Abstract
Tumor susceptibility gene 101 (Tsg101) was identified in a random mutagenesis screen for potential tumor suppressors in NIH 3T3 cells. Altered transcripts of this gene have been detected in sporadic breast cancers and many other human malignancies. However, the involvement of this gene in neoplastic transformation and tumorigenesis is still elusive. Using gene targeting, we generated genetically engineered mice with a floxed allele of Tsg101. We investigated essential functions of this gene in vivo and examined whether the loss of function of Tsg101 results in tumorigenesis. Conventional knockout mice were generated through Cre-mediated excision of the first coding exon in the germ line of mouse mammary tumor virus (MMTV)-Cre transgenic mice. The complete ablation of Tsg101 in the developing embryo resulted in death around implantation. In contrast, mammary gland-specific knockout mice developed normally but were unable to nurse their young as a result of impaired mammogenesis during late pregnancy. Neither heterozygous null mutants nor somatic knockout mice developed mammary tumors after a latency of 2 years. The Cre-mediated deletion of Tsg101 in primary cells demonstrated that this gene is essential for the growth, proliferation, and survival of mammary epithelial cells. In summary, our results suggest that Tsg101 is required for normal cell function of embryonic and adult tissues but that this gene is not a tumor suppressor for sporadic forms of breast cancer.
The tumor susceptibility gene 101 (Tsg101) was first discovered in murine fibroblasts in a screen for potential tumor suppressors by using retroviral gene trap vectors in combination with an antisense strategy (13). A ligand-inducible or constitutive expression of the partial and full-length Tsg101 cDNA in antisense orientation led to instant transformation of NIH 3T3 cells. The transformed cells were capable of forming metastasizing tumors in nude mice. Neoplastic transformation of the NIH 3T3 cells was reported to be reversible when the function of Tsg101 was restored (13). Cloning and sequencing of the human TSG101 cDNA revealed that the mouse and human genes encode proteins with more than 94% similarity at the amino acid level (14). The human TSG101 gene was mapped to chromosome 11 p15.1-p15.2, a site that shows loss of heterozygozity in a subset of sporadic breast cancers and other human malignancies (14). Although somatic mutations or deletions within the TSG101 gene are rare events, aberrant splice forms have been observed frequently in a variety of human cancers (7, 12, 27, 28). After cloning, sequencing, and revising the TSG101 gene structure in both the human and the mouse, we showed that many of the previously described aberrant transcripts in human malignancies were in fact alternative splice forms generated solely by exon skipping (31, 32).
Various biological functions of Tsg101 have been postulated from the predicted protein structure, the intracellular localization of Tsg101, the identification of Tsg101 binding proteins, and the deletion of the putative homologous gene Vsp23p in yeast. These functions include a role in ubiquitination (10, 23), transcriptional regulation (8, 36), endosomal trafficking (1, 6), and cell proliferation (37, 40). Although some of these functions have been verified experimentally in cell culture systems, their relevance in vivo during normal development and neoplastic transformation is still elusive. In addition, it has never been confirmed, using a site-directed targeting approach, that the deletion of both endogenous Tsg101 alleles leads to accelerated growth and neoplastic transformation of nonimmortalized cells and to tumorigenesis in vivo. Based on our earlier findings that (i) Tsg101 is expressed in all tissues of the mouse throughout development and (ii) the 5′ sequence of this gene has common features of housekeeping gene promoters, we hypothesized that a conventional knockout of this gene would result in early embryonic death (32). Therefore, we have chosen the Cre-loxP technology, which allows the conditional deletion of Tsg101 from any given cell type in genetically engineered mice. In this article, we report that Tsg101 is essential for cell growth, cell survival, and normal function of embryonic and adult tissues. Tsg101-deficient cells exhibited a defect in cell cycle regulation and underwent increased cell death. In contrast to an earlier report (13), a Tsg101 knockout did not result in accelerated cell growth and instant neoplastic transformation. Our findings suggest that a null mutation of Tsg101 is not an initiating event for tumorigenesis.
MATERIALS AND METHODS
Gene targeting.
Overlapping BAC clones encompassing the Tsg101 locus were isolated from a mouse 129SvJ genomic library and mapped (31, 32). A 11.5-kb EcoRI fragment was cloned into pZero (Invitrogen, Inc.), sequenced, and used as a template to generate the floxed targeting vector (see Fig. 1A). Three consecutive fragments (2.9, 3.1, and 3.3 kb) approximately 950 bp downstream of the 5′ EcoRI site were amplified with Pfx polymerase (Gibco BRL) and cloned into pZero. The 2.9-kb 5′ fragment was released by EcoRI and subcloned into pLoxpNeo (39), resulting in an additional EcoRI site 5′ of the PGK-neomycin (PGK-neo) expression cassette, which was used to verify the targeting event by Southern blotting. The 3′ homology region of the targeting vector was generated by subcloning the 3.1- and 3.3-kb PCR fragments into pBS64, thereby inserting a loxP sequence and an additional EcoRI site into intron 1. After inserting an oligonucleotide containing an XhoI recognition site into the 5′ end of the homologous region, the entire 6.4-kb fragment was released from the pBS64 vector by using XhoI and NotI and subcloned into the pLoxpNeo vector containing the 2.9-kb 5′ homology region. The final targeting construct, designated pFloxedTSG101, contained a floxed PGK-neo selection marker approximately 3 kb upstream and a third loxP site 230 bp downstream of the first coding exon. The pFloxedTSG101 plasmid was linearized using NotI and electroporated into isogenic RW4 embryonic stem (ES) cells (GenomeSystems, Inc.). Analysis of G418-resistant clones by Southern blotting using 5′ and 3′ diagnostic probes (Fig. 1) indicated that about 7% of the ES cell clones (10 of 150) had undergone correct homologous recombination both in the 5′ region and at the 3′ end downstream of the third loxP site. Of the 10 correctly targeted ES cell clones, 2 (clones 7 and 36) were expanded and injected into C57BL/C blastocysts. Germ line transmission of the Tsg101 floxed allele was achieved from male chimeras of both clones.
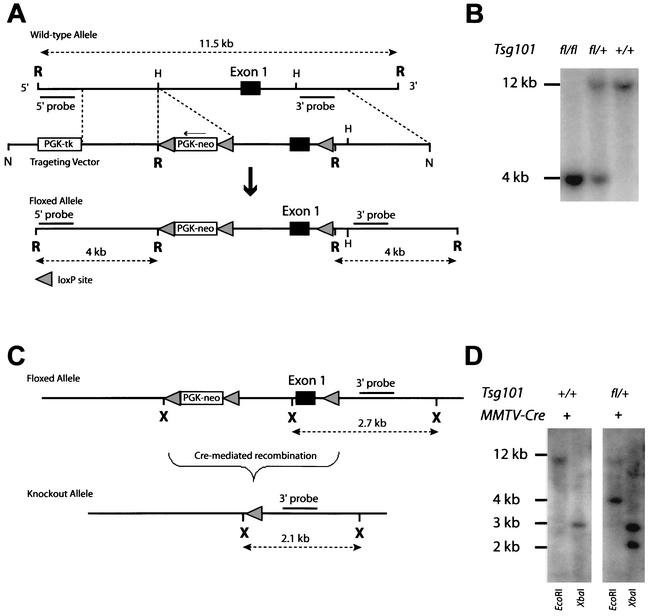
FIG. 1.
