Abstract
The mechanism of DNA hypermethylation-associated tumor suppressor gene silencing in cancer remains incompletely understood. Here, we show by chromatin immunoprecipitation that for three genes (P16, MLH1, and the O6-methylguanine-DNA methyltransferase gene, MGMT), histone H3 Lys-9 methylation directly correlates and histone H3 Lys-9 acetylation inversely correlates with DNA methylation in three neoplastic cell lines. Treatment with the histone deacetylase inhibitor trichostatin A (TSA) resulted in moderately increased Lys-9 acetylation at silenced loci with no effect on Lys-9 methylation and minimal effects on gene expression. By contrast, treatment with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5Aza-dC) rapidly reduced Lys-9 methylation at silenced loci and resulted in reactivation for all three genes. Combined treatment with 5Aza-dC and TSA was synergistic in reactivating gene expression through simultaneous effects on Lys-9 methylation and acetylation, which resulted in a robust increase in the ratio of Lys-9 acetylated and methylated histones at loci showing dense DNA methylation. By contrast to Lys-9, histone H3 Lys-4 methylation inversely correlated with promoter DNA methylation, was not affected by TSA, and was increased moderately at silenced loci by 5Aza-dC. Our results suggest that reduced H3 Lys-4 methylation and increased H3 Lys-9 methylation play a critical role in the maintenance of promoter DNA methylation-associated gene silencing in colorectal cancer.
Gene silencing is an epigenetic modification that can be transmitted faithfully over many cellular generations (34). Disruption of epigenetic mechanisms leads to abnormal development and is involved in malignant transformation (2, 17). DNA methylation and histone modifications are the major epigenetic mechanisms that can affect gene expression in mammals (34). Hypermethylation of DNA in promoter CpG islands is related to transcriptional silencing of genes on the inactivated X chromosome, imprinted genes, and exogenous integrated genes (3). In cancer cells, hypermethylation of DNA in the promoter CpG islands of tumor suppressor genes (TSG) is thought to play a crucial role in carcinogenesis (2, 16). For example, inactivation of the cyclin-dependent kinase inhibitor p16 (also known as INK4A) by hypermethylation was observed in many cancers (29), leading to disruption of cell cycle regulation and providing a growth advantage to affected cells. Loss of DNA repair proteins, such as MLH1 or O6-methylguanine-DNA methyltransferase, because of promoter hypermethylation can lead to the accumulation of DNA damage and subsequent mutations in oncogenes and TSG (9).
Histone modifications also play critical roles in epigenetic silencing (34). Histone proteins assemble into nucleosomes, which function as DNA packaging units as well as transcriptional regulators. The amino-terminal tails of histones protrude from the nucleosome and are subject to chemical modifications including phosphorylation, acetylation, and methylation (15). These modifications of histones affect the access of regulatory factors and complexes to chromatin and influence gene expression. Acetylation of lysine residues on histones H3 and H4 leads to the formation of an open chromatin structure. Methylation at lysine 9 (Lys-9) on histone H3 has recently been shown to be a marker of heterochromatin from yeast to mouse (26, 28). Lys-9 methylation is recognized by heterochromatin-associated proteins, such as HP-1, and is required to maintain heterochromatin. Studies have shown that modifications of histone H3 also contribute to euchromatin gene silencing by switching between Lys-9 acetylation and methylation (25). Another histone modification, methylation of histone H3 Lys-4, localizes to sites of active transcription, and this modification may be stimulatory for transcription. These different combinations of histone modifications at different residues may act synergistically or antagonistically to affect gene expression (15).
Evidence linking DNA methylation, methyl-CpG binding proteins, and histone modifications is accumulating. A group of proteins with methyl DNA binding activity has been identified and demonstrates a link between DNA methylation and histone modifications through the recruitment of histone-modifying enzymes (3). The DNA methyltransferase genes may also play a role in direct repression of transcription through cooperation with histone deacetylases (11, 30). TSG silencing associated with DNA methylation in cancer is accompanied by loss of histone acetylation (21, 24). While deacetylation may be required for initial silencing (19), histone deacetylase inhibitors cannot relieve silencing at most of these loci, although they act synergistically with DNA methyltransferase inhibitors in reversing silencing (5). Here, we show that TSG silencing in cancer is accompanied by histone hypermethylation and that inhibition of DNA methylation (but not histone deacetylation) results in a rapid decrease in H3 Lys-9 histone methylation at silenced loci as well as reactivation of gene expression. We propose that histone methylation is a critical modification responsible for maintenance of DNA methylation-related gene silencing in colorectal cancer cells.
MATERIALS AND METHODS
Cell lines and culture conditions.
HCT116 and RKO cells were grown in high-glucose Dulbecco's modified Eagle's medium (Gibco/BRL) plus 10% fetal bovine serum (Gibco/BRL). SW48 cells were grown in L-15 medium (Gibco/BRL) plus 10% fetal bovine serum in plastic tissue culture plates in a humidified atmosphere containing 5% CO2 at 37°C. HCT116 and RKO cells and SW48 cells were split in 10 ml of their respective media at 1.5 × 105 cells and 1.0 × 106 cells per 100-mm-diameter plastic tissue culture dishes, respectively. Cell lines were obtained from the American Type Culture Collection.
TSA and 5Aza-dC treatment of cells.
Cells were split 12 to 24 h before treatment. Cells were then given one of the following treatments. (i) 5-Aza-2′-deoxycytidine (5Aza-dC) (5 μM; Sigma) or phosphate-buffered saline was used for 72 h. Medium containing 5Aza-dC or phosphate-buffered saline was changed every 24 h. (ii) Trichostatin A (TSA) (300 nM; ICN Biomedicals) or an identical volume of ethanol was used for 24 h. (iii) 5Aza-dC (5 μM) was used for 48 h followed by TSA (300 nM) for an additional 24 h. The dose of 5Aza-dC (5 μM) was chosen based on preliminary studies showing optimal reactivation of gene expression. The timing and sequencing of 5Aza-dC and/or TSA was based on similar preliminary studies as well as published studies (5).
Bisulfite-PCR methylation analysis.
