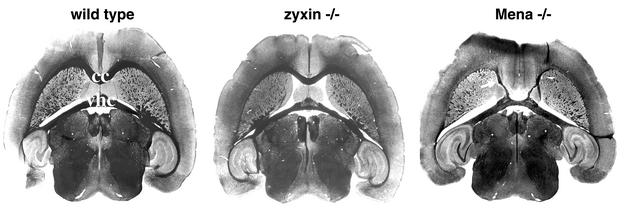
Mice that lack Mena expression are also viable and fertile, but particular deficiencies in brain development were noted in approximately half of the mutant animals examined (29). Specifically, in Mena−/− brains, the corpus callosum and dorsal hippocampal commissure are absent, while the ventral hippocampal commissure is present but reduced in thickness. To evaluate these structures in zyxin-null mice, we compared silver-stained sections of wild-type, zyxin−/−, and Mena−/− brains (Fig. 5). zyxin-null brains did not exhibit defects in any commissure in the brain (ventral or dorsal hippocampal commissures, anterior or posterior commissures, commissures of the superior and inferior colliculi, habenular commissure, or decussation of the superior cerebellar peduncle). Other major fiber tracts such as the internal and external capsules also appeared to be unaffected in zyxin-null brains. Note that the ectopic callosal fibers seen contacting the ventral portion of the ventral hippocampal commissure in Mena−/− brain (arrow) were not seen in the zyxin−/− brain. We did not detect any deficits in axon guidance associated with elimination of zyxin function.
FIG. 5.
Loss of zyxin function does not affect brain development. Horizontal sections from wild-type, zyxin−/−, and Mena−/− adult brains were analyzed by silver staining. The major commissures evident at this plane of section are the corpus callosum (cc) and the ventral hippocampal commissure (vhc). Brain structures, including those affected by loss of Mena function (29), such as the corpus callosum and ventral hippocampal commissure, form normally in the zyxin−/− mice.
Analysis of epidermis.
Transgenic mice expressing a fragment of VASP thought to perturb function in their keratinocytes develop skin blisters that appear to result from perturbation of cell-cell junctions (52). Expression of this mutant VASP disturbs the subcellular distribution of endogenous, full-length Ena/VASP family members (52). Because zyxin plays an important role in the proper subcellular localization of Ena/VASP proteins, it was interesting to consider the possibility that elimination of zyxin might likewise lead to disturbances in the skin by compromising the normal subcellular localization of Ena/VASP family members.
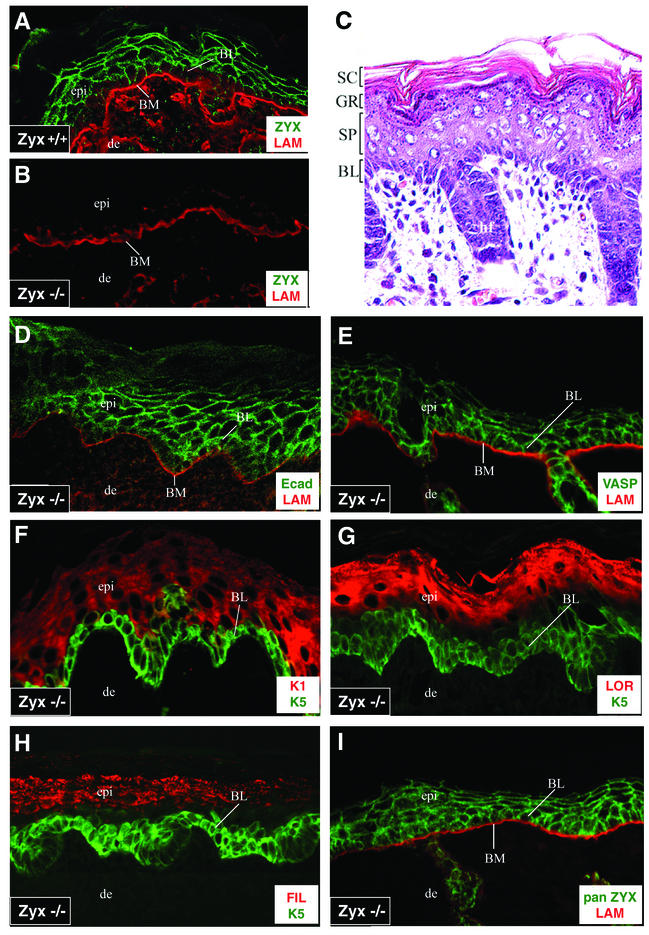
The first step toward evaluating the possibility that zyxin plays a role in cellular adhesions in the skin was to examine the subcellular distribution of zyxin in the skin. As can be seen in Fig. 6A, zyxin was concentrated at the borders of wild-type epidermal keratinocytes. The basement membrane, which is demarcated by laminin, serves to separate the epidermis from the underlying dermis. The intensity of antizyxin labeling was more prominent at the cell borders of suprabasal layers, consistent with the enhancement of intercellular junctions in these layers. As expected, the zyxin-null mice showed no detectable zyxin protein anywhere in the epidermis (Fig. 6B). No visible abnormalities were noted in the skin or hair of the zyxin-null animals, and the skin of these mice also appeared normal at the histological level (Fig. 6C). The epidermis consisted of the standard morphologically distinct layers, and the hair follicles appeared normal.