Targeted deletion of the Tsg101 gene. (A) Targeting strategy to flank the promoter and first coding exon of Tsg101 with loxP sites. The conditional knockout (floxed) allele contains a selectable marker cassette 3 kb upstream and a third loxP site 230 bp downstream of the first coding exon. (B) Southern blot analysis of genomic DNA of mice carrying either two floxed alleles (fl/fl), one floxed and one wild-type allele (fl/+), or two wild-type alleles (+/+). The 11.5-kb EcoRI restriction fragment represents the wild-type allele, whereas homozygous mutants carry only a 4-kb fragment. (C) Cre-mediated deletion of the Tsg101 gene. (D) Southern blot to verify the correct deletion event illustrated in panel C. Genomic DNA was isolated from tail biopsy specimens from transgenic mice carrying the MMTV-Cre transgene and either two Tsg101 wild-type alleles (Tsg101+/+) or one floxed and one wild-type allele (Tsg101fl/+). The 2.1-kb XbaI fragment represents the recombined Tsg101 null allele that is present only when a floxed allele is subject to Cre-mediated recombination. Both the wild-type and unrecombined floxed allele are represented by a 2.7-kb XbaI restriction fragment. R, EcoRI; H, HindIII; N, NotI; X, XbaI; tk, thymidine kinase.
PCR genotyping protocols.
The PCR protocols for genotyping mouse mammary tumor virus (MMTV)-Cre and WAP-Cre mice have been described previously (34). MMTV-Cre and WAP-Cre mice are available from the Jackson Laboratory (stock 003551, 003553, and 003552) and the National Cancer Institute repository in Frederick, Md. For identification of the various Tsg101 alleles by PCR, we used the following primer sets: wild-type allele (5′-GTT CGC TGA AGT AGA GCA GCC AG-3′ and 5′-CAT TTC TGG AGT CCG ATG CGC AG-3′), floxed allele (5′-AGA GGC TAT TCG GCT ATG ACT G-3′ and 5′-TTC GTC CAG ATC ATC CTG ATC-3′), and recombined/null allele (5′-GAT GGT CAT ACC TGG TTA GAA AGC-3′ and 5′-CAT TTC TGG AGT CCG ATG CGC AG-3′). Note that the primer set for the wild-type allele is specific; i.e., it will not recognize the targeted floxed allele or the null allele. All animals used in the described studies were treated humanely and in accordance with federal guidelines and institutional policies.
Southern and Northern hybridization.
Genomic DNA from tissues or cell lines was prepared using standard phenol-chloroform extraction, and 15 μg was digested with EcoRI or XbaI at 37°C overnight. The DNA was separated on a 0.8% agarose gel, blotted onto a nylon membrane (GeneScreen Plus; NEN), and hybridized with a 32P-labeled probe. The 5′ diagnostic probe, which was used to screen targeted ES cells, was generated by PCR using the primer set 5′-TTG CTT GTA TGA GTG CAG GTG CC-3′ and 5′-TAT GAC CTG CTT CTT GCA AAA GCA G-3′. The 3′ internal probe for identification of the various Tsg101 alleles was amplified by PCR using the forward and reverse primers 5′-ATC TTA TCT CCC ATC CTA AGC AGA C-3′ and 5′-TCT CTC ACA TCA CTA AAG CTC AAT G-3′, respectively. Membranes were washed in 0.1× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing sodium dodecyl sulfate (SDS) and exposed for 6 h to a Kodak X-Omat-AR film.
The isolation of total RNA from mammary tissue has been described previously (32). A 20-μg portion of total RNA was separated on a 1.5% formaldehyde gel and transferred to a GeneScreen Plus membrane. Tsg101 transcripts were detected by probing the membranes with 32P-labeled full-length mouse Tsg101 cDNA.
Whole-mount analysis of the mammary gland and other histological techniques.
The preparation and staining of mammary gland whole mounts and detailed protocols for immunohistochemistry and bromodeoxyuridine (BrdU) labeling of proliferating mammary epithelial cells were described previously (35). For immunohistochemistry, mammary gland tissue was fixed for 5 h in 4% (vol/vol) paraformaldehyde or overnight at 4°C in 10% buffered formalin (Fisher Scientific Co.). The specimens were embedded in paraffin by standard methods. Sections were deparaffinized, rehydrated, and treated with antimasking solution from Vector Laboratories, Inc., as specified by the manufacturer. We used a 1:10 dilution of the M30 CytoDEATH stock solution (see detailed protocol by Roche Molecular Biochemicals) to label apoptotic mammary epithelial cells. The Vectastain Elite ABC and peroxidase substrate kit from Vector Laboratories, Inc., was used to visualize labeled cells. The slides were counterstained with hematoxylin. Immunohistochemical detection of BrdU-labeled nuclei was performed by following the Amersham-Pharmacia-Biotech (RPN202) protocol.
Construction of retroviral expression vectors.
The Cre coding sequence from vector pBS185 (a kind gift of B. Sauer, National Institute of Diabetes, Digestive, and Kidney Disease [NIDDK], to L. Hennighausen, NIDDK) was cloned as an XhoI-MluI(blunt) fragment into the EcoRV-XhoI sites of pZero (Invitrogen, Inc.). The pBabe-Cre-puro retroviral vector was constructed by subcloning the Cre recombinase cDNA as an XhoI(blunt)-EcoRI fragment into the BamHI(blunt) and EcoRI sites of pBabe-puro. We have used 293T cells to generate replication-deficient viral particles of pBabe and its Cre-expressing derivative (pBabe-Cre).
Cell culture and immunocytochemistry.
Primary mammary epithelial cell cultures were prepared similarly to methods described previously (16, 20). The epithelial cells were maintained in Dulbecco’s minimal essential medium (DMEM)/F12 supplemented with 2% fetal calf serum, 10 μg of insulin per ml, 10 ng of epidermal growth factor per ml, 10 μg of gentamicin per ml, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. For retroviral infection, cells at passages 2 and 3 were plated at a density of 3 × 105 to 4 × 105 cells per 10-cm culture dish. Infection with pBabe-Cre or control constructs lacking the Cre coding sequence (pBabe) was performed in the presence of 10 μg of polybrene (Sigma) per ml. Forty-eight hours later, the cells were selected in complete medium containing 3 to 7 μg of puromycin (Sigma) per ml.