We performed bisulfite treatment as reported previously (6). Briefly, 2 μg of genomic DNA was denatured with 2 M NaOH for 10 min, followed by incubation with 3 M sodium bisulfite (pH 5.0) for 16 h at 50°C. After treatment, DNA was purified using a Wizard Miniprep column (Promega), precipitated with ethanol, and resuspended in 30 μl of diluted water. Two microliters of the aliquot was used as a template for methylation-specific PCR (MSP) using the oligonucleotide primers shown in Table 1. PCR products were separated by 5% polyacrylamide gel electrophoresis and stained with ethidium bromide.
TABLE 1.
Primers and PCR conditions used in this studya
| Region | Primer sequence | Annealing temp (PCR cycle no.)b |
|---|---|---|
| CHIP assay primers | ||
| P16-1F | AGTTTCGCTCTTGTCTCCCAG | 62 (2), 60 (3), 58 (4), 52 (12) |
| P16-1R | CATGGCGAAACCCTGTCTCTA | |
| P16-2F | GATTATAGACGTGAGCCACCGC | 62 (2), 60 (3), 58 (4), 52 (12) |
| P16-2R | AGGCAGGAGAATCGCTTGAAC | |
| P16-3F | CCCTTCCCCCCTTATAATTACG | 62 (2), 60 (3), 58 (4), 52 (24) |
| P16-3R | GGACGGACTCCATTCTCAAAG | |
| P16-4F | AGACAGCCGTTTTACACGCAG | 62 (2), 60 (3), 58 (4), 52 (21) |
| P16-4R | CACCGAGAAATCGAAATCACC | |
| P16-5F | TAGGAAGGTTGTATCGCGGAGG | 62 (2), 60 (3), 58 (4), 52 (23) |
| P16-5R | CAAGGAAGGAGGACTGGGCTC | |
| P16-6F | CATTCGCTAAGTGCTCGGAGT | 58 (2), 56 (3), 54 (4), 52 (30) |
| P16-6R | CTCCTCTTTCTTCCTCCGGTG | |
| MLH1-1F | GAAACCAAGAACGCTTCCATTT | 62 (2), 60 (3), 58 (4), 52 (20) |
| MLH1-1R | GGCGACCTGAATTTCAGACTTT | |
| MLH1-2F | TCGTGGTCAGTCCAACCATTCT | 62 (2), 60 (3), 58 (4), 52 (20) |
| MLH1-2R | GGTGTAGGCCCTGAGTTGGAG | |
| MLH1-3F | AACGCCTTGCAGGACGCTTA | 62 (2), 60 (3), 58 (4), 52 (20) |
| MLH1-3R | TGAAGAGAGAGCTGCTCGTGC | |
| MLH1-4F | CGAGCAGCTCTCTCTTCAGGA | 58 (2), 56 (3), 54 (4), 52 (24) |
| MLH1-4R | AAGATGGAAGTCGACGAGGCT | |
| MLH1-5F | CTTGCTTCTTTTGGGCGTCAT | 62 (2), 60 (3), 58 (4), 52 (22) |
| MLH1-5R | GGCTTGTGTGCCTCTGCTGA | |
| MLH1-6F | CCCAGCAACCCACAGAGTTGAG | 62 (2), 60 (3), 58 (4), 52 (22) |
| MLH1-6R | CGGAAGTGCCTTCAGCCAATC | |
| MGMT-1F | CCCCATCTCCAAATAAGGTCA | 60 (2), 58 (3), 56 (4), 54 (12), 52 (16) |
| MGMT-1R | CCTAGACACTGCCAGAGCCTG | |
| MGMT-2F | CTCCGGGCTCAGCGTAG | 60 (2), 58 (3), 56 (4), 54 (12), 52 (21) |
| MGMT-2R | GCCTTAGTTTGCCAAATGG | |
| MGMT-3F | AAAAGGTACGGGCCATTTG | 62 (2), 60 (3), 58 (4), 52 (21) |
| MGMT-3R | CAGTCTGCGCATCCTCG | |
| MSP primers | ||
| P16MSPM1 | TTATTAGAGGGTGGGGCGGATCGC | 60 (35) |
| P16MSPM2 | GACCCCGAACCGCGACCGTAA | |
| P16MSPU1 | TTATTAGAGGGTGGGGTGGATTGT | |
| P16MSPU2 | CAACCCCAAACCACAACCATAA | |
| MLH-MSPM1 | GATAGCGATTTTTAACGC | 52 (35) |
| MLH-MSPM2 | TCTATAAATTACTAAATCTCTTCG | |
| MLH-MSPU1 | AGAGTGGATAGTGATTTTTAATGT | |
| MLH-MSPU2 | ACTCTATAAATTACTAAATCTCTTCA | |
| MGMT-MSPM1 | GGTCGTTTGTACGTTCGC | 60 (3), 58 (4), 56 (5), 54 (23) |
| MGMT-MSPM2 | GACCGATACAAACCGAACG | |
| MGMT-MSPU1 | GTAGGTTGTTTGTATGTTTGT | |
| MGMT-MSPU2 | AACCAATACAAACCAAACA | |
| RT-PCR primers | ||
| P16-RTF | CAACGCACCGAATAGTTACGG | 55 (38) |
| P16-RTR | GCGCAGTTGGGCTCCG | |
| MLH1-RTF | AGTGGCTGGACAGAGGAAGA | 55 (38) |
| MLH1-RTR | GATCAGGCAGGTTAGCAAGC | |
| MGMT-RTF | CGAAATAAAGCTCCTGGGCA | 55 (30) |
| MGMT-RTR | GAACTCTTCGATAGCCTCGGG | |
| GAPDH-RTF | TCCCATCACCATCTTCCAG | 55 (25) |
| GAPDH-RTR | ATGAGTCCTTCCACGATACC |
The P16-1 and P16-2 primers amplify Alu sequences that correspond to the upstream region of P16 but may also cross-amplify other Alus.
Temperature is given in degrees Celsius.
CHIP assays.
