Abstract
RNA polymerase II (Pol II) Mediator plays an essential role in both basal and activated transcription. Previously, subunits of the Sin4 Mediator complex (Sin4, Pgd1, Gal11, and Med2) have been implicated in both positive and negative transcriptional regulation. Furthermore, it was proposed that this subcomplex constitutes an activator-binding domain. A yeast nuclear-extract system was used to investigate the biochemical role of the Sin4 complex. In contrast to previous findings, we found at least two general activator-independent roles for the Sin4 complex. First, mutations in sin4 and pgd1 destabilized the Pol II-Med complex, leading to a reduced rate and extent of preinitiation complex (PIC) formation both in the presence and absence of activators. Although reduced in amount compared with the wild type, PICs that are formed lacking the Sin4 complex are stable and can initiate transcription normally. Second, mutation of pgd1 causes partial disruption of the Sin4 complex and leads to a defect in transcription reinitiation. This defect is caused by dissociation of mutant Mediator from promoters after initiation, leading to nonfunctional Scaffold complexes. These results show that function of the Sin4 complex is not essential for transcription activation in a crude in vitro system but that it plays key roles in the general transcription mechanism.
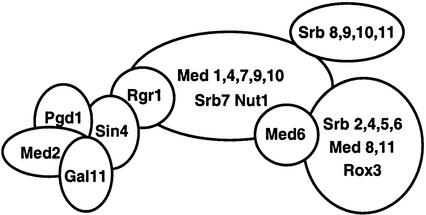
Transcription by Saccharomyces cerevisiae RNA polymerase II (Pol II) requires a number of transcription factors, including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and the Srb/Mediator complex. Yeast Mediator was originally identified genetically for its ability to suppress deletions in the Pol II C-terminal domain (43) and biochemically as a coactivator able to impart activator responsiveness to a minimal reconstituted transcription system (17). The 24 subunits of the yeast Mediator complex are the Srb proteins (Srb2 and Srb4 to Srb11), Med proteins (Med1, Med2, Med4, and Med6 to Med11), Rox3, Nut1, Rgr1, Gal11, Sin4, and Pgd1/Hrs1 (Fig. 1) (10, 17, 24, 25). Complexes of Mediator and Pol II have been purified from yeast as Pol II-Med (25) and as various forms of Pol II holoenzyme containing additional transcription factors (17, 19). Mediator-like complexes with activator binding and coactivator functions have also been isolated from mammalian cells. These complexes, including TRAP/SMCC (13), DRIP (34), ARC (30), hMediator (4), murine Mediator (14), NAT (42), and the smaller CRSP (37) and PC2 (26) complexes, share a number of subunits but are not identical. In addition to a common coactivator function, 20 of the human Mediator subunits have known homologues in yeast Mediator (3, 9).
FIG. 1.
Yeast Mediator. Proposed interactions within the Sin4 complex are shown (8, 16, 29).
Apart from its role in activation, Mediator also plays a general role in transcription. Inactivation of the complex by a temperature-sensitive srb4 mutation results in widespread, rapid elimination of Pol II transcription in vivo (12, 44). In vitro, yeast extracts prepared from an srb2 or srb5 deletion strain or an srb4ts strain have nearly undetectable basal and activated transcription levels (18, 36). Recently, several groups have observed inhibition of basal transcription when TRAP/Mediator is immunodepleted from HeLa nuclear extracts (1, 28). Together, these results indicate that Mediator function is not limited solely to its ability to interact with activators but that it also functions as a general transcription factor.
Biochemical studies have implicated Mediator in preinitiation complex (PIC) formation. In yeast extracts with the weak activator Gal4-AH, PIC formation can be separated into at least three steps by using mutations which block steps in the assembly pathway. TFIID and TFIIA are first recruited to the promoter, followed by cooperative binding of Mediator, Pol II, TFIIF, TFIIB, and TFIIE. TFIIH binds in the final step before initiation. Inactivation of Mediator blocks PIC assembly after the recruitment of TFIID and TFIIA (36). Mediator has also been shown to be critical for transcription in vivo at promoters that do not use the holoenzyme recruitment mechanism. In Drosophila melanogaster, Mediator can be recruited to heat shock promoters at which a stalled Pol II is already bound (33). The yeast HO promoter also uses an alternative, Mediator-dependent recruitment pathway. Mediator is recruited alone to this promoter by the activator SBF. The remainder of the PIC, including Pol II, TFIIB, and TFIIH, is recruited in a later, Cdk1-dependent step (6).
Mediator has also been implicated in reinitiation of transcription, a process distinct from PIC formation and initiation of the first round of transcription. Following transcription initiation, a number of proteins remain bound at the promoter in a complex called Scaffold. This reinitiation intermediate consists of TFIID, TFIIA, TFIIH, Mediator, and the less stable TFIIE. Scaffold can recruit Pol II, TFIIB, and TFIIF back to the promoter for additional rounds of transcription. Reinitiation by this mechanism can occur more rapidly than initiation of the first round of transcription, supporting higher rates of transcription. Some activators can stabilize Scaffold, and this function may help promote high levels of transcription reinitiation during activation (46).
Biochemical and genetic studies have identified two major modules within yeast Mediator, the Srb4 module and the Rgr1 module. The Rgr1 module of Mediator includes Rgr1, Med1, Med4, Med7, Med9, Med10, Srb7, and the Sin4 complex (Sin4, Pgd1, Gal11, and Med2) (22). The Sin4 complex is anchored within Mediator by the Sin4 protein so that deletion of sin4 results in the loss of this entire subcomplex from purified Mediator (Fig. 1). Although Pgd1, Med2, and Gal11 are dependent on each other for their stable association within Mediator, Sin4 association with purified Mediator is only slightly diminished by their loss. Recent in vitro work has implicated the Sin4 complex as an activator-binding module within Mediator. In a reconstituted transcription system, deletion of pgd1, med2, or sin4 almost completely eliminated activation by Gal4-VP16. In contrast, Gcn4 activation was blocked by the sin4 deletion but was only mildly decreased by deletion of pgd1 or med2 (29). These results suggested that the Sin4 complex is essential for the function of certain activators. In addition, Park et al. have shown direct interactions between Gal11 and the activators Gcn4, Gal4, and VP16, as well as an interaction between Gcn4 and Pgd1 (32).
