Abstract
The Drosophila melanogaster Brahma (Brm) complex, a counterpart of the Saccharomyces cerevisiae SWI/SNF ATP-dependent chromatin remodeling complex, is important for proper development by maintaining specific gene expression patterns. The SNR1 subunit is strongly conserved with yeast SNF5 and mammalian INI1 and is required for full activity of the Brm complex. We identified a temperature-sensitive allele of snr1 caused by a single amino acid substitution in the conserved repeat 2 region, implicated in a variety of protein-protein interactions. Genetic analyses of snr1E1 reveal that it functions as an antimorph and that snr1 has critical roles in tissue patterning and growth control. Temperature shifts show that snr1 is continuously required, with essential functions in embryogenesis, pupal stages, and adults. Allele-specific genetic interactions between snr1E1 and mutations in genes encoding other members of the Brm complex suggest that snr1E1 mutant phenotypes result from reduced Brm complex function. Consistent with this view, SNR1E1 is stably associated with other components of the Brm complex at the restrictive temperature. SNR1 can establish direct contacts through the conserved repeat 2 region with the SET domain of the homeotic regulator Trithorax (TRX), and SNR1E1 is partially defective for functional TRX association. As truncating mutations of INI1 are strongly correlated with aggressive cancers, our results support the view that SNR1, and specifically the repeat 2 region, has a critical role in mediating cell growth control functions of the metazoan SWI/SNF complexes.
The coordination between cell proliferation and terminal differentiation is of critical importance during the development of multicellular organisms. The metazoan SWI/SNF-related protein complexes function in both of these processes in a broad range of tissues by altering gene expression at specific promoters through ATP-dependent chromatin remodeling (32). The Saccharomyces cerevisiae SWI/SNF complex is a ∼2-MDa multisubunit assembly capable of ATP-dependent chromatin remodeling both in vitro and in vivo (53). Highly related complexes have also been identified in flies (23, 48) and mammals (73). Among the SWI/SNF complex proteins, none appears to be capable of sequence-specific DNA binding; thus, the precise mechanism for targeting the SWI/SNF complexes to their respective gene targets in vivo is not well understood. The complex can interact with transcription factors to facilitate SWI/SNF function at specific promoters (53). Interactions have been observed with steroid hormone receptors; EKLF (erythroid Kruppel-like factor), which activates the β-globin locus; C/EBPβ, which activates transcription of myeloid genes (35); c-Myc (13, 65); and HRX (56), a homolog of Drosophila melanogaster Trithorax (TRX) that regulates expression of the homeotic genes. Depending on the chromatin context of the binding site, some transcription factors may initially bind to their recognition site and then recruit SWI/SNF to alleviate chromatin repression at the promoter, or conversely, the factor may require SWI/SNF activity for binding. While there is presently very little known about what types of protein interactions might promote such recruitment, several complex subunits may function coordinately to mediate these associations (46). Further, mounting evidence suggests that the various SWI/SNF and related complexes might function in both gene activation and repression (22, 64).
Studies of the fly Brm and mammalian hBrm/Brg1 complexes have revealed widespread and important biological roles in development (45, 66). In addition to transcriptional activation, the complex functions in regulating cell cycle progression through interactions with the retinoblastoma (Rb) protein and/or cyclin-cdk complexes (7).
The purified Brm complex contains at least eight proteins, including BRM, MOR, SNR1, and OSA (15, 48). While brm, mor, and osa were identified in genetic screens as dosage-dependent activators of homeotic gene transcription, counteracting the repressive effects of the Polycomb group of genes (31), none of the other Brm complex genes was identified in any genetic screen. The snr1 gene (SNF5-Related-1) was identified (23) by molecular means based on its similarity to the human INI1/SMARCB1/hSNF5 gene, which encodes a core subunit of the hBrm and Brg1 complexes. Both snr1 and INI1 are strongly conserved with SNF5, which encodes an essential component of the yeast SWI/SNF complex, and SNR1, INI1, and SNF5 are required for full function of their respective complexes in vivo and in vitro (23, 38, 39, 55). While brm, mor, and osa have dosage-sensitive mutant phenotypes in combination with other gene mutations that can be examined in the imaginal tissues, such as leg and wing disks (8, 9, 24, 67), the existing snr1 null mutations do not display significant dosage sensitivity in most genetic assays, apparently inconsistent with SNR1 serving as a core subunit of the Brm complex and making analysis of this essential gene very difficult. Thus, the snr1 gene product is either in excess of that required to assemble the Brm complex, has unidentified functions independent of the complex, or has a limited role at a subset of target genes and thus is dispensable for some Brm complex functions. To help distinguish among these possibilities, we carried out a large-scale noncomplementation chemical mutagenesis screen to identify conditional mutant alleles of snr1 (77), and we report here the molecular and genetic characterization of snr1E1, which exhibits recessive lethality at 29°C and a variety of temperature-dependent phenotypes. Our genetic and biochemical characterizations of snr1E1 reveal that snr1 has essential functions in the development of many tissues including the peripheral nervous system, eye, and wing veins, and surprisingly, it is also required for sustained adult viability. Further, our analyses provide strong links among snr1 function, chromatin remodeling, and the regulation of cell proliferation, confirming its predicted role in restricting cell growth during normal development.
MATERIALS AND METHODS
Fly stocks.
All stocks were maintained on a standard cornmeal medium at 18°C, except for snr1E1/TM6B flies, which were maintained at 29°C. The snr1E1 and isogenic red, e/red, e strains were generated as described elsewhere (76). All other fly strains, markers, and special chromosomes used were as described in Flybase (http://flybase.bio.indiana.edu).
Molecular analysis of the snr1E1 mutation.
Genomic DNA was isolated (23) from snr1E1/snr1E1 adult flies at 18°C. Specific primers flanking the snr1 open reading frame were used for PCR amplification. Products were cloned and fully sequenced on an ABI3700 sequencer (BioResource Center, Cornell University, Ithaca, N.Y.).
Viability and mutant phenotype analyses.
The viability of homozygous snr1E1 flies at 18, 25, and 29°C was determined by crossing balanced heterozygotes and then determining the percentage of nonbalanced flies eclosed relative to siblings. The developmental period at which lethality first occurred was determined by crossing flies for 24 h at 18°C, turning them into fresh bottles, and collecting eggs every 24 h over 3 days. The crosses were then shifted to 29°C for 4 additional days, and eggs were similarly collected. Following hatching (∼24 h after egg laying), first-instar larvae were transferred to vials and allowed to develop at either 18 or 29°C. As controls, progeny from each cross were also maintained at the original temperature. The number of individual larvae and pupae of each genotype was counted at different developmental stages. Homozygous snr1E1 white prepupae raised at 18°C were collected and shifted to 29°C for the remainder of development to determine the precise lethal phase during metamorphosis according to the staging of Bainbridge and Bownes (3). Adult longevity was measured by collecting newly emerged adult flies (<8 h posteclosion at 18°C) and placing them in vials at 29°C. The number of dead flies in each vial was scored every 24 h to determine the percent viability per genotype each day.
Wings were dissected in 100% isopropanol, placed in DPX mounting fluid (Fluka Chemika), and examined on a Zeiss Axioscope microscope at 100× magnification. Adult cuticles were examined by mounting them in Hoyer's medium. For scanning electron microscopy analysis flies were dehydrated in acetone and examined on a JEOL JSM5800LV scanning electron microscope at 10 kV.
Biomass differences in weight, size, and cell number were determined from progeny raised at 18°C from self-crosses, including (i) snr1E1/TM3, (ii) snr1E1/red, e, and (iii) w− ; P[snr1+]/CyO; snr1E1/TM3. Male progeny were collected each day and aged an additional 3 days, and then pools of 10 flies (>100 flies/genotype) were weighed to determine the average weight per fly. Wing size and cell number determinations were made for five wings of each genotype. For wing area measurements, 40× digital images were outlined with Adobe Photoshop (v5.0), and the number of pixels corresponding to each wing was determined and converted into square millimeters by dividing the number of pixels in the wing by the known number of pixels in a square millimeter at the same magnification. Digital images were generated at 200× magnification from the same five wings, and the number of hairs in a constant premeasured area of each wing was counted to determine cell number (42).
Temperature shift analysis.
snr1E1/TM6B flies were crossed at either 18 or 29°C and turned into fresh vials every 24 h. Eggs laid within each 24-h period were aged at the original incubation temperature for the appropriate length of time in 24-h increments and then shifted to the opposite temperature and allowed to develop. Alternatively, flies were raised continuously at 18°C and then shifted to 29°C for 24 h at specific intervals prior to downshifting to 18°C and allowing for the completion of development. The number of snr1E1/snr1E1 adult flies was scored relative to balanced siblings for each 24-h interval to determine percent lethality. The number of wings showing ectopic veins in snr1E1/+ flies was scored and compared to the total number of snr1E1/+ wings at each time point. Developmental staging at 18°C was as follows: embryogenesis, 0 to 24 h; first larval instar, 24 to 72 h; second larval instar, 72 to 120 h; third larval instar, 120 to 216 h; pupation, 216 to 456 h; eclosion (adults), >456 h. The relative developmental staging at 29°C was 0 to 24, 24 to 48, 48 to 72, 72 to 120, 120 to 192, and >192 h. As snr1E1/snr1E1 larvae have a 48- to 72-h developmental delay during the third-larval-instar stage, viability data from each shift during and after that stage were accordingly adjusted.
