Abstract
Ligand-dependent transcriptional activation by nuclear receptors involves the recruitment of various coactivators to the promoters of hormone-regulated genes assembled into chromatin. Nuclear receptor coactivators include histone acetyltransferase complexes, such as p300/CBP-steroid receptor coactivator (SRC), as well as the multisubunit mediator complexes (“Mediator”), which may help recruit RNA polymerase II to the promoter. We have used a biochemical approach, including an in vitro chromatin assembly and transcription system, to examine the functional role for Mediator in the transcriptional activity of estrogen receptor α (ERα) with chromatin templates, as well as functional interplay between Mediator and p300/CBP during ERα-dependent transcription. Using three different approaches to functionally inactivate Mediator (immunoneutralization, immunodepletion, and inhibitory polypeptides), we find that Mediator is required for maximal transcriptional activation by ligand-activated ERα. In addition, we demonstrate synergism between Mediator and p300/CBP-SRC during ERα-dependent transcription with chromatin templates, but not with naked DNA. This synergism is important for promoting the formation of a stable transcription preinitiation complex leading to the initiation of transcription. Interestingly, we find that Mediator has an additional distinct role during ERα-dependent transcription not shared by p300/CBP-SRC: namely, to promote preinitiation complex formation for subsequent rounds of transcription reinitiation. These results suggest that one functional consequence of Mediator-ERα interactions is the stimulation of multiple cycles of transcription reinitiation. Collectively, our results indicate an important role for Mediator, as well as its functional interplay with p300/CBP-SRC, in the enhancement of ERα-dependent transcription with chromatin templates.
Nuclear receptors (NRs) and their cognate ligands (e.g., steroids, retinoids, thyroid hormone, and vitamin D3) are critical regulators of diverse physiological processes, including growth, development, metabolism, homeostasis, and reproduction. The NRs comprise a large superfamily of DNA-binding regulatory proteins that control the transcription of distinct subsets of genes in the chromatin environment of the nucleus (10, 33). Most NRs share a conserved structural and functional organization, with distinct domains for DNA binding, ligand binding, and transcriptional activation (10, 33). Members of the NR superfamily can be categorized based on their dimerization and DNA binding properties (33). Class I NRs include the steroid hormone receptors, which bind to inverted repeat sequences in DNA primarily as homodimers. Class II NRs include retinoid, thyroid hormone, and vitamin D3 receptors, which bind to direct repeats as heterodimers with the common dimerization partner RXR. Although these two classes of NRs share many structural and functional similarities, differences in their association with corepressors, ability to repress basal transcription, and use of particular classes of coactivators have been observed (reviewed in references 13 and 34).
Ligand-dependent transcriptional activation by NRs is a complex process involving the recruitment of various coactivators to the promoters of hormone-regulated genes assembled into chromatin (12, 28). Two widely studied groups of NR coactivators are: (i) p300 and the highly related CREB-binding protein (CBP) and (ii) the mammalian Mediator complexes (TRAP, DRIP, ARC, CRSP, SMCC, etc.) (13, 31, 34, 41). p300 and CBP are recruited to ligand-activated, DNA-bound NRs by the steroid receptor coactivator (SRC) family of bridging factors (SRC1, -2, and -3) (13, 30, 34). The SRC proteins have receptor interaction domains (RIDs) containing LXXLL motifs that contact a hydrophobic groove on the receptor ligand binding domains (30). p300 and CBP are histone acetyltransferases (HATs) capable of acetylating nucleosomal histones, a covalent modification generally associated with the enhancement of transcription (reviewed in references 13, 14, 34, and 36). Mediator complexes interact with many NRs, as well as RNA polymerase II (RNA pol II), helping to recruit RNA pol II to ligand-activated promoters (31, 41). The precise roles of each type of coactivator, as well as the functional interactions among them, during NR-dependent transcription have not been defined.
The mammalian Mediator complexes are heteromeric complexes containing from 7 to 25 distinct polypeptides, many of which share homology to subunits of the yeast Mediator complex (reviewed in references 3, 31, and 41). For the purposes of this paper, we will refer to the mammalian Mediator complexes collectively as “Mediator” and the individual subunits according to the unifying nomenclature proposed by Rachez and Freedman (e.g., Med220 = TRAP220, DRIP205, and ARC205) (41). Although the original studies with yeast suggested that Mediator is tightly associated with the RNA pol II holoenzyme (reviewed in reference 31), recent studies from yeast and more complex eukaryotes suggest that Mediator may not be an obligatory component of the RNA pol II holoenzyme (2, 4, 8, 40, 49). Like the p300/CBP-SRC complexes, Mediator complexes, as well as some individual Mediator subunits (e.g., Med220 and Med150), have been shown to bind directly to some class I and class II NRs in the presence of their cognate ligands (7, 19, 42, 50, 51). Like the RIDs of the SRC proteins, the Med220 RID contains receptor binding LXXLL motifs that appear to be the primary structural feature of the Mediator complex that contacts NR ligand binding domains (31, 41).
A number of important questions regarding Mediator function with NRs have yet to be addressed in detail, including the role or roles of Mediator during the transcription process and whether Mediator interacts functionally with other distinct classes of coactivator proteins, such as the histone-modifying coactivator p300/CBP. Herein, we used a biochemical approach, including an in vitro chromatin assembly and transcription system, to examine the specific roles of Mediator and p300/CBP-SRC complexes in the transcriptional activity of estrogen receptor α (ERα), a class I NR. We find that both Mediator and p300/CBP-SRC are required for maximal transcriptional activation by ligand-activated ERα and that synergism between the two coactivators is important for promoting the formation of a stable transcription preinitiation complex leading to the initiation of transcription. Interestingly, we find that Mediator has an additional distinct role during ERα-dependent transcription not shared by p300/CBP-SRC: namely, to promote preinitiation complex formation for subsequent rounds of transcription reinitiation. Collectively, our results indicate an important role for Mediator, as well as its functional interplay with p300/CBP-SRC, in the enhancement of ERα-dependent transcription with chromatin templates.
MATERIALS AND METHODS
Plasmids.
2ERE-pS2 and pERE contain two or four tandem copies of an estrogen response element (ERE) upstream of the human pS2 promoter or adenovirus E4 promoter, respectively (27). 2Gal-pS2 and 2Gal-E4 contain two tandem copies of a Gal4 UAS site upstream of the pS2 and adenovirus E4 promoter, respectively. pGLM-ENH contains the human monocyte chemoattractant protein 1 (MCP-1) promoter and NF-κB enhancer (48).
Synthesis and purification of recombinant proteins.
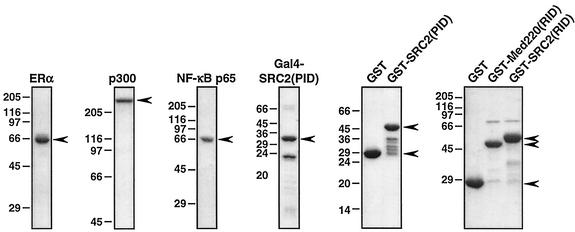
Full-length Flag-tagged human ERα, full-length six-His-tagged human p300, and full-length six-His-tagged NF-κB p65 were synthesized in Sf9 cells by using a baculovirus expression system and purified as described previously (25). The six-His-tagged fusion of the Gal4 DNA binding with SRC2(PID) [referred to as Gal4-SRC2(PID)] was expressed in Escherichia coli and purified by standard nickel-nitrilotriacetic acid (NTA) affinity chromatography (23). The fusions of glutathione S-transferase (GST) with the RID of SRC2 (amino acids 563 to 767), the p300/CBP interaction domain (PID) of SRC2 (amino acids 1010 to 1130), or the RID of Med220 (amino acids 508 to 695) were expressed in E. coli and purified by standard glutathione-agarose affinity chromatography (23). Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses of the purified proteins are shown in Fig. 1.
