Abstract
Sphingolipid metabolism is implicated to play an important role in apoptosis. Here we show that dihydrosphingosine (DHS) and phytosphingosine (PHS), two major sphingoid bases of fungi, have potent fungicidal activity with remarkably high structural and stereochemical specificity against Aspergillus nidulans. In fact, only naturally occurring DHS and PHS are active. Further analysis revealed that DHS and PHS induce rapid DNA condensation independent of mitosis, large-scale DNA fragmentation, and exposure of phosphatidylserine, all common morphological features characteristic of apoptosis, suggesting that DHS and PHS induce apoptosis in A. nidulans. The finding that DNA fragmentation requires protein synthesis, which implies that an active process is involved, further supports this proposition. The induction of apoptosis by DHS and PHS is associated with the rapid accumulation of reactive oxygen species (ROS). However, ROS are not required for apoptosis induced by DHS and PHS, as scavenging of ROS by a free radical spin trap has no effect. We further demonstrate that apoptosis induced by DHS and PHS is independent of metacaspase function but requires mitochondrial function. Together, the results suggest that DHS and PHS induce a type of apoptosis in A. nidulans most similar to the caspase-independent apoptosis observed in mammalian systems. As A. nidulans is genetically tractable, this organism should be an ideal model system for dissecting sphingolipid signaling in apoptosis and, importantly, for further elucidating the molecular basis of caspase-independent apoptosis.
In recent years, mounting evidence has indicated that sphingolipid metabolism is an important cell signaling system. Rapid and transient changes in sphingolipid metabolism are closely associated with a wide range of cellular activities, including the stress response, apoptosis, inflammation, cell cycle regulation, and cancer development (19, 20, 33). For instance, stress signals rapidly and transiently elevate the level of cellular ceramide; conversely, growth factors stimulate the rapid, transient generation of sphingosine-1-phosphate (S-1-P). Importantly, the treatment of cells with short-chain ceramide analogs or the expression of sphingomyelinases reproduces most of the effects of endogenous ceramide, particularly in the stress response and apoptosis (19, 20, 33). Furthermore, S-1-P, as a high-affinity ligand of the edg family of G-protein-coupled receptors, promotes cell survival and proliferation by antagonizing ceramide-mediated apoptosis (52, 61). Thus, ceramide, as an important regulatory component in the stress response and apoptosis, and S-1-P, involved in cell survival and proliferation, have attracted enormous scientific interest in recent years. In particular, a large body of evidence has now accumulated to implicate an important role of ceramide in the stress response and apoptosis. It is proposed that the relative levels of cellular ceramide and S-1-P determine whether cells undergo apoptosis or continue to proliferate (61).
The sphingoid long-chain bases dihydrosphingosine (DHS) and sphingosine are central components of the sphingolipid metabolic pathway (20). They are substrates for the synthesis of both ceramide and sphingoid base phosphates (S-1-P and DHS-1-phosphate [DHS-1-P]). Sphingosine is also the product of ceramide and S-1-P metabolism. It has been shown that sphingosine is also a highly bioactive molecule (20). However, compared to the situation for ceramide and S-1-P, very few studies have been directed toward characterizing the cellular functions of sphingosine. Interestingly, a recent study showed that, paralleling the change in ceramide levels, a rapid, transient elevation of sphingosine levels is also observed in the stress response and apoptosis (9). It was further shown that treatment of cells with exogenous C2-ceramide not only increases the level of endogenous ceramide but also leads to a marked increase in the level of cellular sphingosine (9). The biological significance of such an increase in the cellular sphingosine level during the stress response and apoptosis is not well understood. These studies do raise the possibility that, like ceramide, sphingosine may also play a role in the stress response and apoptosis. This possibility is further supported by the fact that exogenous sphingosine is a potent elicitor of apoptosis (9, 10, 30). Most importantly, sphingosine regulates multiple key signal transduction factors, such as protein kinase C, 3-phosphoinositide-dependent kinase 1 (PDK1), and p21-activated kinase 1 (PAK1) (4, 18, 32).
The basic chemical structure, biosynthesis, and metabolism of sphingolipids are conserved in mammalian and fungal systems (15). In Saccharomyces cerevisiae, an important role for sphingolipids in the stress response was originally proposed based on the observation that SLC (suppressor of long-chain bases) mutants were unable to grow under many types of stress (55). A role for sphingolipids in the yeast stress response to heat now has been well established (28, 71). Interestingly, a detailed analysis of changes in the sphingolipid composition showed that upon heat stress, the levels of the sphingoid long-chain bases DHS and phytosphingosine (PHS) rapidly and transiently increase severalfold within 10 to 20 min (28). Although the level of cellular ceramide also increases severalfold upon heat stress, the increase occurs much more slowly, within 1 to 2 h (28). Furthermore, it was demonstrated that the rapid elevation of the levels of cellular DHS and PHS but not of ceramide is responsible for transient G1 arrest upon heat stress (27) and that heat resistance requires DHS-1-P and PHS-1-phosphate (PHS-1-P) produced by sphingoid base kinase-phosphatase activities (31, 34, 43, 44). Exogenous PHS has also been shown to inhibit yeast growth in a very specific manner that does not involve ceramide (8) but occurs in part through the regulation of ubiquitin-dependent proteolysis (7) and nutrient uptake (59). Moreover, in S. cerevisiae, PHS plays an important role in endocytosis and proper actin cytoskeleton organization by regulating Pkc1 activity via Pkh1/2 (11, 65). Together, these studies strongly suggest that sphingoid bases play a specific signaling role in growth regulation and the stress response in S. cerevisiae.
We are using Aspergillus nidulans, a filamentous fungus, as a model genetic system to study the cellular functions of sphingolipids. Recently, Cheng et al. showed that sphingolipids are essential for the establishment and maintenance of cell polarity via control of the actin cytoskeleton and that the accumulation of cellular ceramide causes cell cycle arrest in G1 (6). In this study, we investigated the cellular functions of DHS and PHS by analyzing the physiological and cellular effects of treating A. nidulans cells with exogenous DHS and PHS. Both DHS and PHS are able to diffuse readily between cell membranes and, most importantly, are able to fully complement the lack of serine palmitoyltransferase (SPT) activity, the first committed and rate-limiting step of the sphingolipid biosynthesis pathway (6). We found that DHS and PHS have potent antifungal activity with remarkable structural and stereochemical specificity. We demonstrate that the antifungal activity of DHS and PHS does not involve ceramide and intriguingly acts through the rapid induction of metacaspase-independent apoptosis. Furthermore, we show that fungal apoptosis is remarkably similar to that of mammalian cells morphologically.
MATERIALS AND METHODS
Strains and general techniques.
The A. nidulans strains used in this study were A773 (pyrG89 pyroA4 wA3), a cell cycle wild-type pyrG+ strain (pyrG89 pyroA4 pyrG+ wA3), lcbA3′Δ-263 (pyrG89 alcA::lcbA pyrG+ lcbA3′Δ wA3), SO182 (nimT23 pyrG89 pabaA1 chaA1), A781 (nimA5 wA2), SO65 (nimXcdc2Y206H riboA1 pyroA4 wA3), smcBcut143′Δ-6 (pyrG89 pyroA4 alcA::smcBcut14 smcBcut143′Δ pyrG+ wA3), casAΔ-6 (pyrG89 pyroA4 casAΔ pyrG+ wA3), and casAOE (pyrG89 pyroA4 alcA::casA pyrG+ wA3). Media, transformation, nimT23 block and release experiments, and general techniques for culturing A. nidulans cells, 4′,6′-diamidino-2-phenylindole (DAPI) staining, indirect immunofluorescence staining, and microscopy were as previously described (6). A minimum of 300 cells were examined to score cellular phenotypes. Protein extraction, immunoprecipitation, and p34cdc2/cyclin B and NIMA kinase assays were carried out as previously described (73). The anti-phosphorylated histone H3 marker was obtained from Upstate Biotechnology Inc. and used under the conditions previously described (14).
Labeling and analysis of sphingolipids.
All chemicals were reagent grade. Sphingolipids were purchased from Avanti Polar Lipids (Alabaster, Ala.). Radiochemicals were purchased from American Radiolabeled Chemicals (St. Louis, Mo.). All the organic solvents were high-pressure liquid chromatography (HPLC) grade and were purchased from Fisher Scientific (Cincinnati, Ohio).
Pulse-labeling of sphingolipids with [4,5-3H]DHS-1-P and [4,5-3H]DHS, extraction, and analysis by thin-layer chromatography were performed as previously described (6, 21, 51). For analysis of cellular ceramide by HPLC, lipids were extracted with solvent system III B (95% ethanol-water-diethyl ether-pyridine-ammonium hydroxide [15:15:5:1:0.018, vol/vol]) (21) and then reextracted with solvent A (chloroform-methanol [1:1, vol/vol]) as described by McNabb et al. (47). The chloroform phase was dried under vacuum and resuspended in chloroform-methanol (9:1 [vol/vol]). Separation of ceramide was performed as described by McNabb et al. (47) with an HPLC system consisting of a Waters 2690 apparatus, Millenium software, and a 3-μm silica column (4.5 by 150 mm) (all from Waters, Inc., Milford, Mass.) and with a mobile phase of chloroform-ethanol-triethylamine-formic acid (90:10:1:2 [vol/vol]). The flow rate was 0.5 ml/min. A Sedex 75 ELSD (Sedere, Alfortville, France) was used with nitrogen gas pressure at 3.2 bars (1 bar = 105 Pa), a drift tube temperature of 70°C, and a gain setting at 6. Under these conditions, ceramide elutes with a retention time of 3 min.
Cloning of condensin subunit smcBcut14 and metacaspase casA.