For immunocytochemistry, cells were fixed in 70% ethanol, washed twice in 1× phosphate-buffered saline (PBS) and incubated for 20 min in blocking solution (1× PBS, 1% bovine serum albumin, 0.1% Tween 20). The slides were treated for several hours with a 1:500 to 1:1,000 dilution of the primary antibody in blocking solution (anti-proliferating cell nuclear antigen [PCNA] [PC-10], anti-cyclin A [C-19] [both from Santa Cruz], or anti-pan-cytokeratin [Sigma]). The primary PCNA and anti-pan-cytokeratin antibodies were fluorescein isothiocyanate (FITC)-conjugated. After an additional washing step, cyclin A was visualized with an FITC-conjugated secondary antibody (sc-2012 [Santa Cruz]; 1:1,000 dilution). The slides were washed repeatedly in 1× PBS and mounted with Vectashield containing 1.5 μg of 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc.).
Western blot analysis.
Cell pellets were lysed on wet ice for 30 min in 1× PBS-1 % NP-40-0.5% sodium deoxycholate-0.1% SDS, 1 mM phenylmethylsulfonyl fluoride-0.4 U of aprotinin per ml-1 mM NaF-0.1 mM sodium orthovandate. Protein was quantified using a Bradford assay (Pierce). Protein (20 to 50 μg per lane) was resolved by SDS-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked for 1 h in 1× TBS-0.1% Tween 20-5% dry milk. They were then incubated with primary antibodies in blocking buffer at 4°C overnight, washed three times for 15 min in washing buffer (1× TBS-0.1% Tween 20), and incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies in blocking buffer. The membranes were washed again three times in washing buffer and once for 15 min in 1× TBS without Tween 20. Protein bands were detected using the ECL chemiluminescence kit for Western blot analysis (Amersham) as specified by the manufacturer. Membranes were stripped using 0.2 M NaOH for consecutive detection of various proteins. The anti-ActB (I-19) and anti-Tsg101 (C-2) antibodies from Santa Cruz Biotechnology were used at a 1:1,000 dilution. Horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology and used at a 1:1,000 dilution.
RESULTS
Generation of genetically engineered mice with a conditional knockout allele of Tsg101.
A conditional knockout (floxed) allele of the Tsg101 gene was generated through homologous recombination in mouse ES cells (Fig. 1A). The PGK-neo selectable marker was placed 3 kb upstream of exon 1 to avoid interference with transcriptional regulation of the Tsg101 locus. A third loxP site was introduced into the first intron immediately following exon 1. Homozygous mutant mice that carry two floxed Tsg101 alleles (Fig. 1B) developed normally until adulthood. Both males and females were fertile, and they exhibited no obvious phenotypic abnormalities. Therefore, the insertion of the selectable marker upstream of Tsg101 had no effect on the transcriptional regulation of Tsg101 and the floxed allele was phenotypically indistinguishable from its wild-type counterpart. Excision of the promoter and the first coding exon was achieved through Cre-mediated recombination (Fig. 1C). The presence of the recombined (null) Tsg101 allele was verified by Southern blotting using XbaI restriction digestion of genomic DNA and an internal genomic probe that recognized all alleles (the wild-type, floxed, and knockout alleles) simultaneously. To verify the accuracy of the predicted recombination event, the Tsg101 floxed mice were crossed with MMTV-Cre transgenic animals that exhibit high expression of Cre recombinase in many organs including the female germ line (33, 34). In cells expressing Cre recombinase, the floxed allele in MMTV-Cre Tsg101fl/+ mice was converted into a null allele with the expected 2.1-kb length of the XbaI restriction fragment, demonstrating that the loxP sites were functional and the knockout strategy was technically successful (Fig. 1D).
A complete knockout of Tsg101 is deleterious at a very early stage during embryonic development.
To test the hypothesis that Tsg101 is indispensable for embryonic development (32), we generated mice with a complete null mutation of Tsg101 by converting the floxed gene into a complete null allele in the female germ line of MMTV-Cre Tsg101fl/+ mice. Line A, which is one of several MMTV-Cre transgenic strains described previously (33, 34), exhibits Cre expression in developing oocytes. This feature makes this particular line useful to generate a complete null allele from any floxed locus. MMTV-Cre (line A) Tsg101fl/+ females were mated with wild-type males to generate Tsg101 heterozygous null mice (Tsg101+/−) and to segregate out the MMTV-Cre transgene. Tsg101+/− mice of both genders were selected from the resulting offspring. These heterozygous knockouts were maintained as a separate population and crossed among each other to generate Tsg101 homozygous null mutants (Tsg101−/−). Initially, 50 mice from six independent litters of heterozygous knockout intercrosses were genotyped. We were unable to identify a single Tsg101 knockout mouse. Surviving pups were either Tsg101+/+ (18 of 50) or Tsg101+/− (32 of 50). These results represented the Mendelian ratio of mutant and wild-type alleles when Tsg101 deficiency results in embryonic death. This observation further suggested that the wild-type Tsg101 locus was not imprinted.
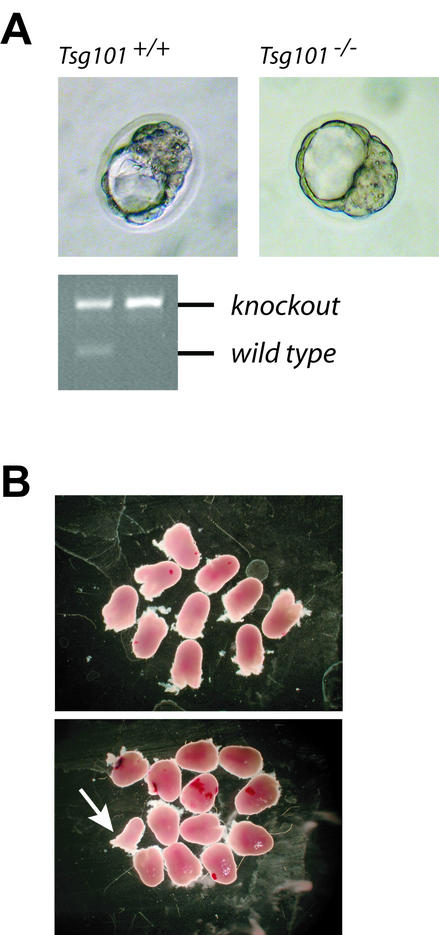
Determination of the time point of embryonic death is important for the design of rescue experiments where viable tissues from a knockout can be transplanted into wild-type recipients to study the loss of function of important tumor suppressor genes in adult animals. To answer this question, we isolated and genotyped embryos at various time points (days E3.5, E6.5, and E12.5) from Tsg101 heterozygous intercrosses. We were able to identify Tsg101−/− blastocysts on day E3.5 which were indistinguishable in their overall morphology from wild-type littermates (Fig. 2A). However, we failed to clearly identify Tsg101 homozygous mutants after implantation. We isolated deciduae on day E6.5, and all embryos, even those few with an abnormal appearance (Fig. 2B, arrow), contained embryos that were identified as wild type or heterozygous mutants. Therefore, death in our animal model occurs at an earlier time point, most probably around implantation or very shortly thereafter. In summary, this experiment confirmed our hypothesis that Tsg101 is crucial for early embryonic development. Since the generation of Tsg101-deficient ES cells and the cultivation of the inner cell mass (ICM) of Tsg101 knockout blastocysts were unsuccessful (see Discussion), it is virtually impossible to rescue viable tissues from conventional Tsg101 knockout embryos and to generate somatic mutants through chimera production or transplantation of cells into adult recipients.