The protocols for chromatin immunoprecipitation (CHIP) have been described previously (20). Briefly, cells are treated with 1% formaldehyde for 8 min to cross-link histones to DNA. After washing, the cell pellets are resuspended in 550 μl of Lysis buffer (150 mM NaCl, 25 mM Tris-Cl [pH 7.5], 5 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) and sonicated seven times for 8 s each. The lysate (500 μl) is then divided into three fractions; the first and second ones (230 μl each) are diluted in 260 μl of lysis buffer, and the third one (40 μl) is used for input control. The first lysate is incubated with 10 μl of anti-Lys-9 acetylated histone H3 antisera (gift from Sharon Dent), 10 μl of anti-Lys-9 methylated histone H3 antibody, or 10 μl of anti-Lys-4 methylated histone H3 antibody (Upstate Biotechnology) at 4°C overnight. The second lysate is incubated with mock preimmune sera (10 μl) or Tris-EDTA buffer (10 μl) at 4°C overnight as a negative control. To collect the immunoprecipitated complexes, protein A-Sepharose beads (Pharmacia Biotech) are added and incubated for 1 h at 4°C. After washing, the beads are treated with RNase (50 μg/ml) for 30 min at 37°C and then proteinase K overnight. The cross-links are then reversed by heating the sample at 65°C for 6 h. DNA is extracted by the phenol-chloroform method, ethanol precipitated, and resuspended in 100 μl of water. PCR amplification of DNA is carried out on diluted DNA aliquots, using the oligonucleotide primers shown in Table 1. The PCR products are visualized by agarose gel electrophoresis and quantitated by capillary electrophoresis using the Agilent 2100 Bioanalyzer (Agilent Technologies). To ensure that PCR amplification was in the linear range, each reaction was initially set up at different dilutions of DNA for various amplification cycle numbers, and we selected the final PCR conditions accordingly. The assays were typically done in duplicate or triplicate. The average fragment size after sonication was 500 bp.
RNA extraction and RT-PCR.
Total cellular RNA was extracted with TriZOL (Gibco/BRL) according to the manufacturer's protocol. RNA was resuspended in diethyl pyrocarbonate-treated water and was quantitated by spectrophotometer. Reverse transcription (RT) reactions were done on 2 μg of total RNA and were performed using Moloney murine leukemia virus RT (Roche) according to the manufacturer's protocol. cDNA was amplified by PCR using oligonucleotides shown in Table 1. PCR products were quantitated with the Agilent 2100 Bioanalyzer. We initially confirmed that PCR amplifications were performed in the linear range, and all reactions included negative controls where reverse transcriptase was omitted.
Statistics.
All CHIP assays on untreated cells were performed three separate times, and the results were expressed as average ± standard error of the mean calculated using Microsoft Excel. All other CHIP assays were done at least in duplicate. There was an excellent correlation (R = 0.7 to 0.8) between measurements obtained in duplicate experiments. In selected cases, we averaged the measurements obtained using different primers in a given gene to simplify comparisons. These values were expressed as mean ± standard deviation (SD) calculated using Microsoft Excel.
RESULTS
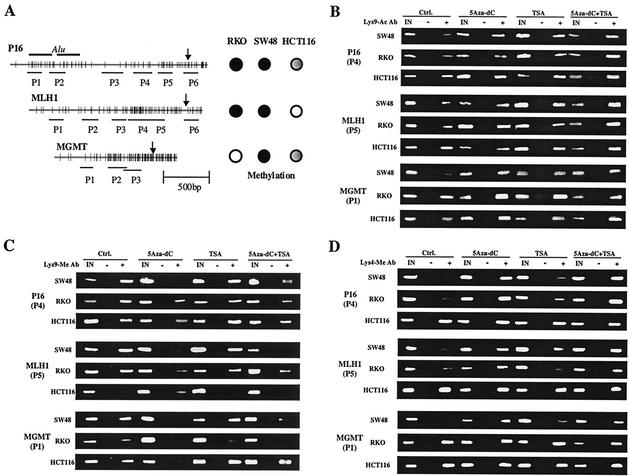
We examined DNA methylation, histone H3 Lys-9 acetylation, and histone H3 Lys-9 methylation in different regions of the promoters of P16, MLH1, and the O6-methylguanine-DNA methyltransferase gene, MGMT, in three colorectal cancer cell lines, SW48, RKO, and HCT116. These three TSG promoters were previously well characterized for methylation status in colorectal cancer, and this hypermethylation is closely related to gene silencing (10, 13, 14). Figure 1A shows a CpG map of each promoter with the location of primers used in this study. These three cell lines have a characteristic DNA methylation status in each promoter region (summarized in Fig. 1A). P16 is densely methylated in RKO and SW48 and partially (one allele only) methylated in HCT116. MLH1 is densely methylated in RKO and SW48 but is not methylated in HCT116. MGMT is densely methylated in SW48, partially methylated in HCT116, and not methylated in RKO. We used CHIP to study histone modification in these various regions. Examples of the results from these assays are shown in Fig. 1B and C. The ratios of PCR from immunoprecipitated DNA versus input DNA are shown in Fig. 2 for each region. Similar results were obtained when we used glyceraldehyde-3-phosphate dehydrogenase instead of input DNA as a DNA loading control (data not shown).
FIG. 1.
Examples of histone H3 lysine 9 CHIP assays. (A) Schema of the P16, MLH1, and MGMT promoter regions. The distribution of CpG sites is represented by vertical bars. Two Alu sequences are upstream of the P16 promoter region; arrows indicate transcription initiation sites. Lines shown below each promoter indicate regions amplified by each set of PCR primers. To the right of each gene, methylation status is indicated by filled circles (fully methylated), partially filled circles (partially methylated), and unfilled circles (unmethylated) in the three cell lines studied. (B) Examples of CHIP assays using anti-acetylated histone H3 Lys-9 antibody. (C) Examples of CHIP assays using anti-methylated histone H3 Lys-9 antibody. (D) Examples of CHIP assays using anti-methylated histone H3 Lys-4 antibody. In these assays, DNA combined with acetylated or methylated histone H3 Lys-9 antibody is immunoprecipitated and detected by PCR amplification. In panel B, note that the histone deacetylase inhibitor TSA induces moderate increased Lys-9 acetylation while the combination of the DNA methyltransferase inhibitor 5Aza-dC and TSA induces remarkable Lys-9 hyperacetylation. In panel C, note that 5Aza-dC and a combination of 5Aza-dC and TSA remarkably decrease Lys-9 methylation. In panel D, note that 5Aza-dC and a combination of 5Aza-dC and TSA remarkably increase Lys-4 methylation. IN, input DNA from whole-cell lysate. The intensities of the bands of PCR products were quantitated by densitometry or using the Agilent 2100 Bioanalyzer.