A homology search with the cut14 gene of Schizosaccharomyces pombe identified a few expressed sequence tags (EST) from the Fungal Genetic Stock Center (Kansas City, Kans.) A. nidulans cDNA database (http://www.genome.ou.edu/asper_blast.html). Based on the EST, a PCR approach was used to screen a single-copy genomic cosmid library (Fungal Genetic Stock Center) grown in 96-well titer plates. Cosmids L05F06 and L28D12 were identified as containing EST. Sequencing of cosmid L05F06 by walking in both directions starting from the EST revealed an open reading frame encoding a protein of 1,179 amino acids and having a high overall level of homology to S. pombe cut14 (45% identity and 61% similarity). This cut14 homolog of A. nidulans was designated smcBcut14.
The A. nidulans metacaspase gene, casA, was also similarly cloned by PCR screening of the single-copy genomic cosmid library after an EST search with S. cerevisiae YOR197w. A single cosmid clone, W15B06, was identified to contain a casA gene encoding a 404-amino-acid protein and having an overall identity of 50% and a similarity of 61% to that of yeast YOR197w.
Because of the presence of introns, to further confirm the protein sequence predictions for smccut14 and casA, we also cloned full-length cDNAs for these two genes by rapid amplification of cDNA ends-PCR as previously described (6).
Generation of alcA::smcBcut14-dependent and casAΔ strains.
Since condensin activity is essential for mitosis, as demonstrated in other model organisms, such as S. cerevisiae and S. pombe (58, 63), we anticipated that smcBcut14 of A. nidulans would also be essential. Thus, to study the potential role of condensin activity in A. nidulans apoptosis induced by sphingoid bases, we created alcohol-dependent smcBcut14 strains as previously described (6). Briefly, we cloned a 2.1-kb fragment of smcBcut14 with a 3′ truncation (encoding 528 amino acids) under the control of the promoter of the A. nidulans catabolic alcohol dehydrogenase gene, alcA, which is alcohol inducible and glucose repressible (69), into pBluscript II SK(+) (Stratagene), with the pyrG gene of A. fumigatus as the selection marker to give rise to plasmid psmcB3′Δ. Circular psmcB3′Δ DNA was then used to transform strain A773. A single homologous recombination event for psmcB3′Δ at the smcBcut14 locus results in the 3′ truncation of the endogenous smcBcut14 gene and brings the full-length smcBcut14 gene under the control of the alcA promoter. Screening for and identification of the alcA::smcBcut14-dependent strains were done as previously described (6).
A casA disruption construct was made by three-step cloning with pBluescript II SK(+). A 1.5-kb 3′-end flanking sequence was amplified from A. nidulans genomic DNA by PCR with a pair of primers, 5′-AAAACAAAAACTGCAGCACCACCACCATCAACAGCCG-3′ and 5′-AAACAAAGAATTCGTTTAAACGCTATCGTATGAACGTATATTATGAACG-3′, and cloned into the PstI and EcoRI sites of pBluescript II SK(+). Then, the A. fumigatus pyrG gene was cloned into the plasmid as a 1.9-kb SpeI-BamHI fragment. Finally, a 2.2-kb 5′-end casA flanking sequence was amplified with the primers 5′-AAACAAAACAAAAGCGGCCGCAGTGGCAGATCATCTACAACAAGCTCGAG-3′ and 5′-AAAACAAACAAACACTAGTTTGTCTGAGGAAGCGGGGAC-3′and cloned into the NotI and SpeI sites to generate pcasΔ, in which the casA open reading frame was disrupted by A. fumigatus pyrG. A linear 5.6-kb NotI-EcoRI fragment from pcasΔ was used to replace the endogenous casA gene by transformation of strain A773. PCR analysis of transformants was used to screen for casAΔ strains as previously described (6).
Analysis of apoptotic markers.
For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) with fluorescein isothiocyanate (FITC)-conjugated dUTP, A. nidulans spores were germinated on coverslips in yeast extract (YG) medium. Fixation, washing, and cell wall digestion were carried out as previously described (6, 49), except that 3 mM EDTA was added to inhibit DNase activity. Cell permeabilization and the TUNEL reaction were carried out as previously described for S. cerevisiae (40) with an in situ cell death detection kit (POD; Boehringer Mannheim, Indianapolis, Ind.). Following the TUNEL reaction, cells were stained with DAPI.
An annexin V binding assay and propidium iodide staining of protoplasts were carried out as described by Madeo et al. (40). A. nidulans spores (109) were germinated at 32°C for 5.5 h in 50 ml of suspension culture before treatment with PHS. Protoplasts were then generated as previously described for A. nidulans transformation (49).
For pulsed-field gel electrophoresis (PFGE), DNA was isolated from A. nidulans as previously described (13). PFGE was carried out with a 1% agarose gel in 0.5× Tris-borate-EDTA buffer (50 mM Tris, 45 mM boric acid, 0.5 mM EDTA, pH. 8.4) by using a CHEF II PFGE system (Bio-Rad, Hercules, Calif.). The pulse time was 1 to 6 s for 11 h at 6 V/cm. The temperature was maintained at 14°C, and the angle was 120°. DNA was visualized after staining with ethidium bromide.
The production of reactive oxygen species (ROS) by A. nidulans cells was detected with dihydrorhodamine 123 (DHR123) (Sigma, St. Louis, Mo.). The free radical spin trap reagent 3,3,5,5,-tetramethyl-pyrroline N-oxide (TMPO) (Sigma-Aldrich) was used to scavenge intracellular ROS as previously described (40). Oligomycin (Sigma), a specific inhibitor of mitochondrial F0F1-ATPase, was directly added to germinating spores at final concentrations of 10 and 20 μM to inhibit mitochondrial function as described in a previous study of S. cerevisiae (46).
Nucleotide sequence accession numbers.
The nucleotide sequences of the smcBcut14 gene and the casA gene have been deposited in GenBank under accession numbers AF488722 and AF528964, respectively.
RESULTS
Potent antifungal activity of naturally occurring sphingoid long-chain bases.
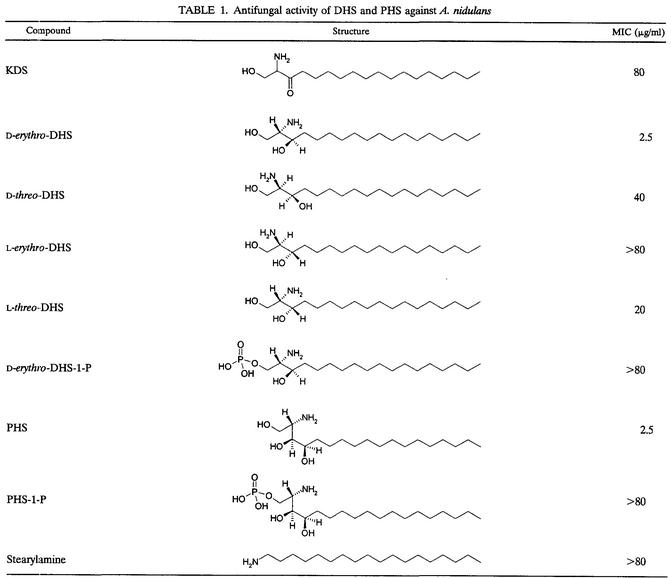
To study the functions of sphingoid bases in A. nidulans, we first determined their antifungal activities. We found that DHS and PHS possess equally potent activities against A. nidulans, with an MIC of 2.5 μg/ml. In contrast, 3-keto-DHS (KDS) had very little antifungal activity (Table 1). Furthermore, the antifungal activity of DHS showed remarkable structural and stereochemical specificity. Only naturally occurring d-erythro-DHS showed potent antifungal activity, whereas all of the nonnatural stereoisomers of DHS, the structurally related compound stearylamine, and DHS-1-P exhibited either no or very weak antifungal activity. PHS-1-P also lacked antifungal activity.
TABLE 1.
Antifungal activity of DHS and PHS against A. nidulans
In S. cerevisiae, PHS specifically inhibits the transport of various nutrients, such as uracil and amino acids (7, 8, 59). As a result, auxotrophic strains for these nutrients are much more sensitive to PHS inhibition than the wild-type strain. Moreover, yeast cells growing in defined medium containing ammonium ions as the nitrogen source are more resistant to PHS than those growing in complex rich medium. However, unlike the situation for yeast cells, we found that PHS is equally potent against A. nidulans cells growing in either defined minimal medium or complex rich medium. Furthermore, nutrient-auxotrophic A. nidulans cells carrying the pyrG89 mutation are not more sensitive to PHS inhibition than wild-type cells.
It is possible that the lack of antifungal activity of KDS, DHS-1-P, and PHS-1-P is a result of a lack of uptake of these compounds by A. nidulans cells. We thus tested their ability to complement the lethality of a null mutant strain lacking SPT activity, as SPT catalyzes the first step of sphingolipid biosynthesis, the formation of KDS through the condensation of serine and palmitoyl coenzyme A. Cheng et al. previously showed that exogenous DHS and PHS fully complement null mutant A. nidulans cells lacking SPT (6). One of the major metabolic routes of DHS-1-P and PHS-1-P is dephosphorylation to DHS and PHS, respectively, by sphingoid base phosphatases (15, 20). Interestingly, genetic studies of S. cerevisiae have shown that the incorporation of exogenous sphingoid bases into sphingolipids requires phosphorylation followed by dephosphorylation upon uptake (45). We therefore reasoned that if cells efficiently take up KDS, DHS-1-P, and PHS-1-P and efficiently dephosphorylate DHS-1-P and PHS-1-P to DHS and PHS, then KDS, DHS-1-P, and PHS-1-P should all complement the lack of SPT function. Indeed, KDS fully complemented the lethality of A. nidulans cells lacking SPT at concentrations as low as 2 μg/ml, demonstrating that the lack of antifungal activity of KDS is not caused by a lack of uptake. On the other hand, DHS-1-P and PHS-1-P failed to complement the null mutant cells lacking SPT. To further analyze the uptake of DHS-1-P and PHS-1-P by A. nidulans cells, we monitored the uptake of radiolabeled DHS and DHS-1-P and their subsequent incorporation into sphingolipids. As shown in Fig. 1A, both DHS and DHS-1-P were efficiently taken up by A. nidulans cells. However, much less radiolabeled DHS-1-P was subsequently incorporated into sphingolipids compared to DHS (Fig. 1A). Thus, the failure to complement cells lacking SPT was likely due to a slow conversion of DHS-1-P to DHS by sphingoid base phosphatases. The results thus suggest that KDS, DHS-1-P, and PHS-1-P indeed lack antifungal activity.
