Abstract
The basic helix-loop-helix protein BETA2/NeuroD activates transcription of the secretin gene and is essential for terminal differentiation of secretin-producing enteroendocrine cells. However, in heterodimeric complexes with its partner basic helix-loop-helix proteins, BETA2 does not appear to be a strong activator of transcription by itself. Mutational analysis of a proximal enhancer in the secretin gene identified several cis-acting elements in addition to the E-box binding site for BETA2. We identified by expression cloning the zinc finger protein RREB-1, also known to exist as a longer form, Finb, as the protein binding to one of the mutationally sensitive elements. Finb/RREB-1 lacks an intrinsic activation domain and by itself did not activate secretin gene transcription. Here we show that Finb/RREB-1 can associate with BETA2 to enhance its transcription-activating function. Both DNA binding and physical interaction of Finb/RREB-1 with BETA2 are required to potentiate transcription. Thus, Finb/RREB-1 does not function as a classical activator of transcription that recruits an activation domain to a DNA-protein complex. Finb/RREB-1 may be distinguished from coactivators, which increase transcription without sequence-specific DNA binding. We suggest that Finb/RREB-1 should be considered a potentiator of transcription, representing a distinct category of transcription-regulating proteins.
The expression of secretin, a peptide hormone, is restricted to S-type enteroendocrine cells present primarily in the small intestine and in the colon. The gene is also transiently expressed in normal β cells of developing pancreatic islets as well as in several pancreatic and intestinal endocrine cell lines (38). It was previously shown that 1.6 kb of 5′-flanking sequence of the secretin gene conferred tissue-specific, developmentally regulated expression of a reporter gene in transgenic mice (19).
Analysis of the secretin gene promoter by transient expression assays identified an enhancer between −174 and −53 bp 5′ of the transcription initiation site. The enhancer was necessary and sufficient for high levels of reporter gene expression in secretin-expressing cell lines but had no activity in cell lines that did not express their endogenous secretin gene (38). Examination of a series of 5′- and 3′-deletion mutants within the enhancer revealed a stepwise loss of activity with deletions of increasing size, suggesting that the enhancer was comprised of multiple regulatory elements.
A consensus E box sequence at −130 was required for full promoter activity. BETA2/NeuroD, a basic helix-loop-helix (bHLH) transcription factor that was originally identified as a transactivator of the insulin gene (25) and as a neurogenic factor in Xenopus embryos (15), was subsequently shown to bind the secretin gene E-box as a heterodimer with the ubiquitously expressed bHLH protein E12 or E47 (22).
BETA2 is expressed in a very limited number of tissues, including neurons, the anterior pituitary, pancreatic islets, and enteroendocrine cells. The insulin, secretin, and pro-opiomelanocortin (POMC) genes are well established targets of BETA2 (23, 25, 29). BETA2 may transactivate the genes encoding glucagon (2) and PDX-1 (33). The secretin gene is the only gene identified thus far that is absolutely dependent on the presence of BETA2 for expression in vivo. Secretin-expressing enteroendocrine cells are totally absent in BETA2 null mice (24). In the endocrine pancreas, BETA2 null mice show moderate reduction in the number of beta cells and a slight reduction in alpha cell number (24), with essentially normal POMC expression in the corticotrophs of the anterior pituitary (16). In the pancreas, BETA2 null mice show marked abnormalities in islet morphogenesis, with reduced numbers of insulin-expressing β cells and severe neonatal diabetes. Examination of the small intestine also revealed the absence of cholecystokinin-expressing enteroendocrine cells in BETA2−/− mice (24). Although BETA2 appears to be expressed in all enteroendocrine cell types (32), the remaining populations appeared to be unaffected by the absence of BETA2, suggesting that BETA2 does not play a major role in maintaining expression of other gut hormones.
Breeding BETA2−/− animals into a different genetic background produces some mice that survive the neonatal period and exhibit neurological phenotypes (17, 18, 21). The specific genes that are activated by BETA2 in the nervous system have yet to be identified. Although insulin gene expression appears to be normal, these animals remain unable to develop mature, healthy-looking islets (8). Adult animals still exhibit the complete absence of secretin-expressing enteroendocrine cells, implying that their absence in newborn BETA2 null mice did not result merely from delayed development (A. Leiter, unpublished observations). The differences in the role of BETA2 in regulating the insulin and secretin genes are not understood. Such a cell type difference in function could arise from the interaction between BETA2 and other factors. BETA2 associates with the homeodomain protein PDX-1 to synergistically increase insulin gene transcription (26). PDX-1 is specifically expressed in pancreatic β cells but not enteroendocrine cells in adult animals. Other non-bHLH proteins that bind to the secretin gene enhancer that could potentially modify the activity of BETA2 have not been characterized yet.
The goal of the present work was to further identify the cis-elements and their trans-acting factors in the secretin gene enhancer that are important for secretin gene transcription. We conducted a detailed mutational analysis of the secretin enhancer and demonstrated that three GC-rich cis-acting elements, in addition to the E-box, are important for its activity. Two of the GC-rich regions, located downstream from the E-box, bind to members of the Sp1 family of proteins. The third GC-rich region, located upstream of the E-box, appears to be a novel site. We have isolated a cDNA encoding a zinc finger protein that binds to the upstream element. This protein is homologous to two previously identified proteins, RREB-1 and Finb, and appears to require the presence of the protein redox factor 1 to stabilize DNA-binding activity. Finb/RREB-1 cannot transactivate the secretin gene by itself but appears to enhance transactivation by BETA2. The observed potentiation required both site-specific DNA binding of Finb/RREB-1 and direct physical interaction with BETA2.
MATERIALS AND METHODS
Cell culture.
The hamster insulin tumor cell line HIT-15 M2.2.2 (3) and the human cervical carcinoma cell line C33A (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (4.5 g of glucose/liter) supplemented with 10% fetal calf serum and 100 U of penicillin, 100 μg of streptomycin, and 292 μg of l-glutamine per ml. Cells were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Plasmid constructions.
The secretin-luciferase reporter gene constructions for transient expression assays consisted of sequences from −209 to +32 of the rat secretin gene (38) with or without mutations cloned upstream of the structural gene encoding firefly luciferase. We generated a series of transversion mutants (A↔C and G↔T) in the secretin promoter by site-directed mutagenesis (12) and confirmed the presence of the desired mutations by DNA sequencing. Expression plasmids for RREB-1 (pMV1-317), Finb (pME-Finb), and Ref-1 (40) were provided by Barry D. Nelkin (37), Tadashi Yamamoto (5), and Tom Curran, respectively. An RREB-1 expression plasmid was generated by subcloning an EcoRI-XbaI cDNA fragment containing the entire coding sequence from pMV1-317 into the same restriction sites in pcDNA3.1 (+) (Invitrogen). The same cDNA fragment was also subcloned into pGEM7Z (+) in EcoRI and XbaI sites. The amino-terminal deletion of RREB-1 was made by excising the part of the cDNA flanked by NcoI sites, followed by blunt-end ligation. The EcoRI-XbaI fragment from this derivative was further subcloned into pcDNA3.1 (+). The EcoRI-XhoI fragment containing the RREB-1 cDNA fused in frame with the FLAG epitope at the C terminus (provided by Barry D. Nelkin) was subcloned into pcDNA3.1 (+). A unique BstEI restriction site at the C terminus was used to construct the N-terminal deletion of RREB-1/FLAG in the same vector. The entire full-length Finb coding sequence from the pME-Finb plasmid was subcloned into pcDNA3.1 (+). A mutant construct expressing Finb without the 125 amino acids at the C terminus (FinbΔC) was made by excising the fragment containing two BamHI sites followed by ligation. Finb or RREB-1 cDNA was also subcloned into pM (Clontech) to create an in-frame fusion with the Gal4 DNA-binding domain (DBD) (amino acids 1 to 147). The bacterial expression plasmid for making glutathione S-transferase (GST) fusion protein with BETA2 was previously described (23).
Transient transfections.
Cells were subcultured at a density of 106 cells in each well of six-well plates (Costar) 18 h prior to transfection. A total of 10 μg of DNA, consisting of 3.3 μg of secretin reporter plasmid and 1 μg of metallothionein-human growth hormone plasmid or Rous sarcoma virus-β-galactosidase plasmid and sheared salmon testis carrier DNA, was introduced into cells by a standard calcium phosphate coprecipitation method, followed 4 h later by a 2-min treatment with 15% glycerol. Cell lysates were harvested 16 h later for the luciferase assay. To account for variations in transfection efficiency, luciferase activity in cell lysates was normalized to the amount of secreted human growth hormone in the culture medium. For some experiments, the luciferase activity was normalized to the Renilla luciferase activity using the dual-luciferase reporter assay as described by the manufacturer.