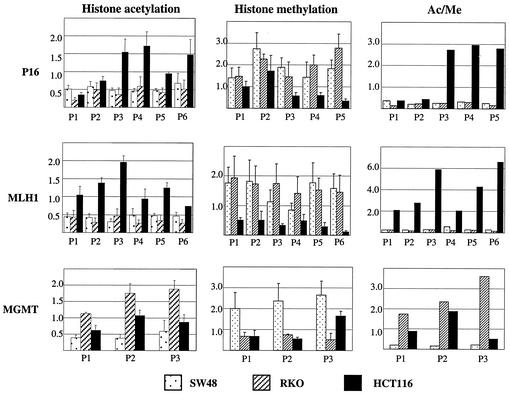
FIG. 2.
Summary of quantitative analysis of histone H3 lysine 9 CHIP assays. Ratios of precipitated DNA over input DNA were used to calculate relative precipitated fold enrichment shown on the y axis. Ac/Me, ratio of acetylation over methylation. The assays were done in triplicate. Error bars represent standard errors of the means.
As can be seen in Fig. 1 and 2, the downstream part of the promoter region of the P16 gene (P3 to P6) shows a higher degree (two- to fourfold) of H3-Lys-9 acetylation in HCT116 (which has only one allele methylated) compared to RKO and SW48 (which have dense biallelic methylation in this region). By contrast, H3-Lys-9 acetylation status was low and almost the same among the three cell lines in the upstream Alu sequences (P1 and P2). Similar results were seen for the MLH1 gene (Fig. 2), which shows a low degree of H3 Lys-9 acetylation in all parts of the promoter region in RKO and SW48 (both of which have dense promoter DNA methylation at this locus). By contrast, a twofold- to fourfold-higher degree of H3 Lys-9 acetylation was detected in HCT116 (which has no MLH1 DNA methylation) at all MLH1 regions studied. Finally, the promoter region of the MGMT gene had relatively low levels of histone Lys-9 acetylation in SW48 (where dense methylation is present), intermediate levels of acetylation in HCT116 (where partial DNA methylation is seen), and the highest degree of acetylation in RKO (which has no DNA methylation in this promoter). Thus, Lys-9 histone H3 acetylation in different regions of the promoters studied correlated very well with the DNA methylation status of each gene.
We next analyzed H3 Lys-9 methylation in these same regions using CHIP. As seen in Fig. 2, Lys-9 histone H3 methylation was almost exactly inversely correlated with Lys-9 acetylation. Thus, in the downstream part of the promoter region of the P16 gene, Lys-9 methylation was higher for SW48 and RKO (which have dense DNA methylation there) than for HCT116 (monoallelic DNA methylation). By contrast, the Alu sequences (P1 and 2) had similarly elevated degrees of Lys-9 methylation in all three cell lines. In particular, HCT116 showed higher degrees of H3 Lys-9 methylation in the Alu sequences than in the downstream area. Similar results were seen at MLH1. HCT116, which has no promoter DNA methylation at this locus, shows a uniformly low degree of H3 Lys-9 methylation. SW48 and RKO, which show DNA methylation-associated silencing at this locus, have higher degrees of H3 Lys-9 methylation across the promoter. In this promoter, as in P16, SW48 and RKO gave nearly identical results, which serves to validate the CHIP assay results. Also, all six MLH1 regions studied (P1 to P6), which span 2 kb, showed nearly identical results. The third gene studied, MGMT, also showed this strong association between promoter DNA methylation and Lys-9 histone methylation. Thus, SW48, which has the highest degree of DNA methylation, has higher H3 Lys-9 methylation at all three regions studied than does RKO, which has no DNA methylation at this locus. HCT116, which has partial DNA methylation at MGMT, also shows intermediate degrees of H3 Lys-9 methylation at this locus. Thus, for all genes, in all cell lines, DNA methylation, Lys-9 histone H3 hypoacetylation, and Lys-9 histone H3 hypermethylation correlated very well. Because acetylation and methylation are mutually exclusive at H3 Lys-9, a relative ratio of acetylation to methylation was used to compare the various genes. As shown in the right panel of Fig. 2, this ratio closely mirrors the DNA methylation status of each gene in each cell line.
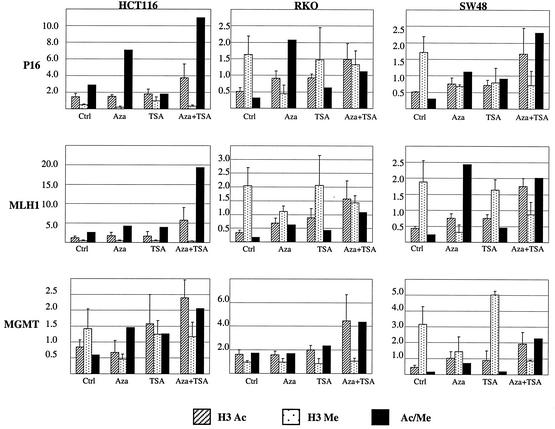
To determine the relative contributions of Lys-9 acetylation and methylation to gene silencing, we next examined the effects of treatment with the histone deacetylase inhibitor TSA and/or the DNA-methyltransferase inhibitor 5Aza-dC on Lys-9 histone H3 acetylation, Lys-9 histone H3 methylation, and gene expression and reactivation. The results of the CHIP studies were almost identical in the different regions of each promoter. For this reason, we averaged the values for each gene in presenting the data. We used the values of P3 to P6 to make the average for P16, because P1 and P2 correspond to Alu sequences. Examples of these results are shown in Fig. 1, and the data are summarized in Fig. 3. We started by examining Lys-9 acetylation. For the P16 gene, treatment with TSA alone had no effect on Lys-9 acetylation in the HCT116 line (partial DNA methylation) but slightly increased Lys-9 acetylation in the silenced cell lines RKO and SW48. Identical results were seen for MLH1. For MGMT, TSA had no effect on the unmethylated cell line RKO and moderately increased Lys-9 acetylation in HCT116 (partial DNA methylation) and SW48 (dense DNA methylation). 5Aza-dC moderately increased Lys-9 acetylation at loci with dense DNA methylation (see for example P16 and MLH1 in RKO) but had no effect on loci with partial or no DNA methylation. However, the combination of 5Aza-dC and TSA was quite effective at increasing Lys-9 acetylation at all loci regardless of DNA methylation status. Thus, TSA and 5Aza-dC had nearly identical effects on histone acetylation. However, as shown later, they had markedly different effects on gene expression, suggesting that factors other than acetylation mediate these effects on silencing.