FIG. 1.
Sphingolipid analysis. (A) Autoradiogram of stable sphingolipid bases labeled with [3H]DHS and [3H]DHS-1-P and separated by chromatography on a Silica Gel 60 thin-layer chromatography plate. Stable sphingolipid bases were extracted from cells after pulse-labeling with [3H]DHS or [3H]DHS-1-P for 1 h. Note the efficient uptake of both [3H]DHS and [3H]DHS-1-P by A. nidulans cells and the much lower level of incorporation of [3H]DHS-1-P than of [3H]DHS into sphingolipids. (B) Levels of cellular ceramide in rapidly growing control cells (C) and cells treated for 1 h with 5 μg of AbA/ml, 4 μg of DHS/ml, or 4 μg of PHS/ml. Error bars indicate standard deviations.
DHS and PHS are substrates for ceramide synthesis. Cheng et al. previously showed that the accumulation of cellular ceramide causes cell cycle arrest in G1 in A. nidulans (6). In S. cerevisiae, PHS has been shown to inhibit growth in a manner independent of ceramide (8). To determine whether DHS and PHS inhibit A. nidulans growth through ceramide, we measured the cellular ceramide level in cells treated with DHS and PHS by HPLC. Cheng et al. previously showed that the inhibition of inositol phosphorylceramide synthase with the selective inhibitor aureobasidin A (AbA) markedly increases the level of cellular ceramide (6). Therefore, AbA-treated cells were used as a positive control. As shown previously, AbA treatment markedly increased the level of cellular ceramide (Fig. 1B). In contrast, DHS and PHS treatment did not significantly increase the level of cellular ceramide (Fig. 1B), thus demonstrating that, as in S. cerevisiae (8), the antifungal activity of DHS and PHS in A. nidulans also does not involve ceramide.
To further determine whether DHS and PHS are fungicidal, we treated A. nidulans germlings growing on coverslips with DHS and PHS and then removed DHS and PHS from the growth medium at various times after treatment. We observed whether the treated cells could resume growth after the removal of DHS and PHS. Remarkably, we found that a brief treatment with DHS or PHS, as short as 4 min, could irreversibly inhibit A. nidulans growth (data not shown). Thus, DHS and PHS are fungicidal against A. nidulans.
DHS and PHS induce rapid DNA condensation independent of mitosis.
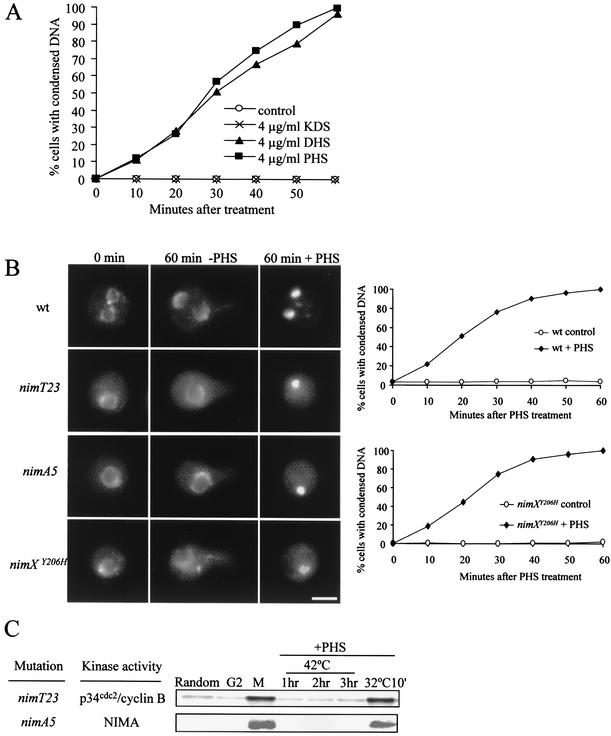
To investigate how DHS and PHS so potently inhibit A. nidulans growth, we examined the phenotypes of cells treated with DHS and PHS. To our surprise, we found that DHS and PHS induced rapid DNA condensation, as revealed by DAPI staining. Mitosis in A. nidulans is a very rapid process and is normally completed within a few minutes (48). Thus, normally <5% of A. nidulans cells in a rapidly growing culture are observed in mitosis with condensed DNA at any time. However, within minutes after treatment with DHS or PHS, a marked increase in the number of cells containing condensed DNA was observed. By 60 min, 100% of the treated cells showed highly condensed DNA (Fig. 2A). In close agreement with their antifungal activity, DHS and PHS were equally effective in inducing DNA condensation, whereas KDS had no effect (Fig. 2A). Because the results from studies done with either DHS or PHS were indistinguishable, we describe only results obtained with PHS in the following sections.
FIG. 2.
Mitosis-independent DNA condensation induced by sphingoid long-chain bases. (A) Time course showing rapid DNA condensation induced by DHS and PHS but not by KDS. Spores of a wild-type strain were germinated on coverslips in YG medium for 5.5 h at 32°C. The germinated spores were transferred to YG medium containing KDS, DHS, or PHS. At 10-min intervals after the transfer, cells were fixed and stained with DAPI to visualize DNA. (B and C) PHS induces DNA condensation in the absence of mitosis-promoting kinase activities. (B) Representative micrographs of DAPI-stained cells showing DNA condensation independent of mitosis-promoting kinase functions and kinetics of DNA condensation in wild-type (wt) and nimXcdc2Y206H mutant cells in the presence of PHS (4 μg/ml). (C) p34cdc2/cyclin B kinase activity and NIMA kinase activity were assayed in nimT23 and nimA5 G2-arrested cells, respectively. Early-log-phase cultures of mutant cells (lane Random) were rapidly upshifted to 42°C from 32°C to inactivate mitosis-promoting kinases and then incubated at 42°C for 2 h to block cells at G2 (lane G2). A portion of the cells was released from the G2 block into mitosis at 32°C for 10 min (lane M). PHS was added to the rest of the cells at a final concentration of 4 μg/ml. The cultures were incubated at 42°C for an additional 3 h in the presence of PHS. Samples were taken at hourly intervals as indicated. Finally, a portion of the PHS-treated cells was also downshifted to 32°C for 10 min after 3 h in the presence of PHS (lane 32°C10′). (D) DNA condensation induced by PHS is not associated with serine-10 phosphorylation of histone H3. nimT23 mutant spores were germinated on coverslips for 6 h at 42°C to cause G2 arrest before PHS addition. Cells were allowed to continue to grow at 42°C in the absence (G2) or presence (G2+PHS) of 4 μg of PHS/ml for another hour. As a positive control, G2-arrested cells without PHS treatment were released into mitosis at 32°C for 10 min (M). Cells were fixed and doubly stained with DAPI and a histone H3 serine-10 phosphorylation-specific antibody (α-histone H3 S10-P). (E) Condensin activity has no role in DNA condensation induced by PHS. (a) Interphase nuclear phenotype of cells lacking smcBcut14 function. Wild-type and smcBcut14 deletion mutant (smcBcut143Δ) cells were germinated at 32°C for 8 h. (b) Spores of wild-type and smcBcut14 deletion mutant strains were germinated at 32°C for 6 h before PHS addition and then allowed to continue to grow in the absence (−PHS) or presence (4μg/ml PHS) of PHS at 4 μg/ml for another hour. Cells were fixed and stained with DAPI to visualize DNA. Bars, 5 μm.
A rapid increase in the number of cells containing highly condensed DNA upon PHS treatment could be a result of a mitotic block (48) or premature entry into mitosis (53, 72). If either of these is the case, then PHS should not cause DNA condensation in interphase-arrested cells with inactive mitosis-promoting kinases. In A. nidulans, the activation of two mitosis-promoting protein kinases, the universally conserved p34cdc2/cyclin B kinase and the NIMA kinase, is required for entry into mitosis (54). To test the hypothesis, we used nimT23 and nimA5 temperature-sensitive mutations to inactivate p34cdc2/cyclin B or NIMA kinase activities or nimXcdc2Y206H to inactivate both p34cdc2/cyclin B and NIMA kinase activities at a restrictive temperature of 42°C and arrest the cell cycle in G2 before PHS treatment. nimTcdc25 encodes a homolog of fission yeast Cdc25 tyrosine phosphatase which is required for p34cdc2/cyclin B activation by dephosphorylation, and nimXcdc2 encodes Cdc2, the catalytic subunit of p34cdc2/cyclin B kinase. It was previously shown that the nimXcdc2Y206H mutation causes G2 arrest at 42°C with both inactive p34cdc2/cyclin B and inactive NIMA kinases (72).