FIG.6.
Differentiation and architecture of the skin are normal in zyxin−/− animals. Skins from wild-type (+/+) (A) and zyxin-null (−/−) (B to I) newborn mice were prepared for analysis. (A) Wild-type mouse skin stained for laminin (LAM) and zyxin (ZYX) exhibits normal basement membrane (BM) (laminin stain) and epidermal zyxin localization (B71 antizyxin polyclonal rabbit serum). (B) zyxin-null mouse skin stained for laminin and zyxin. (C) zyxin-null mouse skin stained with hematoxylin and eosin. BL, basal layer; SC, stratum corneum; GR, granular layer; SP, spinous layer. (D to I) zyxin-null mouse skin stained for E-cadherin and laminin (D), VASP and laminin (E), keratin 1 (K1) and keratin 5 (K5) (F), loricrin (LOR) and keratin 5 (G), filaggrin (FIL) and keratin 5 (H), and panzyxin (pan ZYX) (B38 antibody that detects both zyxin and LPP) and laminin (I). epi, epidermis; de, dermis.
To determine whether adhesion and VASP localization might be perturbed when zyxin is absent, we assessed the status of E-cadherin and VASP in zyxin-null mouse skin. Interference with VASP function in transgenic mice caused blistering and loss of cell-cell contacts which manifested at the cellular level as double lines of E-cadherin between cells (52). In contrast, the zyxin-null mice exhibited normal E-cadherin and VASP localization (Fig. 6D and 6E, respectively).
To test whether the biochemical program of differentiation was normal in zyxin-null epidermis, we used specific markers for the distinct layers of the stratified epithelium. The protein keratin 5, which is a marker for the basal layer of the epidermis, was unaltered in the zyxin-null skin (Fig. 6F). Likewise, the spinous layers exhibited the normal differentiation program, as keratin 1, an early marker of differentiation, was expressed immediately adjacent to the basal layer. Terminal differentiation markers of epidermal differentiation such as loricrin (Fig. 6G) and filaggrin (Fig. 6H), which are hallmarks of the granular layer, also showed normal expression patterns.
Altogether, these data suggest that both the adhesive and differentiation programs of the epidermis are unaffected by the lack of zyxin. One possible explanation may be that the absence of phenotypic abnormalities is due to the presence of other zyxin family members which mask the consequences of loss of zyxin function. Indeed, we determined that both an LPP-specific antibody (38) and a panzyxin antibody that recognizes both zyxin and LPP (Fig. 6I) labeled the cell borders of zyxin-null mouse skin, demonstrating the presence of LPP in the epidermal cell junctions.
DISCUSSION
Zyxin is an evolutionarily conserved protein that has been implicated in cell adhesion, cell motility, and signaling. A number of zyxin binding partners have been identified by in vitro biochemical approaches. The roles of many of these partners have been elucidated by gene ablation studies in the mouse. For example, targeted disruption of the genes encoding p130Cas and Lats-1 results in significant lethality (26, 49). Disturbance of either Mena or VASP function does not lead to inviability but does affect particular cell functions (3, 21, 29). In this study, we generated mice in which the zyxin gene was disrupted. Here we explored the possibility that elimination of the zyxin gene will have consequences similar to what occurs when Mena or VASP coding sequences are ablated.