FIG. 3.
Average of histone H3 Lys-9 acetylation, methylation, and acetylation-to-methylation ratio in each cell line treated with 5Aza-dC, TSA, or both. Shown are the data averaged across each promoter for P16, MLH1, and MGMT. We used the values of P3 to P6 to make the average for P16, because P1 and 2 correspond to Alu sequences. H3 Ac, histone acetylation; H3 Me, histone methylation. Error bars represent the SD of the averaged values for each gene calculated from the values of different regions.
We next analyzed H3 Lys-9 methylation using identical methods. These results are presented in detail in Fig. 3. TSA alone had no effect on H3 Lys-9 methylation, regardless of DNA methylation. However, by contrast to TSA treatment, 5Aza-dC had a marked effect on Lys-9 methylation at silenced loci. For example, 5Aza-dC dramatically reduced histone methylation in the promoter region of P16 and in the promoter of MLH1 in RKO. These changes were specific to promoters showing partial or high degrees of DNA methylation. The combination of 5Aza-dC and TSA had effects on histone Lys-9 methylation similar to those of 5Aza-dC. When we examined the acetylation/methylation ratio for Lys-9 loci with dense DNA methylation, we found that TSA had minimal effects on the ratio, 5-Aza-dC had strong effects on the ratio, and the combination had dramatic effects on this ratio. Interestingly, while TSA or 5Aza-dC alone had little effect on the acetylation/methylation ratio at two loci (MLH1 in HCT116 and MGMT in RKO) where no DNA methylation was observed, the combination treatment did synergistically increase this ratio at these loci.
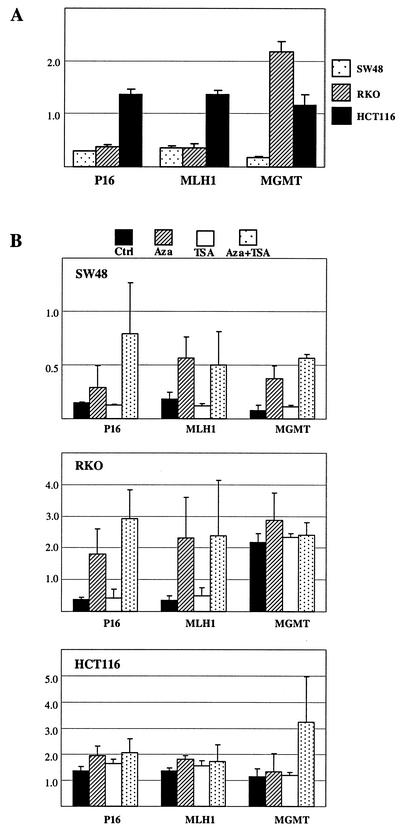
5Aza-dC is a strong inhibitor of DNA methyltransferases, and our data on histone H3 Lys-9 methylation raised the possibility of a nonspecific effect on all trans-methylation reactions. To evaluate the effects of 5Aza-dC and/or TSA on other histone modifications, we examined the status of histone H3 Lys-4 methylation (normally associated with gene activation) at these loci. As shown in Fig. 4, using CHIP, we found a higher degree of Lys-4 methylation in HCT116 than in SW48 and RKO on p16 and MLH1 genes. HCT116, which has partial DNA methylation at MGMT, also shows intermediate degrees of H3 Lys-4 methylation at this locus. Thus, Lys-4 methylation inversely correlates with DNA methylation at all three loci very well. Consistent with its lack of effect on histone Lys-9 methylation, TSA did not affect Lys-4 methylation. On the other hand, 5Aza-dC, or the combination of 5Aza-dC and TSA, increased H3 Lys-4 methylation at all silenced loci in SW48 and RKO, which is precisely opposite to its effect on H3 Lys-9 methylation in these cell lines. Little change in H3 Lys-4 methylation was observed in the HCT116 cell line, where only partial or no methylation is observed at these gene loci. Thus, the effects of 5Aza-dC on histone methylation are not due to nonspecific effects on trans-methylation reactions.
FIG. 4.
Average of histone H3 Lys-4 methylation in the SW48, RKO, and HCT116 cell lines. (A) Histone H3 Lys-4 methylation averaged across each promoter for P16, MLH1, and MGMT without treatment. Note that SW48, which shows dense DNA methylation at all three promoters, has much lower levels of Lys-4 methylation than HCT116, which has no DNA methylation at MLH1 and only partial DNA methylation at P16 and MGMT. (B) Dynamics of Lys-4 methylation in SW48, RKO, and HCT116 treated with 5Aza-dC (Aza), TSA, and both. Note that TSA has no effect by itself and that 5Aza-dC increases Lys-4 methylation in all three genes in SW48 and in P16 and MLH1 in RKO (where dense DNA methylation is present), with a much less pronounced effect in HCT116 (where DNA methylation is absent or partial at these loci and where Lys-4 methylation is already elevated at baseline, compared to SW48 and RKO. Error bars represent the SD of the averaged values for each gene calculated from the values of different regions.