The wild-type and cell cycle mutant cells were germinated at 42°C for 5.5 h. By this time, wild-type cells typically had undergone one round of nuclear division, whereas mutant cells were uniformly arrested at G2 with a single large interphase nucleus (Fig. 2B). Cells were then treated with PHS at 42°C, and DNA condensation was examined at various times after PHS treatment. As shown in Fig. 2B, PHS was still able to rapidly induce DNA condensation in G2-arrested cells with kinetics similar to those observed in wild-type cells (Fig. 2A). To rule out the possibility that PHS somehow activated mutant mitosis-promoting kinases, we assayed p34cdc2/cyclin B and NIMA kinase activities in G2-arrested cells after PHS treatment. As shown previously, p34cdc2/cyclin B kinase in nimT23 mutant cells and NIMA kinase in nimA5 mutant cells were inactive at 42°C (Fig. 2C) (72). Although DNA had become highly condensed after PHS treatment, these two mitosis-promoting kinases remained inactive in the mutant cells (Fig. 2C). Interestingly, p34cdc2/cyclin B and NIMA kinases in PHS-treated mutant cells could still be rapidly activated upon return to a permissive temperature, just as in those cells without PHS treatment, suggesting that molecular mechanisms for the activation of these protein kinases remained intact after PHS treatment, despite the presence of highly condensed DNA. We thus conclude that DNA condensation induced by PHS is independent of mitosis-promoting kinases.
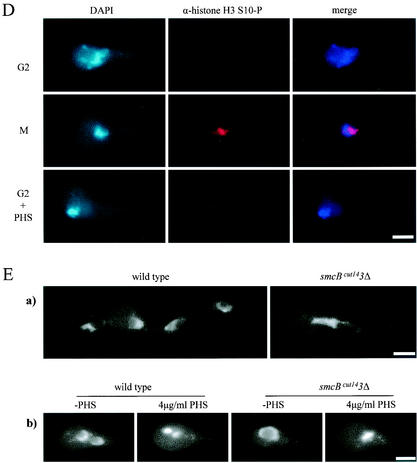
DNA condensation during mitosis requires histone H3 phosphorylation at serine-10 by the highly conserved aurora kinase and condensin activity (23, 25). In A. nidulans, serine-10 phosphorylation of histone H3 during mitosis has been shown to be NIMA kinase dependent (14). Perhaps PHS treatment in A. nidulans bypasses the requirement of NIMA for the activation of aurora kinase in histone H3 phosphorylation to promote DNA condensation. To address this possibility, we monitored histone H3 phosphorylation by indirect immunofluorescence microscopy with a specific antibody against the serine-10 phosphoepitope. As previously reported (14), mitotic DNA condensation during nimT23 block and release was associated with histone H3 phosphorylation (Fig. 2D). In contrast, PHS-induced DNA condensation occurred in the absence of histone H3 phosphorylation (Fig. 2D).
We next investigated whether PHS-induced DNA condensation requires condensin activity. To that end, we cloned the A. nidulans homolog of fission yeast cut14, designated smcBcut14, which encodes an essential and highly conserved subunit of the condensin complex, and generated a conditional mutant strain dependent on the expression of smcBcut14 under the control of the alcA promoter. alcA promoter activity is induced by alcohol as a sole carbon source and is tightly repressed by glucose. When germinated in glucose-containing medium, which tightly represses smcBcut14 expression from the alcA promoter, smcBcut14 deletion mutant cells were unable to undergo nuclear division and were terminally arrested with a single nucleus and uncondensed DNA; these results are consistent with smcBcut14 function being required for DNA condensation (Fig. 2E, panel a). To determine whether PHS could induce DNA condensation in cells lacking smcBcut14 function, we germinated cells for 6 h in glucose-containing medium to inactivate smcBcut14 before we added PHS. The addition of PHS induced rapid DNA condensation in both wild-type and smcBcut14 deletion mutant cells (Fig. 2E, panel b). Therefore, PHS-induced DNA condensation does not require condensin activity. Together, the results further demonstrate that rapid DNA condensation induced by PHS is indeed independent of mitosis.
PHS-treated cells exhibit morphological markers characteristic of apoptosis.
DNA condensation independent of mitosis is observed in cells undergoing apoptosis (50). Perhaps PHS induces apoptosis in A. nidulans and the DNA condensation observed in PHS-treated cells is in fact a manifestation of apoptosis. Apoptosis is a genetically controlled type of active cell death and is characterized by a set of distinctive morphological changes. The most prominent morphological changes include DNA condensation, DNA fragmentation, and exposure of phosphatidylserine (PS) on the cell surface (70). These morphological features are widely used as diagnostic markers of apoptosis. To determine whether PHS indeed induces apoptosis in A. nidulans cells, we next assayed DNA fragmentation and PC externalization.
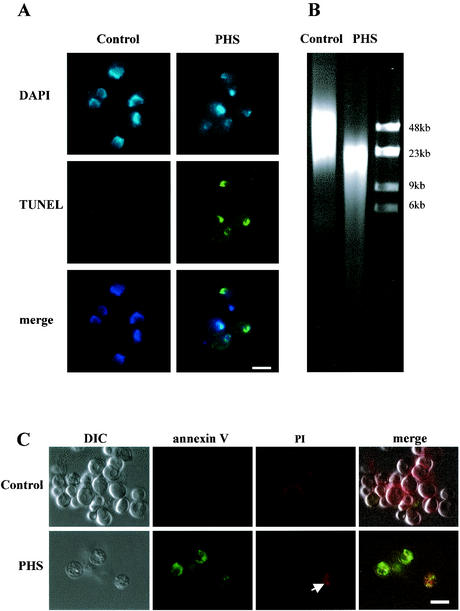
We first used TUNEL staining to detect DNA fragmentation, as DNA breaks labeled with FITC-conjugated dUTP via terminal deoxynucleotidyltransferase can be directly visualized by fluorescence microscopy. To minimize the labeling of DNA breaks occurring during normal DNA replication, DNA damage repair, and mitotic DNA recombination, we used cells synchronized in late G2, because at late G2 cells have completed DNA replication, have repaired DNA damage, and are ready for entry into mitosis. To generate cells synchronized in late G2, we used the nimT23 temperature-sensitive mutation as described for the experiment shown in Fig. 2B. Initially we tried to detect DNA fragmentation by TUNEL with a time course similar to that used for DNA condensation up to 60 min. We observed a very low percentage of PHS-treated cells labeled by TUNEL during this time period. However, we did notice that the number of TUNEL-positive cells increased in a time-dependent manner. Thus, we extended PHS treatment up to 5 h. By 3 h after PHS treatment, the majority of PHS-treated cells showed TUNEL staining (Fig. 3A; also see Fig. 6B). To independently confirm DNA fragmentation detected by TUNEL, we compared the sizes of DNAs isolated from control and PHS-treated cells by PFGE. Indeed, DNA isolated from PHS-treated cells was considerably smaller than DNA isolated from control cells (Fig. 3B). However, PHS treatment did not produce oligonucleosomal DNA ladders typical of caspase-dependent apoptosis. Instead, PHS induced the so-called large-scale DNA fragmentation associated with the caspase-independent apoptosis recently reported for humans and Dictyostelium discoideum (3, 64). The results thus show that PHS treatment induces DNA fragmentation in A. nidulans and that, compared to DNA condensation, DNA fragmentation is a relatively late event.
FIG. 3.
Morphological changes characteristic of apoptosis of cells treated with PHS. (A) nimT23 mutant cells arrested in G2 after germination at 42°C for 5.5 h were treated with PHS (4 μg/ml) for 3 h or left untreated (control). DNA condensation and DNA fragmentation were then visualized by double staining with DAPI and TUNEL. (B) PFGE of DNA isolated from nimT23 G2-arrested cells with or without PHS treatment as described for panel A. (C) Protoplasts derived from PHS-treated cells bind to annexin V strongly. Protoplasts were made from germinating spores with or without PHS treatment (4 μg/ml) and then stained with both FITC-conjugated annexin V and propidium iodide (PI). Spores were germinated for 5.5 h at 32°C before PHS treatment for 1 h. The arrow indicates a cell that was apparently damaged during the protoplast formation process, as it shows general uptake of PI. Bars, 5 μm.
FIG. 6.
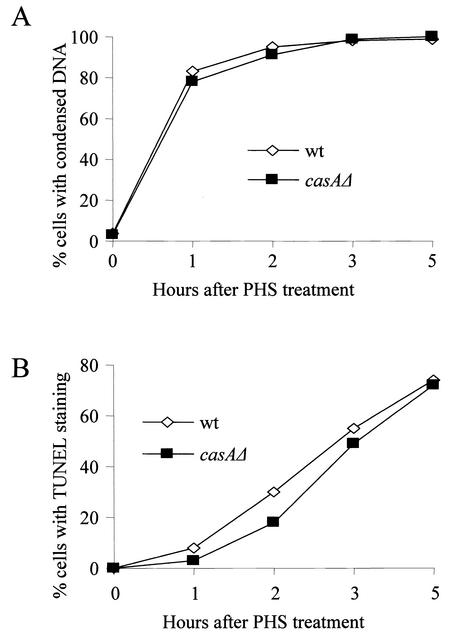
The metacaspase casA has no role in PHS-induced apoptosis. We assayed DNA condensation (A) and DNA fragmentation (B) in the presence or absence of casA. Spores of a wild-type (wt) strain and a casAΔ strain were germinated, treated with PHS, and then processed for DAPI and TUNEL staining as described in the legend to Fig. 3.
PS is predominantly located in the inner leaflet of the plasma membrane of eukaryotic cells. During apoptosis, the plasma membrane loses this asymmetry, and PS becomes externalized to the outer leaflet. In animals, PS so exposed on the cell surface is recognized by PS receptors in phagocytes to facilitate the removal of apoptotic cells by phagocytosis (22, 24). PS exposed on the cell surface can be detected by using annexin V, which binds to PS specifically with a high affinity in the presence of Ca2+.