FIG. 1.
SDS-PAGE analysis of the purified recombinant proteins used in this study. Recombinant Flag-tagged, His-tagged, and GST-fused proteins were expressed in insect cells or in E. coli and were purified by using anti-Flag M2, nickel-NTA, and glutathione-agarose affinity chromatography, respectively, as described in Materials and Methods. Aliquots of the purified proteins were run on acrylamide-SDS gels with subsequent staining with Coomassie brilliant blue R-250. The sizes of molecular mass markers are shown.
Immunoneutralization and immunodepletion of HNEs.
HeLa cell nuclear extracts (HNEs) were prepared according to the method of Dignam et al. (9) without an ammonium sulfate precipitation step. For immunoneutralization of Mediator, 100-μl aliquots of HNE containing approximately 1 mg of total protein were incubated with 4 μg of either anti-Med220, anti-Med33, or control antibodies (final concentration, 40 ng/μl), or 2 μg each of anti-Med220 and anti-Med33 antibodies in combination (20 ng/μl each), in the presence of 45 μg of recombinant protein G (450 ng/μl; Sigma) for 4 h at 4°C with gentle mixing. The extracts were then used in the in vitro transcription reactions described below.
For immunodepletion of Mediator, 250-μl aliquots of HNE containing approximately 2.5 mg of protein were incubated with 5 μg each of anti-Med220 and anti-Med33 antibodies (20 ng/μl each) or 10 μg of control antibody (40 ng of goat anti-mouse immunoglobulin G per μl) in the presence of 45 μl (packed volume) of protein G agarose resin (containing 450 ng of protein G per μl; Gibco BRL) for 4 h at 4°C with gentle mixing. The resin was collected by centrifugation at 1000 × g in a microcentrifuge at 4°C, and the supernatant was saved for immunoblotting and transcription experiments. After removal of the supernatant, the resin was washed three times with a wash buffer containing 20 mM Tris-HCl (pH 7.9), 200 mM NaCl, 0.2 mM EDTA, 10% glycerol, and 0.1% NP40 and boiled in 1× SDS gel loading buffer. Control and Mediator-depleted extracts, as well as the immunoprecipitated material, were analyzed by immunoblotting with a panel of antibodies to various Mediator subunits, coactivators, chromatin remodelers, and components of the basal transcription machinery. Antibodies to the following proteins were obtained from the sources listed: Med220, Med78, Med34, Med33, cdk8, p300, CBP, SRC1, SRC3, Brg1, Brm, TBP, TFIIB, TFIIEβ, TFIIF (RAP74 subunit), and RNA pol II (large subunit) were from Santa Cruz Biotechnology, Inc.; Med150 and Med130 were provided by L. Freedman; SRC2 was provided by P. Chambon; and PCAF was provided by Y. Nakatani.
In vitro chromatin assembly and transcription reactions.
Chromatin assembly reactions were performed with a chromatin assembly extract derived from Drosophila embryos as described previously (5, 25). ERα, 17β-estradiol (E2), NF-κB p65, and Gal4-SRC2(PID) were added during the chromatin assembly reactions, whereas p300 and the GST fusions were added after chromatin assembly was complete. Unless noted otherwise, the final concentrations of factors in the transcription reactions were as follows: ERα, 4.5 nM; E2, 30 nM; NF-κB p65, 200 nM; Gal4-SRC2(PID), 5.5 nM; p300, 30 nM; and the GST fusions, 180 or 225 nM. Multiple- and single-round in vitro transcription reactions were performed with chromatin or naked DNA templates as described previously (25) by using control or Mediator-depleted HNEs. The RNA products were analyzed by primer extension (25). All reactions were performed in duplicate, and each experiment was performed a minimum of three times to ensure reproducibility. The data were analyzed with a Molecular Dynamics Phosphorimager.
RESULTS
Immunoneutralization of Mediator in HNEs impairs ERα-dependent transcription with chromatin templates.
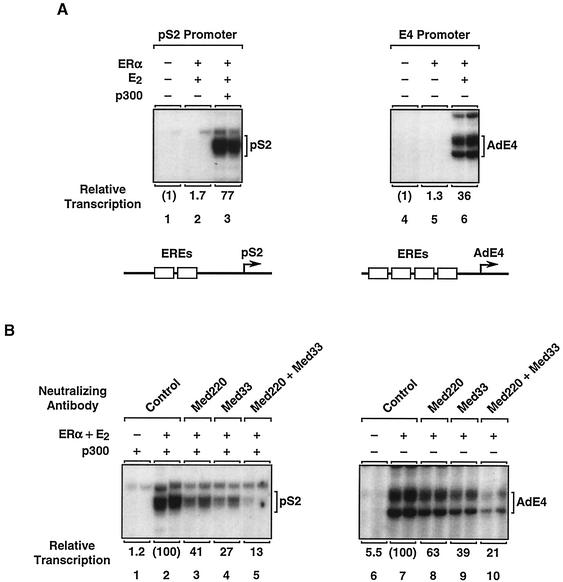
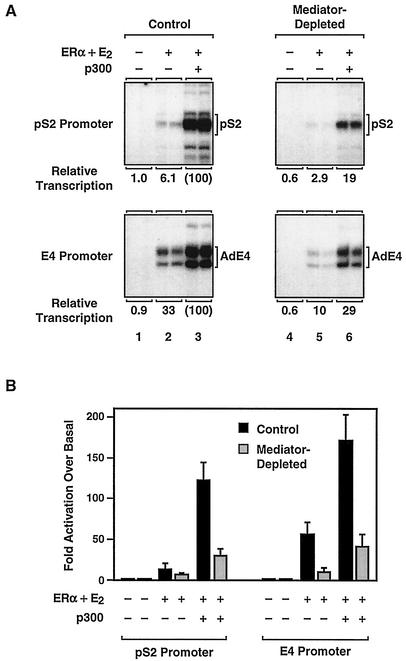
To examine the functional interplay between distinct classes of coactivators during ERα-dependent transcription, we used a biochemical approach, including a previously characterized in vitro chromatin assembly and transcription system (26). Plasmid templates containing EREs located upstream of the human pS2 promoter or the adenovirus E4 promoter (see schematics in Fig. 2A) were assembled into chromatin with a Drosophila embryo extract (S190) (5). The templates were then transcribed in the presence of purified recombinant human ERα (Fig. 1) by using HNE (9). ERα- and 17β-estradiol (E2)-dependent transcription was observed with both templates (Fig. 2A and data not shown). Efficient activation of the E4 template by ERα occurred with the endogenous p300/CBP in the HeLa extract, while efficient activation of the pS2 template required the addition of purified recombinant p300, as we have shown previously (Fig. 2A) (26, 27).
FIG. 2.