Among zyxin's binding partners, the interaction with Ena/VASP family members is the best characterized. As discussed above, numerous studies have illustrated that zyxin serves as a docking site for Ena/VASP family members and is important for the proper subcellular targeting of these proteins to particular subcellular locations, most notably focal adhesions (1, 14, 15, 19, 34, 42). Therefore, it was anticipated that elimination of zyxin might disturb Ena/VASP function by ablating a key Ena/VASP docking site within cells. Based on that perspective, one might anticipate that elimination of zyxin would have similar consequences as elimination of Mena or VASP.
As was observed of individual targeted VASP and Mena gene disruptions in the mouse, mice that lack zyxin function are viable and fertile. However, mutations in either Mena or VASP or expression of a VASP deletion construct revealed deficiencies in platelet activation, neuronal pathfinding, and epithelial cell-cell adhesion (3, 21, 29, 52). Here we examined the consequences of zyxin gene disruption for each of these processes. Despite the established contribution of zyxin to the docking of Mena/VASP proteins at cell adhesion sites (14), VASP−/− and Mena−/− phenotypes are not recapitulated in the zyxin−/− animals. Ena/VASP family members are complex, multidomain proteins. One possible explanation for the observation that zyxin−/− animals do not display the deficiencies observed in VASP−/− and Mena−/− mice is that phenotypes resulting from loss of Mena and VASP functions are independent of zyxin. For example, it is established that the localization, and therefore presumably the function, of Ena/VASP family members at the distal tips of lamellipodial extensions and at sites of T-cell receptor clustering are independent of zyxin (27, 45).
Alternatively, the consequences of losing zyxin could be masked by the expression of related proteins. Indeed, since initiating our efforts to target the zyxin gene, two proteins that are closely related to zyxin, LPP and TRIP6, were identified (33, 38, 53, 55). We detected no upregulated expression of these proteins in the zyxin−/− mice, so it does not appear that the lack of dramatic phenotype can be attributed to compensatory changes in LPP or TRIP6 expression. However, zyxin family members display similar functional domains and subcellular distributions, and the three zyxin family members exhibit similar tissue distribution patterns (11, 13, 31, 38, 39, 54, 55). Importantly, in the cases of blood platelets, brain, and skin, three tissues which we examined closely for a zyxin-null phenotype, we and others have detected expression of multiple zyxin family members (Fig. 4C and 6I) (13, 31, 39).
This is the first report describing the ablation of a gene in the zyxin family. Although no basal phenotype has yet been observed in the zyxin−/− mice, consequences of eliminating the zyxin gene may emerge if these animals are challenged by physiological stress. In order to assess further the role of the zyxin family in vivo, it will be of interest to disrupt the genes that encode other zyxin family members and to generate compound mutations. There now exist many examples in which deletion of a single member of a gene family has minimal consequences but simultaneous elimination of multiple members has a severe impact on development or adult physiology (17, 22, 50). The availability of a genetic model for studying zyxin function is a critical first step toward elucidating the role of zyxin.
Acknowledgments
We are grateful to Kirk Thomas (University of Utah Transgenic Core) for targeting vector constructs, the genomic phage library, and substantial technical advice. We thank Marleen Petit and Wim Van de Ven for anti-LPP serum (MP-2), Susanne Kloeker for anti-TRIP6 serum (B65), and Matthias Krause and Jürgen Wehland for antizyxin monoclonal antibody (164D4). We appreciate the contributions of Bob Schackmann (University of Utah DNA and Peptide Core Facility; NCI CA42014), Wayne Green (University of Utah Health Sciences Center Flow Cytometry Core Facility), and the University of Utah Animal Resource Center. Diana Lim provided invaluable assistance with graphic design.
This work was supported by funds provided by the National Institutes of Health to M.C.B. (GM50877) and to F.B.G. (GM58801), the Huntsman Cancer Foundation to M.C.B., a postdoctoral fellowship award by the Cancer Research Institute (New York) to B.B., a predoctoral training grant by the National Institutes of Health to D.N. (T32-CA09602), and the Fonds der Chemischen Industrie to R.F.