The stability and accumulation of SNR1E1 were measured following each shift by Western blotting with extracts prepared from pupae prior to eclosion that correlated with the shift points (18 to 29°C) or from pupae raised continuously at 18 or 29°C. Fractionated proteins were probed with affinity-purified anti-SNR1, with chemiluminescent detection with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Labs, West Grove, Pa.).
Protein interaction studies.
Immunoprecipitations were carried out with embryonic or pupal extracts (23). Following precipitation with affinity-purified rabbit anti-SNR1 antibodies, proteins were fractionated on sodium dodecyl sulfate-4 to 12% polyacrylamide gels, transferred to nitrocellulose, and probed with affinity-purified rabbit anti-BRM (77), mouse anti-OSA (15), or rat anti-SNR1 (23). Detection was performed with appropriate horseradish peroxidase-conjugated secondary antibodies with enhanced chemiluminescence (Supersignal; Pierce Chemical).
Yeast interaction trap assays were used to define the regions of SNR1 that associated with the SET domain of TRX. Full-length SNR1 (amino acids [aa] 44 to 370) and deletion derivatives were produced as fusions to the GAL4 activation domain (GAL4AD) in pACTII and verified by Western blotting with anti-GAL4 antibodies (Babco Inc.). A C-terminal portion of TRX (aa 3534 to 3759), including the entire SET domain (aa 3610 to 3759), was fused to the GAL4 DNA binding domain in pAS1-CYH (Clontech, Palo Alto, Calif.). The SNR1E1 (G256D) fusion protein was generated by replacing a portion of the wild-type snr1 gene (BamHI-SalI) with the corresponding fragment cloned from snr1E1. Yeast strains of opposite mating types (Y187, Y190) carrying each fusion were mated to produce diploids. Interactions were assayed by growth on His(−) medium supplemented with 25 mM 3-aminotriazole and measurement of lacZ activity (Bio-Rad, Hercules, Calif.). Each combination was tested a minimum of three times, and the results were averaged. Nonbiased PCR mutagenesis of full-length GALAD-SNR1 was carried out as described previously (72) followed by gap repair to identify residues important for SET domain contacts. Noninteracting GAL4AD-SNR1* fusions were identified by an inability to grow on His(−) plates after the appropriate matings. Selected isolates were assayed for lacZ activity, tested for production of full-length SNR1 protein by Western blotting, and then subcloned and sequenced.
RESULTS
Identification of a snr1 conditional allele.
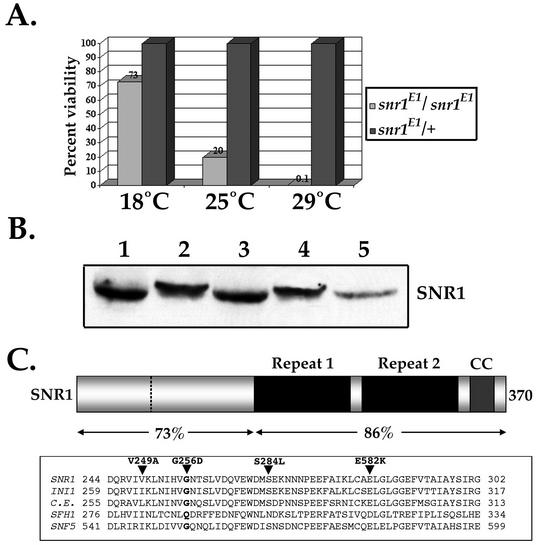
We previously identified P-element-induced snr1 null mutations (snr1R3, snr1SR21) that truncated the protein at aa 131, removing the region conserved among all SNF5 family members (23, 77). We also carried out a large-scale chemical mutagenesis noncomplementation screen and identified four new alleles of snr1 including one temperature-sensitive (TS) mutant (snr1E1) that displayed nearly complete recessive lethality at the 29°C restrictive growth temperature, an intermediate lethality at 25°C, and viability at 18°C (Fig. 1A) (Table 1). Homozygous snr1E1 flies exhibited a 48-h developmental delay at both the permissive and restrictive temperatures that appeared in the third-larval-instar stage and persisted throughout the remainder of development. A wild-type genomic transgene copy of snr1 fully restored viability to both the snr1 null mutations (77) and snr1E1 homozygotes at 29°C, demonstrating that the temperature sensitivity was due to disruption of normal snr1 function (data not shown). However, unlike null alleles, full rescue of snr1E1 required two copies of the transgene. SNR1 protein levels and stability were identical in both wild-type and snr1E1 mutants at both the permissive and restrictive temperatures as determined by immunoblot analysis with pupal extracts (Fig. 1B). We therefore conclude that the snr1E1 mutation functions as an antimorph. A single guanine-to-adenine transition was the only detectable mutation associated with the snr1E1 allele (Fig. 1C), which resulted in a glycine-to-aspartic acid alteration (G256D) within the highly conserved (∼90%) repeat 2. A glycine at this position is found in all SNF5-related proteins except SFH1 (Fig. 1C), which is a core component of the RSC complex (11, 12).
FIG. 1.
Temperature sensitivity and molecular characterization of snr1E1. (A) Percent viability of snr1E1/snr1E1 and snr1E1/+ sibling flies at different incubation temperatures. (B) Western blot of pupal-stage extracts (100 μg). SNR1 (lanes 1 and 3), SNR1E1 (lanes 2 and 4), and SNR1R3 (lane 5) at 18°C (lanes 1 and 2) and 29°C (lanes 3 to 5) were probed with anti-SNR1 antibodies. The SNR1E1 protein displayed a reduced mobility relative to that of the wild type. (C) SNR1 protein structure. Conserved domains include repeat 1, repeat 2, and a coiled-coil region (CC). The dashed line indicates the intron location. The amino acid similarity between SNR1 and INI1 is indicated below the diagram. Boxed is an alignment of repeat 2 from SNF5-family proteins, including SNR1 (D. melanogaster), INI1 (Homo sapiens), CeSNF5 (Caenorhabditis elegans), and SNF5 and SFH1 (S. cerevisiae). Shown are the known positions of affected repeat 2 residues in SNR1E1 (G256D) and mutations in INI1 (S284L), SNF5 (E582K), and SFH1 (D317K).
TABLE 1.
Lethal-phase analysis of snr1E1 homozygous and heterozygous flies
| Genotype (maternal, paternal) and temp (°C)a | No. observed at each stage:
|
|||
|---|---|---|---|---|
| Egg | Larva | Pupa | Adult | |
| +/+, +/+ | ||||
| 18 | 185 | 164 | 156 | 154 |
| 29 | 95 | 76 | 70 | 69 |
| snr1E1/snr1E1, +/+ | ||||
| 18 | 80 | 61 | 57 | 57 |
| 29 | 124 | 101 | 87 | 87 |
| +/+, snr1E1/snr1E1 | ||||
| 18 | 121 | 92 | 89 | 88 |
| 29 | 191 | 80 | 77 | 77 |
| snr1E1/snr1E1, snr1E1/snr1E1 | ||||
| 18 | 213 | 104 | 73 | 15 |
| 29 | 172 | 17 | 17 | 0 |
| snr1E1/+, snr1E1/+ | ||||
| 29 | 246 | 217 | 204 | 136 |
Incubation temperature at which the progeny were raised.
snr1 function is required throughout development.
Expression of the snr1 gene is widespread, with ubiquitous nuclear distribution of the encoded protein in early syncytial embryos that becomes more restricted to late-dividing and differentiating tissues, including the central nervous system and brain (23). There is also a strong maternal contribution of mRNA, and zygotic expression showed modest fluctuations in accumulation throughout development. Null alleles of snr1 exhibit early larval lethality with no obvious defects, presumably owing to high levels of snr1 mRNA deposited into the oocyte that provide sufficient snr1 product to complete embryogenesis.
We determined the developmental stage at which the TS snr1E1 allele caused recessive lethality. Balanced or homozygous flies were mated at 18°C for 3 days and then shifted to 29°C for an additional 4 days. Embryos were collected and allowed to develop at the temperature at which they were laid (18 or 29°C). Although homozygous snr1E1 females were fertile, males exhibited a reduced fertility at 29°C. Progeny from homozygous parents exhibited embryonic lethality at both temperatures, though the effect was more significant at 29°C (Table 1). Mutant embryos were examined, and most had secreted a larval cuticle, indicating late embryonic lethality. Moreover, the embryonic lethality was rescued with a wild-type paternal copy of snr1 at both temperatures. Homozygous snr1E1 progeny produced from heterozygous parents exhibited late-pupal-stage lethality when raised at 29°C. A significant proportion of snr1E1 homozygous flies raised at 18°C exhibited late-pupal lethality when both parents were homozygous for the snr1E1 allele. As the snr1E1 mutant exhibits biphasic lethality, including critical periods during embryogenesis and pupal development, and an snr1-null mutant is lethal during larval growth, we can conclude that snr1 and Brm complex function is required throughout development.