To examine the function of the Sin4 complex in more detail, we used yeast nuclear extracts in combination with an immobilized promoter. The immobilized-template system allows direct assay of PIC and Scaffold formation under conditions which can be directly compared with in vitro transcription. Because transcription factors are derived from unfractionated nuclear extracts, this system allows us to analyze the role of the Sin4 complex in a system that is likely more representative of the array of nuclear proteins present in vivo. In contrast to those of earlier studies, our results show that the Sin4 complex does not function solely as an activator-binding site within Mediator. Instead, the complex also has a general function in Mediator and Pol II-Med stability, PIC formation, and the formation of the Scaffold complex.
MATERIALS AND METHODS
Yeast strains.
The wild-type strain used was BY4705 (MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0). The deletion strains were created by replacing pgd1 and sin4 with the trp1 marker (5).
Nuclear extract.
Nuclear extracts were prepared from 3-liter cultures as described at the website for the Hahn laboratory (www.fhcrc.org/labs/hahn). EDTA was added to a final concentration of 20 mM during the 15-min incubation (at 30°C in 50 mM Tris [pH 7.5]-30 mM dithiothreitol [DTT]). This was necessary because the flocculent deletion strains did not form spheroplasts well otherwise.
Plasmid transcription.
Plasmid transcription reactions were performed essentially as described at the Hahn laboratory website. Transcription reactions were carried out in a 25-μl volume. Nuclear extract (120 μg) was incubated with 150 ng of pSH515 plasmid with or without activator (24 ng of Gal4-VP16, 30 ng of Gal4-AH, or 30 ng of Gal4-Gcn4) in transcription mix containing 1× transcription buffer (10 mM HEPES, 100 mM potassium glutamate, 10 mM magnesium acetate, 5 mM EGTA, 3.5% glycerol), 2.5 mM DTT, 192 μg of phosphocreatine, 0.2 μg of creatine phosphokinase, and 10 U of RNase inhibitor (Amersham) for 40 min to form PICs. To initiate transcription, nucleoside triphosphates (NTPs) were added to a final concentration of 100 μM each. The reaction was incubated at room temperature for 2.5 or 40 min and then stopped with 180 μl of stop mix (100 mM sodium acetate, 10 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 17 μg of tRNA/ml). Samples were extracted with phenol-chloroform and precipitated with ethanol. Transcripts were analyzed by primer extension (35), and quantitation was performed with a PhosphorImager (Molecular Dynamics).
Immobilized-template transcription.
The immobilized-template protocols are described at the Hahn laboratory website. Biotinylated template was prepared by PCR from pSH515 plasmid and immobilized on M-280 streptavidin Dynabeads (Dynal) as previously described (36). Transcription reactions were performed in a 50-μl volume. PICs were assembled in the presence of 0.05% NP-40 as previously described (36), except that PICs were not washed (washed PICs are not able to support multiround transcription). NTPs were added to unwashed PICs to a final concentration of 100 μM each, and transcription was stopped after 2.5 or 40 min by the addition of 360 μl of stop mix. The supernatant was removed from the beads, extracted with phenol-chloroform, and ethanol precipitated. Transcripts were analyzed by primer extension, with actinomycin C1 (15 μg/ml) being added during the extension reaction.
To determine Scaffold function, Scaffold complexes were prepared as described below, except that 100 μM ATP was used in place of NTPs. Templates were washed and resuspended in transcription mix plus 0.5 μg of HaeIII-digested Escherichia coli competitor DNA and 120 μg of nuclear extract from the srb2 deletion strain (Δsrb2 extract). NTPs were immediately added, and transcription was stopped after 2.5 min and analyzed as described above. Residual PIC transcription was determined by resuspending washed PICs in transcription mix plus competitor DNA alone. Scaffold-independent transcription by the Δsrb2 extract was determined by adding transcription mix plus competitor DNA and Δsrb2 extract to immobilized templates that had been preincubated with Gal4-VP16 only.
Analysis of transcription intermediates.
To analyze transcription intermediates, reactions were performed in a 100-μl volume as described at the Hahn laboratory website. Where indicated, some reaction mixtures with mutant nuclear extracts were scaled up to 200 or 400 μl for more accurate quantitation. PICs were formed as described above and then washed three times in 1× transcription buffer containing 0.05% NP-40 and 2.5 mM DTT. PICs were immediately digested with 60 U of PstI (New England Biolabs) for 30 min at 37°C with agitation. Promoter-bound proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting. Quantitation was performed using the LI-COR Biosciences Odyssey infrared imaging system. Bands were visualized with IRDye800- and Cy5.5-labeled secondary antibodies. This system produced Western blots that were linear over a 9- to 16-fold range, depending on the primary antibodies. Odyssey application software was used to quantitate the bands and generate standard curves from serially diluted wild-type PICs. To determine PIC stability, washed PICs were resuspended in transcription mix containing 1 μg of HaeIII-digested E. coli DNA competitor and incubated for 40 min. Templates were washed once, and the samples were then digested and analyzed as described above.
Scaffold complexes were formed by resuspending washed PICs in transcription mix and then adding 100 μM concentrations of NTPs for 3 min. The templates were washed once and then digested and analyzed as described above. To test Scaffold stability, washed Scaffold complexes were resuspended in transcription mix containing 1 μg of HaeIII-digested E. coli competitor DNA, incubated for 40 min, and washed once before the PstI digestion.
Immunoprecipitation.