Immunoneutralization of Mediator in HNEs inhibits ERα-dependent transcription. (A) Ligand- and p300-dependent transcription by ERα with chromatin templates in vitro. The plasmid template 2ERE-pS2 containing two EREs upstream of the human pS2 promoter (bottom, left) or the plasmid template pERE containing four EREs upstream of the adenovirus E4 promoter (bottom, right) was assembled into chromatin in the presence of recombinant ERα, 17β-estradiol (E2), and p300, as indicated. The chromatin templates were transcribed with an HNE, and the resulting RNA products were analyzed by primer extension. Relative levels of transcription are indicated. (B) Effect of Mediator immunoneutralization on ERα-dependent transcription with chromatin templates. HNEs were incubated with anti-Med220 or anti-Med33 antibodies (40 ng/μl), a combination of both antibodies (20 ng/μl each), or a control antibody (40 ng/μl), as indicated. After the formation of immune complexes, the variously treated HNEs were tested for their ability to support ERα-dependent transcription with chromatin templates as described in panel A.
To analyze the contribution of Mediator complexes to ERα-dependent transcription with chromatin templates, we used an in vitro Mediator immunoneutralization protocol conceptually similar to cell-based antibody microinjection protocols described previously (21, 24). Antibodies against two Mediator subunits, Med220 and Med33, were incubated alone (at 40 ng/μl) or in combination (at 20 ng/μl each) with HNE, and immune complexes were allowed to form. Note that either Med220 or Med33 (more typically both) has been found in all of the subunit-defined mammalian Mediator complexes identified to date (31, 41). Control immunoneutralizations were performed under identical conditions with a nonspecific antibody. Aliquots of the antibody-treated extracts were then tested in transcription assays with chromatin templates.
Immunoneutralization with the Med220 and Med33 antibodies in combination caused a five- to sixfold reduction in ERα-dependent transcription with the pS2 and E4 promoters, while the addition of each antibody individually caused about a two- to threefold reduction (Fig. 2B). Thus, under conditions in which Mediator complexes are functionally neutralized, but other coactivators and components of the transcriptional machinery are present, ERα transcriptional activity is reduced. These results suggest that Mediator complexes serve an important coactivator function for ERα-dependent transcription with chromatin templates.
Immunodepletion of Mediator from HNEs.
To explore further the role of Mediator in ERα-dependent transcription, we used an immunodepletion protocol to remove Mediator complexes from the HNE. Immunodepletion of Mediator from transcription extracts has proven to be a useful and reliable approach for studying the activity of the complex (see references 1, 32, 35, 39, and 40, for example). For these experiments, antibodies against Med220 and Med33 were incubated with the extract, and the immune complexes were collected with protein G agarose resin. Control depletions were performed under identical conditions with a nonspecific antibody. Aliquots of the control and Mediator-depleted extracts, as well as the immunoprecipitated material, were analyzed for the presence of various Mediator subunits by immunoblotting.
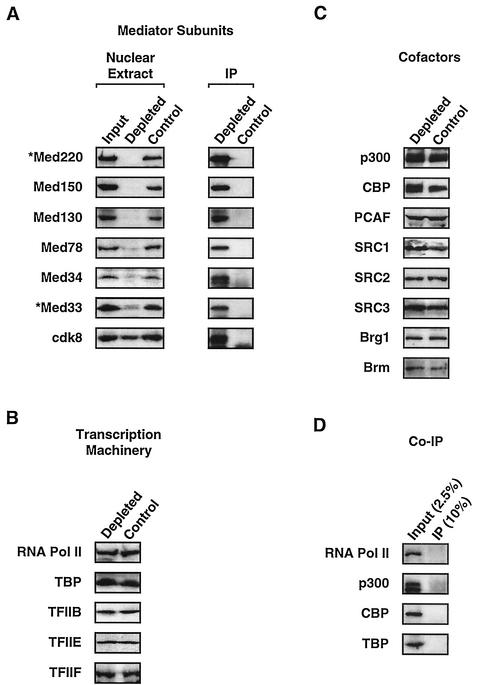
As shown in Fig. 3A, immunodepletion with the Med220 and Med33 antibodies in combination resulted in efficient depletion of Med220 and Med33, both of which were found in the immunoprecipitated material. In contrast, control antibody did not deplete Med220 or Med33. We also assayed for the presence of four other Mediator subunits that are nearly ubiquitous in mammalian Mediator complexes: namely, Med150, Med130, Med78, and Med34 (31, 41). Although antibodies specific for these subunits were not used for immunodepletion, the subunits were efficiently depleted by the antibodies specific for Med220 and Med33 (Fig. 3A), indicating that the immunodepletion protocol was effectively depleting the Mediator complexes, not simply the Med220 and Med33 subunits. As expected, depletion of the Med150, Med130, Med78, and Med34 subunits was not observed with the control antibody, and the same subunits were found in the specifically immunoprecipitated material (Fig. 3A). Quantitative immunoblotting comparing various dilutions of the control- and Mediator-depleted extracts indicated that about 90 to 95% of each Mediator subunit, with the exception of Med33 (about 75 to 80%), was removed by this protocol (data not shown). We also assayed the control and depleted extracts for the presence of cdk8, a protein found in some, but not all, Mediator complexes, as well as some non-Mediator complexes (31, 41, 47). As expected, immunodepletion of the HeLa cell extract with Med220 and Med33 antibodies depleted some (about half), but not all of the cdk8 (Fig. 3A). Together, these results indicate that immunodepletion with Med220 and Med33 antibodies generates an HNE that has dramatically reduced levels of Mediator complexes.
FIG. 3.
Immunodepletion of Mediator from HNEs. HNEs were immunodepleted with a mixture of antibodies to Med220 and Med33 (depleted) or mock depleted with control antibody (control). Aliquots of the original extract (input), Mediator-depleted extract, or control-depleted extract were analyzed by immunoblotting with an array of antibodies to Mediator subunits (Med220, Med150, Med130, Med78, Med34, Med33, and Cdk8) (A, left panel), components of the RNA pol II transcriptional machinery (the large subunit of RNA pol II, TBP, TFIIB, TFIIE p34, and TFIIF p74) (B), and coactivators and chromatin remodelers (p300, CBP, PCAF, SRC1, SRC2, SRC3, Brg1, and Brm) (C). The immunoprecipitated or coimmunoprecipitated material (IP or Co-IP, respectively) was also analyzed by immunoblotting for the presence of Mediator subunits (A, right), as well as RNA pol II, p300, CBP, and TBP (D). Note that the Mediator subunits are designated according to the unified nomenclature proposed by Rachez and Freedman (41).
Since interactions between Mediator and other transcription-related factors could result in their codepletion, we determined the levels of various other factors in the Mediator-depleted nuclear extracts. Previous results have suggested that Mediator can interact with RNA pol II (reviewed in reference 31), as well as some basal transcription factors, including TBP, TFIIB, TFIIE, and TFIIF (39). However, depletion of Mediator had no detectable effect on the levels of these factors in the extracts compared to the levels in the controls (Fig. 3B), and the same factors did not coimmunoprecipitate with the Mediator complex (Fig. 3D) (data not shown). The negative coimmunoprecipitation results with RNA pol II are consistent with recent reports indicating that Mediator complexes may exist free of the RNA pol II holoenzyme (2, 4, 8, 40, 46, 49), but they do not exclude functional interactions between Mediator and RNA pol II.