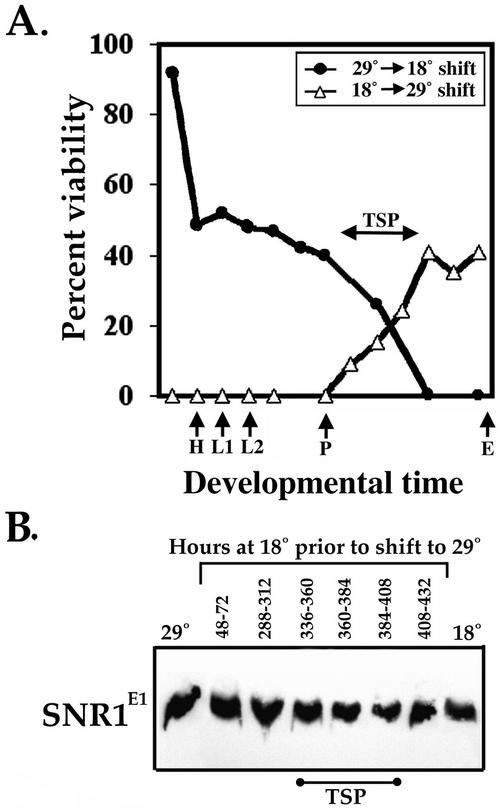
To more precisely define the temporal requirements for snr1, we performed a series of temperature shift experiments. Heterozygous and homozygous snr1E1 embryos were collected, reared at either 18 or 29°C, and later shifted to the opposite temperature at different stages for the remainder of development. Figure 2A shows the percentage of snr1E1 homozygous adult flies that emerged following the shift at each developmental stage relative to their heterozygous siblings. The data show that there is a continuous requirement for snr1 function throughout development, with critical points during late embryogenesis and again during pupal growth. Shifts to 29°C at any point until midpupal stages resulted in almost complete lethality of snr1E1 homozygotes. In contrast, snr1E1 flies shifted from 29 to 18°C at any time prior to the midpupal stage had viability restored. We examined the accumulation and stability of the SNR1E1 protein following temperature shift by Western blotting with extracts prepared from late-staged pupae (Fig. 2B). As controls, we also examined SNR1E1 from pupae raised continuously at 18 or 29°C and found that the SNR1E1 protein was stable and produced at wild-type levels following the shift at all stages examined. Thus, the SNR1E1 protein appears to be defective for function rather than synthesis or stability.
FIG. 2.
TS period and lethal phase of snr1E1. (A) TS periods (TSP) were observed during embryogenesis and pupal stages. H, hatching; L1, first larval instar; L2, second larval instar; P, pupariation; E, eclosion. (B) SNR1E1 synthesis and stability following temperature shift. Shown are results of Western blotting with anti-SNR1 against equivalent extract amounts from homozygous snr1E1 pupae raised at 18 or 29°C or shifted at the times indicated. Time points corresponding to the TSP shown in panel A are indicated by the bar.
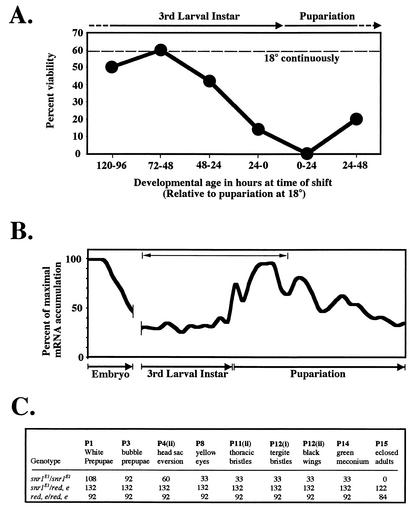
Homozygous snr1E1 larvae raised continuously at 18°C were shifted to 29°C for 24-h intervals and then downshifted to 18°C to complete development to better define the critical TS period (Fig. 3A); thus, each time point represents the percentage of viable adults that emerged when shifted to the restrictive temperature for a discrete 24-h period. Shifts to 29°C prior to or immediately following pupariation (±24 h) resulted in almost complete lethality (Fig. 3A). This sensitive developmental period coincides with significant increases in wild-type snr1 mRNA accumulation (Fig. 3B) and precisely corresponds with the onset of metamorphosis in flies that leads to the final elaboration of the adult structures. Consistent with these data, we observed a significant (70% of total) pupal lethality during the early stages of pupal metamorphosis subsequent to head eversion when flies were raised continuously at 29°C (Fig. 3C). Importantly, pupae that survived beyond these early stages (P4 to 8) continued to develop, although no adults emerged and dissection of the noneclosed pupae revealed only minor bristle defects (data not shown). The early pupal lethality is significantly later than that observed for strong loss-of-function mutations in other Brm complex genes, including null alleles of snr1, which result in death during late embryogenesis or early larval stages and suggests a previously unknown essential requirement for snr1 function for events that occur during metamorphosis. These pupal stages are associated with the last major cell proliferation and differentiation events that occur prior to eclosion (emergence of adults) and closely correlate with the time of new Brm complex synthesis.
FIG. 3.
Essential function of SNR1 during metamorphosis. (A) Temperature shift of snr1E1 revealed a critically sensitive period during the transition from larval to pupal development. Flies (0- to 24-h collections) were raised at 18°C until the times indicated (expressed as hours of development relative to pupariation), shifted to 29°C for 24 h, and then shifted back to 18°C for the remainder of development. The percentage of emerged snr1E1 homozygous adults relative to control siblings was calculated following each shift point and for flies raised continuously at 18°C (dashed line). The approximate developmental stage (at 18°C) relative to each shift point is indicated above the graph. (B) RNA accumulation of snr1 during development. Northern blot analysis of snr1 was performed with specific riboprobes hybridized to developmentally staged mRNA from wild-type flies raised at 25°C. Hybridization signals were quantified and plotted relative to maximal accumulation observed in 0- to 4-h embryos. The line above the graph represents the approximate developmental time period that correlates with the time points shown in panel A. (C) Homozygous snr1E1 pupae exhibit broad lethality during metamorphosis at 29°C. Prepupae (P1) of the genotypes indicated were collected and allowed to complete development (P15). Values represent the numbers of individuals from the original cohort that displayed the morphological marker characteristic of the pupal stage indicated (3).
Adult requirement for snr1 function.
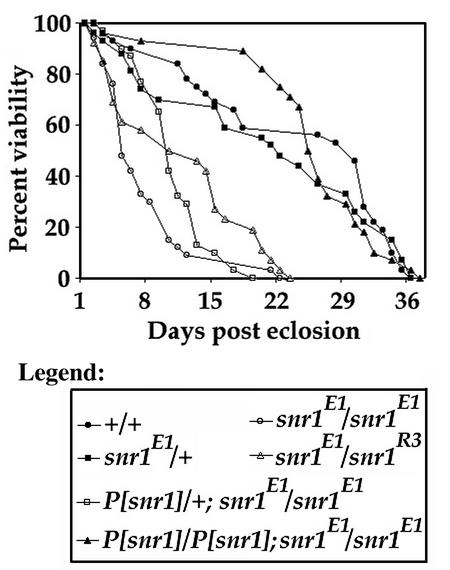
Brm complex components, including snr1, are expressed in both adult males and females; in males there has been no obvious role for the complex, while in females it appears to be required for oogenesis (18, 24). Ectopic expression of snr1 deletions suggested a broad functional role in sustaining adult viability (77). To address this question directly, newly emerged snr1E1/snr1E1 or snr1E1/snr1R3 adult flies raised at 18°C were shifted to 29°C to measure effects on life span (Fig. 4). Homozygous snr1E1 adults and snr1E1/snr1R3 transheterozygotes showed dramatic reductions in longevity compared to both control and snr1E1 heterozygous flies. The shortened life span was apparent immediately following the shift to 29°C with less than 30% viability at 7 days compared with ∼90% for wild-type flies at the same temperature. The viability for snr1E1 homozygous flies maintained at 18°C was 80% at 7 days postemergence (data not shown). Normal 29°C viability was obtained by increasing snr1 dosage with a genomic rescue transgene, with two copies restoring full viability. These results demonstrate a previously unknown requirement for snr1 function in sustained adult viability.
FIG. 4.
snr1 is required for sustained adult viability. Shown are the results of a comparison of longevity between wild-type and snr1 mutant flies. Flies of each indicated genotype were incubated at 29°C, and the number of dead flies was scored every 24 h.
Broad requirement for snr1 function in developing tissues.