Anti-Srb4 polyclonal antibody was bound to protein A Dynabeads (Dynal) at a ratio of 6 μl of antibody to 10 μl of Dynabeads. The antibody was cross-linked to the beads in 0.2 M triethanolamine, 0.003% NP-40, and 20 mM dimethyl pimelimidate and then washed with 1× phosphate-buffered saline, 0.1% bovine serum albumin, and 0.003% NP-40. For the immunoprecipitation, beads were resuspended in nuclear extract and 1× transcription buffer using 24 μl of beads per 960 μg of extract. After incubation at room temperature for 90 min, the beads were washed five times with 1× transcription buffer plus 0.1% bovine serum albumin and 0.05% NP-40. Immunoprecipitated proteins were eluted in 50 mM Tris (pH 8.3), 1 mM EDTA, and 0.15% SDS for 10 min at 37°C. Proteins were analyzed by SDS-PAGE and Western blotting and quantitated as described above.
RESULTS
Sin4 complex mutations result in impaired multiround transcription.
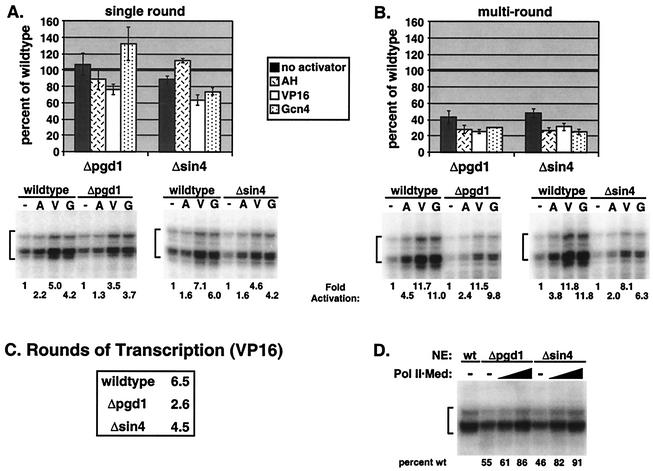
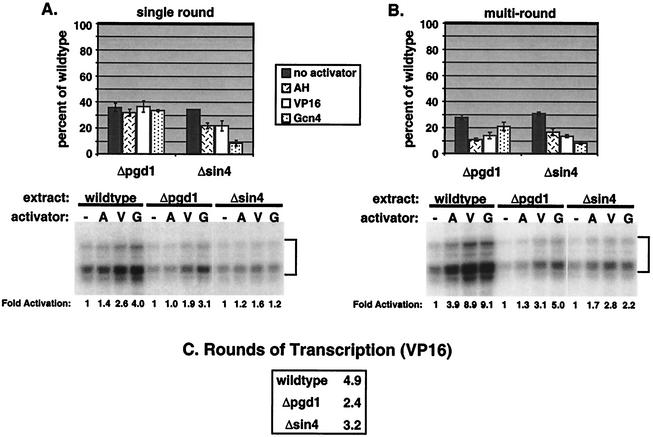
Previous work using purified factors has suggested that the Sin4 complex may play a key role in transcriptional activation. To extend these studies to a crude extract system, sin4 and pgd1 deletion strains were first constructed. As described previously, both deletion strains are viable, flocculent, and not temperature sensitive (29). To determine the effect of the pgd1 and sin4 deletions on in vitro transcription, nuclear extracts from the wild-type and mutant strains were incubated for 40 min with pSH515 plasmid template (which contains a modified HIS4 promoter with a single Gal4 binding site) and activator (Gal4-AH, Gal4-VP16, or Gal4-Gcn4). Forty minutes is sufficient time for maximum PIC formation in wild-type extracts (36). NTPs were then added for either 2.5 or 40 min to assay single-round or multiround transcription, respectively. For single-round transcription (Fig. 2A), neither of the mutants showed strong transcription defects, either in the overall level of transcription or in the level of activation.
FIG. 2.
Δpgd1 and Δsin4 mutants have a general defect in plasmid multiround transcription. PICs were formed on plasmid template for 40 min, NTPs were added to initiate transcription, and the reactions were stopped after 2.5 (A) or 40 (B) min. Transcripts were assayed by primer extension (gels). Results of typical transcription reactions with wild-type, Δpgd1, and Δsin4 extracts and with no activator (−), Gal4-AH (A), Gal4-VP16 (V), or Gal4-Gcn4 (G) are shown. Levels of activation (n-fold) are indicated below the gels. Data in the graphs are averages from at least four experiments, with the level of mutant transcription calculated as a percentage of wild-type transcription. (C) Rounds of transcription were calculated by dividing multiround transcription by single-round transcription. The results shown are an average of three experiments. (D) VP16-activated multiround transcription was assayed as in panel B with 0, 1, or 2 μl of purified Pol II-Med complex (∼0.04 pmol/μl) added to the mutant nuclear extracts (NEs).
In contrast, disruption of the Sin4 complex significantly decreased levels of multiround transcription (Fig. 2B). Basal and activated transcription levels were similarly decreased to 20 to 50% of the levels seen with the wild type. In contrast to previous results from reconstituted systems (29), the level of activation was only mildly decreased, indicating that the activation domains AH, VP16, and Gcn4 function in the absence of the Sin4 complex. The ability of the mutant nuclear extracts to support activation and the similar defect in basal and activated transcription suggest a general requirement for the Sin4 complex in transcription. The number of rounds of transcription supported by each nuclear extract was determined by comparing single-round and multiround transcription levels (Fig. 2C). With the activator Gal4-VP16, wild-type extract completed an average of 6.5 rounds of transcription during the 40-min transcription assay. In contrast, both mutants supported fewer rounds. Extracts from the pgd1 deletion strain (Δpgd1 extracts) completed only 2.6 rounds, while those from the sin4 deletion strain (Δsin4 extracts) supported 4.5 rounds. These results suggest that the Sin4 complex may play a key role during the reinitiation of multiround transcription.