FIG. 2.
Examination of Tsg101-deficient embryos. (A) Tsg101 knockout embryos are phenotypically indistinguishable from their wild-type littermates at E3.5. PCR was used to verify the absence of the wild-type allele in these embryos. B. Deciduae at E6.5 from two Tsg101 heterozygous knockout crosses. At this stage of development, Tsg101 knockouts could not be identified. The abnormal-looking deciduum (arrow) contained a heterozygous mutant.
To determine whether haploinsuficiency of Tsg101 is involved in tumorigenesis, we maintained a cohort of Tsg101+/− mice for a prolonged period. To date, only 1 of 12 24-month-old heterozygous mutants has developed colon cancer, and this tumor was not caused by the loss of the remaining Tsg101 wild-type allele (data not shown). None of the age-matched wild-type controls or other heterozygous mutants currently more than 21 months old exhibited any palpable lesions. Thus, a heterozygous germ line mutation of Tsg101 was not sufficient to initiate tumorigenesis in this animal model.
Inactivation of both Tsg101 alleles in differentiating mammary epithelial cells results in impaired mammogenesis.
To address whether the loss of Tsg101 function plays a role in normal mammogenesis and the development of mammary tumors, we generated mice that carry a WAP-Cre transgene in addition to two floxed alleles of the Tsg101 locus (WAP-Cre Tsg101fl/fl). The WAP-Cre transgene is specifically active in differentiating alveolar cells during late pregnancy and lactation. However, a significant number of recombined cells bypass apoptotic signals, survive remodeling during involution, and serve as progenitor cells for the alveolar compartment in subsequent pregnancies (30, 34). This feature makes the WAP-Cre strain an important tool to study tumor suppressor function in the mammary gland (39).
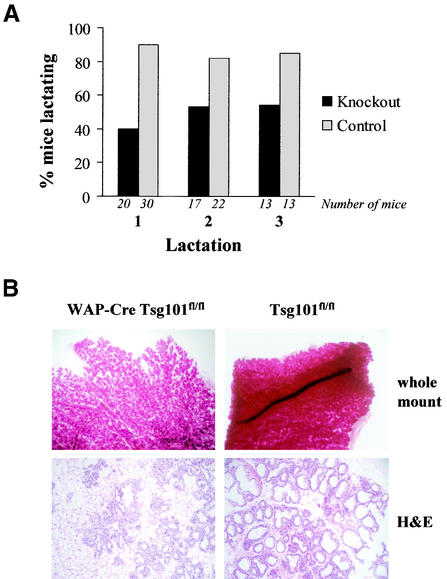
A WAP-Cre-mediated knockout of both alleles of Tsg101 leads to a severe inhibition of normal mammary gland development and the inability of mutant mice to support a litter (Fig. 3). Of 20 conditional knockout mice, 12 (60%) were completely unable to lactate after their first pregnancy, resulting in the loss of their entire litter within 24 h (Fig. 3A). On average, the few lactating conditional knockout mice supported only four or five pups (x = 4.5) by day 12 of lactation. The offspring were often malnourished. A histological examination of mammary tissues revealed that alveolar development was impaired in the Tsg101 conditional knockout mice (WAP-Cre Tsg101fl/fl) compared to their controls (Tsg101fl/fl) (Fig. 3B). It is a common feature of some genetically engineered mouse strains that lactation can be restored in successive pregnancies. To address this issue, we monitored lactation in conditional knockout dams and their controls for three consecutive gestation cycles (Fig. 3A). Interestingly, while some animals were able to establish lactation, almost half of all animals were still unable to nurse their young after the second and third pregnancies. Also, the average litter size that a lactating conditional knockout dam was able to support did not increase (x = 4.5 and 4.0, respectively). Therefore, a complete reversal of the phenotypic abnormalities of the mutant mice was not achieved. This suggested that either normal mammary epithelial cells have only a very limited capacity to bypass Tsg101 gene function or there is selective amplification of alveolar cells that maintain unrecombined alleles of Tsg101 due to WAP-Cre transgene silencing.
FIG. 3.
Analysis of lactation (A) and histological examination of mammary glands (B) from tissue-specific Tsg101 knockout mice (WAP-Cre Tsg101fl/fl) and their controls (Tsg101fl/fl). (A) A mouse is defined as lactating if she can support at least one pup for 12 days. (B) Mammary gland whole mounts were stained in carmine alum (magnification, ×40) and subsequently embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) (magnification, ×100).
WAP-Cre activation, the recombination of the floxed Tsg101 locus, and Tsg101 expression levels in mammary tissues from females during late pregnancy and lactation were monitored by Southern and Northern blot analyses (Fig. 4). Cre-mediated excision of the promoter and first coding exon of Tsg101 could be detected on day 18 of gestation and several hours after delivering the litter. This resulted in a reduced expression of Tsg101 mRNA around parturition (Fig. 4A and C). Based solely on Northern blot analysis, we did not expect to see a complete lack of Tsg101 transcripts in our conditional knockout model since this gene is ubiquitously expressed; i.e., many, if not all, cell types of the mammary stroma have normal Tsg101 mRNA levels. Conditional mutants unable to initiate lactation lost their litter shortly after birth, and the rudimentarily developed alveolar compartment regressed after several days (data not shown). An examination of the mammary glands of the few WAP-Cre Tsg101fl/fl mice that were able to nurse their young over a 12-day lactation period revealed that there was a considerable negative selection against Tsg101-deficient cells. While a few recombined cells were still present at 72 h postpartum (Fig. 4B, lane 2), Tsg101 null cells were virtually absent after 12 days of lactation as determined by Southern blot analysis (lanes 5 to 7). The recombined Tsg101 knockout allele could be detected only by PCR (data not shown). Consequently, the overall expression level of Tsg101 in the mammary gland was unaffected in these animals (Fig. 4D). In the 12-day lactating mammary gland of control mice carrying one floxed allele and one wild-type Tsg101 locus in addition to the WAP-Cre transgene (WAP-Cre Tsg101fl/+), the recombined (null) allele could be detected by Southern blotting (Fig. 4B, lane 4), suggesting that there was no general selection against Cre-expressing or Tsg101 haploinsufficient cells. This result was expected since neither Tsg101 heterozygous knockouts (Tsg101+/−) nor single-transgenic WAP-Cre mice exhibited abnormal mammary phenotypes. In summary, our data suggested that mammary epithelial cells were unable to compensate for the loss of Tsg101. Lactation could only be initiated in animals that exhibited a nonuniform expression of WAP-Cre and a selected amplification of alveolar cells with unrecombined Tsg101 alleles (i.e., negative selection against Tsg101-deficient cells).
FIG. 4.