In additional immunoblot experiments, we analyzed the levels of HAT coactivators (p300, CBP, and PCAF), the SRC family of coactivators (SRC1, SRC2, and SRC3), and the ATPase subunits of mammalian SWI/SNF chromatin remodeling complexes (Brg1 and Brm). As shown in Fig. 3C, depletion of Mediator from the HNE had no detectable effect on the levels of these factors in the extracts compared to those in the controls. Also, p300 and CBP were not found in the coimmunoprecipitates with the Mediator complex (Fig. 3D). These results indicate that our Mediator-depleted extracts contain control levels of selected components of the basal transcriptional machinery, coactivators, and chromatin remodeling complexes.
Basal transcription is moderately reduced with Mediator-depleted nuclear extracts.
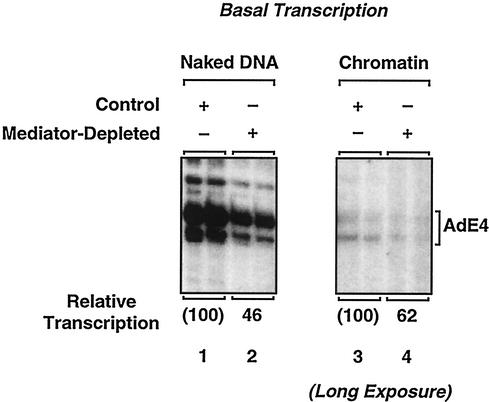
Next, we examined the levels of basal transcription by using the control and Mediator-depleted extracts with the adenovirus E4 promoter-containing plasmid (pERE), either as naked DNA or assembled into chromatin. As shown in Fig. 4, the control extract exhibited considerable basal transcription with the naked DNA template (lane 1), which was reduced about 40-fold by assembling the template into chromatin (lane 3). Basal transcription with Mediator-depleted extract on both the naked DNA and chromatin templates was reduced by about one-third to one-half of the activity observed with the control extracts (lanes 2 and 4). Thus, basal transcription is moderately reduced in Mediator-depleted nuclear extracts, consistent with previous studies that suggested a role for Mediator in basal transcription (1, 32, 35).
FIG. 4.
Immunodepletion of Mediator from HNEs causes a modest reduction in basal transcription. Basal transcription with naked DNA and chromatin templates. The plasmid template pERE, either as naked DNA (left) or assembled into chromatin (right), was transcribed with control or Mediator-depleted HNEs, as indicated. The resulting RNA products were analyzed by primer extension. Note that the chromatin experiment represents a fourfold-longer exposure than the other chromatin transcriptions shown herein, so that the low levels of basal transcription with chromatin can be observed more readily.
Transcriptional activation by ERα is impaired with Mediator-depleted nuclear extracts.
The immunoneutralization experiments shown in Fig. 2 suggested a role for Mediator in ERα-dependent transcription with chromatin templates. To address this issue further, we used the control and Mediator-depleted HNEs in ERα-dependent in vitro transcription assays with the pS2 and E4 promoters shown in Fig. 2A. Maximal ERα-dependent transcription with both promoters was observed in the presence of exogenously added p300 (Fig. 5A, compare lanes 2 and 3 for each promoter). Depletion of Mediator resulted in a three- to fivefold reduction in the maximal level of ERα-dependent transcription with the E4 and pS2 promoters, but it did not abrogate the transcriptional response completely (Fig. 5A, compare control with Mediator-depleted samples for each promoter, especially lanes 3 and 6). When the moderate inhibitory effects of Mediator depletion on basal transcription were included in the analysis from multiple experiments (i.e., expressing the data as fold activation over basal transcription), the effect of Mediator depletion on ERα-dependent transcription was still clearly evident (Fig. 5B). Thus, by two different approaches (i.e., immunoneutralization and immunodepletion, both of which gave similar reductions in ERα-dependent transcription), we observed a role for Mediator in the transcriptional activity of ERα.
FIG. 5.
ERα transcriptional activity with chromatin templates is impaired in Mediator-depleted nuclear extracts. (A) Analysis of ERα-dependent transcription with chromatin templates by using control and Mediator-depleted HNEs. The 2ERE-pS2 (pS2 promoter) and pERE (E4 promoter) plasmid templates were assembled into chromatin in the presence of purified recombinant ERα, E2, and p300 as indicated. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The relative transcription for each promoter is indicated. (B) Summary of multiple experiments like those in panel A showing the effects of Mediator depletion on activated transcription by ERα with the human pS2 and adenovirus E4 promoters. The data are plotted as fold activation over basal to account for the modest effects that Mediator depletion has on basal transcription (Fig. 4). Each bar represents the mean + the standard error from three or more separate determinations.
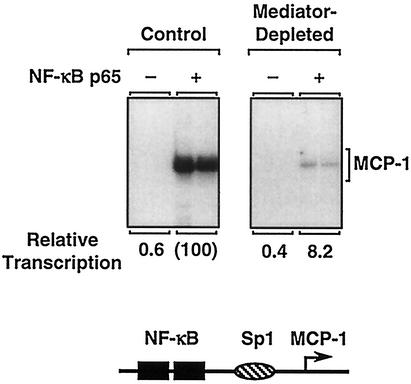
In control experiments to verify our immunodepletion approach, we tested the control and Mediator-depleted extracts with another activator, namely the p65 subunit of NF-κB, the activity of which has been shown previously to be dependent on Mediator (15, 38). p65 strongly activated transcription of an NF-κB-responsive MCP-1 enhancer-promoter construct assembled into chromatin (Fig. 6). Depletion of Mediator resulted in a 14-fold reduction in p65-dependent transcription. Again, when the moderate inhibitory effects of Mediator depletion on basal transcription were included in the analysis, an eightfold reduction in p65-dependent transcription was observed (Fig. 6) (data not shown). Thus, our immunodepletion protocol reproduces the previously characterized contribution of Mediator to the transcriptional activity of p65. The observed differences between the magnitudes of effects with ERα and p65 upon Mediator depletion may reflect activator- or promoter-specific differences in the requirement for Mediator.
FIG. 6.
NF-κB p65 transcriptional activity with chromatin templates is impaired in Mediator-depleted nuclear extracts. Analysis of NF-κB p65-dependent transcription was performed with chromatin templates by using control and Mediator-depleted HNEs. The MCP-1 promoter/enhancer construct pGLM-ENH was assembled into chromatin in the presence of purified recombinant NF-κB p65 as indicated. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The relative transcription for each condition is indicated.
In add-back or complementation experiments with partially immunopurified Mediator fractions, only a partial restoration of ERα-dependent transcriptional activity with the Mediator-depleted extracts was observed (20 to 30% recovery) (data not shown). The lack of a full restoration of transcriptional activity in the add-back experiments is likely due to immunoneutralization of the added Mediator fractions by anti-Mediator antibodies left behind in the depleted extract (see the immunoneutralizing effects of the antibodies in Fig. 2B). (Note that the Med220 subunit of Mediator is required for ERα-Mediator interactions.). However, the immunoneutralization experiments (Fig. 2B) and the inhibitory polypeptide experiments (described below) provide alternate lines of evidence to support the assertion that depletion or inactivation of the Mediator complex, not the inadvertent removal of other factors required for transcription, underlies the reduction in ERα-dependent transcription in these experiments.
Mediator is required for maximal p300/CBP-dependent transcription with chromatin templates.