To show that the transcription phenotypes observed are a direct effect of the deletion of pgd1 and sin4, nuclear extracts were first probed by Western blotting to determine the concentration of other transcription factors. Compared with the extract from the wild type, both mutant extracts contained similar amounts of all examined factors, including TATA-binding protein (TBP), Mediator, Pol II, TFIIB, TFIIA, TFIIH, TFIIE, and TFIIF (data not shown). A Mediator-specific transcription defect in these extracts was also demonstrated by adding back purified Pol II-Med (25) to rescue multiround transcription in the Δpgd1 and Δsin4 extracts. As shown in Fig. 2D, increasing amounts of Pol II-Med restored transcription in the Δpgd1 and Δsin4 extracts to 86 and 91% of wild-type levels, respectively. These experiments showed that the phenotypes discussed in this paper are the direct result of the deletion of pgd1 and sin4 and are not due to secondary effects.
Sin4 complex mutations decrease the rate and extent of PIC formation.
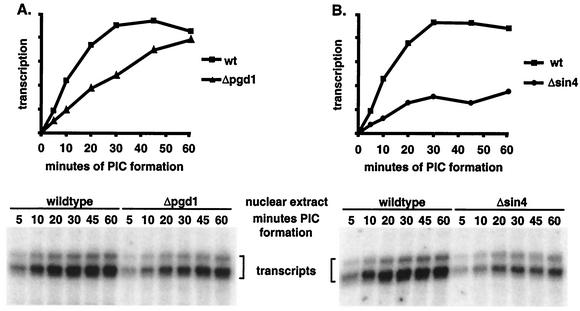
In the single-round transcription assays (Fig. 2A), PICs were formed for 40 min before the addition of NTPs. This method may not necessarily reveal a kinetic defect in PIC formation caused by the Sin4 complex deletions. To examine whether Pgd1 and Sin4 have any role during PIC formation, the rates of PIC formation in wild-type and mutant extracts were measured (Fig. 3). Extract, template, and Gal4-VP16 activator were mixed and incubated at room temperature for up to 60 min. At the intermediate time points shown in Fig. 3, samples were removed and PIC formation was assayed by the addition of NTPs for a single round of transcription. As reported previously, wild-type PICs peaked by 40 min (36). Deletion of pgd1 slowed the initial PIC formation rate by an average of twofold, although the number of PICs formed eventually reached levels near that of the wild type. Deletion of sin4 had a more severe effect. The initial PIC formation rate was one-third of that of the wild type, and PIC formation in the Δsin4 extract reached only about half of the wild-type level after 40 min, consistent with the results shown in Fig. 2A. Thus, the Sin4 complex plays a role in increasing the rate and extent of PIC formation.
FIG. 3.
Sin4 and Pgd1 promote efficient PIC formation. Wild-type extract and Δpgd1 (A) or Δsin4 (B) extract were incubated with Gal4-VP16 activator and plasmid template in transcription mix for the indicated times. PIC formation was assayed by adding NTPs for a single 2.5-min round of transcription.
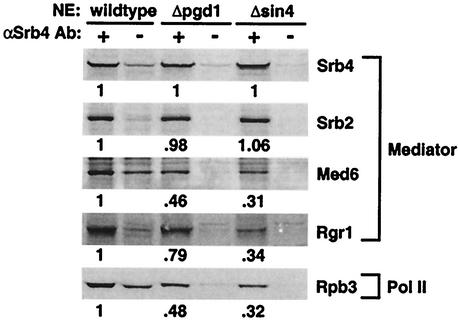
Since the decrease in the rate and extent of PIC formation could be due to a loss of stable, intact Pol II-Med in the mutant extracts, nuclear extracts were immunoprecipitated with anti-Srb4 antibody to isolate the Pol II-Med complex (Fig. 4). Srb4 and Srb2 were coimmunoprecipitated with equal efficiency from mutant and wild-type extracts. However, other components of Mediator (Rgr1 and Med6) and Pol II (Rpb3) were partially lost from the complexes immunoprecipitated from mutant nuclear extracts. This defect was strongest in the Δsin4 extract, which also had the strongest defect in the rate of PIC formation. Since the concentrations of Srb4 and Rpb3 in the wild-type and mutant nuclear extracts were similar (data not shown), these results indicate that the Pol II-Med complex is destabilized by the loss of Sin4 complex components.
FIG. 4.
Sin4 complex stabilizes Pol II-Med. Nuclear extracts (NEs) were incubated with protein A Dynabeads either with (+) or without (−) bound anti-Srb4 polyclonal antibody. Immunoprecipitated proteins were analyzed by SDS-PAGE and Western blotting. After subtracting nonspecific binding (−), the band intensities were normalized to that of the wild type. The ratio of Pol II-Mediator components to Srb4 is shown.
An immobilized-promoter assay was used to directly examine the role of the Sin4 complex during PIC formation and reinitiation. In this assay, a linear fragment of the pSH515 HIS4 promoter was attached via a streptavidin-biotin linkage to a magnetic bead. A single PstI digestion site upstream of the Gal4 binding sites allows liberation of promoter-bound transcription intermediates from the beads for Western blot analysis (36, 46). Single- and multiround transcription assays were repeated using this system to determine whether the Sin4 complex mutants behaved similarly to what was seen when plasmid templates were used (Fig. 5). In contrast to the results with the plasmid template, the mutants assayed on immobilized templates showed marked decreases in single-round transcription, with levels ranging from 3- to 10-fold lower than those seen with the wild type (Fig. 5A). This difference was traced to the inclusion of 0.05% NP-40 in the immobilized transcription reactions. Addition of the same concentration of NP-40 in the single-round plasmid transcription assay specifically decreased mutant transcription to an extent similar to that seen on immobilized templates (data not shown). NP-40 is necessary to reduce nonspecific protein binding to beads and also to prevent sticking of beads to the reaction tubes. Since it has no effect on transcription with extracts from the wild-type yeast strain, this low concentration of detergent likely enhances the above-described instability of mutant Mediator and Pol II-Med complexes. In addition to the general activator-independent defect observed, loss of activation by Gcn4 was seen in single-round transcription with the Δsin4 extracts on the immobilized template. This was the only condition that resulted in a near-complete loss of transcriptional activation.
FIG. 5.