Gel shift assays.
Gel shift assays were performed as described earlier (4) with nuclear extracts prepared from different cell lines by the method of Dignam et al. (1). The binding reaction contained the binding buffer (25 mM HEPES [pH 7.9], 0.1 mM ZnCl2, 1 mM dithiothreitol [DTT], 40 mM KCl, and 5% glycerol) with 32P-labeled DNA probe (30,000 cpm), 2 μg of poly(dI-dC), 5 μg of nuclear protein, and cold competitor when desired in a total volume of 20 μl. Each reaction was initiated by the addition of protein and incubated for 30 min at room temperature, and then 10 μl was electrophoresed in 4% polyacrylamide gels. For immunodetection of proteins in DNA-protein complexes, the extracts were incubated on ice for 1 h with different antibodies, including anti-BETA2 (25), anti-E47 (6), anti-Sp1, and anti-Sp3 (Santa Cruz Biotechnology). For some experiments, the extracts were also incubated with anti-RREB-1 (see below) or anti-Ref-1 (Santa Cruz Biotechnology) antibody.
The following oligonucleotides were used in the gel shift experiments as double-stranded sequences (uppercase) with 4-bp single-stranded overhangs (lowercase), with mutations shown in bold type: wild type −179/−146, tcgaCAGTTGAGGGGCGCCAACACGGCGGTAGGGACAG; wild type −179/−161, gatcCAGTTGAGGGGCGCCAACA; GCI (−124 to −111), tcgaGGGGGGCGGCCCTG; GCII (−78 to −54), tcgaGCGCGGAGCCGGGGCGGTGCCGGAG; Sp1, ATTCGATCGGGGCGGGGCGAGC; Mut −168/−165, tcgaCAGTTGAGGGGATAAAACACGGCGGTAGGGACAG; Mut −154/−151, tcgaCAGTTGAGGGGCGCCAACACGGCGGGCTTGACAG; Mut-170, tcgaCAGTTGAGGTGCGCCAACACGGCGGTAGGGACAG; GC-MT, tcgaGCGCGGAGCCGGGTCTGTGCCGGAG.
At the 5′ end of each nucleotide a SalI or BamHI site was added to facilitate cloning or Klenow fill-in labeling with [α-32P]dCTP.
Methylation interference assay.
A HindIII-EcoRI fragment containing the promoter sequence −174 to −161 or −179 to −146 (as described above) was end labeled at the EcoRI site (sense strand) or the HindIII site (antisense strand), partially methylated with dimethyl sulfate, and used as a probe in a gel shift assay. The bound and the unbound (free) DNA probes were eluted from the gel, cleaved with piperidine, and then analyzed in an 8% sequencing gel as described previously (22).
Purification of recombinant Ref-1.
A His-tagged Ref-1 bacterial expression plasmid (pQE30-Ref-1) was a gift from Tom Curran (St. Jude Children's Hospital, Department of Developmental Neurobiology, Memphis, Tenn.). Recombinant Ref-1 was expressed as a histidine fusion protein in Escherichia coli and purified by nickel chelation chromatography.
Purification of the upstream element-binding factor.
All protein purification was performed at 4°C. Approximately 15 mg of nuclear extract (5 mg/ml) derived from C33A cells (5 × 108 cells) was diluted to 0.1 M KCl by the gradual addition of buffer A (20 mM HEPES [pH 7.6], 0.1 mM EDTA, 0.02 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], and 10% glycerol), applied onto a heparin-agarose column (12 mg of protein per ml), and washed with loading buffer. Bound proteins were eluted with buffer A containing 0.25 or 0.75 M KCl. The DNA-binding activity of each fraction was assayed by a gel shift assay with a DNA probe containing the upstream element. DNA-binding activity, eluted with buffer A containing 0.25 M KCl, was pooled and dialyzed against buffer B (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 0.2 mM PMSF, 0.1% NP-40, and 10% glycerol) containing 0.1 M KCl for 1.5 h at 4°C. The dialysate (3 mg of protein per ml) was then applied to either a DE52 column pre-equilibrated with buffer B containing 0.1 M KCl or a sequence (−174 to −161)-specific DNA-affinity column (10) pre-equilibrated with buffer C (20 mM HEPES [pH 7.9], 0.1 mM ZnCl2, 5 mM DTT, 0.1% NP-40, 1 mM PMSF, and 10% glycerol) containing 0.1 M KCl. Bound proteins from the DE52 column were eluted with a three-bed volume of buffer B with either 0.25 or 0.35 M KCl. The bound proteins from the DNA-affinity column were eluted with buffer C containing either 0.25 or 0.75 M KCl.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto a nitrocellulose membrane. Nonspecific binding to the membrane was blocked with a 3% milk solution in phosphate-buffered saline (PBS) containing 0.1% NP-40 prior to incubation with an anti-Ref-1 antibody (Santa Cruz) or anti-FLAG (M2) antibody (Sigma). An enhanced chemiluminescence (ECL) kit (Amersham) was used for detection of protein.
Immunofluorescent cell staining.
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, followed by permeabilization with 0.2% Triton X-100 in PBS. Cells were washed five times in PBS, blocked with 2% horse serum in PBS for 30 min, and incubated with anti-FLAG M2 antibody (1:400) in PBS containing 0.2% horse serum for 45 min. After washing, cells were incubated with Cy3-conjugated anti-mouse immunoglobulin G (IgG) (1:400; Jackson Immunoresearch) in PBS containing 0.2% horse serum for 45 min.
cDNA isolation.
Proteins were expressed from a HIT λgt11 cDNA library (27) and screened with a single labeled copy of the upstream element binding site (−179 to −146). Approximately 2 × 106 recombinant phage clones were examined by standard methods (34), with some modification. The binding buffer contained 10 μg of poly(dI-dC) per ml, 10 mM DTT, and 7.5 μg of recombinant Ref-1 per ml.
Additional sequence at the 5′ end of the identified partial cDNA was obtained by 5′ rapid amplification of cDNA ends (RACE) of the synthesized cDNA from HIT cell RNA using the SMART RACE cDNA amplification kit (Clontech). Gene-specific primers were designed for amplification of the cDNA generated by RACE. The forward primer was designed from the sequence of a conserved nucleotide sequence in the mouse and human Finb genes and the reverse primer was designed from the sequence of cloned hamster cDNA. The primer sequences were as follows: forward, 5′-CAACCAGGTGTTTGCCTTCTCCGGG-3′; and reverse, 5′-CCTCTTCTCAGTTGTGCCTTCCCCATC-3′.
Production of an antipeptide antibody against RREB-1.
The peptide LQDLTRHMRSHTGERPYK was commercially synthesized for rabbit immunization and for affinity purification (New England Peptide, Inc., Fitchburg, Mass.).
Binding analysis with GST fusion proteins.
Bacterially expressed GST fusion proteins were adsorbed to glutathione-Sepharose beads (Pharmacia) and washed. The beads were then incubated with [35S]methionine-labeled in vitro-transcribed and -translated proteins synthesized using the TNT-reticulocyte lysate system (Promega). The beads were extensively washed with the binding buffer (PBS with 0.1% NP-40), and the bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by autoradiography.
RESULTS
Identification of multiple cis-active regulatory elements in the secretin enhancer.
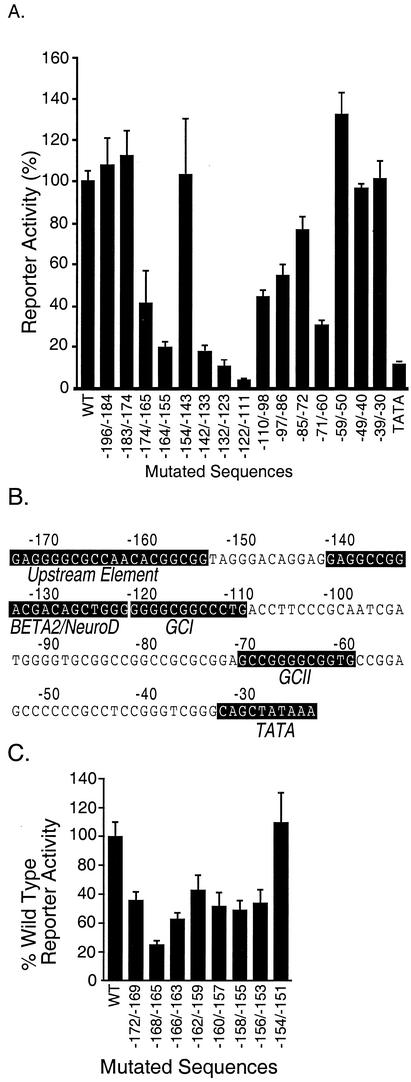
Deletion analysis from the 5′ end of the secretin gene enhancer at −174 revealed a stepwise loss of transcriptional activity, suggesting the presence of multiple cis-regulatory elements (38). The cis-regulatory elements in this region were further localized by introducing a series of 10- to 13-bp block transversions into secretin-luciferase reporter genes between −196 and −20. In secretin-expressing HIT cells, three mutationally sensitive domains in addition to the TATA box were identified by the substantial loss of luciferase activity with six block substitution mutants spanning −174 to −165, −164 to −155, −142 to −133, −132 to −123, −122 to −111, and −71 to −60 (Fig. 1A).