Adult snr1E1 homozygous flies raised at 18°C exhibited a range of mutant phenotypes, including eye and wing disruptions and an increased body size. Eye morphology was frequently affected (12%), with reduced ommatidia, duplicated interommatidial bristles, and general head tissue reduction (Fig. 5B to D). The eye phenotypes displayed a broad expressivity, ranging from a weak disruption of the posterior portion of the eye and head tissues to significant reductions of eye size (Fig. 5B). The frequency and severity of these disruptions were increased (26%) when snr1E1 was heterozygous over a null allele (snr1R3), and these phenotypes were completely rescued by the presence of a genomic transgene, indicating a functional requirement for snr1 during normal proliferation and differentiation of eye tissue.
FIG. 5.
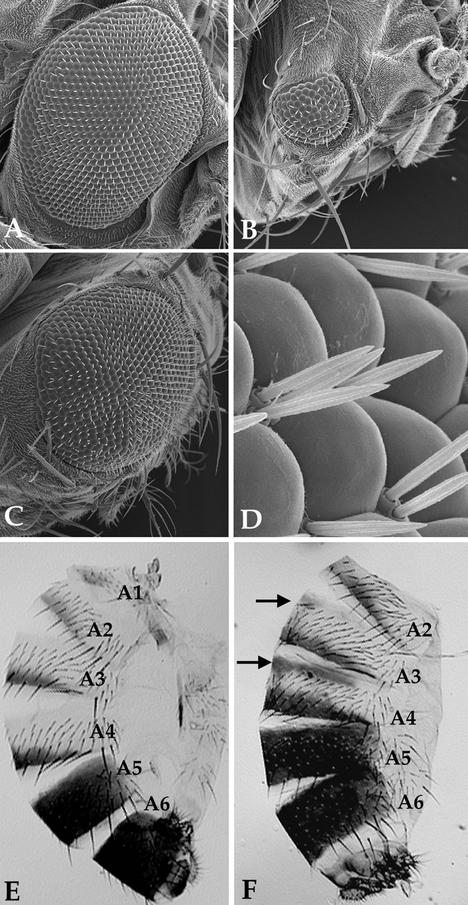
snr1 is required for proper eye and abdomen development. Scanning electron micrographs (150×) of adult eyes from flies raised at 18°C are shown. (A) Wild type; (B) snr1E1/snr1E1; (C) snr1E1/snr1R3. Note the reduced eye size, irregularly shaped ommatidia, and fused or missing interommatidial bristles in panels B and C. (D) Magnified (2,000×) view of an eye from an snr1E1/snr1E1 fly showing duplicated and fused interommatidial bristles. (E and F) Abdominal pigmentation in similarly aged adult males. Shown are lateral views of wild-type (E) and snr1E1/snr1R3 (F) flies raised at 25°C with segments A2 to A6 indicated. Arrows in panel F indicate ectopic pigmentation in the anterior portions of the A3 and A4 tergites.
Adult snr1E1 homozygous flies also frequently displayed duplication of dorsal thoracic macrochaetae (79%) and ectopic mechanosensory bristles along the L1 and L3 longitudinal wing veins (data not shown). These phenotypes were similar to the bristle duplications observed in the eyes and in somatic clones produced with a null allele (77) and were rescued by the presence of a single wild-type copy of snr1, confirming a critical role for snr1 in peripheral nervous system development. Another frequently observed snr1E1 phenotype was ectopic anterior A4 pigmentation in adult snr1E1/snr1R3 male flies (67%; Fig. 5F), consistent with disruptions in homeotic gene expression. This phenotype was also rescued by increasing snr1 dosage and completely suppressed in combination with a null allele of brm (data not shown). In addition, snr1E1/snr1E1 and snr1E1/snr1R3 adults were flightless regardless of incubation temperature, suggesting an inability to coordinate flight muscle movement.
Heterozygous snr1E1 adult flies showed a temperature-dependent ectopic vein phenotype (Fig. 6B). The penetrance ranged from 33% among flies reared at 18°C (n = 482) to 71% at 29°C (n = 464). The penetrance was significantly enhanced in snr1E1/snr1E1 mutant flies (85% at 18°C, n = 159), including incomplete longitudinal veins posterior to both the L4 and L5 veins (Fig. 6C). The highest penetrance and expressivity were observed for heterozygous snr1E1/snr1R3 flies reared at 29°C (98%, n = 187) (Fig. 6D). Importantly, the vein phenotypes were fully rescued by increasing the snr1 gene dosage (Fig. 6E and F) (Table 2), indicating that the extra veins were caused by the loss of functional snr1.
FIG. 6.
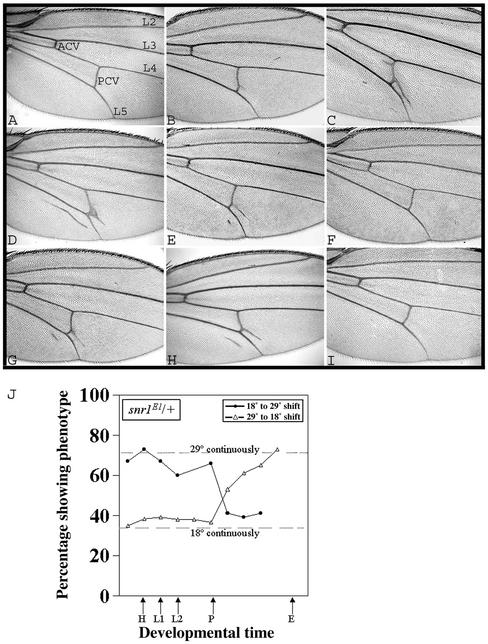
snr1 is required for proper wing vein development. (A to I) Wings (100× magnification) shown are from wild-type (A), snr1E1/+ (B), snr1E1/snr1E1 (C), snr1E1/snr1R3 (D), P[snr1+]/+; snr1E1/snr1R3 (E), P[snr1+]/P[snr1+]; snr1E1/snr1R3 (F), snr1E1/mor1 (G), snr1E1/osa2 (H), and snr1E1/brm2, snr1R3 (I) flies. Wings in panels A to C were from flies raised at 18°C, and those in panels D to I were from flies raised at 29°C. Longitudinal wing veins L2, L3, L4, and L5; anterior cross vein (ACV); and posterior cross vein (PCV) are indicated in panel A. Note the ectopic veins emanating from the posterior cross vein and posterior to L5 in panels B to E, G, and H. (J) Temperature shift showing a critical requirement for SNR1 function during early pupal development coincident with vein formation. Heterozygous snr1E1/+ flies were shifted to 18 or 29°C at the stages indicated (Fig. 2).
TABLE 2.
Enhancement and suppression of snr1E1 wing vein phenotypes by members of the Brm complex
| Genotype | No. of wings examined | Severity of phenotypea
|
||
|---|---|---|---|---|
| − | + | ++ | ||
| snr1E1/+ | 391 | 33 | 67 | 0 |
| snr1E1/snr1R3 | 268 | 1 | 1 | 98 |
| P[snr1+]/+; snr1E1/snr1R3 | 129 | 14 | 50 | 36 |
| P[snr1+]/P [snr1+]; snr1E1/snr1R3 | 147 | 66 | 26 | 8 |
| snr1E1/osa1 | 58 | 19 | 60 | 21 |
| snr1E1/osa2 | 100 | 18 | 47 | 35 |
| snr1E1/osa00090-eld | 116 | 39 | 61 | 0 |
| snr1E1/mor1 | 80 | 4 | 63 | 33 |
| snr1E1/Df(3R)sbd105, pp, Ubxbx−1, sr1, es [mor] | 86 | 21 | 79 | 0 |
| snr1E1/Df(2L)cl-h3 [E(brm)25D-26B] | 40 | 0 | 0 | 100b |
| snr1E1/Df(3L)ZN47 [E(brm)64E1-65C] | 38 | 42 | 32 | 26 |
| snr1E1/brm2 | 278 | 91 | 9 | 0 |
| snr1E1/brm2, snr1R3 | 82 | 10 | 40 | 50 |
| snr1E1, brm2/snr1R3 | 72 | 4 | 31 | 65 |
| brmK804R/+; snr1E1/+ | 88 | 24 | 76 | 0 |
| brmK804R/brmK804R; snr1E1/+ | 70 | 99 | 1 | 0 |
| snr1E1/Df(3L)brm11 | 174 | 21 | 79 | 0 |
| Df(1)c246; snr1E1 [BAP60] | 64 | 47 | 53 | 0 |
Phenotypes (at 29°C) were scored as follows: two or more ectopic veins, ++; one ectopic vein, +; and wild-type vein pattern, −. Numbers are expressed as percentages of the total wings examined for each genotype.
Df(2L)cl-h3/+ heterozygotes exhibit a fully penetrant ectopic wing vein phenotype. In combination with snr1E1, the ectopic wing vein phenotype was strongly enhanced relative to control sibling flies, with the strongest effects posterior to the L5 longitudinal vein.
The wing veins are formed from the imaginal disk epithelium during the late larval and early pupal stages, coincident with the TS lethal period for the snr1E1 allele and the time of new Brm complex synthesis. As the snr1E1 vein phenotype is temperature dependent, we carried out shift experiments to determine the precise time of snr1 function in regulating vein development. We observed a short sensitive period from early to midpupal development (Fig. 6J), indicating that snr1 acts on wing vein development during much of the early pupal period.