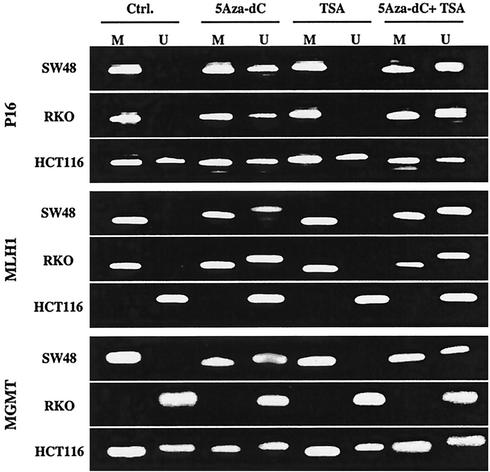
We next used MSP to evaluate the effect of 5Aza-dC and/or TSA treatment on each promoter DNA methylation status (Fig. 5). As expected, 5Aza-dC and the combination of 5Aza-dC and TSA treatment resulted in demethylation of P16 and MLH1 in SW48 and RKO, and of MGMT in SW48, in which the gene is silenced in association with promoter region hypermethylation. By contrast, TSA alone did not affect the DNA methylation status in any gene.
FIG. 5.
MSP analysis in each promoter region after treatment. The presence of PCR products in lane M indicates the presence of methylated alleles, and that in lane U indicates unmethylated alleles. Cells were treated with 5Aza-dC, TSA, a combination of 5Aza-dC and TSA, or no drug as a control (Ctrl.). After 5Aza-dC and the combination of 5Aza-dC and TSA treatment, unmethylated bands were detected for P16 and MLH1 in SW48 and RKO and for MGMT in SW48. By contrast, TSA did not affect the methylation status for any gene.
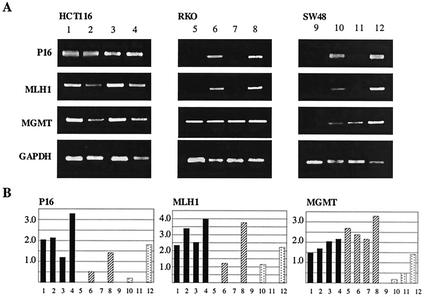
We next used RT-PCR to determine whether changes of histone modification status after 5Aza-dC and TSA treatment are reflected in gene expression (Fig. 6). P16 was expressed in HCT116 at baseline and minimally affected by treatment. This gene was silenced in RKO and SW48. In both of these cell lines, TSA was not able to activate gene expression, even though acetylation increased. By contrast, 5Aza-dC alone reactivated expression of the gene in these two silenced cell lines. This was achieved with the same degree of histone acetylation change as that achieved by TSA. Essentially similar results were obtained for MLH1, which was expressed in HCT116 but silenced in RKO and SW48. Again, TSA had no effect on gene expression, while 5Aza-dC reactivated the gene, and the combination of 5Aza-dC and TSA synergistically increased gene expression. MGMT gene expression behaved a little differently. MGMT was expressed in both RKO (unmethylated) and HCT116 (partially methylated) but was silenced in SW48 (fully methylated). In SW48, both TSA and 5Aza-dC alone were able to reactivate gene expression, and the combination of 5Aza-dC and TSA also synergistically increased gene expression there. Combining these observations with the effects of the drugs on histone modifications, one can see that TSA increases histone acetylation in highly methylated promoters, with little effect on gene expression. 5Aza-dC achieves substantial gene reactivation with the same degree of increased histone acetylation. However, TSA and 5Aza-dC differ markedly with regards to histone Lys-9 methylation, which is unaffected by the former but substantially inhibited by the latter. Thus, gene expression restoration correlates much better with decreased histone Lys-9 methylation (and concomitant increased Lys-4 methylation) than with increased Lys-9 acetylation, which leads us to conclude that histone methylation is a critical mechanism used to achieve maintenance of gene silencing at these loci. The synergy between TSA and 5Aza-dC can then be explained by simultaneous action on Lys-9, as evidenced by the markedly increased acetylation/methylation ratio achieved by the combination.
FIG. 6.
RNA expression of each gene in each cell line was examined by RT-PCR. (A) Examples of RT-PCR results. Cells were treated with 5Aza-dC (lanes 2, 6, and 10), TSA (lanes 3, 7, and 11), a combination of 5Aza-dC and TSA (lanes, 4, 9, and 12), and no drug as a control (lanes 1, 5, and 9). The name of each gene amplified is indicated on the left of each column. (B) The ratio of each gene of interest to glyceraldehyde-3-phosphate dehydrogenase is shown on the y axis. Lane numbers correspond to the lanes in panel A.
DISCUSSION
We studied the dynamics of histone acetylation and methylation in the promoter regions of the P16, MLH1, and MGMT genes. Histone H3 Lys-9 methylation correlated, and histone H3 Lys-9 acetylation and H3 Lys-4 methylation inversely correlated, very well with DNA methylation in the three genes. Our data on histone acetylation are consistent with previous reports linking DNA methylation and transcriptional silencing (18, 24) and now extend these observations to histone methylation. Methylation at Lys-9 on histone H3 that is specifically catalyzed by Suv39h has recently been shown to be a marker of heterochromatin (26, 28). H3 Lys-9 methylation contributes to establishing and maintaining heterochromatin through providing a binding site for the HP-1 protein. Our data indicate that H3 Lys-9 methylation is also closely related to DNA methylation and acts as an epigenetic mark of silencing in the tumor suppressor genes, P16, MLH1, and MGMT. Recent reports noted that H3 Lys-9 methylation can be regulated by Suv39h-HP1-independent pathways and occurs in facultative heterochromatin on the inactive X chromosome (4, 27). Given similarities between X inactivation and TSG silencing in cancer, H3 Lys-9 methylation in the promoters of the three genes we studied may also contribute to form folded states of chromatin. In contrast to histone H3 Lys-9 methylation, histone H3 Lys-4 methylation may be associated with activation of transcription. Recently it was reported that Lys-4 methylation prevents the association of histone H3 with NuRD and displaces the deacetylase activity of the NuRD repressor leading to the acetylation of lysine 9 (36). Our data showing that H3 Lys-4 methylation directly correlates with H3 Lys-9 acetylation and gene expression in P16, MLH1, and MGMT suggest that these three genes can also be regulated in a histone H3 Lys-4 methylation-dependent manner.