FIG. 1.
Mutational analysis of the secretin promoter. HIT cells were transiently transfected with a secretin-luciferase reporter gene (Materials and Methods) without (wild type [WT]) or with transversion mutations (10 to 13 bp) at different positions (shown by the numbers under each bar) including the TATA box (A) or with 4-bp transversion mutations at different positions (C). Luciferase activity was measured in cell extracts 16 h later. Results are expressed aspercentages relative to the wild-type reporter and are the means ± standard errors of the means (SEM) normalized for transfection efficiency for at least three independent experiments. (B) The mutationally sensitive sequences present in the promoter region are highlighted with a black background.
We previously showed that sequences between −142 and −123, which include an E-box sequence, comprise the binding site for the bHLH protein BETA2 together with one of its bHLH heterodimeric partners, E12 or E47 (22). Examination of the nucleotide sequences in the mutationally sensitive elements −122 to −111 and −71 to −60 revealed the presence of two GC-rich sequences (GCI and GCII) homologous to Sp1 binding sites (Fig. 1B).
Sequences at the 5′ end of the enhancer between −174 and −155 did not conform to any transcription binding sites in the Tfsites database. We generated a series of secretin-luciferase reporter genes containing 10 consecutive 4-bp transversion mutations within this region to more precisely map the sequence elements important for transcriptional activity of the enhancer (Fig. 1C). A mutation spanning −168 to −165 showed the greatest reduction of luciferase activity (75%). Other mutations flanking this region also reduced reporter gene expression to a lesser extent, indicating that these sequences may in part influence the activity of this upstream element.
Identification of factors binding to the mutationally sensitive elements of the enhancer.
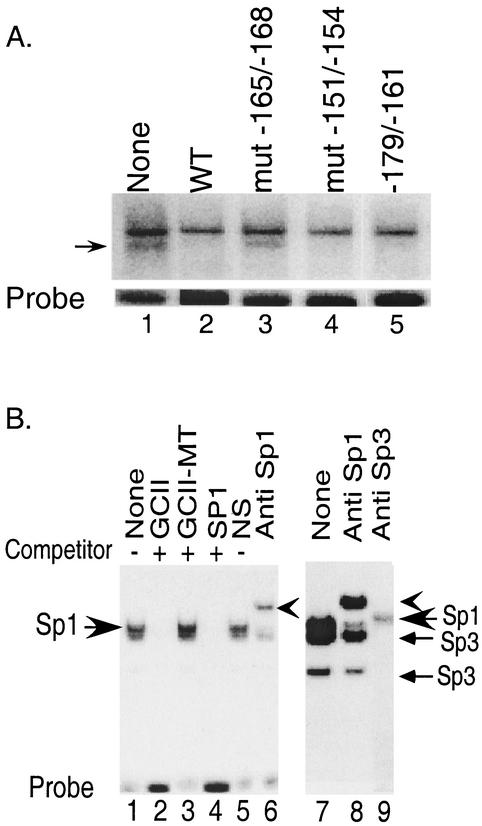
We examined nuclear extracts from HIT cells by electrophoretic mobility shift assays for the presence of factors that bind to the upstream element defined by scanning substitution mutagenesis. Several DNA-protein complexes were generated with a labeled DNA probe containing sequence between −174 and −146 (Fig. 2A, lane 1). The faster-migrating complex (marked by an arrowhead) was competed out by an excess of unlabeled wild-type oligonucleotide, indicating sequence specificity of binding (Fig. 2A, lane 2).
FIG. 2.
Characterization of factors binding to mutationally sensitive elements in the secretin gene enhancer. (A) A HindIII-EcoRI fragment containing sequence −174 to −146 was used as a probe in a gel shift assay with HIT cell nuclear extract. Arrowhead, specific DNA-protein complex. Lanes 2 to 5 included a 100-fold molar excess of unlabeled competitor from −174 to −146, with the indicated 4-bp block mutations in lanes 3 and 4. Lane 5 included a 100-fold molar excess of a competitor spanning −174 to −161. WT, wild type. (B) Left panel, HIT nuclear extract was incubated with a GCI probe in the absence (lane 1) or presence of different competitors at 100-fold molar excess (lanes 2 to 4) or in the presence of a nonimmune IgG (lane 5) or anti-Sp1 antibody (lane 6). The smaller arrow shows the slower mobility complex supershifted by the Sp1 antibody. Right panel, experimental conditions were as described for the left panel except that the electrophoresis was carried out to separate the individual complexes. Extracts were treated with anti-Sp1 (lane 8) or anti-Sp3 (lane 9) antibody.
A similar oligonucleotide containing the 4-bp transversion mutation −168 to −165 failed to compete for the probe, suggesting that these sequences in the native gene were required for binding. Moreover, the same mutation reduced the transcriptional activity of the reporter gene by 75% (Fig. 1C), suggesting that the formation of this complex is required for full transcriptional activity. Downstream regions such as −154 to −151 do not appear to participate in complex formation, since an oligonucleotide containing the transversion −154 to −151 competed for the specific DNA-protein complex and reporter genes containing a mutation retained full activity. An oligonucleotide containing sequences from −174 to −161 also efficiently competed for the same complex at the same molar excess as the longer competitor (lane 5). Thus, functional binding and transcriptional activity require minimal sequences from −174 to −161 (upstream element). The relatively GC-rich upstream element did not appear to bind to members of the Sp1 family since Sp1 consensus sequences failed to compete for the complex (not shown).
Proteins binding to the mutationally sensitive sequences, −122 to −111 (GCI) and −71 to −60 (GCII), were examined using a gel shift assay with a radiolabeled DNA probe containing the nucleotide sequence −122 to −111 (GCI) (Fig. 2B). Incubation with HIT cell nuclear extracts generated two closely migrating DNA-protein complexes, the slower of which may represent a doublet (Fig. 2B, lane 1). All complexes were competed out by an excess of unlabeled, wild-type oligonucleotide for GCII (lane 2), GCI (not shown), or an oligonucleotide with an Sp1 binding site (lane 4) that is very similar to either GC box. Thus, the two GC boxes appear to bind to similar proteins. Competitors containing a double point mutation designed to disrupt Sp1 binding in the Sp1 consensus sequences in the GC box did not compete for any of the complexes (lane 3), indicating a loss of binding activity. A polyclonal Sp1 antibody (Fig. 2B, lanes 6 and 8) but not the control antibody (lane 5) supershifted the slower-mobility complex in the doublet, indicating that Sp1 is a part of the DNA-protein complexes formed with the GC-rich sequences. The ability to form the lower complex in the doublet as well as the faster-moving complex did not appear to be affected by an anti-Sp1 antibody (Fig. 2B, lane 8), whereas an anti-Sp3 antibody completely blocked the formation of the two faster-moving complexes and partially inhibited formation of the slow-moving complex, indicating the presence of Sp3 in all three complexes.
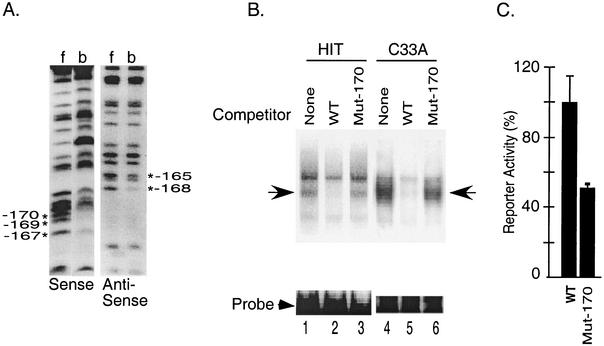
To further characterize the nucleotide sequences in the upstream element that participate in the formation of the complex, we next identified DNA-protein contact points in the complex using methylation interference assays (Fig. 3A). Methylation of G residues at −170, −169, and −167 (sense strand) and −168 and −165 (antisense strand) interfered with the formation of the DNA-protein complex. Introduction of a single G-to-T substitution at −170 (Mut −170) disrupted the binding of the complex in nuclear extracts from both HIT and C33A cells (Fig. 3B). Although an excess of wild-type unlabeled sequence displaced the bound protein (lanes 2 and 5), a 100-fold excess of unlabeled oligonucleotides containing the −170 mutation failed to compete (lanes 3 and 6), suggesting that this single base change disrupts binding of proteins to the upstream element. The proteins binding to the upstream element appear to be present in nuclear extracts from C33A cervical carcinoma cells as well as HIT cells (lanes 1 and 4), indicating that these proteins are also expressed in nonendocrine cells. The −170 single point mutation also reduced luciferase activity in transient expression assays by approximately 50%, indicating that the loss of binding activity correlates with loss of transcriptional activity (Fig. 3C).