The FITC-conjugated annexin V binding assay for PS exposure in A. nidulans was carried out after removal of the cell wall by using cell wall hydrolytic enzymes in a manner similar to that previously described for S. cerevisiae (40, 41). By 1 h after PHS treatment, >50% of protoplasts derived from treated cells bound to annexin V strongly, whereas <3% of protoplasts derived from untreated control cells bound to annexin V (Fig. 3C). Propidium iodide staining showed that the plasma membranes of protoplasts derived from both treated and control cells remained largely intact, as the general uptake of propidium iodide was observed in <3% of protoplasts (Fig. 3C). Furthermore, control protoplasts which bound to annexin V usually also took up propidium iodide, indicating that their plasma membranes were damaged, likely during the protoplast formation process.
In summary, PHS treatment of A. nidulans cells induces rapid mitosis-independent DNA condensation, DNA fragmentation, and PS externalization. These morphological changes observed in PHS-treated cells are features characteristic of apoptosis. Together, the results strongly suggest that PHS induces apoptosis in A. nidulans.
Apoptosis is not a general response to antifungal compounds.
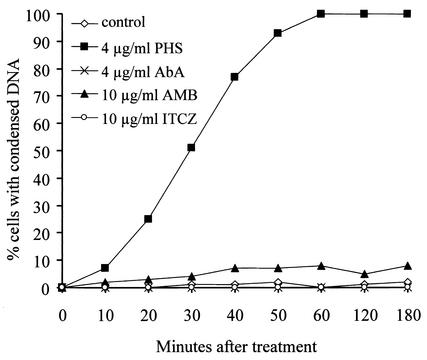
As shown in Table 1, DHS and PHS have potent antifungal activity, and their antifungal activity was subsequently shown to be associated with their ability to induce apoptosis. We next examined whether apoptosis in A. nidulans is specific to the action of DHS and PHS or is a general response to antifungal agents. To distinguish these two possibilities, we compared the abilities of PHS and several different chemical classes of antifungal agents, which all target membrane functions, to induce rapid DNA condensation; included in this analysis was AbA, which specifically inhibits sphingolipid synthesis. All of these antifungal agents rapidly inhibited A. nidulans cell growth after addition. However, DAPI staining of treated cells revealed that only PHS induced DNA condensation (Fig. 4). Since DNA condensation is one of the established and necessary morphological markers of apoptosis, we thus conclude that apoptosis induced by DHS and PHS is not a general response to antifungal activity. Rather, DHS and PHS play a direct and specific signaling role in bringing about apoptosis. This proposition further explains why the antifungal activity of DHS and PHS shows such high structural and stereochemical specificity.
FIG. 4.
Specificity of PHS in inducing DNA condensation. Spores of a wild-type strain were germinated at 32°C for 6 h before treatment with PHS and several potent antifungal compounds: amphotericin B (AMB), itraconazole (ITCZ), and AbA. DNA condensation was determined by DAPI staining at the indicated times following compound treatment.
DNA fragmentation but not DNA condensation requires protein synthesis.
Although many key regulators of apoptosis, such as caspases, are activated through posttranslational processing, apoptosis in many cell types requires protein synthesis (16). Recently, it was shown that in S. cerevisiae, apoptosis-like cell death induced by oxygen stress could be prevented by the protein synthesis inhibitor cycloheximide; consequently, cycloheximide treatment was shown to significantly increase the rate of survival of yeast cells under oxygen stress (41). To explore whether apoptosis induced by PHS in A. nidulans also requires protein synthesis, we analyzed the consequences of pretreating cells with cycloheximide to inhibit protein synthesis for the subsequent induction of DNA condensation and DNA fragmentation by PHS.
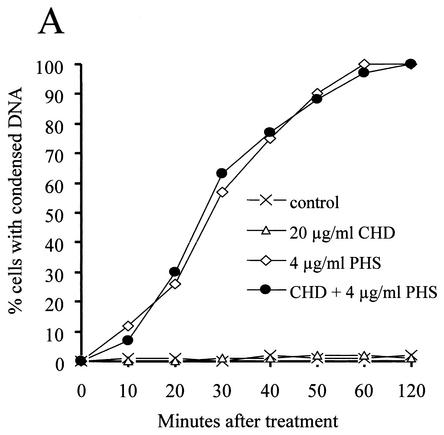
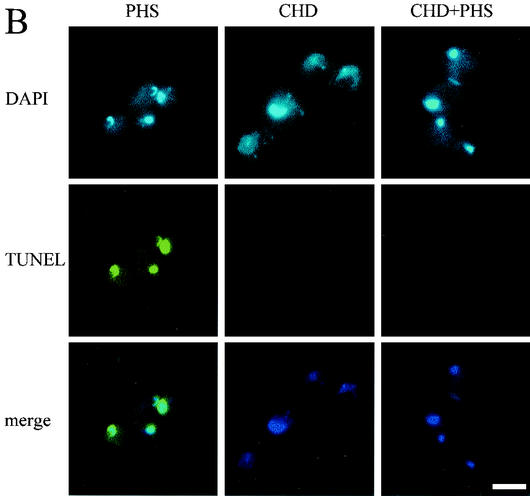
Again, we carried out the experiment with G2-arrested cells and with the nimT23 temperature-sensitive mutation as described for the experiments shown in Fig. 2 and 3. To find an appropriate concentration of cycloheximide for this experiment, we first determined that cycloheximide has an MIC against A. nidulans of 10 μg/ml. We actually used 20 μg of cycloheximide/ml in this experiment, because the addition of cycloheximide at this concentration to G2-arrested cells effectively inhibited further growth. As shown in Fig. 5, the addition of cycloheximide alone to G2-arrested cells had no effect on nuclear morphology. However, the addition of PHS rapidly induced DNA condensation in cells with or without cycloheximide pretreatment with identical kinetics (Fig. 5). Interestingly, pretreatment of cells with cycloheximide completely prevented DNA fragmentation (Fig. 5B). The data thus demonstrate that DNA fragmentation but not DNA condensation requires protein synthesis. These results help explain why DNA condensation occurs much more rapidly than DNA fragmentation upon PHS treatment. The data further show that although DNA condensation and DNA fragmentation in apoptosis normally occur close together, they are in fact separate events.
FIG. 5.
Effect of inhibiting protein synthesis on DNA condensation and DNA fragmentation induced by PHS. (A) Time course showing rapid DNA condensation induced by PHS in either the presence or the absence of the protein synthesis inhibitor cycloheximide. After germination at 42°C for 5.5 h, nimT23 mutant cells were pretreated with 20 μg of cycloheximide (CHD)/ml for 30 min before PHS addition. At various times after PHS treatment, cells were fixed and stained with DAPI to visualize DNA. (B) Pretreatment of cells with cycloheximide prevents DNA fragmentation induced by PHS. nimT23 mutant spores were germinated and treated with cycloheximide and PHS as described for panel A, except that PHS treatment lasted for 3 h. Cells were then fixed and stained with DAPI and TUNEL. PHS, PHS treatment alone; CHD, cycloheximide treatment alone; CHD+PHS, pretreatment with cycloheximide followed by treatment with PHS. Bar, 5 μm.
The highly conserved fungal metacaspase is not required for PHS-induced apoptosis.
The activation of caspases plays a central role in the execution of apoptosis (5, 62). However, as revealed by genome sequencing, fungi have no canonical caspases, except for one highly conserved fungal cysteine protease distantly related to caspases. Molecular phylogenic analysis suggests that this fungal cysteine protease may be at the evolutionary root of caspases (68). Recently, metacaspase YCA1 of S. cerevisiae was indeed shown to have a caspase-like enzymatic activity (42). We cloned the metacaspase gene, designated casA, from A. nidulans based on sequence homology to YCA1 encoded by YOR197w. Preliminary experiments showed that the overexpression of casA inhibited A. nidulans growth and caused morphological changes consistent with apoptosis (J. Cheng and X. S. Ye, unpublished data). To address the question of whether this metacaspase plays a role in PHS-induced apoptosis, we generated a casAΔ strain and then analyzed the effect of casAΔ on apoptosis induced by PHS treatment. The casA gene is not essential, as the casAΔ strain grows and sporulates equally as well as the parental strain. PHS treatment of casAΔ cells induced rapid DNA condensation and subsequent DNA fragmentation just as in wild-type cells with similar kinetics (Fig. 6), indicating that casA does not play a role in apoptosis induced by PHS.
A role for mitochondria but not ROS in PHS-induced apoptosis.
The production of ROS is closely associated with many forms of apoptosis (60). Furthermore, in plant and yeast cells, ROS are shown to be necessary and sufficient to induce certain forms of apoptosis (2, 41). We thus examined whether PHS treatment also produces ROS in A. nidulans.
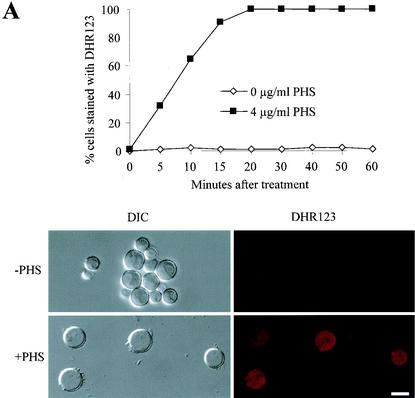
To assay ROS production, we preloaded cells with DHR123 before PHS treatment. In the presence of ROS, DHR123 is oxidized to rhodamine, which can then be visualized under a fluorescence microscope. As shown in Fig. 7A, actively growing A. nidulans cells produced very little ROS, as these cells showed very weak fluorescence of rhodamine. In contrast, PHS-treated cells rapidly showed intense rhodamine fluorescence (Fig. 7A), indicating a rapid generation of ROS. Indeed, kinetic analyses of ROS production and DNA condensation indicated that ROS generation preceded DNA condensation.