Brm complex interactions in wing patterning.
Expression of dominant-negative versions of brm, osa (15, 24), and snr1 (77) can affect wing vein patterning, and the overexpression phenotypes are sensitive to the dosage of other Brm complex genes. We therefore examined the effect of decreasing the dosage of known Brm complex genes on the snr1E1 wing vein phenotype. Consistent with a dominant-negative phenotype, the snr1E1 phenotype was strongly enhanced by reduction in snr1 gene dosage with a null allele (snr1R3), with almost every fly showing extensive ectopic veins (Table 2). Heterozygous hypomorphic mutations in osa and mor also enhanced the severity of the snr1E1 extra vein phenotype (Table 2) (Fig. 6G and H). While point mutations in osa (osa1 and osa2) enhanced the wing phenotype, an insertion that truncates the protein (osaeld or osa13) (70) had no strong effect. It was previously suggested that the point mutations in osa allowed for defective or nonfunctional proteins to be produced that could competitively interfere with wild-type OSA in the Brm complex (70). Similarly, allele-specific interactions were observed with heterozygous mor1, but not a deficiency of mor, perhaps due to the removal of other genes in the deficiency that modified the phenotype. Although there are no available specific alleles of genes encoding the Brm complex subunits BAP60 and BAP111, heterozygous deficiencies that removed these genes also showed only modest effects on the snr1E1 wing phenotype (Table 2). Importantly, deficiencies previously identified as enhancers of the BRMK804R dominant-negative lethality [E(brm)25D-26B and E(brm)64E1-65C] strongly enhanced the snr1E1 phenotype. These deficiencies are believed to include genes essential for brm function in vivo (48).
Surprisingly, both a brm2 null allele and the brmK804R dominant-negative (24) allele suppressed the vein phenotype of snr1E1, as well as the strong ectopic veins associated with the snr1E1/snr1R3 combination at 29°C (Table 2) (Fig. 6I). The suppression was comparable when brm2 was recombined onto either the snr1E1 or snr1R3 chromosome. The ability of the brmK804R transgene to suppress the snr1E1 phenotype was dependent on the dosage of the transgene (Table 2). The BRMK804R protein is efficiently incorporated into the Brm complex, disrupting the ATPase function of BRM, though not complex formation or stability, and thus functions as a dosage-sensitive dominant-negative (15, 24, 49). Our results indicate that reduced brm function in the wing could compensate for the reduced snr1 function, suggesting that the snr1E1 wing phenotype was possibly due to derepression of Brm complex activity in non-vein-forming regions. These data suggest that SNR1 might serve to restrict Brm complex ATP-dependent activities in specific cells while assisting these activities in other cells.
The SNR1E1 protein is present in Brm complexes.
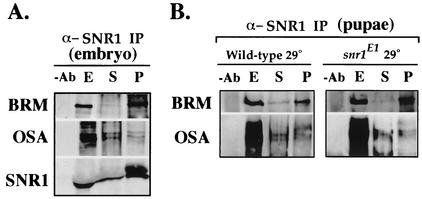
As reduced brm function could suppress the mutant phenotypes of both snr1E1/+ and snr1E1/snr1R3 flies, one likely possibility was that the SNR1E1 protein functioned as a dominant-negative to interfere with normal Brm complex function at specific targets. We addressed this possibility by first determining whether SNR1E1 was incorporated into the Brm complex. Our highly specific affinity-purified SNR1 antibodies (10, 23, 76) were capable of efficiently coprecipitating BRM and OSA from wild-type embryo extracts, confirming that the SNR1 antibodies were capable of precipitating the Brm complex (Fig. 7A). As the SNR1E1 protein is stable, we performed coimmunoprecipitation experiments with extracts prepared from both wild-type and snr1E1 homozygous pupae raised at 29°C. We found that the SNR1E1 protein was incorporated into the Brm complex as determined by coimmunoprecipitation of both BRM and OSA (Fig. 7B). Consistent with our genetic data indicating that mutant alleles of other Brm complex genes could affect the snr1E1 phenotypes, our coimmunoprecipitation data revealed that SNR1E1 functions as a weak dominant-negative, impinging on SNR1-dependent functions of the Brm complex.
FIG. 7.
SNR1E1 stably associates with components of the Brm complex. (A) Immunoprecipitation (IP) with affinity-purified anti-SNR1 antibodies (C. B. Zraly et al., submitted for publication) demonstrates association with BRM and OSA in wild-type embryo extracts. Lanes: −Ab, G-Sepharose control with no primary antibody (lane represents 50% of pelleted material); E, 100 μg of extract; S, supernatant fraction (lane represents 20% of input protein extract); P, pellet fraction (lane represents 50% of coimmunoprecipitated proteins). Western blots were probed with affinity-purified rabbit anti-BRM (77), mouse monoclonal anti-OSA (15), or rat anti-SNR1 (23). (B) Immunoprecipitation (IP) with anti-SNR1 from extracts prepared with wild-type or snr1E1 homozygous pupae raised at 29°C. Lanes are as indicated for panel A.
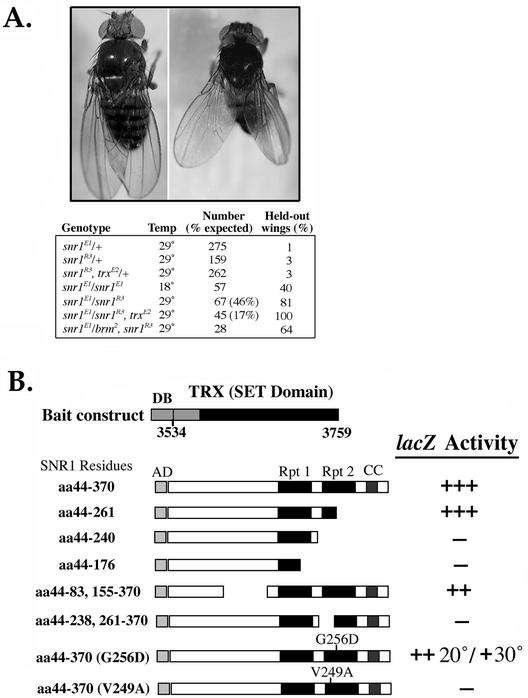
Although little is known regarding the identity of bona fide cellular targets of the Brm complex, the homeotic genes are strong candidates, possibly via cooperation with TRX (8, 9, 28, 31, 67, 70, 77). We previously showed with the snr1R3 null allele that snr1 interacted with trx in the abdomen (23). To determine whether the snr1E1 phenotypes were related to disrupted trx function, we generated a trxE2, snr1R3 recombinant and examined interactions with snr1E1 (Fig. 8A). snr1E1/snr1R3 flies exhibited a held-out wing phenotype similar to those for brm/mor and brm/osa interactions, which resemble a loss of Antp function (9). A mutation in trx amplified the snr1 held-out wing phenotype, and there was a significantly enhanced lethality (17% of expected progeny); although we did not observe enhancement of a trx abdomen phenotype, nor did we observe any effects on sex comb number on the male prothoracic leg with snr1E1, which is often used as an assay for trx function (31, 66). This allele-specific interaction suggested that the SNR1E1 protein might affect localized protein interactions, as SNR1 was shown previously to be capable of forming direct contacts with the SET domain of TRX (42).
FIG. 8.
SNR1E1 is partially defective for associations with TRX. (A) Genetic interactions between snr1E1 and trx. The snr1E1 allele was maternally provided in every cross. Adult flies of each genotype were scored for viability and held-out wings at 18 or 29°C. Values are expressed as the percentages of total adults observed versus number expected for combinations involving snr1E1 and trxE2 and percentages of total adults observed with held-out wings. (B) Yeast two-hybrid interaction tests between SNR1 derivatives fused to the GAL4AD and the SET domain (TRX) fused to the GAL4 DNA binding domain. The amino acid regions of SNR1 and TRX included in each fusion are indicated. Interaction ability was determined by lacZ activity (+++, 80 to 100% of full length; ++, 50 to 80%; +, 30 to 50%; −, <30%).
To further explore the allele-specific genetic interactions between snr1E1 and trx, we examined the association between SNR1 and TRX in more detail by yeast interaction trap analyses (Fig. 8B). A full-length SNR1 protein (aa 44 to 370) fused to the GAL4AD was capable of strong and specific interactions with the SET domain of TRX (aa 3534 to 3759). Deletions of SNR1 revealed that the repeat 2 region was necessary for the observed association. The full-length (aa 44 to 370) SNR1E1 protein containing the G256D change within repeat 2 was expressed in yeast fused to GALAD and verified by immunoblotting (data not shown). The SNR1E1 protein showed temperature-dependent association with the isolated SET domain (20 > 30°C); even at the lower temperature, the interaction was modestly diminished relative to the wild-type SNR1 protein. Though interactions between SNR1 and SET are reduced by the G256D change, they are only moderately affected, suggesting that other nearby residues might provide more direct contacts or stabilize the interaction. To help identify specific SNR1 residues important for the interaction, we carried out a nonbiased PCR mutagenesis screen. As a result of several rounds of screening, we identified a single valine-to-alanine conversion at aa 249 within repeat 2 that completely abolished the two-hybrid interaction between SNR1 and the SET domain. The mutant protein was fully stable by Western blot analysis and functional for control interactions (data not shown). The critical V249 residue resides within 7 aa of the G256D in SNR1E1, supporting our view that the repeat 2 region was necessary to mediate specific associations and that SNR1 can form direct associations with TRX.