FIG. 3.
Characterization of the factors binding to the upstream element in the secretin enhancer. (A) Partially methylated probes were incubated with HIT cell nuclear extract in an electrophoretic mobility shift assay. Following electrophoresis, free probe (f) and probe bound to proteins in the upstream element complex (b) were eluted from the gel, cleaved with piperidine, and resolved on an 8% sequencing gel to generate G ladders. *, G residues that are important for complex formation. (B) The upstream element probe was incubated in a gel shift reaction with the nuclear extract from HIT cells or C33A cells in the absence (lane 1) or presence of a 100-fold molar excess of the wild type (WT) (lane 2) or the Mut-170 (lane 3) competitor. Arrow, specific DNA-protein complex. (C) HIT cells were transiently transfected with a secretin-luciferase reporter plasmid (WT) or the same reporter with a point mutation at −170 (Mut-170). Luciferase activity was measured in cell extracts 20 h later. Results are expressed as percentages relative to the wild-type reporter and are the means ± SEM normalized for transfection efficiency for at least three independent experiments.
Proteins binding to the upstream element require an additional cellular factor, Redox factor 1 (Ref-1), for DNA binding.
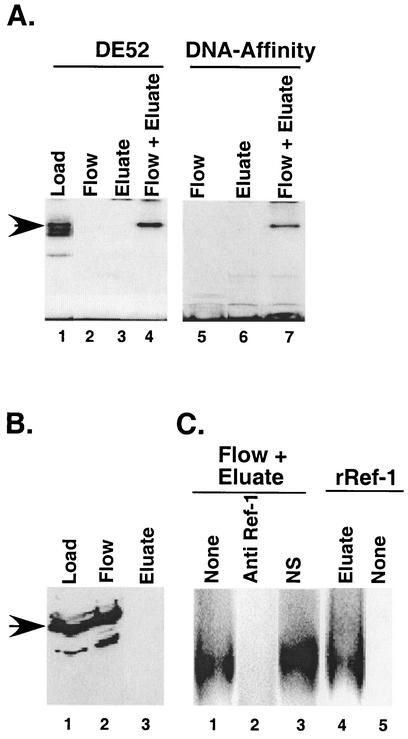
To further characterize the upstream element binding proteins, we fractionated nuclear extracts from C33A cells on heparin-agarose columns followed by either a DE-52 agarose or DNA oligonucleotide affinity column, testing fractions for DNA-binding activity using an upstream element probe. Most of the DNA-binding activity was eluted from the heparin-agarose column with 0.25 M KCl (Fig. 4A, lane 1) and was further fractionated through either a DE52 column or an oligonucleotide affinity column. After either of these column steps, no DNA-binding activity was detected in either the material not retained on the column (flowthrough fractions) or in proteins eluted in the presence of salt (Fig. 4A, lanes 2, 3, 5, and 6).
FIG. 4.
Stabilization of DNA-binding activity of the upstream element binding factor by Ref-1. (A) The DNA-binding activity was purified from C33A nuclear extracts. Aliquots from a heparin-agarose column salt eluate (lane 1) as well as the flowthrough and salt-eluted fractions of subsequent columns, either a DE52 column (lanes 2 to 4) or a DNA-affinity column (lanes 5 to 7), were tested for upstream element DNA-binding activity in a gel shift assay. The lost DNA-binding activity (lanes 2, 3, 5, and 6) was restored by mixing the two fractions (flow + eluate) (lanes 4 and 7). (B) Immunoblot analysis of proteins in the DE52 load, flow, and eluate fractions with anti-Ref-1 antibody. Proteins present in an equivalent volume of each column fraction were separated as described in Materials and Methods. Arrow, 38-kDa protein that reacts with the Ref-1 antibody. (C) An aliquot of the DE52 eluate fraction was mixed with the DE52 flow fraction that was untreated (lane 1), the flow fraction pretreated with 0.4 μg of anti-Ref-1 antibody (lane 2), a control antibody (lane 3), or a recombinant Ref-1 protein (∼200 ng) (lane 4) and was assayed for DNA-binding activity. Ref-1 alone was devoid of DNA-binding activity (lane 5).
The apparent loss of the DNA-binding activity suggested that the components necessary for DNA binding might have separated during the DE52 and affinity column fractionation. This was confirmed by mixing proteins from the flowthrough with fractions eluted with 0.25 M KCl for the DE52 column or 0.75 M KCl for the affinity column to reconstitute binding activity (Fig. 4A, lanes 4 and 7). We also observed that the reconstitution in the mixing experiments was most efficient in the presence of DTT (not shown). The DNA-binding activity of a number of transcription factors, including AP1, NF-κB, c-Myb (40), HIF-1α (14), Pax-5 (36), Pax-8 (35), and p53 (9), is stabilized under reducing conditions by the cellular protein Ref-1. The stabilization of DNA-binding activity probably resulted from maintaining a reducing environment for the cysteine residues near the DBD.
To determine if Ref-1 was required for reconstitution of DNA-binding activity of factors binding to the upstream element, we examined fractions from the DE52 column by Western blotting for the presence of Ref-1 (Fig. 4B). We observed that most of the 38-kDa Ref-1 immunoreactive protein loaded onto the DE52 column (lane 1) was not retained on the column, came out in the flow (lane 2), and was undetectable in the eluate (0.25 M KCl) (lane 3). Pretreatment of the flow fractions with Ref-1 antibody completely blocked reconstitution of DNA-binding activity in the salt-eluted fractions (Fig. 4C, compare lanes 1 and 2), whereas normal control IgG had no effect on reconstitution (Fig. 4C, lane 3). The ability of anti-Ref-1 antibody to neutralize reconstitution by the flow fractions suggested that Ref-1 might be essential for maintaining DNA-binding activity of proteins retained on the column. The role of Ref-1 was confirmed by the ability of purified His-tagged recombinant Ref-1 to substitute for flow fractions in reconstituting the DNA-binding activity of proteins eluted from the DE52 column (Fig. 4C, lane 4). Like the column flow fractions, the recombinant Ref-1 showed no intrinsic binding to DNA (Fig. 4C, lane 5), suggesting that it functioned by stabilizing a separate protein in the salt eluted fractions.
Identification of a protein binding to the upstream element.
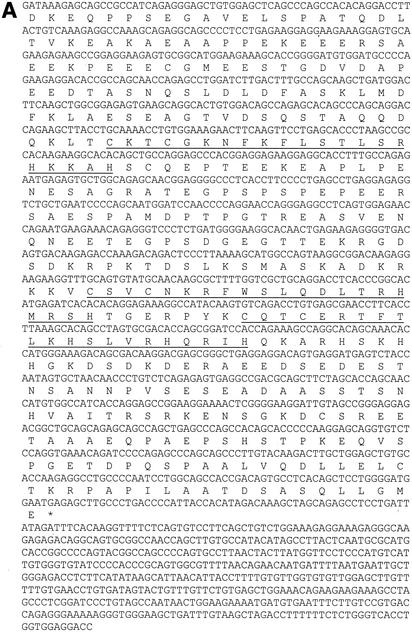
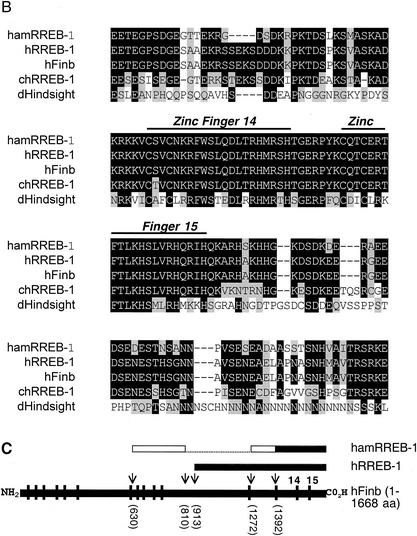
Initial screening of proteins expressed from a bacteriophage cDNA library made from HIT cells failed to identify any clones that expressed proteins that bound to a single-copy labeled binding site probe containing sequences −179 to −146. Based on the requirement for Ref-1 to stabilize the DNA-binding activity of the proteins binding to the upstream element, we subsequently screened the library in the presence of bacterially expressed, purified, histidine-tagged Ref-1. After screening 2 million clones, we isolated several cDNA clones with an open reading frame of 281 amino acids containing two Kruppel-type C2H2 zinc fingers (Fig. 5A). A BLAST search against multiple databases revealed this protein to be the hamster homologue of two related if not identical human proteins, Finb (finger protein in nuclear bodies) (5) and RREB-1 (Ras-responsive element binding protein 1) (37), as well as chicken RREB-1 (20). These two larger proteins extend beyond the amino terminus of the hamster protein with 15 and 4 zinc fingers, respectively.