Mutants have a general defect in single-round and multiround transcription on an immobilized template. PICs were formed on an immobilized template for 40 min. As described for Fig. 2, NTPs were added to initiate transcription and the reactions were stopped after 2.5 (A) or 40 (B) min. Transcripts were assayed by primer extension (gels). Results of typical transcription reactions with wild-type, Δpgd1, and Δsin4 extracts with no activator (−), Gal4-AH (A), Gal4-VP16 (V), or Gal4-Gcn4 (G) are shown. Levels of activation (n-fold) are indicated below the gels. Data shown in the graphs are the averages from at least two experiments, with the level of mutant transcription calculated as a percentage of wild-type transcription. (C) Rounds of transcription were calculated by dividing multiround transcription by single-round transcription. The results shown are an average of two experiments.
Consistent with the results presented in Fig. 2, multiround transcription defects were greater than those seen for single-round transcription (Fig. 5B). Multiround defects were also about twofold greater in the presence of activators. Reinitiation occurs poorly in the absence of an activator but can contribute to the high levels of transcription induced by activators (11). Therefore, defects in reinitiation due to Sin4 complex deletions could be amplified during multiple rounds of transcription to produce the twofold activation defect observed.
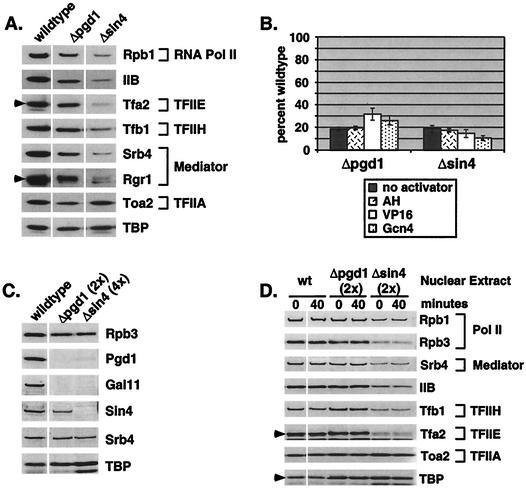
Although the experiments presented in Fig. 3 and 4 showed a kinetic defect in functional PIC formation that was attributable to decreased Pol II-Med stability, loss of the Sin4 complex might also impair a step in transcription initiation after PIC formation. To test this possibility, the immobilized-template system was used to directly examine the number of PICs formed in wild-type and mutant extracts prior to transcription initiation. Following 40 min of PIC formation, immobilized templates were washed to remove unbound proteins, and promoter-bound proteins were examined by Western blotting. Representative PICs, formed with the activator Gal4-VP16, are shown in Fig. 6A. For each mutant extract, all PIC components assayed were similarly reduced, with the exception of TBP and TFIIA. This was expected, since previous experiments using a Δsrb2 extract showed that TBP and TFIIA can bind to a promoter independently of Mediator (36). The experiment was repeated without activator and with Gal4-AH and Gal4-Gcn4. Both mutant extracts formed fewer PICs than did the wild-type extract with all activators used (Fig. 6B). The defects in PIC formation closely mirrored the defects in single-round transcription. These results and the PIC formation time course shown in Fig. 3 suggest that the major cause of the single-round transcription defect is the requirement of the Sin4 complex for efficient PIC formation.
FIG. 6.
Pgd1 and Sin4 are required for optimal PIC formation. (A) PICs were formed in the presence of Gal4-VP16 for 40 min. Promoter-bound proteins were assayed by SDS-PAGE and Western blotting. (B) The experiment described in panel A was repeated with no activator, Gal4-AH, Gal4-VP16, and Gal4-Gcn4. Mutant PIC formation is shown as a percentage of wild-type PIC formation, based on the presence of a complete PIC (including Pol II, TFIIB, TFIIH, and Mediator). The results are an average of at least two experiments. (C) Identical to panel A but showing Sin4 complex subunits. (D) Washed PICs were formed as described for panel A, and PICs were then resuspended in transcription mix plus competitor DNA. After 0 or 40 min, PICs were washed once and bound proteins were visualized by SDS-PAGE and Western blotting.
Sin4 complex composition is similar between purified and crude systems.
The differences in transcription activation results between the nuclear extract transcription system used here and the reconstituted transcription system used previously could be due to a difference in the composition of the Sin4 complexes of the two systems. To determine whether the composition is affected by the different methods used to obtain Mediator, PICs were probed for components of the Sin4 complex (Fig. 6C). In agreement with the previously described composition of purified Mediator (29), Med2 (data not shown), Pgd1, and Gal11 were all lost from promoter-bound Mediator in a Δpgd1 extract. In a Δsin4 extract, all four components of the subcomplex (Pgd1, Med2, Sin4, and Gal11) were absent from the promoter-bound Mediator. Thus, the Mediator recruited to promoters in our system has the same Sin4 complex composition as the purified Mediator used in the reconstituted transcription system.
Once formed, PICs do not require the Sin4 complex for stable association with DNA.
Based on the on-rate defect shown in Fig. 3, the Sin4 complex was found to function during transcription factor recruitment in PIC formation. However, these experiments cannot reveal an additional stabilizing role of the complex once PICs have bound at a promoter. For example, the Sin4 complex might interact with proteins and/or DNA after PIC formation to stabilize the PIC. In this case, PICs formed from mutant nuclear extracts would be expected to be less stable than wild-type PICs. Alternatively, the Sin4 complex may be required only during transcription factor recruitment to the promoter but not for PIC stability. To distinguish between these models, PICs were formed as described above, washed, and then resuspended in transcription mix plus nonspecific competitor DNA. Bound transcription factors were isolated after 40 min and analyzed by Western blotting. No significant difference in PIC stability was observed between wild-type and mutant nuclear extracts (Fig. 6D), indicating that the Sin4 complex does not play a significant role in stabilizing PICs once they are formed.
Pgd1 mutants disrupt Scaffold formation.