REFERENCES
- 1.Ahern-Djamali, S. M., A. R. Comer, C. Bachmann, A. S. Kastenmeier, S. K. Reddy, M. C. Beckerle, U. Walter, and F. M. Hoffmann. 1998. Mutations in Drosphila Enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol. Biol. Cell 9:2157-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alwine, J. C., D. J. Kemp, and G. R. Stark. 1977. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. USA 74:5350-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aszódi, A., A. Pfeifer, M. Ahmad, M. Glauner, X.-H. Zhou, L. Ny, K.-E. Andersson, B. Kehrel, S. Offermanns, and R. Fässler. 1999. The vasodilator-stimulated phosphoprotein (VASP) is involved in cGMP- and cAMP-mediated inhibition of agonist-induced platelet aggregation, but is dispensable for smooth muscle function. EMBO J. 18:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bear, J. E., M. Krause, and F. B. Gertler. 2001. Regulating cellular actin assembly. Curr. Opin. Cell Biol. 13:158-166. [DOI] [PubMed] [Google Scholar]
- 5.Bear, J. E., J. J. Loureiro, I. Libova, R. Fassler, J. Wehland, and F. B. Gertler. 2000. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101:717-728. [DOI] [PubMed] [Google Scholar]
- 6.Bear, J. E., T. M. Svitkina, M. Krause, D. A. Schafer, J. J. Loureiro, G. A. Strasser, I. V. Maly, O. Y. Chaga, J. A. Cooper, G. G. Borisy, and F. B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109:509-521. [DOI] [PubMed] [Google Scholar]
- 7.Beckerle, M. 1998. Spatial control of actin filament assembly: lessons from Listeria. Cell 95:741-748. [DOI] [PubMed] [Google Scholar]
- 8.Beckerle, M. C. 1997. Zyxin: zinc fingers at sites of cell adhesion. Bioessays 19:949-957. [DOI] [PubMed] [Google Scholar]
- 9.Brakebusch, C., R. Grose, F. Quondamatteo, A. Ramirez, J. L. Jorcano, A. Pirro, M. Svensson, R. Herken, T. Sasaki, R. Timpl, S. Werner, and R. Fässler. 2000. Skin and hair follicle integrity is crucially dependent on Beta-1 integrin expression on keratinocytes. EMBO J. 19:3990-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capecchi, M. R. 1989. Altering the genome by homologous recombination. Science 244:1288-1292. [DOI] [PubMed] [Google Scholar]
- 11.Crawford, A. W., and M. C. Beckerle. 1991. Purification and characterization of zyxin, an 82,000-dalton component of adherens junctions. J. Biol. Chem. 266:5847-5853. [PubMed] [Google Scholar]
- 12.Crawford, A. W., J. W. Michelsen, and M. C. Beckerle. 1992. An interaction between zyxin and alpha-actinin. J. Cell Biol. 116:1381-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuppen, E., M. van Ham, D. G. Wansink, A. de Leeuw, B. Wieringa, and W. Hendricks. 2000. The zyxin-related protein TRIP6 interacts with PDZ motifs in the adaptor protein RIL and the protein tyrosine phosphatase PTP-BL European J. Cell Biol. 79:283-293. [DOI] [PubMed] [Google Scholar]
- 14.Drees, B., E. Friederich, J. Fradelizi, D. Louvard, M. C. Beckerle, and R. M. Golsteyn. 2000. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 275:22503-22511. [DOI] [PubMed] [Google Scholar]
- 15.Drees, B. E., K. M. Andrews, and M. C. Beckerle. 1999. Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 147:1549-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drees, B. E., and M. C. Beckerle. 1999. Zyxin, p. 95-98. In T. Kreis and R. Vale (ed.), Guidebook to the extracellular matrix, anchor, and adhesion proteins, 2nd ed. Oxford University Press, Oxford, UK.