Southern blot (A and B) and Northern blot (C and D) analyses to determine the recombination efficiency and expression levels of Tsg101 in the mammary glands of Tsg101 conditional mutants (WAP-Cre Tsg101fl/fl) and their controls (Tsg101fl/fl or WAP-Cre Tsg101fl/+). (A and B) The 2.1-kb XbaI fragment represents the recombined Tsg101 null allele. The unrecombined floxed or wild-type alleles are 2.7 kb (see Fig. 1). (A and C) Recombination efficiency and Tsg101 mRNA expression levels in mammary tissues from mice during late pregnancy (day 18 of gestation) and around parturition (16 h postpartum). (B and D) Southern and Northern analyses of mammary glands from lactating mice at 1, 3, and 12 days of lactation. Note the decline in the number of Tsg101-deficient cells in conditional knockout animals that are able to establish lactation (compare lanes fl/fl yes for 18 days and 16 h in panel A with lanes fl/fl yes for 72 h and fl/fl yes for 12 days [3 lanes] in panel B).
To investigate a possible involvement of a Tsg101 null mutation in mammary tumorigenesis, WAP-Cre Tsg101fl/fl mice and their controls were maintained over a period of 2 years and monitored for mammary tumor formation. These animals were continuously mated to (i) repeatedly activate WAP-Cre in the newly developed alveolar compartment and subsequently increase the number of Tsg101-deficient cells temporarily, (ii) increase the overall proliferation rate of mammary epithelial cells in response to higher estrogen and progesterone levels during pregnancy, and (iii) introduce and select for genomic mutations that might cooperate with the lack of Tsg101 toward neoplastic transformation. Thus far, none of the mice, which are currently up to 24 months old, have developed mammary gland lesions. These data suggest that in contrast to a previous report (14), Tsg101 is not a tumor suppressor gene for the initiation of sporadic forms of breast cancer.
Tsg101 deficiency results in the death of mammary epithelial cells.
Since Tsg101 is expressed in all cell types of the mammary gland and throughout mammogenesis, we did not reason that Tsg101 is specifically required for terminal differentiation of the alveolar compartment. In addition, Tsg101 is preferentially deleted from late-differentiating cells expressing the Wap gene (i.e., WAP-Cre-mediated recombination). Therefore, it was not surprising that we observed only slightly lower levels of β-casein and Wap mRNA in the conditional knockouts (data not shown). These marginal differences were probably caused by a smaller number of alveolar cells in the mutant mice than in their littermate controls (Fig. 3B). We postulated that Tsg101 could be important for cell growth and proliferation and that it may be crucial for cell survival since differentiating alveolar cells exhibit a much lower proliferation rate (35). This hypothesis is supported by the fact that there is a substantial negative selection against Tsg101-deficient cells during lactation (i.e., after completion of terminal differentiation). We used BrdU labeling and immunohistochemistry with the M30 CytoDEATH antibody to address whether impaired alveolar development is caused by a block in proliferation, an increase in cell death, or both in the WAP-Cre mediated conditional knockout of Tsg101 (Fig. 5). In particular, we studied the presence of dividing and apoptotic cells shortly after WAP-Cre becomes highly active (days 16 and 17 of gestation) and immediately postpartum, when milk secretion is initiated. Similar numbers of BrdU-labeled cells were observed in the knockouts and the controls at both stages of mammogenesis, suggesting that cell proliferation was not drastically reduced in the mutant mice (Fig. 5A to D). In general, apoptotic epithelial cells were barely visible in the mammary glands of wild-type mice at the onset of lactation, as determined by immunohistochemistry using the M30 CytoDEATH antibody (Fig. 5E). This antibody specifically recognizes a caspase-cleaved epitope of the keratin 18 (K18) protein (4). The cytoDEATH antibody recognizes only dying epithelial cells since K18 is a specific marker for the epithelial compartment of the mammary gland. In contrast to the controls, Tsg101 conditional knockout mice exhibited numerous apoptotic epithelial cells around parturition. The number of dying cells within a developing alveolus seemed to be larger in less well-organized lobular structures (compare Fig. 5F, G, and H). The staining intensity and number of dying cells were, however, irregular throughout entire cross sections, which made it virtually impossible to objectively quantify the exact percentage of apoptotic cells in vivo. Therefore, we reexamined cell growth and cell death in primary mammary epithelial cell cultures, where Cre-expressing cells can be selected and the deletion of Tsg101 can be controlled in a timely manner.
FIG. 5.
Detection of proliferating (arrows in panels B to D) and apoptotic (arrows in panels E to H) cells at various stages of mammary development in Tsg101 mammary-specific knockout mice (WAP-Cre Tsg101fl/fl B, D, and F to H) and their controls (Tsg101fl/fl [A, C, and E]). (A and B) 17 days of gestation (magnification, ×400). (C to H) Shortly after parturition (magnification, ×400). Proliferating cells were detected using BrdU labeling, whereas apoptotic cells were identified by immunohistochemistry using the M30 CytoDEATH antibody. Slides were counterstained with hematoxylin. Note the larger number of dying cells in less organized or collapsed alveolar structures (F to H).
Deletion of Tsg101 from primary mammary epithelial cells reveals an important function of this gene in proliferation and cell survival.
We cultivated primary mammary epithelial cells from Tsg101fl/fl animals and infected them with a retroviral vector, which expresses Cre recombinase under the viral long terminal repeat in addition to a simian virus 40 promoter-driven puromycin resistance gene (Fig. 6A). The presence of mammary epithelial cells in the cultures was verified by immunohistochemistry with an FITC-labeled pan-cytokeratin antibody (Fig. 6B). It was necessary to grow these cells in a defined medium (see Materials and Methods), which contains less than 2% fetal calf serum, to suppress the growth of contaminating fibroblasts. After retroviral Cre infection, selection with puromycin allowed us to enrich for Tsg101-deficient cells. The Cre-mediated excision of the promoter and first coding exon of the Tsg101 gene resulted in a considerable downregulation of the encoded protein as determined by Western blotting (Fig. 6C). We did not detect any smaller protein variants in the conditional knockout cells, suggesting that our targeting strategy led to complete inhibition of the Tsg101 locus. After confirming that Tsg101 had been deleted from our cultures, we monitored cell growth and cell survival for eight consecutive days after infection with the Cre retrovirus. The knockout of Tsg101 led to a severe growth inhibition and cell death in less than 1 week (Fig. 6D). Control cells with two functional Tsg101 loci expressing Cre recombinase (Tsg101+/+; pBabe-Cre) and Tsg101 floxed cells infected with a retrovirus lacking Cre (Tsg101fl/fl; pBabe) exhibited some growth retardation when puromycin selection was initiated. These control cells gradually recovered after a few days, while virtually all Tsg101 deficient cells detached from the dish and died. We noted that the primary mammary epithelial cells seemed to be more sensitive to puromycin treatment than were the embryonic fibroblasts (11). We therefore lowered the puromycin concentration to achieve optimal selection (see Materials and Methods). In contrast to earlier reports (18, 26) a low-level expression of Cre recombinase from a retroviral vector had no direct effect on cell growth in our experimental system. In addition, we showed earlier that expression of Cre in a mouse mammary epithelial cell line (HC11) and in the mammary glands of transgenic animals (WAP-Cre or MMTV-Cre) causes neither cytotoxicity nor phenotypic abnormalities (34).