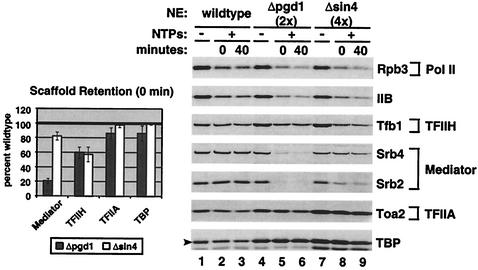
Since the Δpgd1 mutant showed a significant decrease in transcription reinitiation, the role of the Sin4 complex during reinitiation was more closely examined. High rates of reinitiation require the stable association of Scaffold (TFIID, TFIIA, TFIIH, TFIIE, and Mediator) at the promoter following initiation and the recruitment of Pol II, TFIIB, and TFIIF to this Scaffold-bound promoter for additional rounds of transcription (46). Scaffold composition was assayed to determine whether the Sin4 complex is required for Scaffold formation. As shown in Fig. 7, washed PICs were incubated with NTPs for 3 min and then washed to remove any released proteins. Under these conditions, wild-type extract releases Pol II and TFIIB, as well as TFIIF (data not shown), from the promoter, while TFIIH, Mediator, TFIIA, and TBP remain stably bound (Fig. 7, compare lanes 1 and 2). The polymerase remaining at the template is largely nonfunctional, as it can give only very low levels of transcription when incubated with NTPs (see below) (46). Although Pol II and TFIIB were released from a Δpgd1 PIC, only a partial Scaffold formed. Mediator was almost completely lost from the Scaffold, and TFIIH was also slightly reduced (Fig. 7, compare lanes 4 and 5 with lanes 1 and 2). This result is consistent with the low number of rounds of transcription completed by the Δpgd1 extract.
FIG. 7.
Δsin4, but not Δpgd1, nuclear extracts (NEs) can form stable Scaffold complexes. PICs (without NTPs) were formed for 40 min in the presence of the activator Gal4-VP16 and washed. To form Scaffold complexes, washed PICs were resuspended in transcription mix and initiated with NTPs (+) for 3.0 min. Washed promoter-bound Scaffold complexes were either immediately digested from the beads (0 min) or resuspended in transcription mix plus competitor DNA for 40 min before being washed and digested (40 min). For more accurate quantitation, 2× Δpgd1 and 4× Δsin4 reactions were performed. The graph compares the retention of Scaffold components in mutants to their retention in wild-type extracts at time 0. The calculations are based on at least five experiments.
Different results were observed when the remaining component of the Sin4 complex was removed. Following transcription from a Δsin4 PIC, the Scaffold complex remained largely intact (Fig. 7, lane 8 and graph), with only a modest decrease in Mediator and TFIIH. An average of 79% of Mediator from the Δsin4 extract remained bound at the promoter compared with 96% of Mediator from wild-type extract. This result is consistent with the fact that Δsin4 extracts support more rounds of transcription than do Δpgd1 extracts (on a plasmid, 4.5 rounds for Δsin4 extract compared with 6.5 for wild-type extract and only 2.6 for Δpgd1 extract). Since Mediator is largely lost from Scaffold in the Δpgd1 extract but to a lesser extent in the Δsin4 extract, it appears that Sin4 (in the absence of Pgd1, Med2, and Gal11) has a negative effect on the association of Mediator with the Scaffold.
To further examine the reinitiation defect, Scaffold stability was tested for up to 40 min. Yudkovsky et al. have previously shown that the increased stability of Scaffold resulting from the presence of certain activators is associated with higher levels of multiround transcription (46). Since the Δpgd1 and Δsin4 extracts support fewer rounds of transcription than does wild-type extract, it is possible that the Sin4 complex may be required for Scaffold stability over time. However, when we tested this by forming Scaffold complexes and incubating them for up to 40 min in transcription buffer with nonspecific competitor DNA, no significant differences in stability between the wild-type and mutant extracts were observed (Fig. 7, compare the results obtained with the 0- and 40-min incubation times).
Deletion of Pgd1 but not Sin4 renders Scaffold inactive.
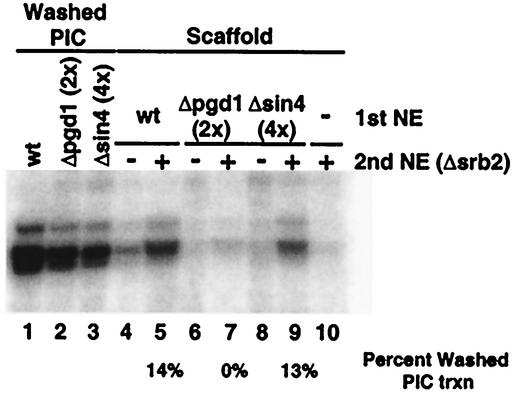
For rapid reinitiation, Scaffold must be capable of recruiting Pol II, TFIIB, and TFIIF to the promoter for additional rounds of transcription. The following experiment was performed to test the effects of Sin4 complex deletions on Scaffold function. PICs were formed with Gal4-VP16 for 40 min and then washed. Single-round transcription was assayed by the addition of NTPs to washed PICs for 2.5 min (Fig. 8, lanes 1 to 3). To test Scaffold function (Fig. 8, lanes 4 to 9), ATP was added to washed PICs to form Scaffold complexes. Scaffold complexes formed by the addition of ATP are essentially identical to those formed by NTPs (45) (data not shown). Scaffold complexes were then washed and resuspended in transcription mix containing either no second nuclear extract or Δsrb2 extract. The Δsrb2 extract is nearly inactive in transcription (Fig. 8, lane 10) and PIC formation but contains active Pol II, TFIIB, and TFIIF that can be recruited by Scaffold for a round of transcription. NTPs were immediately added, and transcription was stopped after 2.5 min. To calculate transcription specific to the Scaffold, two controls were included. In case any functional PICs remained after Scaffold formation, transcription was performed on Scaffold complexes without the addition of a second nuclear extract. As shown in Fig. 8, lanes 4, 6, and 8, transcription in the absence of a second nuclear extract was very low. In addition, Δsrb2 extract was added to a Scaffoldless promoter (Fig. 8, lane 10) to measure Scaffold-independent transcription.
FIG. 8.