Many transcriptional activators use distinct classes of coactivators, such as p300/CBP and Mediator. Thus, a fundamental question in transcriptional regulation is whether these distinct factors interact functionally. In Fig. 5A, we showed that stimulation of ERα-dependent transcription by exogenously added p300 was reduced by about 60% upon Mediator depletion with the pS2 promoter construct (16-fold stimulation by p300 with control extract versus 7-fold stimulation by p300 with Mediator-depleted extract; compare lanes 2 and 3 with lanes 5 and 6), suggesting a functional link between the activities of Mediator and p300 with ERα in the context of the pS2 promoter. A similar effect of Mediator depletion was not observed with the E4 promoter in the presence of exogenously added p300 (i.e., the p300 response was the same with or without Mediator depletion [Fig. 5A]). However, additional studies described below using more physiological levels of p300/CBP (i.e., the endogenous levels found in the HNE without exogenously added p300) demonstrate a functional interaction between Mediator and p300/CBP during ERα-dependent transcription with the E4 promoter (see Fig. 8B). These initial studies prompted us to explore in more detail the functional interplay between Mediator and p300/CBP during transcription by RNA pol II with chromatin templates.
FIG. 8.
Mediator and p300/CBP synergistically activate ERα-dependent transcription with chromatin templates. (A) Effect of p300/CBP inhibition on Mediator-stimulated ERα transcriptional activity with chromatin templates. ERα transcriptional activity with the pERE reporter template assembled into chromatin was assayed with control or Mediator-depleted HNEs in the presence or absence of the p300/CBP inhibitor GST-SRC2(PID) as indicated. The resulting RNA products were analyzed by primer extension. The relative transcription for each condition is indicated. GST-SRC2(PID), which blocks interactions between the endogenous p300/CBP and SRC proteins in the HNEs, was added at a 40-fold excess relative to the concentration of ERα. (B) Summary of selected conditions from experiments like those shown in panel A. The data are plotted as fold activation over basal and show the requirements for both Mediator and p300/CBP during ERα-activated transcription with chromatin templates. Each bar represents the mean + standard error from three or more separate determinations.
We used an assay system that bypasses the need for ERα so that we could focus on functional interactions between Mediator and p300/CBP in the absence of an activator protein. For p300/CBP-dependent activation, we used a fusion of the PID from SRC2 with the DNA binding domain from yeast Gal4 [referred to as Gal4-SRC2(PID)], as well as pS2 and E4 promoter constructs containing two Gal4 binding sites. We showed previously that the Gal4-SRC2(PID) fusion protein potently activates transcription with chromatin templates in a p300/CBP-dependent manner (23), which likely occurs through direct interactions between the SRC2(PID) and the endogenous p300/CBP in the HNE (Fig. 7A and B). In chromatin transcription experiments, the transcriptional activity of Gal4-SRC2(PID) with Mediator-depleted HNE was reduced by about 50% relative to control extract (Fig. 7B), even when the moderate inhibitory effects of Mediator depletion on basal transcription were included in the analysis (Fig. 7C). These results provide additional support for functional interactions between p300/CBP and Mediator during transcription by RNA pol II with chromatin templates.
FIG. 7.
Activity of a p300/CBP-dependent transcriptional activator is reduced in Mediator-depleted nuclear extracts. (A) Schematic representation of the mechanism of transcriptional activation by Gal4-SRC2(PID). Gal4-SRC2(PID) directly recruits endogenous p300/CBP in HNE to promoters assembled into chromatin via the PID of SRC2. (B) Analysis of transcriptional activation by Gal4-SRC2(PID) with control and Mediator-depleted HNEs. The 2Gal-pS2 (top panel) and 2Gal-E4 (bottom panel) plasmid templates containing two Gal4 UAS sites upstream of the pS2 and E4 promoters, respectively, were assembled into chromatin in the presence or absence of Gal4-SRC2(PID), as indicated. The chromatin samples were subjected to in vitro transcription analysis, and the resulting RNA products were analyzed by primer extension. The relative transcription for each promoter is indicated. (C) Summary of multiple experiments like those in panel B showing the effects of Mediator depletion on activated transcription by Gal4-SRC2(PID) with the human pS2 and adenovirus E4 promoters. The data are plotted as fold activation over basal to account for the modest effects that Mediator depletion has on basal transcription (Fig. 4). Each bar represents the mean + the standard error from three or more separate determinations.
Mediator and p300/CBP-SRC function synergistically during ERα-dependent transcription with chromatin templates, but not naked DNA.
To investigate further the interplay between Mediator and p300/CBP-SRC complexes during ERα-regulated transcription with chromatin templates, we used a previously characterized dominant-negative inhibitor of interactions between p300/CBP and SRC coactivators. The inhibitor, GST-SRC2(PID), is a fusion of GST with the SRC2(PID) described above. This fusion polypeptide blocks ligand-dependent recruitment of p300/CBP to ERα by inhibiting interactions between p300/CBP and SRC proteins (23). As shown in Fig. 8A, left panel, GST-SRC2(PID) was a potent inhibitor of ERα-dependent transcription when added to in vitro transcription reactions with chromatin templates and control (i.e., Mediator containing) HNE (in this case, with the E4 promoter; compare lanes 2 and 4). Addition of GST-SRC2(PID) had no effect on ERα-dependent transcription with naked DNA templates (Fig. 9, compare lanes 2 and 4), as expected when blocking the activity of a histone-modifying enzyme such as p300/CBP. The inhibition of ERα-dependent transcription with chromatin templates by GST-SRC2(PID) was relieved by the addition of recombinant p300 (Fig. 9B, compare lanes 3 and 4), demonstrating specificity for the inhibitor. Together, these results indicate that p300/CBP-SRC interactions are required for ERα-dependent transcription with chromatin templates even in the presence of Mediator.
FIG. 9.
Inhibition of ERα-dependent transcription by GST-SRC2(PID) does not occur with naked DNA templates and is relieved by the addition of purified recombinant p300. (A) Effect of p300/CBP inhibition on Mediator-stimulated ERα transcriptional activity with naked DNA templates. ERα transcriptional activity with the pERE reporter template as naked DNA was assayed with HNE in the presence or absence of the p300/CBP inhibitor GST-SRC2(PID) as indicated. The resulting RNA products were analyzed by primer extension. The relative transcription for each condition is indicated. GST-SRC2(PID), which blocks interactions between the endogenous p300/CBP and SRC proteins in the HNEs, was added at a 40-fold excess relative to the concentration of ERα (compare to Fig. 8A). (B) Relief of GST-SRC2(PID)-mediated inhibition of ERα transcriptional activity with chromatin templates by exogenously added p300. ERα transcriptional activity with the pERE reporter template assembled into chromatin was assayed with HNE in the presence or absence of GST-SRC2(PID) and purified recombinant p300, as indicated. The resulting RNA products were analyzed by primer extension. The relative transcription for each condition is indicated.