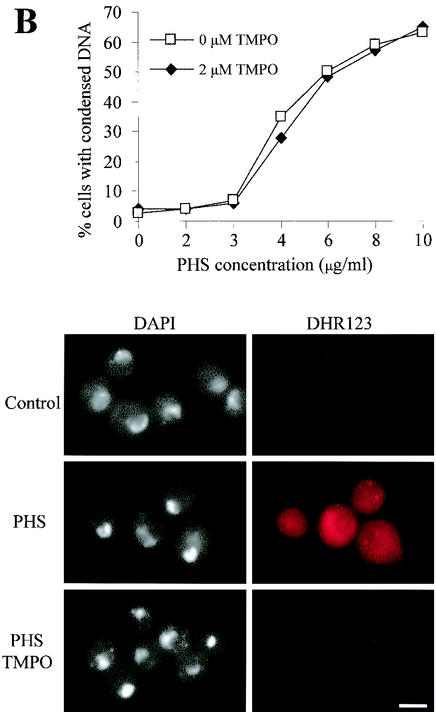
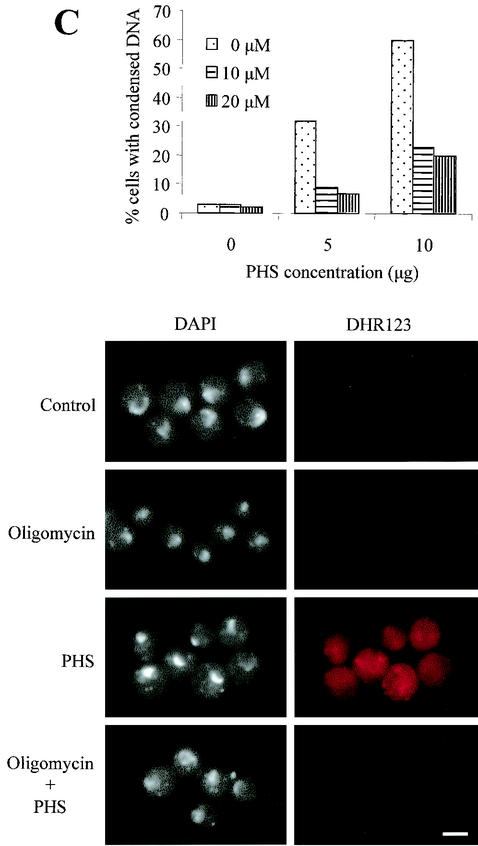
FIG. 7.
Requirement for mitochondrial function in apoptosis induced by PHS. (A) Rapid production of ROS in nimT23 G2-arrested cells after PHS treatment. nimT23 mutant spores were germinated at 42°C for 4 h and then loaded with DHR123 (5 μg/ml) for 2 h before PHS treatment. Shown in the lower panel are cells stained with DHR123 in the presence or absence of PHS for 20 min. DIC, differential interference contrast microscopy. (B) Scavenging of ROS by TMPO has no effect on DNA condensation induced by PHS. nimT23 mutant spores were germinated in the absence or presence of 2 μM TMPO at 42°C for 4 h and then loaded with DHR123 as described for panel A before the addition of various concentrations of PHS for 30 min. Cells were fixed and stained with DAPI to visualize DNA. Micrographs of corresponding cells show DAPI and DHR123 staining. (C) Inhibition of mitochondrial function by oligomycin prevents DNA condensation induced by PHS. nimT23 mutant spores were germinated for 4 h at 42°C in the presence or absence of 10 or 20 μM oligomycin and then loaded with DHR123 for 2 h before PHS treatment as described for panel B. Cells were fixed and stained with DAPI. Micrographs of corresponding cells show DAPI and DHR123 staining. Bars, 5 μm.
To further determine whether ROS are required for PHS-induced apoptosis, we analyzed the effect of scavenging of ROS by the free radical spin trap TMPO on DNA condensation. Cells were first loaded with TMPO for 4 h and then treated with various concentrations of PHS for 30 min. As expected, the number of cells with condensed DNA increased as a function of PHS concentration (Fig. 7B). However, pretreatment with TMPO to scavenge ROS had no effect on DNA condensation induced by PHS (Fig. 7B). We also included DHR123 to demonstrate the effectiveness of TMPO in scavenging of ROS. The lack of rhodamine fluorescence upon PHS treatment in cells pretreated with TMPO indicated that TMPO, as expected, very effectively scavenged ROS (Fig. 7B). The results thus demonstrate that the production of ROS is not required for PHS-induced apoptosis.
Mitochondria play essential roles in both caspase-dependent apoptosis and caspase-independent apoptosis (1, 29). It is also known that the mitochondrial F0F1-ATPase proton pump is required for Bax (a proapoptotic protein)-induced cell death in both S. cerevisiae and mammalian cells (46). To explore a potential role of mitochondria in PHS-induced apoptosis, we inhibited the normal mitochondrial function of nimT23 mutant cells arrested in G2 at 42°C as described above with oligomycin, a specific inhibitor of mitochondrial F0F1-ATPase. Pretreatment with oligomycin markedly inhibited DNA condensation caused by subsequent treatment with PHS (Fig. 7C). Oligomycin also prevented ROS generation, as indicated by a lack of DHR123 oxidation (Fig. 7C). Oligomycin did not kill A. nidulans cells during the duration of this experiment, as the cells were able to rapidly enter synchronous mitosis in the presence of oligomycin upon release from the G2 block at a permissive temperature of 32°C (data not shown). Together, the results show that mitochondria play an essential role in PHS-induced apoptosis.
DISCUSSION
Metabolites derived from the sphingolipid metabolic pathway are highly bioactive, and a large body of evidence now suggests that they play important roles in signaling the stress response and apoptosis. However, the molecular mechanisms through which sphingolipids regulate apoptosis are still not well understood and remain highly controversial (17, 19, 33, 67). Recently, Cheng et al. showed that sphingolipid biosynthesis plays an important role in cell cycle progression through G1 in A. nidulans and that the accumulation of cellular ceramide causes G1 arrest (6). In this study, we characterized the cellular activities of the sphingoid long-chain bases DHS and PHS in A. nidulans. Both DHS and PHS were found to be potent inducers of apoptosis in A. nidulans. Furthermore, apoptosis induced by DHS and PHS in A. nidulans has all the major morphological hallmarks characteristic of caspase-independent apoptosis in mammalian cells.
Apoptosis is a type of genetically and physiologically controlled cell death and is associated with a series of stereotypical morphological changes at the cellular level. These morphological changes include loss of plasma membrane asymmetry, resulting in PS externalization, DNA condensation, and DNA fragmentation. Indeed, DHS and PHS rapidly induce all of these major morphological changes that define apoptotic cell death in A. nidulans. In addition to these morphological changes that are characteristic of apoptosis, the contention that DHS and PHS induce bona fide apoptosis in A. nidulans is further supported by the following observations. DHS and PHS are highly potent fungicidal molecules, suggesting that they cause irreversible cell death. In fact, exposure of A. nidulans cells to DHS and PHS for as little as a few minutes was enough to initiate the irreversible process of cell death. This result indicates that DHS and PHS act as triggers. Once the cell death process is initiated, their presence is no longer required. These findings imply that A. nidulans cells actively participate in the dying process. Indeed, we showed that DNA fragmentation induced by DHS and PHS requires protein synthesis, as pretreatment with cycloheximide, a specific inhibitor of protein synthesis, completely abolished TUNEL staining, thus providing direct evidence for DNA fragmentation as an inducible and active process. Furthermore, we showed that mitochondria play a central role in DHS- and PHS-induced apoptosis. On the basis of these data, together with the demonstration of large-scale DNA fragmentation by PFGE and no requirement for metacaspase, we propose that DHS and PHS induce caspase-independent apoptosis in A. nidulans via a molecular mechanism likely very similar to that of the evolutionarily conserved caspase-independent apoptosis observed in mammalian systems and D. discoideum (3, 29, 38, 64). It is worthwhile to note that caspase-independent apoptosis is often induced by stress factors, such as ceramide. As a role for sphingolipid metabolism in the stress response has been well established, it is thus conceivable that sphingolipids may play an important, evolutionarily conserved role in caspase-independent apoptosis. This hypothesis is consistent with recent reports that showed the induction of apoptosis by sphingosine and PHS in animal cells (10, 36). Indeed, a role for sphingosine has been implicated in mitochondrion-dependent Fas-induced apoptosis (9).
One of the most dramatic and rapid cellular changes occurring in response to DHS and PHS treatment is DNA condensation. DNA becomes highly condensed within minutes after the addition of these sphingoid bases. To determine whether this rapid DNA condensation is cell cycle dependent, we used temperature-sensitive cell cycle mutants to inactivate mitosis-promoting p34cdc2/cyclin B and NIMA kinases and to cause cell cycle arrest in G2 before the addition of DHS and PHS. The results showed that DNA condensation induced by DHS and PHS occurs independent of mitosis-promoting kinases. Furthermore, we showed that this DNA condensation also does not require histone H3 phosphorylation and condensin activity, as does normal mitotic DNA condensation. Together, these results demonstrate that DNA condensation induced by DHS and PHS is independent of mitosis. As DNA condensation independent of mitosis is a hallmark of apoptosis in animals, the results further support the conclusion that DHS and PHS induce apoptosis in A. nidulans. In animal cells, apoptotic DNA condensation has been shown to be dependent on the cleavage of nuclear lamins A and B by caspases, as the expression of noncleavable forms of lamins prevents DNA condensation (50, 56, 57). However, as revealed by genome sequencing, fungi and plants lack homologs of nuclear lamins. At present, how DHS and PHS induce such rapid and dramatic DNA condensation in A. nidulans in the absence of lamins is not understood. Furthermore, how the nuclear structure is normally organized in fungi and plants is also not known, although their nuclei undergo dramatic morphological changes during the cell division cycle, just as animal cell nuclei do. We believe that further elucidation of how DHS and PHS induce DNA condensation in genetically tractable A. nidulans will provide insights into the organization of the nucleus in fungi and perhaps also in plants.