- 17.Esteban, L. M., C. Vicario-Abejón, P. Fernández-Salguero, A. Fernández-Medarde, N. Swaminathan, K. Yienger, E. Lopez, M. Malumbres, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fradelizi, J., V. Noireaux, J. Plastino, B. Menichi, D. Louvard, C. Sykes, R. M. Golsteyn, and E. Friederich. 2001. ActA and human zyxin harbour Arp2/3-independent actin polymerization activity. Nat. Cell Biol. 3:699-707. [DOI] [PubMed] [Google Scholar]
- 19.Gertler, F. B., K. Niebuhr, M. Reinhard, J. Wehland, and P. Soriano. 1996. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87:227-239. [DOI] [PubMed] [Google Scholar]
- 20.Goh, K. L., L. Cai, C. L. Cepko, and F. B. Gertler. 2002. Ena/VASP proteins regulate cortical neuronal positioning. Curr. Biol. 12:565-569. [DOI] [PubMed] [Google Scholar]
- 21.Hauser, W., K.-P. Knobeloch, M. Eigenthaler, S. Gambaryan, V. Krenn, J. Geiger, M. Glazova, E. Rohde, I. Horak, U. Walter, and M. Zimmer. 1999. Megakaryocyte hyperplasia and enhanced agonist-induced platelet activation in vasodilator-stimulated phosphoprotein knockout mice. Proc. Natl. Acad. Sci. USA 96:8120-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heber, S., J. Herms, V. Gajic, J. Hainfellner, A. Aguzzi, T. Rülicke, H. Kretzschmar, C. v. Kock, S. Sisodia, P. Tremml, H.-P. Lipp, D. P. Wolfer, and U. Müller. 2000. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members J. Neurosci. 20:7951-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirota, T., T. Morisaki, Y. Nishiyama, T. Marumoto, K. Tada, T. Hara, N. Masuko, M. Inagaki, K. Hatakeyama, and H. Saya. 2000. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149:1073-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hobert, O., J. W. Schilling, M. C. Beckerle, A. Ullrich, and B. Jallal. 1996. SH3 domain-dependent interaction of the proto-oncogene product Vav with the focal contact protein zyxin. Oncogene 12:1577-1581. [PubMed] [Google Scholar]
- 25.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 26.Honda, H., H. Oda, T. Nakamoto, Z. Honda, R. Sakai, T. Suzuki, T. Saito, K. Nakamura, K. Nakao, T. Ishikawa, M. Katsuki, Y. Yazaki, and H. Hirai. 1998. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat. Genet. 19:361-365. [DOI] [PubMed] [Google Scholar]
- 27.Krause, M., A. S. Sechi, M. Konradt, D. Monner, F. B. Gertler, and J. Wehland. 2000. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J. Cell Biol. 149:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lanier, L., M. Gates, W. Witke, A. S. Menzies, A. Wehman, J. Macklis, D. Kwiatkowski, P. Soriano, and F. Gertler. 1999. Mena is required for neurulation and commissure formation. Neuron 22:313-325. [DOI] [PubMed] [Google Scholar]
- 30.Laurent, V., T. P. Loisel, B. Harbeck, A. Wehman, L. Gröbe, B. M. Jockusch, J. Wehland, F. B. Gertler, and M.-F. Carlier. 1999. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 144:1245-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macalma, T., J. Otte, M. E. Hensler, S. M. Bockholt, H. A. Louis, M. Kalff-Suske, K.-H. Grzeschik, D. von der Ahe, and M. C. Beckerle. 1996. Molecular characterization of human zyxin. J. Biol. Chem. 271:31470-31478. [DOI] [PubMed] [Google Scholar]
- 32.Mansour, S. L., K. R. Thomas, C. Deng, and M. R. Capecchi. 1990. Introduction of a lacZ reporter gene into the mouse int-2 locus by homologous recombination. Proc. Natl. Acad. Sci. USA 87:7688-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy, K. K., K. Clark, Y. Fortin, S.-H. Shen, and D. Banville. 1999. ZRP-1, a zyxin-related protein, interacts with the second PDZ domain of the cytosolic protein tyrosine phosphatase hPTP1E. J. Biol. Chem. 274:20679-20687. [DOI] [PubMed] [Google Scholar]
- 34.Niebuhr, K., F. Ebel, R. Frank, M. Reinhard, E. Domann, U. D. Carl, U. Walter, F. B. Gertler, J. Wehland, and T. Chakraborty. 1997. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 16:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nix, D. A., and M. C. Beckerle. 1997. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138:1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nix, D. A., J. Fradelizi, S. Bockholt, B. Menichi, D. Louvard, E. Friederich, and M. C. Beckerle. 2001. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 276:34759-34767. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Alvarado, G. C., C. Miles, J. W. Michelsen, H. A. Louis, D. R. Winge, M. C. Beckerle, and M. F. Summers. 1994. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat. Struct. Biol. 1:388-398. [DOI] [PubMed] [Google Scholar]