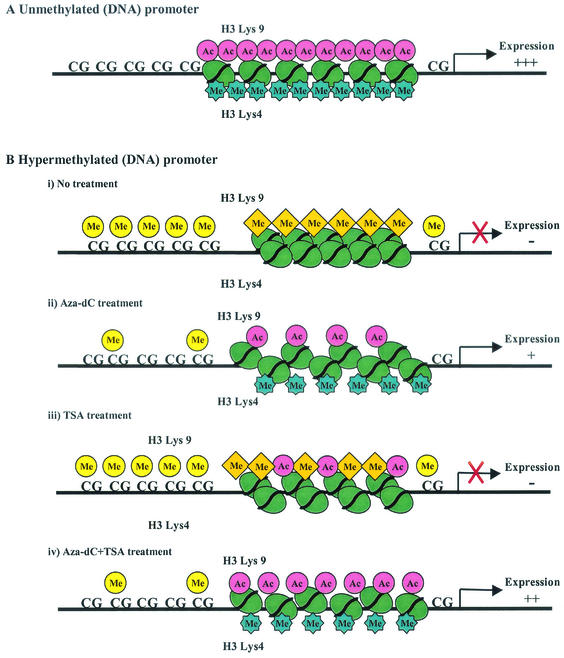
It has previously been reported that histone deacetylation is important to the establishment of initial silencing at loci with promoter methylation (3). However, once silencing is established, inhibition of histone deacetylation can generally not result in activation of gene expression (5, 7, 8), suggesting other factors than acetylation status in maintaining silencing at these loci. Our data are entirely consistent with these observations. Both TSA and 5Aza-dC moderately increased the Lys-9 acetylation status of P16 and MLH1 to similar levels, but only 5Aza-dC reactivated expression of these genes. Our observation that the DNA-methyltransferase inhibitor 5Aza-dC decreased Lys-9 methylation in both genes while TSA had no effect on Lys-9 methylation now identify this alternate histone modification as responsible for maintaining the silenced state at these loci. In the present study, we still cannot address which modification is more important in gene silencing between histone H3 Lys-9 acetylation and methylation. It is possible that both methylation and lack of acetylation are needed for silencing rather than one or the other. The combination of 5Aza-dC and TSA resulted in markedly increased Lys-9 acetylation and moderately decreased Lys-9 methylation in all three cell lines and was most effective at restoration of gene expression as previously reported (5). The mechanism of this synergistic activity on gene expression may now be explained by a dual activity on histone H3 Lys-9 acetylation and histone methylation. In Fig. 7, we summarize the present data and propose a model for the role of histone modifications in silencing of P16, MLH1, and MGMT in colorectal cancers. Intriguingly, the synergistic effect of 5Aza-dC and TSA on the Lys-9 acetylation/methylation ratio is also apparent in unmethylated promoters, suggesting the possibility of an action of 5Aza-dC that is separate from induction of promoter demethylation. It is possible that 5Aza-dC-related DNA methyltransferase degradation disrupts complexes between DNA methyltransferases and histone-modifying proteins (11, 30), with resultant effects on transcription that are partly independent of DNA methylation. This could also explain 5Aza-dC-induced activation of unmethylated genes, such as P21 (23) and APAF1 (31).
FIG. 7.
A schematic representation of the proposed mechanism in the role of histone modifications in unmethylated and methylated promoters. (A) Unmethylated CpG promoter. Acetylation in histone H3 Lys-9 and methylation in Lys-4 result in an unfolded chromatin structure leading to gene expression. (B) Methylated CpG promoter. (i) Methylation in Lys-9 and no methylation in Lys-4 cause a repressive folded chromatin structure, leading to gene silencing. (ii) 5Aza-dC, which is a DNA demethylating agent, decreases Lys-9 methylation dramatically, increases Lys-9 acetylation slightly and Lys-4 methylation moderately, and reactivates gene expression. (iii) TSA increases Lys-9 acetylation but has no effect on Lys-9 methylation or Lys-4 methylation and does not relieve transcriptional silencing. (iv) The combination of 5Aza-dC and TSA decreases Lys-9 methylation and increases Lys-9 and Lys-4 methylation. The acetylation level for Lys-9 is higher than that observed when 5Aza-dC is used alone. The synergy between TSA and 5Aza-dC, which reactivate gene expression most effectively, can be explained by the markedly increased acetylation/methylation ratio achieved by the combination.
To clarify whether 5Aza-dC and TSA exclusively affect histone H3 Lys-9, we examined the dynamics of histone H3 Lys-4 methylation after treatment with 5Aza-dC and TSA. 5Aza-dC and the combination of 5Aza-dC and TSA increased Lys-4 methylation, whereas TSA alone had no effect. These results are also consistent with the observations that 5Aza-dC or the combination of 5Aza-dC and TSA, but not TSA alone, reactivate gene expression at silenced loci. Thus, 5Aza-dC can affect two distinct histone H3 lysines in different ways, increasing methylation at Lys-4 and decreasing methylation at Lys-9, in concordance with its effects on gene expression (Fig. 7). This finding rules out the possibility that effects of 5Aza-dC on histone methylation relate to nonspecific inhibition of all trans-methylation reactions. Overall, these findings are consistent with the proposed hypothesis that combinations of histone modifications at different residues act synergistically or antagonistically to affect gene expression (15).
The mechanism underlying modifications of histone methylation by 5Aza-dC is not known. One simple possibility is that promoter methylation is essential to maintain histone methylation. However, the observation is also consistent with previous reports of 5Aza-dC inducing uncoiling of chromatin, particularly in constitutive heterochromatin (12, 22). DNMT1, which is predominantly a maintenance DNA methyltransferase, localizes to replication foci, interacts with histone deacetylases in vitro, and can repress transcription in transient-transfection assays, suggesting the possibility that it is involved in repression of transcription in cooperation with histone deacetylases at replication forks (1, 30). It is possible that DNMT1 and other DNA methyltransferases make complexes not only with histone deacetylases but also with histone methyltransferases. Decreased expression of DNMT1 induced by 5Aza-dC could then lead to histone demethylation via disruption of these silencing complexes.