FIG. 5.
The protein binding to the upstream element is the hamster homologue of human Finb/RREB-1. (A) The nucleotide and deduced amino acid (single-letter code) sequences of the identified cDNA clone are shown. The first 420-nucleotide sequence was obtained by 5′-RACE, whereas the rest was obtained from the identified cDNA clone. The zinc finger domains are underlined and an asterisk indicates the stop codon. (B) ClustalW alignment of hamster RREB-1/Finb (hamRREB-1) (residues 183 to 330) with the region containing zinc fingers 14 and 15 of human Finb (hFinb), human RREB-1 (hRREB-1), chicken RREB-1 (chRREB-1), and the Drosophila Hindsight protein (dHindsight). Identical amino acids are shown as white text on a black background, conservative substitutions are shown as black text on a gray background, and nonconserved residues are shown as black text on a white background. Gaps (-) were created to maximize the alignment. (C) Structural organization of hamster RREB-1, human RREB-1, and human Finb. Horizontal black bars denote sequenced (nucleotide) regions of each protein. The open bars indicate the homologous region of hamster RREB-1 identified by 5′-RACE and the dotted line areas have not been sequenced. Vertical bars indicate positions of zinc fingers in full-length human Finb. Arrows and numbers in parentheses show the positions of amino acids in each protein relative to the full-length protein.
Multiple sequence alignment of the deduced amino acid sequence of the hamster protein with the sequences of human Finb, human RREB-1, and chicken RREB-1 shows that a region spanning 148 amino acids, including the two zinc fingers, is highly homologous to each of the other three proteins (Fig. 5B and C). In addition, the two zinc fingers show considerable homology with the protein product of the Drosophila melanogaster hindsight gene. A high level of homology between the hamster cDNA and the two corresponding human transcripts (75 to 84%) was also observed in the nucleic acid sequences. Using available mouse and human RREB-1 genomic sequences, we obtained additional hamster nucleotide sequence by 5′-RACE extending beyond the original clone that was homologous with the human Finb sequence (Fig. 5A). The 5′ end of the RACE amplified fragment was also highly homologous to the human transcript 5′ of the start of RREB-1 (not shown). A hamster cDNA probe hybridized to a 9.4-kb transcript in HIT cell RNA (not shown) that was comparable in size to the human mRNA (37). Thus, the protein identified in the present work appears to be encoded by the hamster gene for Finb/RREB-1.
Finb/RREB-1 binding to the secretin enhancer is required for full transcriptional activity.
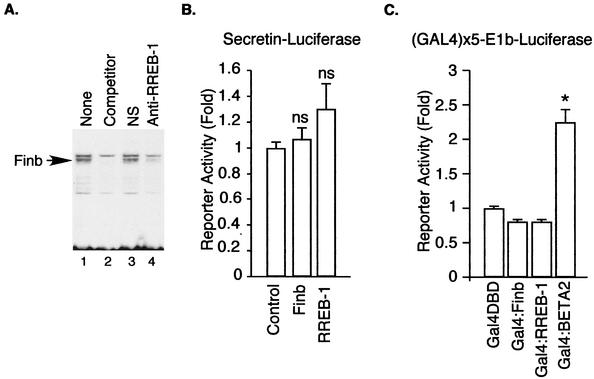
To determine whether Finb/RREB-1 participates in binding to the upstream element, we generated an antipeptide antibody against a short sequence, LQDLTRHMRSHTGERPYK, in the zinc finger region of hamster RREB-1 conserved in the human protein. HIT cell nuclear extracts were treated with the antipeptide antibody before performing gel shift experiments with the upstream element probe (Fig. 6A). Treatment of nuclear extracts from HIT cells with the anti-RREB-1 but not control antiserum prevented formation of the complex, indicating the presence of the cloned protein in the DNA-protein complexes binding to the upstream enhancer element (Fig. 6A).
FIG. 6.
Functional role for RREB-1/Finb. (A) Immunodepletion of RREB-1 diminishes the DNA-protein complex at the upstream element. Nuclear extracts from HIT cells were treated with protein A-Sepharose bound to either anti-RREB-1 (lane 4) or control IgG (lane 3). Note the loss of the specific band denoted by an arrow (lane 1). The specific band is competed out by unlabeled wild-type competitor (lane 2). (B) C33A cells were cotransfected with an equal amount (1 μg) of Finb or RREB-1 expression plasmid or empty vector (control) and a secretin reporter plasmid (0.25 μg). Results shown are means ± SEM for at least seven separate experiments. ns, not significantly different from control. (C) C33A cells were cotransfected with an equal amount (0.5 μg) of Gal4-Finb, Gal4-RREB-1, or Gal4-BETA2 expression plasmid or Gal4 DBD alone and an E1b-luciferase reporter plasmid containing five GAL4 binding sites. Results are shown as the means ± SEM for at least five separate experiments. *, significantly different (P < 0.001) from Gal4 DBD.
Previous studies raised questions about whether RREB-1/Finb directly activates transcription upon DNA binding. To determine whether RREB-1/Finb directly activates transcription from the binding site identified in the present work, we cotransfected C33A cells with a Finb/RREB-1 expression plasmid and a secretin-luciferase reporter plasmid. Neither Finb nor RREB-1 significantly increased the activity of the reporter gene, suggesting that Finb and RREB-1 are not direct transcriptional activators, as was similarly noted previously with the calcitonin (37) and c-erbB2 genes (5) (Fig. 6B). Expression of Finb or RREB-1 as a GAL4 fusion protein failed to increase transcription of a reporter gene with multiple GAL4 binding sites above that seen with GAL4(1-147) alone (Fig. 6C), in contrast to a positive control (GAL4-BETA2).
RREB-1/Finb physically and functionally interacts with BETA2/NeuroD.
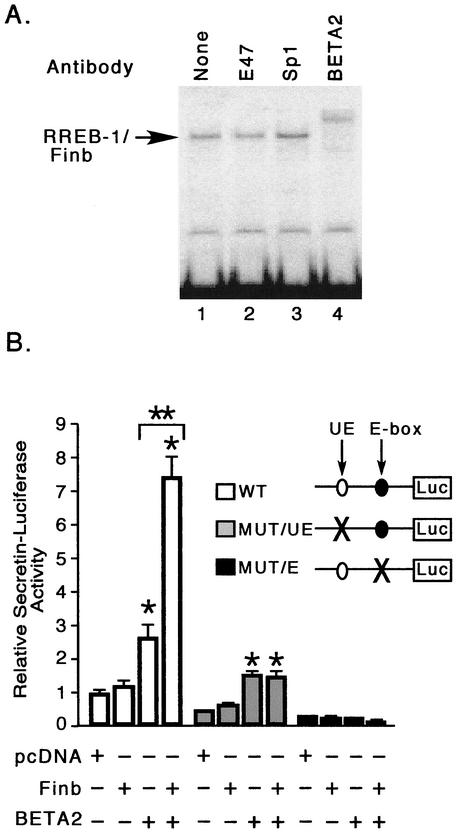
The requirement for Finb/RREB binding to the secretin enhancer for full transcriptional activity despite the lack of intrinsic transcription-activating function by these proteins suggested that Finb or RREB-1 might interact with other proteins binding to the secretin enhancer. We therefore examined DNA-protein complexes containing Finb in gel shift assays for the presence of other proteins known to bind to the enhancer (Fig. 7A). An anti-BETA2/NeuroD antibody supershifted the Finb-containing complex bound to the upstream element probe, indicating the presence of BETA2 in this complex (lane 4). In contrast, antibodies against E47 (lane 2) or Sp1 (lane 3) had no effect on the Finb-DNA complex. The presence of BETA2 in DNA-protein complexes with the upstream element probe, which lacks an E-box sequence, suggested that BETA2 might be recruited to the complex by associating with RREB-1/Finb rather than by direct binding to DNA. A mixture of in vitro-translated BETA2 and E12 did not retard the mobility of the upstream element probe, confirming that these bHLH proteins do not directly bind to the upstream element (not shown).
FIG. 7.