FIG. 6.
Tsg101 deletion in primary epithelial cultures. (A) Experimental design. (B) Pan-cytokeratin staining to verify the presence of epithelial cells (magnification, ×400). (C) Tsg101 protein expression in uninfected (no virus) and Cre expressing (pBabe-Cre) cultures. The blot was reprobed with an antibody against β-actin to verify equal loading between lanes. (D) Declining amounts of viable cells in Tsg101-deficient cultures (Tsg101fl/fl; pBabe-Cre) between 3 and 7 days after Cre expression. Tsg101fl/fl cultures infected with pBabe or wild-type cells expressing Cre (Tsg101+/+; pBabe-Cre) served as controls.
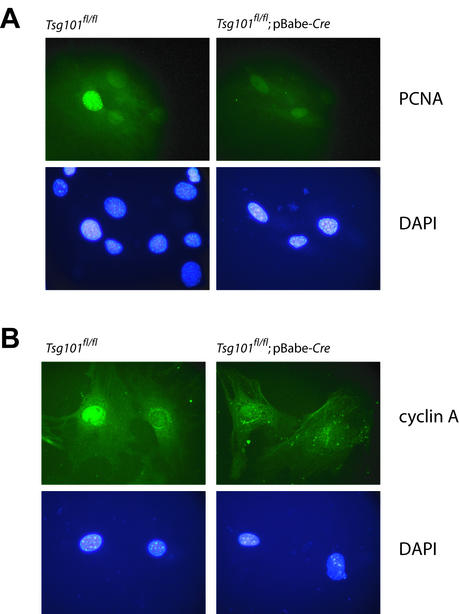
The deletion of Tsg101 from primary cells in culture confirmed our in vivo observations that the protein encoded by this gene is indispensable for normal cell growth and cell survival. It was suggested previously that Tsg101 may play a role in proliferation (24, 40). However, we did not observe a considerable reduction of proliferating cells in the mammary glands of WAP-Cre Tsg101fl/fl females. Since Wap is expressed mainly in differentiating cells exhibiting reduced proliferation, the animal model might not entirely recapitulate a phenotype that suggests an additional role for Tsg101 in cell proliferation. Therefore, we studied primary cells in vitro to determine whether Tsg101 deficiency results in perturbation of the cell cycle before cell death is initiated. More than 7.2% of the control cells (Tsg101fl/fl) expressed PCNA, whereas only 1.0% of the Tsg101-deficient cells exhibited positive staining (Fig. 7A). These in vitro data indicated that Tsg101-deficient primary cells have a reduced rather than an increased proliferative capacity. Since expression and nuclear accumulation of cyclin A could not be detected in the conditional knockout cells (Fig. 7B), these findings suggest that Tsg101-deficient cells arrest in the G1 phase and eventually die before the S phase is initiated. Results of the immunohistochemical examination of primary cells are in accordance with our observations in Tsg101-deficient mouse embryonic fibroblasts (MEFs). These MEFs exhibited a sustained G1/S arrest as determined by fluorescence-activated cell sorter and Western blot analysis of all major cyclins. Tsg101-deficient MEFs had an inactive cyclin-dependent kinase 2 (Cdk2)/cyclin E complex (11). In summary, the conditional deletion of Tsg101 from primary cells confirmed that (i) this gene is essential for both proliferation and cell survival and (ii) Tsg101 deficiency does not result in neoplastic transformation.
FIG. 7.
Immunocytochemistry for PCNA (A) and cyclin A (B) in Tsg101-deficient cultures (Tsg101fl/fl; pBabe-Cre) and their controls (Tsg101fl/fl). Nuclei were counterstained with DAPI. Note the presence of replicating cells and the expression and nuclear accumulation of cyclin A in the controls, which indicates that these cells, in contrast to the Tsg101 knockouts, progress normally into S phase.
DISCUSSION
Tsg101 and development.
Based on the promoter sequence and the expression profile, we postulated that a conventional knockout of Tsg101 would result in early embryonic death (32). The generation and analysis of embryos lacking Tsg101 completely confirmed this hypothesis. In our Tsg101Δ exon 1−/− model, deficiency of this gene resulted in embryonic death around the time of implantation. This assumption was also verified recently in a different mouse model carrying a deletion of exons 8 and 9, which encode the coiled-coil domain near the C-terminal end of Tsg101 (24). These mice die between E5.5 and E6.5 due to a defect in cell proliferation and mesoderm formation. An increase in cell death was not observed in these mutants. In the Tsg101Δ exon 1−/− model, we were unable to identify homozygous mutant embryos at day E6.5. This suggests that there might be marginal differences in the phenotypes of the two mouse models depending on which functional domains were deleted from the Tsg101 locus. Since neither the generation of Tsg101-deficient ES cells nor the cultivation of the ICM from Tsg101Δ exon 8/9−/− blastocycts was successful (24), it is virtually impossible to rescue Tsg101-deficient cells by transplantation.
To investigate the biological effects of Tsg101 deficiency on the proliferation and differentiation of breast epithelial cells and to verify the proposed function of this gene as a tumor suppressor for sporadic forms of breast cancer, we developed a knockout mouse with a mammary cell-specific deletion of Tsg101. Deficiency of Tsg101 in differentiating mammary epithelial cells resulted in impaired mammary development. One consequence was the inability of the mutant mice to lactate normally. This phenotype was caused mainly by apoptosis of differentiated cells, which normally exhibit a low proliferation rate (35). This phenomenon seems not to be a unique feature of mammary epithelial cells or of differentiating cells in general. We observed cell death as a consequence of Tsg101 deficiency in a variety of proliferating cell types including MEFs (11). In agreement with our findings, Garrus et al. (6) reported a slight growth inhibition of 293 cells when Tsg101 was transiently knocked down for 72 h by using the small interfering RNA approach. In addition, the introduction of Tsg101 siRNAs into human breast cancer cells (MCF-7) was highly toxic when these cells were treated repeatedly over time. Whereas Tsg101-deficient MCF-7 cells died, transfection of other siRNA control vectors had little or no effect on the growth of these cells (X. Lin and W. Nelson, John-Hopkins University, personal communication). In summary, these observations suggest that Tsg101 is important for the survival of nonimmortalized (e.g., mammary epithelial cells and MEFs) as well as immortalized and transformed (e.g., 293T and MCF-7 cells) cell lines. Our further studies of transformed Tsg101fl/fl MEFs, which were able to form tumors in nude mice, seemed to support this assumption. The Cre-mediated deletion of Tsg101 in explanted tumor cells resulted in instant cell death (M. D. Henry, A. Krempler, and K.-U. Wagner, unpublished data).
Tsg101 and cell cycle regulation.