In subsequent experiments, we used GST-SRC2(PID) in conjunction with Mediator-depleted HNEs (Fig. 8A, right panel) to examine ERα transcriptional activity under conditions in which Mediator, p300/CBP, or both were depleted or functionally inactivated. Selected conditions from experiments like the one shown in Fig. 8A are summarized graphically in Fig. 8B as “fold activation over basal,” thus taking into account the effects of Mediator depletion on basal transcription. The results indicate that p300/CBP and Mediator function synergistically during ERα-dependent transcription with chromatin templates. This conclusion is best demonstrated by comparing the levels of activity of ERα under conditions that can be roughly approximated as −Mediator and p300 (condition 5; ∼5% of full activity), + Mediator (condition 4; ∼5% of full activity, no enhancement), + p300 (condition 3; ∼30% of full activity, a 5-fold enhancement), and + Mediator and p300 (condition 2; 100% of full activity, a 20-fold enhancement). Thus, neither Mediator nor p300/CBP alone is sufficient for maximal ERα-dependent transcription with chromatin templates, and the activity with both coactivators together is greater than the sum of activities with either coactivator alone.
Both Mediator and p300/CBP-SRC are required for the efficient stimulation of preinitiation complex formation by liganded ERα.
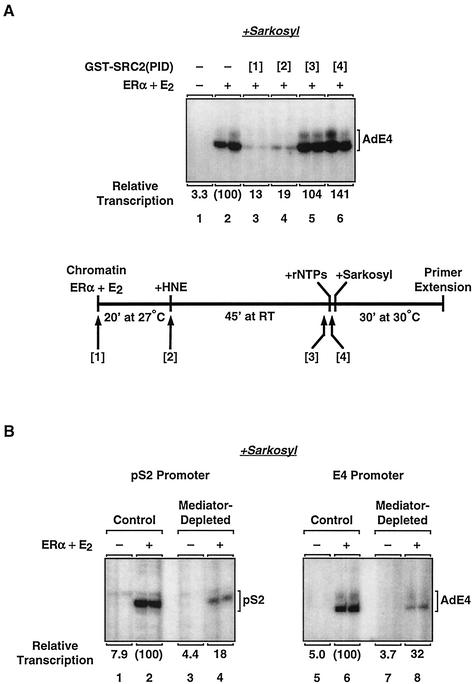
The contribution of distinct coactivator complexes (i.e., Mediator and p300/CBP-SRC) to ERα-dependent transcription raises a question regarding the specific roles that each of these coactivators play in the transcription process. To explore the role of p300/CBP-SRC complexes in ERα-dependent transcription in more detail, we performed order-of-addition experiments with the p300/CBP inhibitor, GST-SRC2(PID). In these experiments, the effects of inhibiting p300/CBP activity at various points in the process of ERα-dependent transcription in the presence of Mediator were examined. As shown in Fig. 10A, GST-SRC2(PID) was added as follows: [1] to the chromatin, before the addition of the HNE; [2] simultaneously with the HNE; [3] after preinitiation complex formation, just prior to the addition of ribonucleoside 5′-triphosphates (rNTPs); or [4] after transcription initiation (Fig. 10A, bottom). For these experiments, transcription was limited to a single round by the detergent Sarkosyl, which was added after the formation of transcription preinitiation complexes and the initiation of transcription (i.e., by the addition of rNTPs). Sarkosyl inhibits the assembly of transcription preinitiation complexes, but not elongation by transcriptionally engaged RNA pol II, thus allowing the examination of events associated with transcription initiation in the absence of subsequent rounds of reinitiation (17, 18).
FIG. 10.
Both Mediator and p300/CBP-SRC are required for the efficient stimulation of preinitiation complex formation by liganded ERα. (A) Effect of p300/CBP inhibition at various points in the process of ERα-dependent transcription in the presence of Mediator. (Top) The pERE (E4 promoter) template was assembled into chromatin in the presence of purified recombinant ERα and 17β-estradiol (E2) as indicated. The p300/CBP inhibitor GST-SRC2(PID) was added at the time points shown in the schematic diagram of the experimental setup (bottom). The samples were then subjected to in vitro transcription analysis, with the addition of the detergent Sarkosyl (0.2% [wt/vol]) after the initiation of transcription (i.e., 10 s after the addition of rNTPs). Under these conditions, Sarkosyl inhibits transcription reinitiation, but not elongation, and thus a single round of transcription is obtained. The resulting RNA products were analyzed by primer extension. Note that transcription initiates primarily from the most 3′ start site of the E4 promoter in experiments in which Sarkosyl is added (26). (Bottom) Experimental setup. GST-SRC2(PID) was added as follows: [1] to the chromatin, before the addition of the HNE; [2] simultaneously with the HNE, [3] after preinitiation complex formation, just prior to the addition of rNTPs; or [4] after transcription initiation. RT, room temperature. (B) Analysis of ERα-dependent transcription with chromatin in a single round by using control and Mediator-depleted HNEs. The 2ERE-pS2 (pS2 promoter) and pERE (E4 promoter) templates were assembled into chromatin in the presence of purified recombinant ERα and E2 as indicated. The chromatin samples were then subjected to in vitro transcription analysis, with the addition of Sarkosyl after the initiation of transcription as described in panel A. The resulting RNA products were analyzed by primer extension.
As shown in Fig, 10A, inhibition of p300/CBP activity before the formation of a stable preinitiation complex (i.e., conditions 1 and 2, shown in lanes 3 and 4, respectively) led to an inhibition of ERα-dependent transcription. In contrast, inhibition of p300/CBP activity after the formation of a stable preinitiation complex (i.e., conditions 3 and 4, shown in lanes 5 and 6, respectively) had no effect. Thus, p300/CBP-SRC is required for the formation of a stable preinitiation complex, even in the presence of Mediator. These results are consistent with those of our previous studies suggesting a role for p300/CBP-SRC interactions in the ERα-dependent formation of stable preinitiation complexes leading to the initiation of transcription with chromatin templates (23, 26).
To explore the role of Mediator complexes in ERα-dependent transcription in more detail, we examined the possibility that Mediator might also participate in the formation of a stable preinitiation complex. To address this issue, we performed single-round transcription experiments with control and Mediator-depleted nuclear extracts (Fig. 10B). Again, transcription was limited to a single round by the detergent Sarkosyl to allow examination of events associated with preinitiation complex formation and transcription initiation in the absence of transcription reinitiation. Depletion of Mediator in a single round of transcription (i.e., under conditions in which transcription of each template in the population is limited to one initiation event) resulted in a three- to fivefold reduction in ERα-dependent transcription with the pS2 and E4 promoters (Fig. 10B, compare control with Mediator-depleted samples for each promoter). These results suggest a role for Mediator in the formation of a stable preinitiation complex leading to the stimulation of transcription initiation by liganded ERα with chromatin templates. Thus, both Mediator and p300/CBP-SRC are required for the efficient stimulation of preinitiation complex formation by liganded ERα.
Mediator and p300/CBP-SRC have distinct roles during ERα-dependent transcription with chromatin templates.
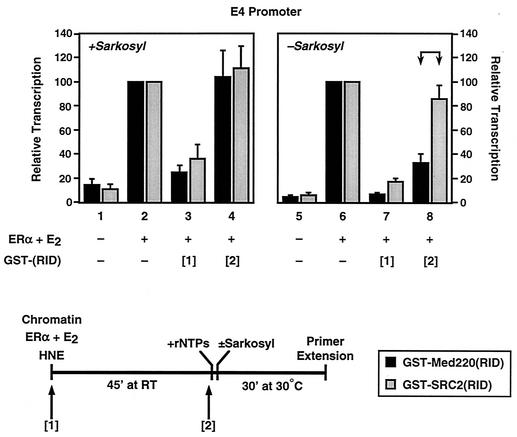
To explore the possibility that Mediator or p300/CBP-SRC might have additional distinct roles during ERα-dependent transcription with chromatin templates, in addition to their common roles in preinitiation complex formation, we performed a set of experiments with two additional dominant-negative polypeptide inhibitors. The inhibitors GST-Med220(RID) and GST-SRC2(RID) are fusions of GST with the RIDs of Med220 and SRC2, respectively. Both inhibitors bind to the ligand binding domain of ERα in an E2-dependent manner (data not shown) and block ERα-dependent transcription with chromatin templates (Fig. 11, compare bar 2 with bar 3 and bar 6 with bar 7).