Finb/RREB-1 interacts with BETA2/NeuroD. (A) Heparin-agarose-purified binding activity from HIT cell nuclear extract was examined in a gel shift assay in the absence (lane 1) or presence of anti-E47 (lane 2), anti-Sp1 (lane 3), or anti-BETA2 (lane 4) antibody. (B) C33A cells were cotransfected with an equal amount (1 μg) of Finb expression plasmid or pcDNA (empty vector) in the absence or presence of BETA2 expression plasmid (0.05 μg) and a reporter plasmid (0.25 μg) containing the wild-type secretin promoter (white bars), the mutant promoter lacking the functional upstream element (gray bars), or the E-box element (black bars). Results are shown as the means ± SEM for at least three separate experiments. *, significantly different (P < 0.001) from the respective control (pcDNA); **, significantly different (P < 0.001) from BETA2 alone.
To examine whether Finb functionally interacts with BETA2 to regulate secretin gene transcription, we cotransfected C33A cells with both Finb and BETA2 with a secretin-luciferase reporter gene containing one binding site for each protein. This cell line does not express BETA2, yet the secretin promoter retains sensitivity to transactivation by exogenously expressed BETA2, thus allowing us to assay BETA2-dependent transcription (31). As shown earlier (Fig. 6B), Finb did not increase basal expression of the reporter in the absence of BETA2. BETA2 alone increased reporter expression approximately 2.7-fold. Cotransfection of Finb with BETA2 increased expression 7.5-fold, suggesting that Finb potentiates transcriptional activation of BETA2 despite the lack of intrinsic activation function (Fig. 7B). Finb failed to increase BETA2-dependent transcription with a reporter gene containing a mutation in the Finb/RREB-1 binding site, suggesting that potentiation cannot occur unless Finb is bound to DNA (Fig. 7B). Expression of a reporter containing an E-box mutation did not increase with Finb as well, indicating that recruitment of BETA2 by protein-protein interactions with Finb could not explain potentiation by the latter protein (Fig. 7B). Thus, Finb can potentiate BETA2 only when both proteins are bound to DNA.
We examined the ability of RREB-1 to interact with BETA2 in an in vitro binding assay to determine whether BETA2 and RREB-1 associate directly. Both in vitro-translated and -labeled Finb and RREB-1 were retained by a GST-BETA2 affinity resin (Fig. 8A, lanes 3 and 9) but not by GST alone (lanes 1 and 7) bound to the resin, indicating that Finb and RREB-1 directly interact with BETA2. To localize the BETA2 interaction domain in Finb/RREB-1, we synthesized several deletion mutants of RREB-1/Finb and tested their ability to bind to GST-BETA2 agarose beads. Deletion of the C-terminal 125 residues of RREB-1/Finb had no effect on the interaction with BETA2 (Fig. 8A, lane 6), whereas a mutant with 285 residues removed from the N terminus of RREB-1 did not bind to the GST-BETA2 resin (Fig. 8A, lane 11). This observation suggests that BETA2 interacts with the region of Finb/RREB-1 spanned by amino acids 912 to 1196 of Finb, near the N terminus of RREB-1.
FIG. 8.
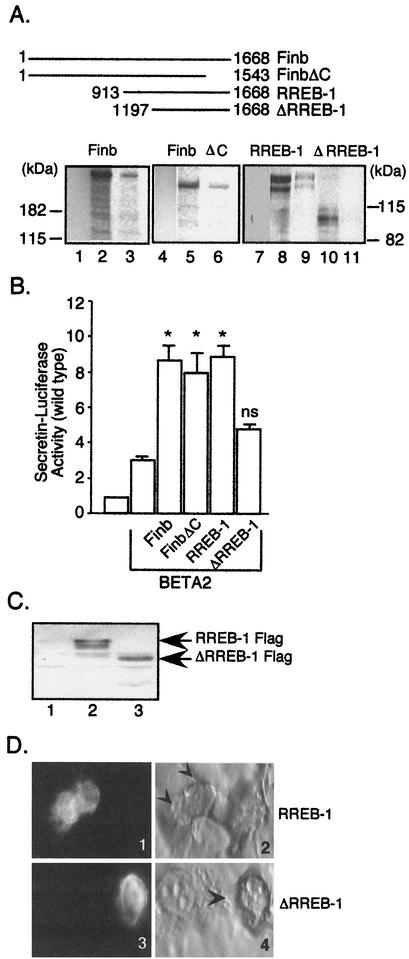
Functional cooperation between Finb/RREB-1 and BETA2 requires a BETA2 interaction domain in Finb. (A) The top panel shows the Finb and RREB-1 protein truncations relative to the full-length protein. Numbers indicate the positions of amino acids in each protein. The bottom panel shows in vitro-translated, 35S-labeled Finb/RREB-1 proteins examined for the ability to bind to BETA2 expressed as a GST fusion protein. Lanes 3, 6, 9, and 11, radiolabeled proteins captured by GST-BETA2 proteins; lanes 1, 4, and 7, proteins captured by GST alone. Lanes 2, 5, 8, and 10 show approximately 10% of the input protein applied to the affinity matrix. (B) C33A cells were cotransfected with a BETA2 expression plasmid, wild-type secretin reporter plasmid, and either Finb, FinbΔC, RREB-1, or ΔRREB-1 expression plasmid. Results are shown as the means ± SEM for at least five separate experiments normalized for transfection efficiency. ns, not significantly different versus BETA2 alone; *, P ≤ 0.001 versus BETA2 alone. (C) C33A cells were transfected with an equivalent amount of expression plasmid for RREB-1-FLAG (lane 2), ΔRREB-1-FLAG (lane 3), or pcDNA (lane 1). The expressed proteins were detected in the cell lysates by immunoblotting with a monoclonal anti-FLAG M2 antibody (Sigma). (D) Immunofluorescent staining of FLAG-tagged, transfected RREB-1. Staining of transfected RREB-1 (panel 1) and ΔRREB-1 (panel 3) is shown. The corresponding bright field views are shown in panels 2 and 4, respectively, with FLAG-stained cells denoted by arrowheads.
We examined the ability of Finb, FinbΔC, RREB-1, or ΔRREB-1 to potentiate the activity of BETA2 to determine whether physical interaction of Finb and RREB-1 was required for potentiation. We transiently cotransfected C33A cells with an expression plasmid for Finb, FinbΔC, RREB-1, or ΔRREB-1 plus BETA2 and a wild-type secretin-luciferase reporter (Fig. 8B). As shown earlier for Finb and RREB-1 (Fig. 6B), FinbΔC and ΔRREB-1 themselves had no effect on expression of a secretin-luciferase reporter (not shown). FinbΔC, like Finb and RREB-1, potentiated the BETA2-induced activation of the reporter more than 2.5-fold (Fig. 8B). In contrast, the effect of ΔRREB-1, which lacks the BETA2 interaction domain, did not significantly increase BETA2-dependent transcription. The loss of potentiation did not result from the failure to express the ΔRREB-1 mutant, as immunoblotting for transfected RREB-1 containing a FLAG epitope tag clearly shows that the mutant protein was expressed (Fig. 8C). Immunostaining for the transfected RREB-1 protein with FLAG antibody showed similar nuclear staining for both the wild-type and mutant protein, indicating that the loss of potentiation did not arise from a difference in intracellular localization of the mutant protein (Fig. 8D). Our results suggest that potentiation of BETA2 activity on the secretin gene enhancer by Finb/RREB-1 requires that both Finb and BETA2 interact with their respective DNA-binding sites and physically associate with each other.
DISCUSSION
The expression of the secretin gene is mainly restricted to a subpopulation of enteroendocrine cells in adult animals and to a fraction of pancreatic β cells during fetal development (38). Cell-specific expression of many genes relies on the interaction of both cell-type specific and ubiquitously expressed transcription factors with cis-acting elements of the promoters. Transcription of the secretin gene in secretin-expressing cell lines depends on the presence of an enhancer within 175 bp of the transcription start site (38). We previously showed that the enhancer activity of the secretin promoter is highly dependent upon the presence of an E-box that binds a cell-specific bHLH protein, BETA2, together with one of its ubiquitously expressed heterodimeric partners, E12 or E47 (22).
In this paper, we have further characterized the secretin enhancer and identified three additional mutationally sensitive elements. These include two GC-rich sequences that bind to Sp1 and related proteins, including Sp3, as well as the Finb/RREB-1 binding site. The widespread distribution of Sp1, Sp3, and Finb/RREB-1 in most tissues makes it unlikely that they direct cell type-specific expression of secretin in enteroendocrine cells based on their selective expression in those cells. BETA2/NeuroD is the only factor binding to the secretin enhancer with a relatively restricted expression pattern limited to enteroendocrine cells, pancreatic islet cells, developing neurons, and the corticotrophs of the anterior pituitary. The secretin gene is the only target gene for BETA2 identified thus far that is known to bind to Finb/RREB-1. The combination of these factors binding to the enhancer as an enhanceosome complex may contribute to cell type-specific expression of the secretin gene, potentially along with other yet-to-be-identified factors that bind at some distance from this proximal enhancer.