SNR1 functions to restrict cellular proliferation.
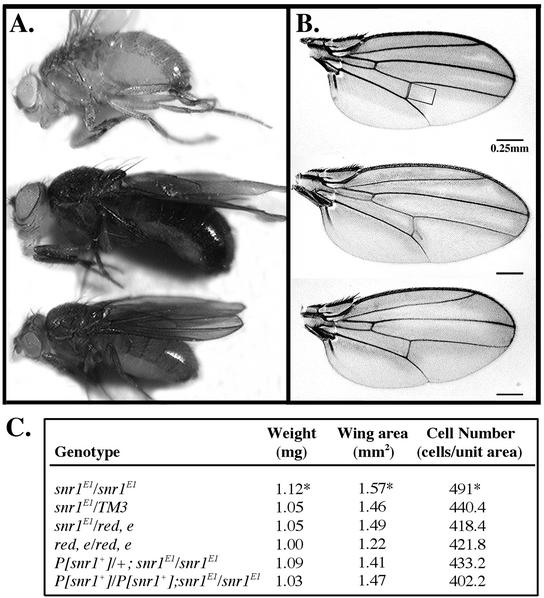
Homozygous snr1E1 flies raised at 18°C were significantly larger than either their heterozygous siblings or control flies (Fig. 9). Male flies generated from independent crosses were examined and scored for body mass and cell number. Body mass was calculated from at least 100 similarly aged male flies of each genotype, and cell number was scored by counting cells in a defined area of the wing, as each wing cell is known to secrete only one hair during development (42). As controls, heterozygous siblings were scored from crosses that included either a balancer chromosome or the parental chromosome on which the snr1E1 allele was generated and crosses in which either one or two copies of the snr1 rescue transgene were included. Both body mass and cell number were significantly (P < 10−4) greater among the snr1E1 flies than among controls, and this effect was ameliorated in a dosage-dependent manner by introducing wild-type copies of snr1. The significantly greater cell number observed in the wings of homozygous snr1E1 flies can be extrapolated to the increased biomass associated with flies of the same genotype and suggests that the larger size of snr1E1 mutant flies is likely due to increased cell proliferation. Hence, a normal function of snr1 appears to be restricting cell growth in the wing and possibly acting to repress cell proliferation in the entire organism.
FIG. 9.
snr1 functions to restrain cell growth and proliferation. (A) Male snr1E1/snr1E1 flies (middle) are larger than either the parental red, e (top) or rescued P[snr1+];snr1E1/snr1E1 (bottom) flies raised at 18°C. Similarly aged flies were photographed in the same optic field for comparison. (B) Wing size comparisons among flies of the same genotypes shown in panel A. The boxed region in the wild-type red, e wing (top) represents the area examined from all wings for cell number analyses. The wings shown were photographed at a 40× magnification. (C) Biomass differences among snr1E1 flies with various gene doses. The average weight, wing area, and cell number for sibling male flies and standard deviations were determined. *, values for snr1E1/snr1E1 in all categories are significantly different from those for all other genotypes in the same category. Probability values for snr1E1/snr1E1 flies relative to parental red, e flies ranged from P < 10−3 to P < 10−4 in the same category.
DISCUSSION
We report here the characterization of the only identified conditional mutation among genes encoding the metazoan SWI/SNF chromatin remodeling complexes. A TS recessive lethal allele of the highly conserved Drosophila snr1 gene has allowed new insights into the developmental functions of the Brm chromatin remodeling complex, including strong in vivo evidence that SNR1 has an essential role in restricting normal cell growth. The biological importance of snr1, and its human counterpart INI1, appears to be critical, as both are essential for viability and normal development. Our present studies have identified previously unknown functions for snr1 during late stages of embryogenesis, for tissue patterning events including proper eye and wing vein formation and for sustained adult viability. Recently it was shown that INI1, but no other mammalian hBrm or Brg1 complex gene, is disrupted in nearly every confirmed case (∼90%) of aggressive rhabdoid tumors that appear most often in early childhood (58, 71), and INI1 deletions are frequently associated with chronic myeloid leukemia (29). Further, murine INI1 is required for embryogenesis as early as the peri-implantation stage (E3.5), and embryonic stem cell-derived heterozygous adult mice are predisposed (>30%) to soft tissue and nervous system carcinomas (33). Thus, INI1 is a tumor suppressor and both INI1 and snr1 have unique and possibly direct roles in restricting cell growth, either independently or as components of their respective Brm complexes.
The snr1E1 mutation genetically functions as an antimorph. The mutant phenotypes associated with snr1E1 are consistent with diminished snr1 function and likely reflect altered Brm complex activities. Our conclusions are based on several lines of evidence: (i) the SNR1E1 protein is stable and associates with other Brm complex subunits even at the restrictive temperature; (ii) snr1E1 heterozygous phenotypes can be modified by allele-specific interactions with mutations in genes encoding other Brm complex components; (iii) the snr1E1 phenotypes are sensitive to snr1 dosage, being more severe in combination with a null allele and suppressed by increasing snr1 dosage by using rescue transgenes; and (iv) both the embryonic and pupal lethal phases associated with snr1E1 are significantly different from null mutants of snr1, which die during early larval development. The embryonic lethality was observed only when both parents were homozygous for snr1E1, consistent with a disruption of Brm complex functions in late embryos (8, 9, 24, 70), whereas the late lethality of snr1E1 homozygotes derived from heterozygous parents implicates essential Brm complex functions during metamorphosis, coinciding with the last major proliferation and differentiation events prior to the emergence of adults.
Our coimmunoprecipitation data revealed that other Brm complex subunits, BRM and OSA, were also stable in an snr1E1 mutant and that both could be coprecipitated with SNR1 antibody at equal efficiency in either wild-type or snr1E1 mutant backgrounds. Moreover, the snr1E1 mutant phenotypes could be strongly suppressed by a brmK804R dominant-negative transgene, whose product is stably incorporated into the Brm complex (24). While it is formally possible that snr1E1 results in reduced complex stability, our studies are consistent with the view that snr1E1 affects a postassembly step of Brm complex functions rather than disruption or destabilization of complex assembly. This contrasts with studies of SNF5, the yeast homolog of SNR1, showing that conditional mutations affecting the repeat 2 region (snf5-51ts; E582K), but not the repeat 1 region (snf5-83bts; D475N), affected complex assembly as measured by both coprecipitation of other subunits and gel fractionation (27). In snf5-51ts mutant extracts, SWI/SNF complex stability was significantly affected at both the permissive and restrictive temperatures and SNF2/SWI2 was not efficiently coprecipitated with the SNF5E582K protein. Further, diminished stability of yeast SWI3 (a homolog of Drosophila MOR) was observed in a strain carrying disruptions of SWI1 and SWI2 (52). However, similar to the brmK804R mutation, disrupting the ATPase function of the yeast SWI/SNF complex with a dominant-negative SWI2 mutant has no significant effect on complex assembly (51).
Developmental functions of SNR1 and the Brm complex.
brm is required for embryogenesis (8), and the expression patterns of brm and snr1 are nearly identical (23, 24). snr1-null mutants die during larval growth, suggesting that SNR1 is not required for all Brm complex functions in embryos. However, our analyses of snr1E1 showed that snr1 was required during embryogenesis, as are conserved components of the Brm complex, brm, mor, and osa. We conclude, therefore, that snr1 is required for a subset of embryonic functions of the complex and that the maternal contribution of snr1 is normally sufficient to complete these functions.
Brm complex components are widely expressed throughout development, with elevated levels observed during pupal stages. Late requirements for Brm complex functions in specific tissue patterning have been inferred from the use of overexpression of dominant-negative transgenes and mosaic clone analyses. Our present studies provide strong evidence for a sustained essential role of snr1 in late cell growth and patterning events. Some of the late snr1E1 mutant phenotypes are consistent with loss of steroid hormone activity. Pupal case defects observed in snr1E1 homozygous pupae (our unpublished data) are similar to those observed in some ecdysone receptor mutants (4) as well as early ecdysone-regulated genes, such as E74 (25). In addition, fluctuating RNA levels of snr1 generally coincide with rising titers of ecdysone. In light of observed interactions between mammalian hormone receptors and the SWI/SNF complexes (47, 53), it is likely that the Brm complex assists in regulating events during Drosophila metamorphosis, possibly in collaboration with the ecdysone receptor. Consistent with this view, we have observed interactions between snr1E1 and some ecdysone receptor mutants suggestive of reduced receptor functions (our unpublished observations). Homozygous snr1E1 flies raised at 29°C rapidly lose viability during early metamorphosis (pupariation), and transient temperature pulse-shifts (18 to 29 to 18°C) show strong sensitivity coincident with the transition from larval to pupal development. As homozygous adults at 18°C are unable to fly and dissected pupae (29°C) show ectopic sensory bristles, it is likely that snr1 (and presumably the Brm complex) is required for aspects of nerve or muscle cell function or development, supporting results for heterozygous INI1 chimeric mice that revealed a strong predisposition (∼30%) to develop tumors affecting the central nervous system (33).