Our studies of Tsg101-deficient primary cells revealed that in addition to cell survival, Tsg101 is important for cell proliferation. The retroviral Cre-mediated conditional knockout of Tsg101 resulted in cell cycle arrest and death before the cells entered the S phase. These observations were consistent with our findings about the role of Tsg101 in proliferating embryonic fibroblasts (11). Tsg101-deficient MEFs did not incorporate BrdU and lacked expression of cyclins A and B. In addition, their cyclin E/Cdk2 complex was inactive, which clearly suggested a sustained G1/S arrest before Tsg101-deficient cells died. The mechanism responsible for the inactivation of Cdk2 in the conditional knockout cells needs to be identified. In a recent report, Oh et al. (22) showed that exogenous levels of Tsg101 in differentiating keratinocytes can positively regulate p21WAF-1/C1P-1, a suggested tumor suppressor protein that mediates G1 arrest in a p53-dependent manner (2, 5, 19). In proliferating keratinocytes, which exhibit stable levels of p21, overexpression of Tsg101 exerts growth suppression through modulation of p21 binding to the cyclin E/Cdk2 complex. It is possible that the mechanisms, which mediate growth arrest in Tsg101-deficient cells and the Tsg101 overexpression model, are the same. However, we did not observe a significant change in p27Kip1 and p21WAF-1/CIP-1 protein levels in the conditional knockouts, suggesting that Tsg101 is not essential for the stability of these major Cdk2 inhibitors in MEFs (11). Preliminary studies of Tsg101−/− MEFs showed a larger amount of phosphorylated Cdk2 (A. Krempler and K.-U. Wagner, unpublished data), and we are currently investigating whether inhibiting phosphorylations mediate inactivation of the cyclin E/Cdk2 complex in Tsg101-deficient cells.
The role of Tsg101 as a positive regulator of p21 and/or Cdk2 activity would provide a mechanism for the suggested function of this protein as an inhibitor of the cell cycle and a potential tumor suppressor. In contrast, Li et al. (15) reported a role for Tsg101 as a positive regulator of Mdm2, which is an oncogene that mediates the degradation of p53. The same group of investigators was able to verify their findings by using a Tsg101-deficient mouse model, which exhibited a cell cycle arrest mediated by upregulation of p53 and p21 (24). Through introduction of a p53-null mutation into the Tsg101Δ exon 8/9−/− background, Ruland et al. (24) were able to prolong the survival of mutant embryos for about 2 to 3 days. Given the suggested importance of Tsg101 as a key regulator of Mdm2, this rescue seemed to be marginal compared to that in Mdm2/p53 double-mutant mice, where a knockout of p53 completely rescued embryonic death caused by Mdm2 deficiency (9, 21). In addition, the suggested function for Tsg101 as a stabilizer of Mdm2 (i.e., an oncogene) is inconsistent with various reports that Tsg101 overexpression leads to cell cycle arrest and cell death (22, 32, 37, 40). The findings that Tsg101 deficiency resulted in cell cycle arrest through upregulation of p53 and p21 therefore contradict the mechanism proposed by Oh et al. (22), in which Tsg101 is a crucial mediator of p21 protein stability and of p21 function as a cell cycle inhibitor on the cyclin E/Cdk2 complex (see above). Our conditional knockout model did not reveal any involvement of p53 and its main effectors in the deleterious phenotype caused by Tsg101 deficiency (11). Various upstream (p19ARF, p16Ink4a, and Mdm2) and downstream (p21WAF-1/CIP-1 and p27Kip1) targets of p53 exhibited no significant change in their protein levels in Tsg101−/− MEFs. Furthermore, neither the functional inhibition of p53 nor the deletion of the p53 gene had a noticeable effect on the deleterious phenotype. Therefore, our findings do not support a biologically relevant function of Tsg101 in regulating the Mdm2-p53 feedback loop, as previously suggested (15, 24). On the other hand, Tsg101 deficiency causes a cell cycle arrest in addition to cell death. While p53 is not essential for mediating cell death in Tsg101−/− MEFs (11), more detailed analysis of whether p53 plays a role in cell cycle arrest observed in the Tsg101 conditional knockout model is needed.
Tsg101 and neoplastic transformation.
Li and Cohen (13) reported that a functional knockout of Tsg101 using a conventional antisense approach resulted in instant neoplastic transformation of immortalized fibroblasts. However, the role of the Tsg101 gene as a tumor suppressor in human malignancies is still controversial. Genomic deletions and aberrant splice variants have been implicated in sporadic forms of breast cancer and a variety of other human malignancies (see, for example, references 7, 12, 14, 28, and 29). However, there are also numerous contradictory reports (see, for example, references 3, 17, 27, and 38). To verify whether the loss of function of Tsg101 is involved in neoplastic transformation in vivo, we maintained a large cohort of Tsg101 mutant mice (heterozygous complete and tissue-specific knockouts) for 2 years. These mice were closely monitored for tumor formation. Neither haploinsufficiency of Tsg101 (see also the report by Ruland et al. [24]) nor the deletion of both Tsg101 alleles in mammary epithelial cells resulted in tumor formation. Our data suggest that a loss of function of Tsg101 is insufficient to trigger neoplastic transformation; therefore, our observations contradict earlier reports by Li et al. (13, 14). Generally, in vitro models such as NIH 3T3 cells, which were used to identify Tsg101 and its tumor suppressive properties, are less reliable since these cells are immortal and carry numerous mutations in other tumor suppressor loci such as p53, p19Arf, or p16Ink4a (25). Specifically, the original NIH 3T3 cell line and its tumorigenic derivative (SL6 cells) described by Li and Cohen (13) are deficient in p19Arf and p16Ink4a (Krempler and Wagner, unpublished). More importantly, we showed recently that the antisense strategy used by Li et al. did not completely inhibit gene expression. SL6 cells still express a significant amount of the Tsg101 protein (11), and therefore it needs to be established (i) whether a knockdown triggers a different phenotype from a null mutation and (ii) whether antisense constructs target other genes with partial sequence similarity to Tsg101. In summary, the results of our studies on Tsg101 conditional knockout mice demonstrate that it is very important to generate genetically engineered mouse models with defined targeted mutations to verify a proposed tumor-suppressive function of genes identified in random mutagenesis screens in immortalized cell lines.
Acknowledgments
We thank Stanley Cohen (Stanford) for providing the Tsg101 cDNA and SL6 cells. We are grateful to Alan Diehl (University of Pennsylvania) for his valuable suggestions and for providing the pBabe retroviral vector and 293T cells.
This work was supported by Public Health Service grant CA-93797 from the National Cancer Institute to K.U.W. The embryonic stem cell work was funded in part by the NIH Breast Cancer Think Tank. A.K. receives a stipend from the Deutsche Forschungsgemeinschaft (DFG, KR 2107/1-1).