RREB-1 was first identified as a 756-amino-acid protein containing four zinc fingers that bound to a distal Ras-responsive element of the human calcitonin gene (37). However, an open reading frame extended upstream from the methionine residue assigned as the first codon, suggesting that RREB-1 was part of a larger protein. The isolation of a cDNA encoding the much larger 1,668-amino-acid protein Finb, containing 15 zinc finger domains, confirmed that RREB-1 was contained within a larger protein. Finb was cloned from a cDNA expression library from human breast carcinoma cell lines by virtue of its ability to bind to a regulatory element of the human c-erbB2 gene promoter (5). Differences between several Finb clones suggested that the mRNA might undergo alternate splicing with the generation of multiple forms of the protein.
The identified DNA sequences of the RREB-1/Finb binding site exhibit considerable divergence. The DNA-binding sites for Finb/RREB-1 in the calcitonin and c-erbB2 genes share only 8 of 14 nucleotides with the minimal sequence in the secretin gene upstream element and 3 of 14 bases with each other (Table 1). Similarly, the secretin gene upstream element shared only 4 of 10 positions with a consensus binding site sequence identified by cyclic amplification and selection of target sequences. One possible explanation for the divergence in binding sites is that DNA-binding specificity is context specific depending on the surrounding sequences and/or the presence of other nearby DNA-bound proteins. The two zinc fingers nearest to the C terminus of RREB-1 bound with similar affinities to two different sequences, an RREB-1 binding site and the sequence GGTCCT (42). The isolation of the hamster homologue of Finb containing the same two C-terminal zinc finger motifs further suggested that the DBD of Finb resides within these two zinc fingers. Of note, these two zinc fingers were highly conserved in the Drosophila zinc finger protein encoded by the hindsight gene (41).
TABLE 1.
Comparison of Finb/RREB-1 binding sites in different promotersa
| Gene | Binding site |
|---|---|
| Secretin | GAGGGGCGCCAACA |
| Calcitonin | --GGTCCCCCACCA |
| c-erbB2 | GACCGGAGAAACCA |
| RREB-1 consensus | ----CCCCAAACCA |
The minimal Finb binding site in the rat secretin promoter (−174 to −161) was aligned with the binding sites in the human calcitonin and c-erbB2 promoters as well as the published RREB-1 consensus binding site (37). The conserved nucleotides are in bold. The common nucleotides in each binding site are underlined.
The function of Finb is not known. However, transcripts for this gene have been identified in all tissues examined in both mammals and avian species with the exception of the brain (5, 20). Hindsight, a related zinc finger protein in Drosophila, is also expressed in several tissues during fly development (41). Interestingly, analyses of flies with mutant hindsight alleles revealed that expression of the protein, particularly in aminoserosal cells adjacent to the epidermis, was required to direct epidermal cells to undergo cell shape changes driving germ band retraction (13). Similarly, hindsight expression in all tracheal cells during tracheal morphogenesis was required to maintain epithelial integrity and direct assembly of the extracellular structures and to prevent apoptosis (39).
Although Finb and RREB-1 cDNAs were isolated on the basis of their ability to bind to the c-erbB2 or calcitonin promoters, respectively, their role in gene transcription was unclear. Neither Finb nor RREB-1 stimulated transcription of reporter genes containing the c-erbB2 or calcitonin promoter sequences. In the case of the calcitonin gene, transcriptional activation occurred only in the presence of activated Ras or Raf. These results suggested that Finb and RREB-1 might not be direct transcriptional activators. Although Finb increased expression of metallothionein and thymidine kinase reporter genes, it is not known whether the observed activation results from the binding of Finb to these promoters or from the regulation of other transcription factors (5). The role of redox conditions and Ref-1 on Finb function is unclear in vivo. Overexpression of Ref-1 had no effect on secretin promoter activity either in the absence or the presence of cotransfected Finb (not shown). Ref-1 may function to stabilize the DNA-binding activity of Finb/RREB-1 in vitro by maintaining its conformation during protein fractionation. Using an antipeptide antibody against Finb/RREB-1, we show for the first time the presence of Finb/RREB-1 in DNA-protein complexes generated from unfractionated nuclear extracts. In addition, loss of transcriptional activity in transient transfection experiments upon introduction of mutations into the Finb/RREB-1 binding site suggests that Finb/RREB-1 has a direct role in secretin gene transcription.
Expression of Finb or RREB-1 fused to the DBD of GAL4 failed to activate a GAL4-luciferase reporter, further implying that this protein lacks an intrinsic activation domain. Our results suggest that the ability to potentiate the transcriptional activity of BETA2 may be the major function of Finb/RREB-1 in the regulation of secretin gene expression. This potentiation requires binding of both proteins to the enhancer and the presence of a BETA2 interaction domain in Finb.
The present work suggests that the ability of Finb/RREB-1 to interact with other transcription factors may be more important than its ability to directly activate transcription. RREB-1 could not activate the calcitonin promoter except in the presence of activated Ras or Raf. The dependence on the Ras signaling pathway may indicate that RREB-1 interacts with another transcription factor regulated by protein kinases. However, the nature of the proteins involved in Ras signaling that interact with RREB-1 and the mechanism of transcriptional activation when Ras is activated remain to be elucidated.
We and others have shown that BETA2 functions as a relatively modest transcriptional activator even in the presence of its bHLH partner proteins (23, 29, 30). The insulin and POMC genes are two genes that are regulated by BETA2 for which the activity of BETA2 is enhanced by PDX-1 and Pitx-1, respectively. The present work provides a third example of how yet another DNA-binding protein, Finb/RREB1, interacts with BETA2 to increase transcriptional activation of the secretin gene, one of the targets of BETA2. Finb/RREB-1 is devoid of intrinsic activating function, unlike the two homeodomain proteins PDX-1 and Pitx-1, which are potent transcriptional activators on their own.
The homeodomain protein PDX-1 interacts with BETA2 and synergistically enhances its transcriptional activity on the insulin promoter (7, 26, 30). Additional studies suggest that the p300 coactivator may further enhance the synergy between BETA2 and PDX-1 (30). The functional synergy between PDX-1 and BETA2 does not occur if the binding site for either is disrupted (30).
The synergism between BETA2 and the homeodomain protein Pitx-1 on the POMC gene differs from the potentiation of BETA2 by Finb/RREB-1 in several respects (28). Pitx-1 does not appear to directly interact with BETA2 but instead associates with one of the ubiquitously expressed bHLH dimerization partners of BETA2. A second major difference is that Pitx-1 enhances BETA2-dependent transcription of POMC reporter genes without site-specific DNA binding (28).
The activity of a number of transcription factors is synergistically increased by other transcription factors binding to separate sites on DNA. In these cases, the transcriptional activation in the presence of both factors exceeds the product of the transcriptional activities of either factor alone. Potentiation of BETA2 activity by Finb/RREB-1 differs from the many known examples of synergistic activation between two or more DNA-binding proteins, each with its own intrinsic activation functions.
In the present work, we show that Finb/RREB-1 increases the activity of BETA2 despite its lack of an intrinsic activation domain. Transcriptional activity generated by the interaction of an activator with a nonactivator partner is well established for heteromeric complexes binding to a single DNA element or closely spaced half-sites. Increased transcriptional activity arising from interactions between BETA2 and Finb/RREB-1 binding to distinct sites separated by 19 bp appears to be uncommon.
The zinc finger protein Ikaros appears to potentiate activity of several transcription factors that bind to the thymidine kinase promoter, including Sp1 and CTF, even in the absence of a domain that confers activation to a GAL4 DBD, suggesting that potentiation is not related to an intrinsic activation domain (11). Like Finb/RREB-1, potentiation by Ikaros is absolutely dependent on its ability to bind to DNA. However, the mechanism of transcriptional potentiation by Ikaros appears to differ from that by Finb/RREB-1. Unlike that by Finb/RREB-1, potentiation by Ikaros does not require protein-protein interaction with the transcription factors it potentiates. Instead, the ability of Ikaros protein to potentiate transcription appears to correlate with its localization in pericentromeric heterochromatin.
Finb/RREB-1 appears to modulate the transcription-activating function of the bHLH protein BETA2 by an uncommon mechanism. Most proteins involved in regulation of RNA polymerase II transcription can be classified as general transcription factors, gene-specific transcription factors, or transcription cofactors (coactivators and corepressors). In the present work, we show that Finb/RREB-1 increases the activity of BETA2 yet is not a classical activator of transcription that functions by recruiting its own activation domain to DNA. The function of Finb/RREB-1 is also distinct from that of coactivators, which modulate transcription without site-specific binding to DNA. Thus, we suggest that Finb/RREB-1 should be classified as a potentiator of transcription. Further studies of the regulation of secretin gene transcription and other Finb target genes will provide important new information about the mechanisms controlling eukaryotic gene expression.