PHS is the predominant sphingoid base in S. cerevisiae. Recent studies with this yeast species showed that PHS is an important signaling molecule and has key signaling functions in the heat stress response, endocytosis, and regulation of the actin cytoskeleton. Although several studies have shown that PHS potently inhibits S. cerevisiae growth, it does so primarily through inhibition of nutrient uptake (7, 8, 59). However, PHS appears not to inhibit nutrient uptake in A. nidulans because, unlike those of S. cerevisiae, A. nidulans auxotrophic strains are not more sensitive than the wild type to treatment with PHS or growth in complex rich medium. Instead, PHS rapidly induces apoptosis in A. nidulans.
Why does PHS not induce apoptosis in S. cerevisiae, as in A. nidulans, as shown in this study? We offer one plausible explanation. As discussed above, PHS induces caspase-independent apoptosis in A. nidulans. It has been shown in mammalian systems that a key regulator of caspase-independent apoptosis is the apoptosis-inducing factor (AIF), a mitochondrial intermembrane protein (38). AIF is strongly conserved in nearly all eukaryotic and prokaryotic species, including Aspergillus and S. pombe, but with an ostensible exception for S. cerevisiae, which does not have a homolog of AIF. AIF is normally confined to mitochondria but can be rapidly translocated to the nucleus upon the induction of apoptosis by stress agents, such as ceramide and staurosporine (38). The mitochondrial-nuclear translocation of AIF is shown to coincide with the initial phase of chromatin condensation and large-scale DNA fragmentation via a mechanism independent of caspase activation (29, 64). In addition, AIF also acts on mitochondria and causes the disruption of transmembrane potential. It is thus conceivable that PHS induces caspase-independent apoptosis in A. nidulans by promoting AIF nuclear translocation from mitochondria. This notion may also explain why mitochondria play an essential role in PHS-induced apoptosis in A. nidulans.
Recently, apoptosis-like cell death in S. cerevisiae was reported by several research groups, and a common feature of the apoptosis-like cell death in S. cerevisiae was the accumulation of ROS (12, 35, 37, 39, 40, 41, 42). Indeed, the apoptosis-like cell death induced by a Cdc48 temperature-sensitive mutation or low levels of H2O2 stress can be largely prevented by free radical-scavenging compounds, such as TMPO, or by hypoxia (41), indicating that ROS are essential for yeast apoptosis. Here we show that the accumulation of ROS is also associated with apoptosis induced by DHS and PHS in A. nidulans. However, we found that ROS are only associated with and not required for apoptosis induced by DHS and PHS, as scavenging of free radicals by TMPO had no observable effect on DNA condensation (Fig. 7A and B). ROS production has also been found to be associated with mammalian cell apoptosis (66). However, mammalian cell apoptosis still occurs in the near absence of oxygen (26), suggesting that ROS may also not be essential for certain forms of apoptosis of mammalian cells. As apoptosis-like cell death in S. cerevisiae appears to be mediated through the metacaspase YCA1 (41), ROS may be necessary for the activation of metacaspase to promote apoptosis. Indeed, H2O2 treatment induces apoptosis accompanied by a caspase-like enzymatic activity requiring YCA1 (41). It has also been found that a metacaspase-dependent apoptosis pathway also operates in A. nidulans, as the overexpression of casA, an A. nidulans homolog of the YCA1 gene, induces apoptosis-like cell death (Cheng and Ye, unpublished). However, the fact that the PHS-induced apoptosis that we describe in this work is independent of metacaspase activity helps explain why, although ROS are induced by PHS treatment, ROS are not required to mediate the apoptosis-like events that we have documented in response to PHS.
In summary, our present work explored the biological activities of exogenous sphingoid bases in A. nidulans and made the interesting discovery that DHS and PHS are potent and specific inducers of apoptosis. The ability of DHS and PHS to induce apoptosis is structurally and stereochemically specific. In fact, only the naturally occurring forms of DHS and PHS are able to induce apoptosis and do so without the involvement of ceramide. Moreover, apoptosis is a specific cellular response to DHS and PHS actions, as several different chemical classes of antifungal compounds similarly tested failed to induce apoptosis. Such high structural and stereochemical specificity suggests that DHS and PHS must act on very specific molecular targets in A. nidulans to bring about apoptosis. Thus, identification of the molecular targets of DHS and PHS in future work will provide new insights into sphingolipid signaling mechanisms and caspase-independent apoptosis.
Acknowledgments
We thank Sheng-bin Peng for critically reading the manuscript and Margaret Shaw for technical assistance in MIC assays. We also thank members of the laboratory of X. S. Ye and Doreen Ma for valuable discussions during the course of this work.
Jijun Cheng and Tae-Sik Park were supported by postdoctoral fellowships from Lilly Research Laboratories.
REFERENCES
- 1.Adrain, C., and S. J. Martin. 2001. The mitochondrial apoptosome: a killer unleashed by the cytochrome seas. Trends Biochem. Sci. 26:390-397. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, M. E., R. I. Pennell, P. J. Meijer, A. Ishikawa, R. A. Dixon, and C. Lamb. 1998. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92:773-784. [DOI] [PubMed] [Google Scholar]
- 3.Arnult, D., I. Tatischeff, J. Estaquier, M. Girard, F. Sureau, J. P. Tissier, A. Grodet, M. Dellinger, F. Traincard, A. Kahn, J. C. Ameisen, and P. X. Petit. 2001. On the evolutionary conservation of the cell death pathway: mitochondrial release of an apoptosis-inducing factor during Dictystelium discoideum cell death. Mol. Biol. Cell 12:3016-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokoch, G. M., A. M. Reilly, R. H. Daniels, C. C. King, A. Olivera, S. Spiegel, and U. G. Knaus. 1998. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J. Biol. Chem. 273:8137-8144. [DOI] [PubMed] [Google Scholar]
- 5.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J. J., T. S. Park, A. S. Fischl, and X. S. Ye. 2001. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21:6198-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, N., G. Jenkins, Y. A. Hannun, J. Heitman, and L. M. Obeid. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275:17229-17232. [DOI] [PubMed] [Google Scholar]
- 8.Chung, N., C. Mao, J. Heitman, Y. A. Hannun, and L. M. Obeid. 2001. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 276:35614-35621. [DOI] [PubMed] [Google Scholar]
- 9.Cuvillier, O., L. Edsall, and S. Spiegel. 2000. Involvement of sphingosine in mitochondria-dependent Fas-induced apoptosis of type II Jurkat T cells. J. Biol. Chem. 275:15691-15700. [DOI] [PubMed] [Google Scholar]
- 10.Dalla Libera, L., R. Sabbadini, C. Renken, B. Ravara, M. Sandri, R. Betto, A. Angelini, and G. Vescovo. 2001. Apoptosis in the skeletal muscle of rats with heart failure is associated with increased serum levels of TNF-alpha and sphingosine. J. Mol. Cell Cardiol. 33:1871-1878. [DOI] [PubMed] [Google Scholar]
- 11.deHart, A. K. A., J. D. Schnell, D. A. Allen, and L. Hicke. 2002. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Carratore, R., C. Della Croce, M. Simili, E. Taccini, M. Scavuzzo, and S. Sbrana. 2002. Cell cycle and morphological alterations as indicative of apoptosis promoted by UV irradiation in S. cerevisiae. Mutat. Res. 513:183-191. [DOI] [PubMed] [Google Scholar]
- 13.Denning, D. W., K. V. Clemons, L. A. Hanson, and D. A. Stevens. 1990. Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. J. Infect. Dis. 162:1151-1158. [DOI] [PubMed] [Google Scholar]
- 14.De Souza, C. P., A. H. Osmani, L. P. Wu, J. L. Spotts, and S. A. Osmani. 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102:293-302. [DOI] [PubMed] [Google Scholar]
- 15.Dickson, R. C. 1998. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu. Rev. Biochem. 67:27-48. [DOI] [PubMed] [Google Scholar]
- 16.Gobe, G. C., B. Harmon, J. Leighton, and D. J. Allan. 1999. Radiation-induced apoptosis and gene expression in neonatal kidney and testis with and without protein synthesis inhibition. Int. J. Radiat. Biol. 75:973-983. [DOI] [PubMed] [Google Scholar]
- 17.Green, D. R. 2000. Apoptosis and sphingomyelin hydrolysis: the flip side. J. Cell Biol. 150:F5-F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannun, Y. A., C. R. Loomis, A. H. Merrill, and R. M. Bell. 1986. Sphingosine inhibition of protein kinase C activity and of phorbol dibutyrate binding in vitro and in human platelets. J. Biol. Chem. 261:12604-12609. [PubMed] [Google Scholar]
- 19.Hannun, Y. A., and C. Luberto. 2000. Ceramide in the eukaryotic stress response. Trends Cell Biol. 10:73-80. [DOI] [PubMed] [Google Scholar]
- 20.Hannun, Y. A., C. Luberto, and K. M. Argraves. 2001. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry 40:4893-4903. [DOI] [PubMed] [Google Scholar]
- 21.Hanson, B. A., and R. L. Lester. 1980. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J. Lipid Res. 21:309-315. [PubMed] [Google Scholar]
- 22.Henson, P. M., D. L. Bratton, and V. A. Fadok. 2001. The phosphatidylserine receptor: crucial molecular switch? Nat. Rev. Mol. Cell Biol. 2:627-633. [DOI] [PubMed] [Google Scholar]