REFERENCES
- 1.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 2.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 3.Carney, M. E., G. L. Maxwell, J. M. Lancaster, C. Gumbs, J. Marks, A. Berchuck, and P. A. Futreal. 1998. Aberrant splicing of the TSG101 tumor suppressor gene in human breast and ovarian cancers. J. Soc. Gynecol. Investig. 5:281-285. [DOI] [PubMed] [Google Scholar]
- 4.Caulin, C., G. S. Salvesen, and R. G. Oshima. 1997. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol. 138:1379-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 6.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 7.Gayther, S. A., P. Barski, S. J. Batley, L. Li, K. A. de Foy, S. N. Cohen, B. A. Ponder, and C. Caldas. 1997. Aberrant splicing of the TSG101 and FHIT genes occurs frequently in multiple malignancies and in normal tissues and mimics alterations previously described in tumours. Oncogene 15:2119-2126. [DOI] [PubMed] [Google Scholar]
- 8.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 10.Koonin, E. V., and R. A. Abagyan. 1997. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat. Genet. 16:330-331. [DOI] [PubMed] [Google Scholar]
- 11.Krempler, A., M. D. Henry, A. A. Triplett, and K. U. Wagner. 2002. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53 independent cell death. J. Biol. Chem. 277:43216-43223. [DOI] [PMC free article] [PubMed]
- 12.Lee, M. P., and A. P. Feinberg. 1997. Aberrant splicing but not mutations of TSG101 in human breast cancer. Cancer Res. 57:3131-3134. [PubMed] [Google Scholar]
- 13.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 14.Li, L., X. Li, U. Francke, and S. N. Cohen. 1997. The TSG101 tumor susceptibility gene is located in chromosome 11 band p15 and is mutated in human breast cancer. Cell 88:143-154. [DOI] [PubMed] [Google Scholar]
- 15.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, M., K. U. Wagner, and P. A. Furth. 2000. Transfection of primary mammary epithelial cells by viral and nonviral methods, p. 233-244. In M. M. Ip and B. B. Ash, (ed.), Methods in mammary gland biology and breast cancer. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 17.Lin, S. F., P. M. Lin, T. C. Liu, J. G. Chang, Y. C. Sue, and T. P. Chen. 2000. Clinical implications of aberrant TSG101 transcripts in acute myeloblastic leukemia. Leuk. Lymphoma 36:463-466. [DOI] [PubMed] [Google Scholar]
- 18.Loonstra, A., M. Vooijs, H. B. Beverloo, B. A. Allak, E. van Drunen, R. Kanaar, A. Berns, and J. Jonkers. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 98:9209-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Caballero, J., J. M. Flores, P. Garcia-Palencia, and M. Serrano. 2001. Tumor susceptibility of p21 (Waf1/Cip1)-deficient mice. Cancer Res. 61:6234-6238. [PubMed] [Google Scholar]
- 20.Medina, D. and F. S. Kittrell. 2000. Establishment of mouse mammary cell lines, p. 137-145. In M. M. Ip and B. B. Ash (ed.), Methods in mammary gland biology and breast cancer. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 21.Montes de Oca, L. R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 22.Oh, H., C. Mammucari, A. Nenci, S. Cabodi, S. N. Cohen, and G. P. Dotto. 2002. Negative regulation of cell growth and differentiation by TSG101 through association with p21Cip1/WAF1. Proc. Natl. Acad. Sci. USA 99:5430-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponting, C. P., Y. D. Cai, and P. Bork. 1997. The breast cancer gene product TSG101: a regulator of ubiquitination? J. Mol. Med. 75:467-469. [PubMed] [Google Scholar]
- 24.Ruland, J., C. Sirard, A. Elia, D. MacPherson, A. Wakeham, L. Li, D. L. P. Luis, S. N. Cohen, and T. W. Mak. 2001. p53 Accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA 98:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 26.Silver, D. P., and D. M. Livingston. 2001. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell 8:233-243. [DOI] [PubMed] [Google Scholar]
- 27.Steiner, P., D. M. Barnes, W. H. Harris, and R. A. Weinberg. 1997. Absence of rearrangements in the tumour susceptibility gene TSG101 in human breast cancer. Nat. Genet. 16:332-333. [DOI] [PubMed] [Google Scholar]
- 28.Sun, Z., J. Pan, G. Bubley, and S. P. Balk. 1997. Frequent abnormalities of TSG101 transcripts in human prostate cancer. Oncogene 15:3121-3125. [DOI] [PubMed] [Google Scholar]
- 29.Turpin, E., B. Dalle, A. de Roquancourt, L. F. Plassa, M. Marty, A. Janin, Y. Beuzard, and H. de The. 1999. Stress-induced aberrant splicing of TSG101: association to high tumor grade and p53 status in breast cancers. Oncogene 18:7834-7837. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, K. U., C. A. Boulanger, M. D. Henry, M. Sgagias, L. Hennighausen, and G. H. Smith. 2002. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development 129:1377-1386. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, K. U., P. Dierisseau, and L. Hennighausen. 1999. Assignment of the murine tumor susceptibility gene 101 (tsg101) and a processed tsg101 pseudogene (tsg101-ps1) to mouse chromosome 7 band B5 and chromosome 15 band D1 by in situ hybridization. Cytogenet. Cell Genet. 84:87-88. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, K. U., P. Dierisseau, E. B. Rucker, G. W. Robinson, and L. Hennighausen. 1998. Genomic architecture and transcriptional activation of the mouse and human tumor susceptibility gene TSG101: common types of shorter transcripts are true alternative splice variants. Oncogene 17:2761-2770. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, K. U., K. McAllister, T. Ward, B. Davis, R. Wiseman, and L. Hennighausen. 2001. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 10:545-553. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, K. U., R. J. Wall, L. St-Onge, P. Gruss, A. Wynshaw-Boris, L. Garrett, M. Li, P. A. Furth, and L. Hennighausen. 1997. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25:4323-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, K. U., W. S. Young, X. Liu, E. I. Ginns, M. Li, P. A. Furth, and L. Hennighausen. 1997. Oxytocin and milk removal are required for post-partum mammary-gland development. Genes Funct. 1:233-244. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, M., Y. Yanagi, Y. Masuhiro, T. Yano, H. Yoshikawa, J. Yanagisawa, and S. Kato. 1998. A putative tumor suppressor, TSG101, acts as a transcriptional suppressor through its coiled-coil domain. Biochem. Biophys. Res. Commun. 245:900-905. [DOI] [PubMed] [Google Scholar]
- 37.Xie, W., L. Li, and S. N. Cohen. 1998. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl. Acad. Sci. USA 95:1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, C. F., J. Greenman, and E. Solomon. 1998. Truncated TSG101 transcripts are present in peripheral blood from both familial breast cancer patients and controls. Eur. J. Cancer 34:1077-1080. [DOI] [PubMed] [Google Scholar]
- 39.Xu, X., K. U. Wagner, D. Larson, Z. Weaver, C. Li, T. Ried, L. Hennighausen, A. Wynshaw-Boris, and C. X. Deng. 1999. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 22:37-43. [DOI] [PubMed] [Google Scholar]
- 40.Zhong, Q., Y. Chen, D. Jones, and W. H. Lee. 1998. Perturbation of TSG101 protein affects cell cycle progression. Cancer Res. 58:2699-2702. [PubMed] [Google Scholar]