We note that the MGMT gene behaves a little differently than P16 and MLH1. MGMT was restored by TSA alone, and this was associated with increased Lys-9 acetylation but not with a change in Lys-9 methylation or Lys-4 methylation. TSA was also reported to activate expression of a silenced ER gene (35). TSA alone is thought to modulate the expression of a minority of cellular genes (33), suggesting that regulatory mechanisms of gene expression by histone acetylation and deacetylation independent of DNA methylation may be important to a subset of genes silenced in cancer. Nevertheless, 5Aza-dC was still effective at restoring MGMT expression, and the combination of 5Aza-dC and TSA was also most effective there, suggesting an important role for histone methylation in this gene as well.
Epigenetic mechanisms involving DNA methylation and histone modifications are clearly closely linked, but the critical initiating events in silencing remain to be defined. In Neurospora, histone methylation may be directing DNA methylation to specific sites (32). Our data showing that inhibition of DNA methylation rapidly changes histone methylation suggest that the situation is more complex in mammalian cells. Our data are most consistent with a model whereby DNA methylation targets histone modifications to gene promoters, with Lys-9 histone methylation being a critical modification for the maintenance of gene silencing. Because Lys-9 acetylation and methylation affect the same residue, histone H3 Lys-9 deacetylation may be a prerequisite for Lys-9 methylation. Moreover, Lys-4 methylation was rapidly modified by DNA methyltransferase inhibition as well, also suggesting that histone H3 methylation could be modified in a DNA methylation-dependent manner. Further studies are clearly needed to understand the relationship between DNA methylation and histone methylation in initial establishment of the silenced state.
Acknowledgments
Yutaka Kondo is supported by the Yasuda Medical Research Foundation and the Nitto Foundation in Japan. We thank Sharon Dent and Madelene Coombes (M. D. Anderson Cancer Center) for helpful discussion and technical assistance.
This work was supported by a Research Grant from the American Cancer Society (RPG9909801MGO) and by the George and Barbara Bush Endowment for Innovative Cancer Research.
REFERENCES
- 1.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S. B., J. G. Herman, J. R. Graff, P. M. Vertino, and J. P. J. Issa. 1998. Alterations in DNA methylation—a fundamental aspect of neoplasia. Adv. Cancer Res. 72:141-196. [PubMed] [Google Scholar]
- 3.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression—belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 4.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 6.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffee, B., F. Zhang, S. T. Warren, and D. Reines. 1999. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 22:98-101. [DOI] [PubMed] [Google Scholar]
- 8.El Osta, A., P. Kantharidis, J. R. Zalcberg, and A. P. Wolffe. 2002. Precipitous release of methyl-CpG binding protein 2 and histone deacetylase 1 from the methylated human multidrug resistance gene (MDR1) on activation. Mol. Cell. Biol. 22:1844-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller, M., G. Gaidano, S. N. Goodman, V. Zagonel, D. Capello, B. Botto, D. Rossi, A. Gloghini, U. Vitolo, A. Carbone, S. B. Baylin, and J. G. Herman. 2002. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J. Natl. Cancer Inst. 94:26-32. [DOI] [PubMed] [Google Scholar]
- 10.Esteller, M., M. Toyota, M. Sanchez-Cespedes, G. Capella, M. A. Peinado, D. N. Watkins, J. P. Issa, D. Sidransky, S. B. Baylin, and J. G. Herman. 2000. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 60:2368-2371. [PubMed] [Google Scholar]
- 11.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 12.Haaf, T. 1995. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol. Ther. 65:19-46. [DOI] [PubMed] [Google Scholar]
- 13.Herman, J. G., A. Merlo, L. Mao, R. G. Lapidus, J. P. Issa, N. E. Davidson, D. Sidransky, and S. B. Baylin. 1995. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 55:4525-4530. [PubMed] [Google Scholar]
- 14.Herman, J. G., A. Umar, K. Polyak, J. R. Graff, N. Ahuja, J. P. J. Issa, S. Markowitz, J. K. V. Willson, S. R. Hamilton, K. W. Kinzler, M. F. Kane, R. D. Kolodner, B. Vogelstein, T. A. Kunkel, and S. B. Baylin. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA 95:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163-167. [DOI] [PubMed] [Google Scholar]
- 17.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 18.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 19.Kass, S. U., N. Landsberger, and A. P. Wolffe. 1997. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 7:157-165. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 21.Magdinier, F., and A. P. Wolffe. 2001. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl. Acad. Sci. USA 98:4990-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malheiro, I., B. Porto, T. Mello-Sampayo, and V. Goyanes. 1995. Specific induction of uncoiling in NORs of human acrocentric chromosomes by 5-azacytidine and 5-azadeoxicytidine. Cytobios 83:17-23. [PubMed] [Google Scholar]
- 23.Milutinovic, S., J. D. Knox, and M. Szyf. 2000. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J. Biol. Chem. 275:6353-6359. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, C. T., F. A. Gonzales, and P. A. Jones. 2001. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 29:4598-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 26.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 27.Peters, A. H., J. E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff, and T. Jenuwein. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77-80. [DOI] [PubMed] [Google Scholar]
- 28.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 29.Rocco, J. W., and D. Sidransky. 2001. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp. Cell Res. 264:42-55. [DOI] [PubMed] [Google Scholar]
- 30.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 31.Soengas, M. S., P. Capodieci, D. Polsky, J. Mora, M. Esteller, X. Opitz-Araya, R. McCombie, J. G. Herman, W. L. Gerald, Y. A. Lazebnik, C. Cordon-Cardo, and S. W. Lowe. 2001. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409:207-211. [DOI] [PubMed] [Google Scholar]
- 32.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 33.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 34.Wolffe, A. P., and M. A. Matzke. 1999. Epigenetics: regulation through repression. Science 286:481-486. [DOI] [PubMed] [Google Scholar]
- 35.Yang, X., A. T. Ferguson, S. J. Nass, D. L. Phillips, K. A. Butash, S. M. Wang, J. G. Herman, and N. E. Davidson. 2000. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 60:6890-6894. [PubMed] [Google Scholar]
- 36.Zegerman, P., B. Canas, D. Pappin, and T. Kouzarides. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 277:11621-11624. [DOI] [PubMed] [Google Scholar]