- 38.Petit, M. M. R., J. Fradelizi, R. M. Golsteyn, T. A. Y. Ayoubi, B. Menichi, D. Louvard, W. J. M. Van De Ven, and E. Friederich. 2000. LPP, an actin cytoskeleton protein related to Zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petit, M. M. R., R. Mols, E. F. P. M. Schoenmakers, N. Mandahl, and W. J. M. Van de Ven. 1996. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics 36:118-129. [DOI] [PubMed] [Google Scholar]
- 40.Raghavan, S., C. Bauer, G. Mundschau, Q. Li, and E. Fuchs. 2000. Conditional ablation of beta-1 integrin in skin: severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J. Cell Biol. 150:1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhard, M., K. Giehl, K. Abel, C. Haffner, T. Jarchau, V. Hoppe, B. M. Jockusch, and U. Walter. 1995. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 14:1583-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhard, M., K. Jouvenal, D. Tripier, and U. Walter. 1995. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc. Natl. Acad. Sci. USA 92:7956-7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinhard, M., J. Zumbrunn, D. Jaquemar, M. Kuhn, U. Walter, and B. Trueb. 1999. An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J. Biol. Chem. 274:13410-13418. [DOI] [PubMed] [Google Scholar]
- 44.Renfranz, P. J., and M. C. Beckerle. 2002. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr. Opin. Cell Biol. 14:88-103. [DOI] [PubMed] [Google Scholar]
- 45.Rottner, K., M. Krause, M. Gimona, J. V. Small, and J. Wehland. 2001. Zyxin is not colocalized with vasodilator-stimulated phosphoprotein (VASP) at lamellipodial tips and exhibits different dynamics to vinculin, paxillin, and VASP in focal adhesions. Mol. Biol. Cell 12:3103-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmeichel, K. L., and M. Beckerle. 1998. LIM domains of cysteine-rich protein 1 (CRP1) are essential for its zyxin-binding function. Biochem. J. 331:885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmeichel, K. L., and M. C. Beckerle. 1994. The LIM domain is a modular protein-binding interface. Cell 79:211-219. [DOI] [PubMed] [Google Scholar]
- 48.Skoble, J., V. Auerbuch, E. D. Goley, M. D. Welch, and D. A. Portnoy. 2001. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J. Cell Biol. 155:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St. John, M., W. Tao, X. Fei, R. Fukumoto, M. L. Carcangiu, D. G. Brownstein, A. F. Parlow, J. McGrath, and T. Xu. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21:182-186. [DOI] [PubMed] [Google Scholar]
- 50.Stein, P. L., H. Vogel, and P. Soriano. 1994. Combined deficiencies of Src, Fyn, and Yes tyrosine kinases in mutant mice. Genes Dev. 8:1999-2007. [DOI] [PubMed] [Google Scholar]
- 51.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasioukhin, V., C. Bauer, M. Yin, and E. Fuchs. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100:209-219. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., J. E. Dooher, M. Koedood Zhao, and T. D. Gilmore. 1999. Characterization of mouse TRIP6: a putative intracellular signaling protein. Gene 234:403-409. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Y., and T. D. Gilmore. 2001. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains Biochim. Biophys. Acta 1538:260-272. [DOI] [PubMed] [Google Scholar]
- 55.Yi, J., and M. C. Beckerle. 1998. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 49:314-316. [DOI] [PubMed] [Google Scholar]
- 56.Yi, J., S. Kloeker, C. C. Jensen, S. Bockholt, H. Honda, H. Hirai, and M. C. Beckerle. 2002. Members of the zyxin family of LIM proteins interact with members of the p130cas family of signal transducers. J. Biol. Chem. 277:9580-9589. [DOI] [PubMed] [Google Scholar]
- 57.Zucker, M. B. 1989. Platelet aggregation measured by photometric method. Methods Enzymol. 169:117-133. [DOI] [PubMed] [Google Scholar]