Mediator is required for Scaffold function. PICs and Scaffold were formed as described in the legend to Fig. 5, except that ATP was used instead of NTPs. In lanes 1 to 3, washed-PIC transcription (trxn) was assayed. To assay Scaffold transcription in lanes 5, 7, and 9, Scaffold complexes were resuspended in transcription mix plus competitor DNA, NTPs, and Δsrb2 nuclear extract (NE) for 2.5 min. As a control, background transcription was determined in lanes 4, 6, and 8 by resuspending Scaffold complexes in transcription mix plus competitor DNA without a second nuclear extract. Scaffold-independent transcription by Δsrb2 nuclear extract was assayed in lane 10 on a template that had previously been incubated only with Gal4-VP16. For the calculations presented in the figure and text, Scaffold-specific transcription was determined by subtracting these two controls from lanes 5, 7, and 9.
As reported previously (46), wild-type Scaffold supported transcription by the Δsrb2 nuclear extract (Fig. 8, lane 5). The Scaffold transcription level in this experiment was only about 14% of the PIC transcription level because the Δsrb2 extract was incubated with Scaffold for only 2.5 min of transcription in order to assay just the first reinitiation events. When Δpgd1 Scaffold-specific transcription was calculated by subtracting background transcription (Δsrb2 extract-independent transcription [Fig. 8, lane 6] and Scaffold-independent transcription [lane 10]) from transcription by the Δpgd1 Scaffold in the presence of Δsrb2 extract (lane 7), the Δpgd1 Scaffold did not support any specific transcription. Since Mediator is missing from the Δpgd1 Scaffold, this result indicates that Mediator is required for Scaffold function in reinitiation. In contrast, a Δsin4 Scaffold (Fig. 8, lane 9) functioned similarly (as a percentage of PIC transcription) to a wild-type Scaffold. Scaffold is therefore able to recruit the transcription machinery for reinitiation in the absence of the Sin4 complex.
DISCUSSION
Based on studies using purified factors, the Sin4 complex has previously been suggested to be an activator-specific component of the yeast Mediator. In contrast, our study has uncovered activator-independent functions for the Sin4 complex in transcription. First, the complex functions during PIC formation to allow stable association of Pol II-Med and its efficient recruitment into the PIC. Second, genetic disruption of a subset of Sin4 complex subunits destabilizes Mediator within the Scaffold, a complex of general factors and Mediator left behind at the promoter after transcription initiation. These biochemical activities can explain some of the contradictory genetic and transcriptional phenotypes observed with subunits of the Sin4 complex.
The Sin4 complex and a role in activated and basal transcription.
In contrast to previous results, deletion of pgd1 and sin4 did not result in the specific loss of transcriptional activation when assayed in yeast nuclear extracts. When single-round transcription was assayed, the defects observed were similar for both unactivated basal transcription and transcription activated by AH, VP16, and Gcn4 activation domains. This demonstrates a fundamental role for the Sin4 complex in transcription, whether or not it is stimulated by an activation domain. Myers et al. and Park et al. observed that a transcription system reconstituted with purified factors was dependent on the Sin4 complex for VP16 and Gcn4 activation (29, 32). In contrast, these activators stimulated transcription in our system in the absence of Sin4 complex components. This was not due to a difference in Mediator composition between the two systems. A likely reason for the different results could be the spectrum of transcription factors present in the two transcription systems. Nuclear extracts contain not only the general transcription factors of purified systems but also coactivators such as TAFIIs and SAGA. These coactivators are targets of some activators and are known to be required for activation at certain promoters (2, 15, 21, 23, 27). If activators have no alternative in the purified system, then they may act solely through the Sin4 complex to stimulate transcription. However, since additional activator targets are available in the crude system, elimination of one activation pathway, such as through the Sin4 complex, may have little or no effect on levels of transcription activation.
Studies with both humans and yeast have identified a general role for Mediator in transcription. Studies with yeast have shown that mutations in the Srb2,4,5,6 complex can eliminate both basal and activated transcription (36, 44). Furthermore, mutations in Srb2, Srb4, or Srb5 were shown to reduce the formation of functional PICs by blocking the recruitment of factors downstream of TFIID and TFIIA to the promoter (36). Depletion of hMediator from HeLa extracts reduces both basal and activated transcription. In previous experiments, hMediator also appeared to act cooperatively with TFIID to promote transcription (1, 28). We found that, within Mediator, the Sin4 complex can play a general role in transcription.
Sin4 complex promotes efficient PIC formation.
Kinetic analysis of the PIC formation rate in pgd1 and sin4 mutants revealed that the Sin4 complex functions to increase both the rate and extent of PIC formation. This was found by assaying PIC function in transcription (Fig. 3) and by directly measuring the number of PICs formed by use of an immobilized-template assay (Fig. 6A). Both the pgd1 and sin4 deletions resulted in a noticeable decrease in the rate of PIC formation and in the amount of intact Pol II-Med but did not noticeably decrease the stability of PICs once formed. This suggests that the Sin4 complex performs its most important function during the promoter binding step when Mediator cooperatively binds with most of the other general factors. Consistent with this model, the defects in PIC formation correlate with the defects in single-round transcription, indicating that PICs formed with or without the Sin4 complex are able to initiate transcription with no apparent defect. Thus, even though fewer PICs are formed in the Sin4 complex mutants, PICs that do form are as stable as those formed in the wild type and can initiate transcription normally.
Role of the Sin4 complex and Mediator in transcription reinitiation.
Mediator has been determined to be a component of the Scaffold complex, although its role within the Scaffold was not clear (46). The results with the Δpgd1 nuclear extracts, in which Mediator is specifically lost from the Scaffold, revealed that Mediator plays a key role in Scaffold function (Fig. 7 and 8). Although all components of Scaffold except Mediator remain associated with the promoter, the Δpgd1 Scaffold is unable to support reinitiation. The loss of Scaffold function can be attributed to the loss of Mediator and not to the slight decrease in TFIIH, because TFIIH association is similarly decreased in the functional Δsin4 Scaffold. Since Mediator has been observed to interact with the C-terminal domain of Pol II both physically and genetically (17, 31, 43), Mediator might be a key factor in the recruitment of Pol II and other factors to the promoter for reinitiation.