A requirement for Brm complex activity to regulate cell growth during eye development has been shown by overexpression of OSA or dominant-negative BRMK804R that resulted in severe reductions in eye size (15, 49). Further, the OSA overexpression phenotype could be enhanced by mutations in snr1 and mor (15). One possibility is that the Brm complex might impact eye development by affecting cell division, as studies have linked snr1 and the Brm complex to regulation of S phases in the eye imaginal disk through interactions with cell cycle regulators (61). It is interesting, therefore, that dominant interference of DmCdk2 function (a cyclin-dependent kinase specific to S-phase progression) also leads to disrupted eyes and duplicated ommatidial bristle phenotypes (34) nearly identical to those seen for our snr1E1 mutants. The duplicated ommatidial bristles and ectopic mechanosensory bristles on the thorax present in snr1E1 mutants also appear similar to the phenotypes associated with somatic loss of both snr1 (77) and brm activities (24), as well as alterations in Notch pathway genes in early pupae (30). Thus, it is possible that the Brm complex is involved in regulation of Notch pathway or other neurogenic genes, and the duplicated bristle phenotypes may not be a direct result of reductions in mitotic divisions but rather of impairment of neurogenic cell fates.
SNR1 can modulate Brm complex activities both positively and negatively.
We and others previously showed that a null allele of snr1 could modestly enhance a brm loss-of-function phenotype in the prothoracic region and that snr1 could function synergistically with other Brm complex genes in eye development, including enhancement of an OSA overexpression phenotype (10, 15, 23). However, there is no identified function for snr1 in leg development (77), unlike brm, mor, and osa (31, 67), consistent with our phenotypic analysis of snr1E1.
The positions of the wing veins are initially determined during the late third-instar larval stage, and this positional information is subsequently refined and patterns are maintained during the late larval and early pupal periods (5, 6, 21). The BRM ATPase activity is required for the formation of the wing veins (15, 24), perhaps analogous to its function in homeotic gene regulation. For example, the Brm complex may be important for the transcriptional activation or maintenance of genes required for vein formation, such as the highly conserved rhomboid gene that encodes a serine protease and is expressed in all vein primordia where it functions to activate the epidermal growth factor receptor signaling pathway (reviewed in reference 41). The ectopic vein phenotype observed in snr1 mutants extends our previous knowledge of Brm complex function in wing development by identifying a critical temporal and position-specific requirement for snr1 function and suggesting that snr1 might function antagonistically to brm in specific cells of the wing. The snr1E1 ectopic vein phenotype is both temperature and snr1 dose dependent, and the TS period precisely corresponds to early pupal wing patterning events. The appearance of ectopic wing veins in an snr1E1 mutant is generally similar to phenotypes caused by ectopic expression of rhomboid (62, 63) or the transforming growth factor β homolog dpp (20, 21). The restricted temporal function is significant in that OSA-containing Brm complexes are required for repression of the fly transforming growth factor β homolog decapentaplegic (dpp) (16), and dpp function is required in early pupal wings for the proper formation of the veins. Thus, a specific functional requirement for SNR1 to regulate dpp expression or other vein-promoting genes in the wing may explain the presence of extra veins in snr1E1 mutants (our unpublished observations).
Reducing the dosage of functional snr1 enhances the snr1E1 vein phenotype while increasing snr1 gene dosage or reducing Brm complex ATP-dependent activity has the effect of suppressing the snr1E1 vein phenotype. Specifically, the ectopic wing vein phenotype associated with heterozygous snr1E1/+ was strongly enhanced by a null allele of snr1 and gene-specific mutations in mor and osa, as well as deficiencies defined as enhancers of a brm dominant-negative (48). Deficiencies or null-allele mutants that reduce the gene dosage of some Brm complex components have little detectable effect on the snr1E1 phenotype, suggesting that modest reduction in Brm complex or individual subunit abundance alone is not sufficiently limiting. There are no reported phenotypes associated with the heterozygous null mutants, although in a few specific cases null mutations in some Brm complex genes can display dosage-sensitive genetic interactions. By contrast, in trans-heterozygous combination with either a brm2 null allele or the brmK804R dominant-negative, we observed strong suppression of the vein phenotype associated with snr1E1. As the SNR1E1 protein appears to be incorporated into the Brm complex, the suppression of the ectopic wing vein phenotype by reducing (though not eliminating) brm suggests that one in vivo function of SNR1 is to restrict or modulate Brm complex activity in specific tissues or cells.
In support of this conclusion, studies of the Drosophila BRM-related ISWI protein revealed that individual subunits of chromatin remodeling complexes can affect the in vitro biochemical properties of the complex distinct from the recombinant ATPase subunit alone (37). Our data are consistent with the view that SNR1 functions antagonistically to restrict Brm complex activity in non-vein-forming wing tissues during early pupal development. One possibility is that SNR1 may serve to regulate Brm complex gene targets in vivo through associations with vein-specific repressors. By analogy, the yeast SWI/SNF complex can be targeted to the histone HTA1-HTB1 promoter in association with the HIRp repressors, possibly through interactions with SNF5 (22). Another possibility is that SNR1 (as well as MOR and OSA) could serve to regulate the gene activation functions of the Brm complex in certain cells. Consistent with this latter possibility, OSA function is normally limiting in the wing, with overexpression giving rise to morphological changes that were suppressed by decreasing BRM levels (15). As SNR1E1 functions as a weak dominant-negative, then decreasing wild-type snr1 should exacerbate the snr1E1 mutant phenotype and decreasing the amount of functional Brm complex through a brm null mutation should suppress the phenotype, precisely as we observed. Additional genetic and biochemical tests will be required to distinguish among these and other possibilities.
While components of the Brm complex are present in male and female adult flies, the only previously known function was a requirement for proper oogenesis. It is interesting, therefore, that snr1E1 females are fertile at 29°C, implying that SNR1 has a limited role in oogenesis, perhaps as a stabilizing subunit of the Brm complex. An unexpected finding was a novel requirement for snr1 function in adult longevity, as snr1E1 adult flies exhibited significantly shortened life spans at 29°C (<7 days). Misexpression of the trx-G group gene ash1 also caused reduced longevity (36), and ash1 genetically interacts with brm (68). ASH1 is a component of a large (∼2-MDa) complex distinct from the Brm complex (48); it is not known whether SNR1 is a component of the Ash1 complex, nor have genetic interactions been identified, perhaps not surprising since null alleles of snr1 weakly interact with brm mutants. As ash1 is important for trx function (57, 68) and SNR1 physically and genetically interacts with TRX, it may be possible that ASH1 can also form transient complexes with SNR1 in vivo. Thus, the decreased life span observed for snr1E1 mutant flies may reflect diminished Brm complex activity or some other complex, such as Ash1, required for sustained adult gene transcription.
SNR1 functions in restricting cell growth.
A primary phenotype of the snr1E1 mutants is an increased mitotic index, suggesting a direct role in controlling cell proliferation. Addition of a wild-type copy of snr1 suppressed the growth defects, arguing against secondary recessive mutations. Further, no obvious growth defects were observed in dissected imaginal tissues, the distribution of SNR1E1 appeared normal, and there was no aberrant mitotic activity or chromosome morphology in larval brain tissues (our unpublished observations). In correspondence with the snr1E1 TS period, the defect in cell growth regulation occurred during pupal metamorphosis, which is characterized by rapid cell division and differentiation of larval tissues into adult structures (26). Further, disruption of INI1, but no other Brm complex gene, is associated with aggressive soft tissue cancers affecting renal and nervous system tissues as well as hematopoietic cell lineages (58, 59). Importantly, no recurrent lesions are found at other loci in these tumors, suggesting that loss of INI1 is primarily responsible for tumorigenesis.
The loss of normal cell proliferation control associated with the snr1E1 mutant raises the possibility that SNR1 and the Brm complex might have a direct role in regulating aspects of the cell cycle. The mammalian Brm complexes directly interact with several transcription factors required for growth and development, as well as cell cycle regulatory proteins including CycE/CDK2, retinoblastoma (Rb), and histone deacetylases (60, 69, 75). In addition, BRG1, one of the two mammalian counterparts to BRM, is required for RAS-mediated transformation of SW13 cells (44). The Drosophila Brm complex can form stable associations with the CycE/CDK2 heterodimer that controls entry into S phase, and mutations in Brm complex genes can suppress the rough eye phenotype associated with a hypomorphic cyclinE allele (10). Thus, the Brm complex appears to function downstream of CycE/CDK2 to restrict S-phase entry. Our present studies demonstrate that SNR1 is important to mediate this aspect of Brm complex function, as the SNR1E1 protein remains tightly associated with the Brm complex even at the restrictive temperature and a single copy of the mutant allele is sufficient to elicit the proliferation defect. Another potential explanation for the snr1E1 growth defects is the disruption of cellular apoptosis, leading to increased cell proliferation. The mammalian GADD34 protein, implicated in DNA damage-induced apoptosis and regulation of cell growth, physically associates with INI1 (2). Although the contact residues involved are unknown and there is no obvious GADD34 counterpart in flies, it is intriguing that members of the SWI2/SNF2 family of ATPases are proteins that have roles in DNA repair and replication (50), suggesting that modification or disruption of protein-DNA interactions may be required for diverse cellular activities and that multiprotein chromatin remodeling complexes may assist these functions. Thus, while the precise role SNR1/INI1 plays in restricting cell growth is unclear, one likely possibility is that loss of SNR1/INI1 function results in the inability to recruit Brm complexes to critical in vivo gene targets.