FIG. 11.
Mediator and p300/CBP-SRC have distinct roles in the ERα-dependent transcription process. The effect of blocking Mediator-ERα and p300/CBP-SRC-ERα interactions at different times during the transcription cycle is shown. The pERE (E4 promoter) template was assembled into chromatin in the presence of purified recombinant ERα and E2 as indicated. The GST-Med220(RID) or GST-SRC2(RID) inhibitors were added at a 50-fold molar excess relative to ERα at the time points shown in the schematic diagram of the experimental setup (bottom) as follows: [1] to the chromatin, before the addition of the HNE and [2] after preinitiation complex formation, just prior to the addition of rNTPs. The samples were then subjected to in vitro transcription analysis with or without the addition of Sarkosyl (0.2% [wt/vol]) after the initiation of transcription (i.e., 10 s after the addition of rNTPs) as described in the legend to Fig. 10. The resulting RNA products were analyzed by primer extension. Each bar represents the mean + standard error from three or more separate determinations.
Although one might expect a priori that these inhibitors would block any ligand-dependent protein-protein interactions involving the ERα ligand binding domain, they in fact show a surprising level of specificity: i.e., GST-Med220(RID) inhibits Mediator activity with ERα, whereas GST-SRC2(RID) inhibits p300/CBP-SRC activity with ERα (unpublished observations). The specificity is clearly illustrated in a variety of assays showing different inhibitory activities for the two GST fusions. For example, GST-Med220(RID) blocks ERα-dependent transcription with naked DNA templates (i.e., under Mediator-dependent, p300/CBP-SRC-independent conditions), whereas GST-SRC2(RID) does not. Conversely, GST-SRC2(RID) blocks the recruitment of p300 HAT activity to chromatin-bound ERα via SRC2, whereas GST-Med220(RID) does not (unpublished observations). Mutant versions of these inhibitors lacking functional LXXLL motifs do not show these activities, demonstrating specificity for receptor binding (data not shown). Thus, these inhibitors are useful tools for studying the individual contributions of Mediator and p300/CBP-SRC activities to ERα-dependent transcription.
We tested the effects of these inhibitory polypeptides on ERα-dependent transcription with chromatin templates in single- versus multiple-round transcription experiments. Preliminary dose-response experiments were used to determine the saturating concentrations for the inhibitors under the transcription conditions shown in Fig. 11 (data not shown); the minimal saturating concentration of the inhibitors (225 nM; a 50-fold molar excess relative to ERα) was then used in all subsequent experiments. In single-round transcription experiments (i.e., with the addition of Sarkosyl), both inhibitors reduced ERα-dependent transcription by about 70% when added before the formation of a stable preinitiation complex (Fig. 11, addition at time point 1 [see bar 3]), but not afterwards (addition at time point 2 [see bar 4]). These results confirmed the results of Fig. 10, which showed a role for both Mediator and p300/CBP-SRC in promoting the formation of a stable preinitiation complex in the presence of liganded ERα.
In multiple-round transcription experiments (i.e., in the absence of Sarkosyl), GST-SRC2(RID) gave a result similar to that observed in the single-round experiment, reducing ERα-dependent transcription by about 80% when added before the formation of a stable preinitiation complex, but not afterwards (Fig. 11, bars 7 and 8). This suggests that p300/CBP-SRC-ERα interactions are required primarily for the formation of stable preinitiation complexes leading to the first round of initiation, but not subsequent rounds of reinitiation. In contrast, in multiple-round transcription experiments, GST-Med220(RID) reduced ERα-dependent transcription by about 75 to 90% when added either before or after the formation of a stable preinitiation complex (Fig. 11, bars 7 and 8). This result suggests that Mediator-ERα interactions are required for the formation of stable preinitiation complexes leading to the first round of initiation, as well as subsequent rounds of reinitiation. Thus, Mediator and p300/CBP-SRC are differentially required for ERα-dependent transcription reinitiation with chromatin templates.
DISCUSSION
Mediator functions as a coactivator for ERα with chromatin templates.
Mediator complexes are important coactivators that enhance the transcriptional activity of NRs and other transcriptional activators (12, 31, 41). For class II NRs (i.e., those that function as heterodimers with RXR), clear biochemical data demonstrating physical interactions of Mediator complexes with the receptors (42, 51), as well as enhancement of receptor transcriptional activity (11, 16, 29, 42), have existed for many years. In contrast, the evidence for physical and functional interactions of Mediator complexes with class I NRs (i.e., a steroid receptor, such as ERα) had been more limited until recently. Although a number of initial studies showed physical interactions of class I NRs with individual Mediator subunits (e.g., Med220, Med150, etc.) (6, 7, 19, 50) and recruitment of Mediator subunits to steroid hormone-regulated promoters in chromatin immunoprecipitation (ChIP) assays (6, 45), interactions of class I NRs with intact Mediator complexes had not been demonstrated until very recently (22).
Kang et al. have recently shown that Mediator complexes can interact directly with ERα and ERβ via the Med220 subunit of the complex and stimulate receptor-dependent transcription with naked DNA templates in vitro (22). We have shown with three different functional assays (immunoneutralization in Fig. 2B, immunodepletion in Fig. 5, and inhibitory polypeptides in Fig. 11) that Mediator is required for maximal ERα-dependent transcription with chromatin templates. Thus, Mediator functions as a bona fide coactivator for ERα and can do so in the context of chromatin, the physiological template for transcription by RNA pol II.
Mediator and p300/CBP function synergistically during ERα-dependent transcription.
Previous studies have demonstrated synergism between Mediator and other transcription-related factors, such as TBP-associated-factors (TAFs) and a SWI/SNF-related chromatin remodeling complex (PBAF) (1, 16, 20, 29, 32, 37, 39). In contrast, a clear demonstration of functional interplay between Mediator and coactivators with histone-modifying activities has been more elusive. For example, recent ChIP experiments have provided inconsistent results regarding the simultaneous recruitment of Mediator and p300/CBP-SRC to estrogen-regulated promoters (6, 45), and they do not provide a functional readout of coactivator activity that might indicate cross talk between these two coactivators.
Our biochemical assays indicate a previously uncharacterized functional interplay between Mediator and p300/CBP during ERα-dependent transcription with chromatin templates. Specifically, we have shown that both Mediator and p300/CBP-SRC are required for maximal ERα transcriptional activity (see, for example, Fig. 5, 8A, and 10) and that they can function synergistically to enhance ERα-dependent transcription with chromatin templates, but not naked DNA (Fig. 8B and 9A). A similar functional interplay between Mediator and p300 has recently been reported with another NR superfamily member (HNF-4) (32), suggesting that this type of synergism might be a common aspect of transcriptional regulation by many, if not all, NRs that use both Mediator and p300/CBP as coactivators. Interestingly, functional interplay between Mediator and p300/CBP-SRC was also observed when the need for a transcriptional activator protein (e.g., ERα) was bypassed by using Gal4-SRC2(PID) (Fig. 7), although synergism was not demonstrated. Thus, synergism between Mediator and p300/CBP-SRC may only occur in the context of a transcriptional activator protein.