Unexpectedly, removal of only a part of the Sin4 complex from Mediator has the most severe effect on Scaffold formation and reinitiation. Deletion of pgd1, which causes the additional loss of the Med2 and Gal11 subunits from Mediator, destabilizes Mediator upon transcription initiation. In contrast, deletion of sin4 removes all of the Sin4 complex subunits but retains the remainder of Mediator in a stable Scaffold complex upon initiation. Together, these results demonstrate that Sin4 can destabilize the Scaffold complex in the absence of Pgd1, Gal11, and Med2. This is consistent with the finding that the Δpgd1 mutant extract gives the fewest number of rounds of transcription reinitiation. While these results are surprising, they may explain several previous studies in which Δpgd1 or Δmed2 strains showed stronger phenotypes than Δsin4 strains. For example, using an in vivo reporter gene activated by Gal4-VP16, Myers et al. found a strong activation defect in a Δmed2 strain (12% of wild-type activation) and a milder defect when sin4 was deleted (63% of wild-type activation). Similarly, Δpgd1 and Δgal11 strains are phenotypically Gal−, while Δsin4 strains are Gal+ (29).
Sin4 has been implicated previously in transcription repression by a number of screens for negative regulators. However, most of these studies screened or selected for mutations that increased transcription under repressive or nonactivating conditions (7, 40, 41). In our experiments, we identified a negative role for Sin4 in Scaffold formation under nonrepressing conditions. This negative function of Sin4, revealed by deletion of pgd1, might be related to the role of Sin4 in transcription repression. Alternatively, these may be two separate functions of Sin4.
As noted earlier, deletions in the Sin4 complex result in a mild, nonspecific activation defect in multiround transcription (Fig. 2B and 5B). It is known that activators can promote multiround transcription and that at least some activators, such as VP16, can stabilize the Scaffold complex (20, 38, 39, 46). If high levels of activated transcription rely on efficient reinitiation, then the reinitiation defect caused by deletions in the Sin4 complex may contribute to the nonspecific activation defect observed. Even if the Sin4 complex is not the exclusive target of activators, its disruption may still affect activation by impairing transcription reinitiation.
Acknowledgments
This work was supported by NIH grant GM53451. W.M.R. is a Howard Hughes Medical Institute Predoctoral Fellow. S.H. is an associate investigator of the Howard Hughes Medical Institute.
We thank L. Hoskins for the Δsrb2 extract. We also thank A. Aguilera for the pT7-7 His(6)Hrs1 plasmid, Y. J. Kim for the pEh-RGR1N plasmid, D. Stillman for the Sin4 antibody, Y. Liu for the Rgr1 antibody and Pol II-Med complex, H. Sakurai for the Gal11 antibody, R. Kornberg for the Tfb1 antibody, and N. Yudkovsky for comments on the manuscript.
REFERENCES
- 1.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boube, M., L. Joulia, D. L. Cribbs, and H.-M. Bourbon. 2002. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 7.Covitz, P. A., W. Song, and A. P. Mitchell. 1994. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics 138:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson, Y. W. Jiang, Y. Li, R. D. Kornberg, and F. J. Asturias. 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97:14307-14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustafsson, C. M., and T. Samuelsson. 2001. Mediator—a universal complex in transcriptional regulation. Mol. Microbiol. 41:1-8. [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson, C. M., L. C. Myers, J. Beve, H. Spåhr, M. Lui, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 1998. Identification of new mediator subunits in the RNA polymerase II holoenzyme from Saccharomyces cerevisiae. J. Biol. Chem. 273:30851-30854. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, S. 1998. Activation and the role of reinitiation in the control of transcription by RNA polymerase II. Cold Spring Harbor Symp. Quant. Biol. LXIII:181-188. [DOI] [PubMed] [Google Scholar]
- 12.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 13.Ito, M., C.-X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z.-Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, Y. W., P. Veschambre, H. Erdhument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K. M., J. Wang, A. Smallwood, C. Arayata, and M. Carey. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16:1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee, and Y.-Y. Kim. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276:42003-42010. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y.-J., S. Björklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 18.Koleske, A. J., S. Buratowski, M. Nonet, and R. A. Young. 1992. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell 69:883-894. [DOI] [PubMed] [Google Scholar]
- 19.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466-469. [DOI] [PubMed] [Google Scholar]
- 20.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y.-J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X.-Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., S. Bjorklund, Y. W. Jiang, Y.-J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Y., J. A. Ranish, R. Aebersold, and S. Hahn. 2001. Yeast nuclear extract contains two major forms of mediator complexes. J. Biol. Chem. 276:7169-7175. [PubMed] [Google Scholar]
- 26.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 27.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 28.Mittler, G., E. Kremmer, H. T. M. Timmers, and M. Meisterernst. 2001. Novel critical role of a human mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers, L. C., C. M. Gustafsson, K. C. Hayashibara, P. O. Brown, and R. D. Kornberg. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Näär, A. M., P. A. Beurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 31.Näär, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J. M., H.-S. Kim, S. J. Han, M.-S. Hwang, Y. C. Lee, and Y.-J. Kim. 2000. In vivo requirement of activator-specific binding targets of Mediator. Mol. Cell. Biol. 20:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y.-J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 34.Rachez, C., Z. Suldan, J. Ward, C.-P. B. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranish, J. A., and S. Hahn. 1991. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 266:19320-19327. [PubMed] [Google Scholar]
- 36.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 38.Sandaltzopoulos, R., and P. B. Becker. 1998. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol. 18:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan, P. L., T. P. Mayall, E. Verdin, and K. A. Jones. 1997. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 11:3327-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song, W., I. Treich, N. Qian, S. Kuchin, and M. Carlson. 1996. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol. Cell. Biol. 16:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stillman, D. J., S. Dorland, and Y. Yu. 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics 136:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73:1361-1375. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, C. M., and R. A. Young. 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. USA 92:4587-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yudkovsky, N. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]