Acknowledgments
This work was supported in part by NIH grants DK43673 and DK52870 to A.B.L. and by GRASP Digestive Disease Center grants P30-DK34928 and T32-DK07542.
We thank Barry Nelkin (Johns Hopkins University, Baltimore, Md.), Tadashi Yamamoto (University of Tokyo, Tokyo, Japan), and Tom Curran (St. Jude Children's Research Hospital, Memphis, Tenn.) for generously providing plasmids as noted under Materials and Methods.
REFERENCES
- 1.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumonteil, E., B. Laser, I. Constant, and J. Philippe. 1998. Differential regulation of the glucagon and insulin I gene promoters by basic helix-loop-helix transcription factors E47 and Beta2. J. Biol. Chem. 273:19945-19954. [DOI] [PubMed] [Google Scholar]
- 3.Edlund, T., M. D. Walker, P. J. Barr, and W. J. Rutter. 1985. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science 230:912-916. [DOI] [PubMed] [Google Scholar]
- 4.Fried, M., and D. Crothers. 1983. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 11:141-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto-Nishiyama, A., S. Ishii, S. Matsuda, J.-I. Inoue, and T. Yamamoto. 1997. A novel zinc finger protein, Finb, is a transcriptional activator and localized in nuclear bodies. Gene 195:267-275. [DOI] [PubMed] [Google Scholar]
- 6.German, M. S., M. A. Blanar, C. Nelson, L. G. Moss, and W. J. Rutter. 1991. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol. Endocrinol. 5:292-299. [DOI] [PubMed] [Google Scholar]
- 7.Glick, E., D. Leshkowitz, and M. D. Walker. 2000. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J. Biol. Chem. 275:2199-2204. [DOI] [PubMed] [Google Scholar]
- 8.Huang, H. P., K. Chu, E. Nemoz-Gaillard, D. Elberg, and M. J. Tsai. 2002. Neogenesis of b-cells in adult BETA2/NeuroD-deficient mice. Mol. Endocrinol. 16:541-551. [DOI] [PubMed] [Google Scholar]
- 9.Jayaraman, L., K. G. K. Murthy, C. Zhu, T. Curran, S. Xanthoudakis, and C. Prives. 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11:558-570. [DOI] [PubMed] [Google Scholar]
- 10.Kadonaga, J. T., and R. Tjian. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83:5889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koipally, J., E. J. Heller, J. R. Seavitt, and K. Georgopoulos. 2002. Unconventional potentiation of gene expression by Ikaros. J. Biol. Chem. 277:13007-13015. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamka, L. M., and D. H. Lipshitz. 1999. Role of the amnioserosa in germ band retraction of the Drosophila melanogaster embryo. Dev. Biol. 214:102-112. [DOI] [PubMed] [Google Scholar]
- 14.Lando, D., I. Pongratz, L. Poellinger, and M. L. Whitelaw. 2000. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and HIF-like factor. J. Biol. Chem. 275:4618-4627. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. E., S. M. Hollenberg, L. Snider, D. L. Turner, N. Lipnick, and H. Weintraub. 1995. Conversion of Xenopus ectoderm into neurons by neuroD, a basic helix-loop-helix protein. Science 268:836-844. [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., C. Lin, A. Glieberman, K. A. Ohgi, T. Herman, H. P. Huang, M. J. Tsai, and M. G. Rosenfeld. 2001. Tbx19, a tissue-selective regulator of POMC gene expression. Proc. Natl. Acad. Sci. USA 98:8674-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, M., F. A. Pereira, S. D. Price, M. Chu, C. Shope, D. Himes, R. A. Eatock, W. E. Brownell, A. Lysakowski, and M. J. Tsai. 2000. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 14:2839-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, M., S. J. Pleasure, A. E. Collins, J. L. Noebels, F. J. Naya, M. J. Tsai, and D. H. Lowenstein. 2000. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl. Acad. Sci. USA 97:865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez, M. J., B. H. Upchurch, G. Rindi, and A. B. Leiter. 1995. Studies in transgenic mice reveal potential relationships between secretin-producing cells and other endocrine cell types. J. Biol. Chem. 270:885-891. [DOI] [PubMed] [Google Scholar]
- 20.Miyake, J. H., D. P. Szeto, and W. E. Stumph. 1997. Analysis of the structure and expression of the chicken gene encoding a homolog of the human RREB-1 transcription factor. Gene 202:177-186. [DOI] [PubMed] [Google Scholar]
- 21.Miyata, T., T. Maeda, and J. E. Lee. 1999. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutoh, H., B. Fung, F. Naya, M.-J. Tsai, J. Nishitani, and A. B. Leiter. 1997. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc. Natl. Acad. Sci. USA 94:3560-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutoh, H., F. J. Naya, M.-J. Tsai, and A. B. Leiter. 1998. The basic helix loop helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes Dev. 12:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naya, F. J., H. Huang, Y. Qiu, H. Mutoh, F. DeMayo, A. B. Leiter, and M.-J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 11:2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naya, F. J., C. M. M. Stellrecht, and M.-J. Tsai. 1995. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9:1009-1019. [DOI] [PubMed] [Google Scholar]
- 26.Ohneda, K., R. G. Mirmira, J. Wang, J. D. Johnson, and M. S. German. 2000. The homeodomain of PDX1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 20:900-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyton, M., L. G. Moss, and M. J. Tsai. 1994. Two distinct class A helix-loop-helix transcription factors, E2A and BETA1, form separate DNA binding complexes on the insulin gene E box. J. Biol. Chem. 269:25936-25941. [PubMed] [Google Scholar]
- 28.Poulin, G., M. Lebel, M. Chamberland, F. W. Paradis, and J. Drouin. 2000. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 20:4826-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulin, G., B. Turgeon, and J. Drouin. 1997. NeuroD1/b2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17:6673-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qui, Y., M. Guo, S. Huang, and R. Stein. 2002. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 22:412-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratineau, C., M. W. Petry, H. Mutoh, and A. B. Leiter. 2002. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J. Biol. Chem. 277:8847-8853. [DOI] [PubMed] [Google Scholar]
- 32.Rindi, G., C. Ratineau, A. Ronco, M. E. Candusso, M. J. Tsai, and A. Leiter. 1999. Targeted ablation of secretin-producing cells in transgenic mice reveals a common differential pathway with multiple enteroendocrine cell lineages in the small intestine. Development 126:4149-4156. [DOI] [PubMed] [Google Scholar]
- 33.Sharma, S., U. S. Jhala, T. Johnson, K. Ferreri, J. Leonard, and M. Montminy. 1997. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol. Cell. Biol. 17:2598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, H., J. H. LeBowitz, A. S. Baldwin, and P. A. Sharp. 1988. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell 52:415-423. [DOI] [PubMed] [Google Scholar]
- 35.Tell, G., L. Pellizzari, D. Cimarosti, C. Pucillo, and G. Damante. 1998. Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun. 252:178-183. [DOI] [PubMed] [Google Scholar]
- 36.Tell, G., A. Scaloni, L. Pellizzar, S. Formisano, C. Pucillo, and G. Damante. 1998. Redox potential controls the structure and DNA binding activity of the paired domain. J. Biol. Chem. 273:25062-25072. [DOI] [PubMed] [Google Scholar]
- 37.Thiagalingam, A., A. D. Bustros, M. Borges, R. Jasti, D. Compton, L. Diamond, M. Mabry, D. W. Ball, S. B. Baylin, and B. D. Nelkin. 1996. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol. Cell. Biol. 16:5335-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler, M. B., J. Nishitani, A. M. J. Buchan, A. S. Kopin, W. Y. Chey, T. Chang, and A. B. Leiter. 1992. Identification of a transcriptional enhancer important for enteroendocrine and pancreatic islet cell-specific expression of the secretin gene. Mol. Cell. Biol. 12:3531-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilk, R., B. H. Reed, U. Tepass, and D. Lipshitz. 2000. The hindsight gene is required for epithelial maintenance and differentiation of the tracheal system in Drosophila. Dev. Biol. 219:183-196. [DOI] [PubMed] [Google Scholar]
- 40.Xanthoudakis, S., G. Miao, F. Wang, Y.-C. E. Pan, and T. Curran. 1992. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 11:3323-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yip, M. L. R., M. L. Lamka, and H. D. Lipshitz. 1997. Control of germ-band retraction in Drosophila by the zinc-finger protein HINDSIGHT. Development 124:2129-2141. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, L., J. Zhao, and H. J. Edenberg. 1999. A human raf-responsive zinc-finger protein that binds to divergent sequences. Nucleic Acids Res. 27:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]