- 23.Hirano, T. 2000. Chromosome cohesion, condensation and separation. Annu. Rev. Biochem. 69:115-144. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, P. R., A. M. deCathelineau, C. A. Ogden, Y. Leverrier, D. L. Bratton, D. L. Daleke, A. J. Ridley, V. A. Fadok, and P. M. Henson. 2001. Phosphatidylserine (PS) induces PS receptor-mediated macropinocytosis and promotes clearance of apoptotic cells. J. Cell Biol. 155:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson, M. D., and M. C. Raff. 1995. Programmed cell death and Bcl-2 protection in very low oxygen. Nature 374:814-816. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins, G. M., and Y. A. Hannun. 2001. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 276:8574-8581. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins, G. M., A. Richards, T. Wahl, C. Mao, L. M. Obeid, and Y. A. Hannun. 1997. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272:32566-32572. [DOI] [PubMed] [Google Scholar]
- 29.Joza, N., S. A. Susin, E. Daugas, W. L. Stanford, S. K. Cho, C. Y. J. Li, T. Sasaki, A. J. Elia, H. Y. M. Cheng, L. Ravagnan, K. F. Ferri, A. Wakeham, R. Hakem, H. Yoshida, Y. Y. Kong, T. W. Mak, J. C. Zuniga-Pflucker, G. Kroemer, and J. M. Penninger. 2001. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410:549-554. [DOI] [PubMed] [Google Scholar]
- 30.Kagedal, K., M. Zhao, I. Svensson, and U. T. Brunk. 2001. Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem. J. 359:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S., H. Fyrst, and J. Saba. 2000. Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156:1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, C. C., F. T. Zenke, P. E. Dawson, E. M. Dutil, A. C. Newton, B. A. Hemmings, and G. M. Bokoch. 2000. Sphingosine is a novel activator of 3-phosphoinositide-dependent kinase 1. J. Biol. Chem. 275:18108-18113. [DOI] [PubMed] [Google Scholar]
- 33.Kolesnick, R. N., and M. Kronke. 1998. Regulation of ceramide production and apoptosis. Annu. Rev. Physiol. 60:645-665. [DOI] [PubMed] [Google Scholar]
- 34.Lanterman, M. M., and J. D. Saba. 1998. Characterization of sphingosine kinase (SK) activity in Saccharomyces cerevisiae and isolation of SK-deficient mutants. Biochem. J. 332:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 36.Lee, J. S., D. S. Min, C. Park, C. S. Park, and N. J. Cho. 2001. Phytosphingosine and C2-phytoceramide induce cell death and inhibit carbachol-stimulated phospholipase D activation in Chinese hamster ovary cells expressing the Caenorhabditis elegans muscarinic acetylcholine receptor. FEBS Lett. 499:82-86. [DOI] [PubMed] [Google Scholar]
- 37.Ligr, M., I. Velten, E. Frohlich, F. Madeo, M. Ledig, K. U. Frohlich, D. H. Wolf, and W. Hilt. 2001. The proteasomal substrate Stm1 participates in apoptosis-like cell death in yeast. Mol. Biol. Cell 12:2422-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenzo, H. K., S. A. Susin, J. Penninger, and G. Kroemer. 1999. Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 6:516-524. [DOI] [PubMed] [Google Scholar]
- 39.Ludovico, P., M. J. Sousa, M. T. Silva, C. Leao, and M. Corte-Real. 2001. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147:2409-2415. [DOI] [PubMed] [Google Scholar]
- 40.Madeo, F., E. Frohlich, and K. U. Frohlich. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. Sigrist, D. H. Wolf, and K. U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 43.Mandala, S. M., R. Thornton, Z. Tu, M. B. Kurtz, J. Nickels, J. Broach, R. Menzeleev, and S. Spiegel. 1998. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl. Acad. Sci. USA 95:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao, C., J. Saba, and L. M. Obeid. 1999. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 342:667-675. [PMC free article] [PubMed] [Google Scholar]
- 45.Mao, C., M. Wadleigh, G. M. Jenkins, Y. A. Hannun, and L. M. Obeid. 1997. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 272:28690-28694. [DOI] [PubMed] [Google Scholar]
- 46.Matsuyama, S., O. Xu, J. Velours, and J. C. Reed. 1998. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol. Cell 1:327-336. [DOI] [PubMed] [Google Scholar]
- 47.McNabb, T. J., A. E. Cremesti, P. R. Brown, and A. S. Fischl. 1999. The separation and direct detection of ceramides and sphingoid bases by normal-phase high-performance liquid chromatography and evaporative light-scattering detection. Anal. Biochem. 276:242-250. [DOI] [PubMed] [Google Scholar]
- 48.Morris, N. R. 1975. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26:237-254. [DOI] [PubMed] [Google Scholar]
- 49.Oakley, B. R., and S. A. Osmani. 1993. Cell cycle analysis using the filamentous fungus Aspergillus nidulans, p. 127-142. In P. Fantes and R. Brooks (ed.), The cell cycle, a practical approach. IRL Press, Oxford, United Kingdom.
- 50.Oberhammer, F. A., K. Hochegger, G. Froschl, R. Tiefenacher, and M. Pavelka. 1994. Chromatin condensation during apoptosis is accompanied by degradation of lamin A+B, without enhanced activation of cdc2 kinase. J. Cell Biol. 126:827-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh, C. S., D. A. Toke, S. Mandala, and C. E. Martin. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272:17376-17384. [DOI] [PubMed] [Google Scholar]
- 52.Olivera, A., and S. Spiegel. 1993. Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature 365:557-560. [DOI] [PubMed] [Google Scholar]
- 53.Osmani, S. A., R. T. Pu, and N. R. Morris. 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53:237-244. [DOI] [PubMed] [Google Scholar]
- 54.Osmani, S. A., and X. S. Ye. 1996. Cell cycle regulation in Aspergillus by two protein kinases. Biochem. J. 317:633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patton, J. L., B. Srinivasan, R. C. Dickson, and R. L. Lester. 1992. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J. Bacteriol. 174:7180-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao, L., D. Perez, and E. White. 1996. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 135:1441-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruchaud, S., N. Korfali, P. Villa, T. J. Kottke, C. Dingwall, S. H. Kaufmann, and W. C. Earnshaw. 2002. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 21:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saka, Y., T. Sutani., Y. Yamashita, S. Saito, M. Takeuchi, Y. Nakaseko, and M. Yanagida. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromsome condensation and segregation in mitosis. EMBO J. 13:4936-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skrzypek, M. S., M. M. Nagiec, R. L. Lester, and R. C. Dickson. 1998. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J. Biol. Chem. 273:2829-2834. [DOI] [PubMed] [Google Scholar]
- 60.Slater, A. F. G., C. Stefan, I. Nobel. D. J. van den Dobbelsteen, and S. Orrenius. 1995. Signaling mechanisms and oxidative stress in apoptosis. Toxicol. Lett. 82/83:149-153. [DOI] [PubMed] [Google Scholar]
- 61.Spiegel, S., and S. Milstien. 2000. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 476:55-57. [DOI] [PubMed] [Google Scholar]
- 62.Strasser, A., L. O'Connor, and V. M. Dixit. 2000. Apoptosis signaling. Annu. Rev. Biochem. 69:217-245. [DOI] [PubMed] [Google Scholar]
- 63.Stunnikov, A. V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromsome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587-599. [DOI] [PubMed] [Google Scholar]
- 64.Susin, S. A., H. K Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, J. Mangion, E. Jacotot, P. Costantini, M. Loeffler, N. Larochette, D. R. Goodlett, R. Aebersold, D. P. Siderovski, J. M. Penninger, and G. Kroemer. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441-446. [DOI] [PubMed] [Google Scholar]
- 65.Sylvie, F., R. Lombardi, T. Schmelzle, M. N. Hall, and H. Riezman. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan, S., Y. Sagara, Y. Liu, P. Maher, and D. Schubert. 1998. The regulation of reactive oxygen species production during programmed cell death. J. Cell Biol. 141:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tepper, A. D., P. Ruurs, T. Wiedmer, P. J. Sims, J. Borst, and van W. J. Blitterswijk. 2000. Sphingomyelin hydrolysis to ceramide during the execution phase of apoptosis results from phospholipids scrambling and alters cell-surface morphology. J. Cell Biol. 150:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uren, A. G., K. O'Rourke, L. Aravind, M. T. Pisabarro, S. Seshagiri, S., E. V. Koonin, and V. M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 5:961-967. [DOI] [PubMed] [Google Scholar]
- 69.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 70.Webb, S. J., D. J. Harrison, and A. H. Wyllie. 1997. Apoptosis: an overview of the process and its relevance in disease. Adv. Pharmacol. 41:1-34. [DOI] [PubMed] [Google Scholar]
- 71.Wells, G. B., R. C. Dickson, and R. L. Lester. 1998. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J. Biol. Chem. 273:7235-7243. [DOI] [PubMed] [Google Scholar]
- 72.Ye, X. S., X. Xu, R. T. Pu, R. R. Fincher, S. L. McGuire, A. H. Osmani, and S. A. Osmani. 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14:986-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye, X. S., L. P. Wu, and S. A. Osmani. 2001. Signal transduction in filamentous fungi: regulation of protein kinases, p. 157-174. In N. Talbot (ed.), Molecular and cellular biology of filamentous fungi: a practical approach. Oxford University Press, Oxford, United Kingdom.