Mediator and p300/CBP-SRC have both shared and distinct roles in ERα-dependent transcription with chromatin templates.
A fundamental question related to the use of distinct coactivators by a transcriptional activator is what specific roles those coactivators play in the transcription process. Our previous studies (23, 26), as well as the work described herein (Fig. 10A and 11), indicate that the recruitment of p300/CBP to liganded ERα via SRC proteins enhances the formation of a stable transcription preinitiation complex leading to the initiation of transcription. However, p300/CBP-SRC-ERα interactions are dispensable once transcription initiation has occurred (Fig. 10A and 11). We find that Mediator also plays a role in stimulating the formation of a stable preinitiation complex in the presence of liganded ERα, as shown by the impairment of ERα transcriptional activity with Mediator-depleted extracts in single-round transcription experiments (Fig. 10B). Thus, both Mediator and p300/CBP-SRC are required for the efficient stimulation of preinitiation complex formation by liganded ERα. Together, our results suggest that enhanced formation of stable transcription preinitiation complexes is one consequence of the functional synergism that occurs between Mediator and p300/CBP.
Interestingly, we find that Mediator has an additional distinct role in the process of ERα-dependent transcription: namely, to promote preinitiation complex formation for subsequent rounds of transcription reinitiation (Fig. 11). These results fit well with our previous observation that ERα plays a dual role in the transcription process, stimulating both preinitiation complex formation leading to an initial round of transcription, as well as stimulating reassembly of the transcription preinitiation complex for subsequent rounds of transcription reinitiation (26). Together, these two observations suggest that a functional consequence of Mediator-ERα interactions is the stimulation of multiple cycles of transcription reinitiation. A role for Mediator in transcription reinitiation has been suggested previously based on in vitro studies with yeast factors and naked DNA templates (52). Thus, this aspect of Mediator function may be conserved from yeast to mammals.
Mechanistically, transcription reinitiation is likely to involve a subset of the transcription machinery that remains at the promoter after initiation has occurred, forming a platform for the subsequent reassembly of a new transcription preinitiation complex (43, 44, 52, 53). In yeast, this reinitiation intermediate has been shown to include Mediator, which may help to stabilize the reinitiation complex (52). Additional studies with yeast indicate that Mediator is not associated with the transcribing RNA pol II (46), consistent with the possibility that Mediator participates in the formation of a “reinitiation platform.” Direct interactions between transcriptional activators and the transcription machinery, including Mediator, are likely required to stabilize the reinitiation intermediate (43, 44, 52). Our results obtained with the GST-Med220(RID) inhibitor (Fig. 11) suggest that this is indeed the case with ERα and Mediator. Together, our studies indicate that different classes of coactivators can function at distinct steps in the transcription process to stimulate ERα-dependent transcription. It will be interesting and informative to define these distinct activities in further detail in future studies.
Acknowledgments
We thank John Lis, Jim Kadonaga, Edwin Cheung, Kathy Lee, and Erik Andrulis for critical reading of the manuscript. We are grateful to Len Freedman, Pierre Chambon, and Pat Nakatani for antibodies; John Hiscott for the NF-κB p65 baculovirus; Teizo Yoshimura for the pGLM-ENH plasmid; Mi Young Kim for the GST-SRC2(PID) and Gal4-SRC2(PID) expression constructs; and Tory Manning for the p300 protein.
This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (DK58110) to W.L.K.
REFERENCES
- 1.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 5.Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5:241-262. [Google Scholar]
- 6.Burakov, D., L. A. Crofts, C. P. Chang, and L. P. Freedman. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 277:14359-14362. [DOI] [PubMed] [Google Scholar]
- 7.Burakov, D., C. W. Wong, C. Rachez, B. J. Cheskis, and L. P. Freedman. 2000. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J. Biol. Chem. 275:20928-20934. [DOI] [PubMed] [Google Scholar]
- 8.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 9.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, R. M. 1988. The steroid and thyroid hormone receptor superfamily. Science 240:889-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman, L. P. 1999. Increasing the complexity of coactivation in nuclear receptor signaling. Cell 97:5-8. [DOI] [PubMed] [Google Scholar]
- 13.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 14.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 15.Guermah, M., S. Malik, and R. G. Roeder. 1998. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell. Biol. 18:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawley, D. K., and R. G. Roeder. 1987. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 262:3452-3461. [PubMed] [Google Scholar]
- 18.Hawley, D. K., and R. G. Roeder. 1985. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem. 260:8163-8172. [PubMed] [Google Scholar]
- 19.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, K. M., J. Wang, A. Smallwood, C. Arayata, and M. Carey. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16:1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 22.Kang, Y. K., M. Guermah, C. X. Yuan, and R. G. Roeder. 2002. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. USA 99:2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, M. Y., S. J. Hsiao, and W. L. Kraus. 2001. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 25.Kraus, W. L., and J. T. Kadonaga. 1999. Ligand- and cofactor-regulated transcription with chromatin templates, p. 167-189. In D. Picard (ed.), Steroid/nuclear receptor superfamily: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 26.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraus, W. L., and J. Wong. 2002. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur. J. Biochem. 269:2275-2283. [DOI] [PubMed] [Google Scholar]
- 29.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 30.Leo, C., and J. D. Chen. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 32.Malik, S., A. E. Wallberg, Y. K. Kang, and R. G. Roeder. 2002. TRAP/SMCC/Mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 22:5626-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 35.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizzen, C. A., and C. D. Allis. 1998. Linking histone acetylation to transcriptional regulation. Cell Mol. Life Sci. 54:6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 39.Park, J. M., B. S. Gim, J. M. Kim, J. H. Yoon, H.-S. Kim, J.-G. Kang, and Y.-J. Kim. 2001. Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol. 21:2312-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 41.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 42.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, S. G., B. Choy, S. S. Walker, Y. S. Lin, and M. R. Green. 1995. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr. Biol. 5:508-516. [DOI] [PubMed] [Google Scholar]
- 44.Sandaltzopoulos, R., and P. B. Becker. 1998. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol. 18:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 46.Svejstrup, J. Q., Y. Li, J. Fellows, A. Gnatt, S. Bjorklund, and R. D. Kornberg. 1997. Evidence for a mediator cycle at the initiation of transcription. Proc. Natl. Acad. Sci. USA 94:6075-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 48.Ueda, A., Y. Ishigatsubo, T. Okubo, and T. Yoshimura. 1997. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene. Cooperation of two NF-kappaB sites and NF-kappaB/Rel subunit specificity. J. Biol. Chem. 272:31092-31099. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G., G. T. Cantin, J. L. Stevens, and A. J. Berk. 2001. Characterization of mediator complexes from HeLa cell nuclear extract. Mol. Cell. Biol. 21:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warnmark, A., T. Almlof, J. Leers, J. A. Gustafsson, and E. Treuter. 2001. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ER alpha and ER beta. J. Biol. Chem. 12:12.. [DOI] [PubMed] [Google Scholar]
- 51.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 53.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]