SNR1 functions to target some Brm complex activities.
The SNF5-related family of proteins share strong similarity within their C termini, with considerable homology among two imperfect repeats, termed repeat 1 and repeat 2. The repeat sequences have been shown previously to contribute to complex stability and function (1, 13, 17, 27, 40, 43, 74). The repeat regions have also been implicated in mediating contacts between the complex and cellular factors, suggesting a possible targeting role for SNF5-family proteins (13, 46, 65). Both SNR1 and INI1 can physically interact with the homologous transcription factors TRX and HRX, which regulate Hox gene expression in flies and mammals (56). Disruption of trx and HRX leads to homeotic phenotypes, and misregulation of HRX function has been strongly linked to the majority of infant acute leukemias (14), possibly through involvement with INI1. While a null allele of snr1 enhanced a trx loss-of-function phenotype in the abdomen (23), the snr1E1 mutant showed only modest dosage-dependent interactions. However, the held-out wing phenotype observed in trans-heterozygotes is strongly reminiscent of homeotic loss-of-function phenotypes and is remarkably similar to brm/osa and brm/mor as well as trans-heterozygous brm allele interactions (8, 70).
While TRX was shown to associate with overexpressed SNR1 in vivo, SNR1 is not a stable component of the embryonic TAC1 complex (TRX acetylation complex), which contains TRX, the acetyltransferase dCBP, and the antiphosphatase protein SBF1 (54). We have shown that SNR1 and TRX can form direct contacts mediated through the TRX SET domain and the highly conserved SNR1 repeat 2 region. Although the contact between the SET domain and SNR1E1 is moderately affected in yeast two-hybrid assays, we were unable to detect any reduction in the in vivo association measured by coprecipitation (our unpublished observations), suggesting that TRX likely interacts with other Brm complex components in vivo. The affected amino acid in SNR1E1 resides within seven residues of a critically important valine (V249) that, when mutated to an alanine, resulted in the complete loss of SNR1 association with SET in a two-hybrid assay. Our results suggest that, at the restrictive temperature, the SNR1E1 protein may compromise interactions with TRX through local alterations in SNR1 protein structure. However, it is not known if conformational changes in SNR1E1 affect interactions among the other Brm complex subunits that might adversely influence the ability of TRX to properly regulate a subset of its targets. Importantly, not all targets of TRX or the Brm complex are affected by SNR1E1, suggesting that SNR1 is essential for some, but not all, Brm complex activities. Although TRX can form some functional associations with the Brm complex in the presence of SNR1E1, the interaction between the SNR1 repeat 2 region and the SET domain is important for normal development. As SNR1 is not a component of the TAC1 complex, this implies that the interactions must be highly regulated, perhaps transiently. In light of this possibility, it is intriguing that SBF1 also contacts TRX through the SET domain, and this association may be important for growth regulation (19). For example, forced expression of mammalian SBF1 in cultured cells resulted in competitive interference of endogenous SET domain interactions with dual-specificity phosphatases (such as myotubularin) leading to oncogenic transformation of NIH 3T3 cells and impaired differentiation of myoblasts (19). While the effect of SNR1E1 on the formation or function of the TAC1 complex is presently unknown, one possibility is that reduced interaction between SNR1E1 and the SET domain of TRX may lead to inappropriate TAC1 function.
Thus, it appears that SNR1/INI1 is necessary to target specific chromatin remodeling functions of the metazoan Brm complexes in vivo through interactions with cellular factors mediated through the repeat regions. Further examination of the response of cellular targets to disruptions of Brm complex activities by SNR1E1 will be important to link chromatin remodeling and specific developmental processes.
Acknowledgments
D.R.M. and C.B.Z. made equivalent contributions to the presented work.
We thank Kevin Fabrizio and Runjhun Nanchal for assistance with the genetic analyses; Robert Hanna (SUNY-ESF) for help with the electron microscopy studies; and Michael Goldberg, Byron Williams, and Bruce Edgar for advice and help with the mitotic analyses. We also thank Dan Garza for communicating unpublished observations on snr1 mRNA accumulation during late development, Jessica Treisman for anti-OSA antibodies, and Christian Muchardt for anti-BRM antibodies.
This work was partly supported by funds from the Ruth Meyer Research Scholar Program (Syracuse University), the March of Dimes (5-FY97-702), and the National Science Foundation (MCB-0221563) to A.K.D.
REFERENCES
- 1.Abrams, E., L. Neigeborn, and M. Carlson. 1986. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:3643-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H. T., R. Chinery, D. Y. Wu, S. J. Kussick, J. M. Payne, A. J. Fornace, and D. C. Tkachuk. 1999. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol. Cell. Biol. 19:7050-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bainbridge, S. P., and M. Bownes. 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66:57-80. [PubMed] [Google Scholar]
- 4.Bender, M., F. B. Imam, W. S. Talbot, B. Ganetzky, and D. S. Hogness. 1997. Drosophila ecdysone receptor mutations reveal functional differences among receptor isoforms. Cell 91:777-788. [DOI] [PubMed] [Google Scholar]
- 5.Biehs, B., M. A. Sturtevant, and E. Bier. 1998. Boundaries in the Drosophila wing imaginal disc organize vein-specific genetic programs. Development 125:4245-4257. [DOI] [PubMed] [Google Scholar]
- 6.Bier, E. 2000. Drawing lines in the Drosophila wing: initiation of wing vein development. Curr. Opin. Genet. Dev. 10:393-398. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142-145. [DOI] [PubMed] [Google Scholar]
- 8.Brizuela, B. J., L. Elfring, J. Ballard, J. W. Tamkun, and J. A. Kennison. 1994. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics 137:803-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brizuela, B. J., and J. A. Kennison. 1997. The Drosophila homeotic gene moira regulates expression of engrailed and HOM genes in imaginal tissues. Mech. Dev. 65:209-220. [DOI] [PubMed] [Google Scholar]
- 10.Brumby, A. M., C. B. Zraly, J. A. Horsfield, J. Secombe, R. Saint, A. K. Dingwall, and H. Richardson. 2002. Drosophila cyclin E interacts with components of the Brahma complex. EMBO J. 21:3377-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 12.Cao, Y., B. R. Cairns, R. D. Kornberg, and B. C. Laurent. 1997. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 17:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, S. W., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 14.Cimino, G., M. C. Rapanotti, T. Sprovieri, and L. Elia. 1998. ALL1 gene alterations in acute leukemia: biological and clinical aspects. Haematologica 83:350-357. [PubMed] [Google Scholar]
- 15.Collins, R. T., T. Furukawa, N. Tanese, and J. E. Treisman. 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18:7029-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, R. T., and J. E. Treisman. 2000. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 14:3140-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig, E., Z.-K. Zhang, K. P. Davies, and G. V. Kalpana. 2002. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: implications for tumorigenesis. EMBO J. 21:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby, M. A., C. Miller, T. Alon, K. L. Watson, C. P. Verrijzer, R. Goldman-Levi, and N. B. Zak. 1999. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui, X., I. De-Vivo, R. Slany, A. Miyamoto, R. Firestein, and M. L. Cleary. 1998. Association of SET domain and myotubularin-related proteins modulates growth control. Nat. Genet. 18:331-337. [DOI] [PubMed] [Google Scholar]
- 20.de Celis, J. F. 1997. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development 124:1007-1018. [DOI] [PubMed] [Google Scholar]
- 21.de Celis, J. F. 1998. Positioning and differentiation of veins in the Drosophila wing. Int. J. Dev. Biol. 42:335-343. [PubMed] [Google Scholar]
- 22.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 23.Dingwall, A. K., S. J. Beek, C. M. McCallum, J. W. Tamkun, G. V. Kalpana, S. P. Goff, and M. P. Scott. 1995. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elfring, L. K., C. Daniel, O. Papoulas, R. Deuring, M. Sarte, S. Moseley, S. J. Beek, W. R. Waldrip, G. Daubresse, A. DePace, J. A. Kennison, and J. W. Tamkun. 1998. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fletcher, J. C., and C. S. Thummel. 1995. The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development 121:1411-1421. [DOI] [PubMed] [Google Scholar]
- 26.Fristrom, D., and J. W. Fristrom. 1993. The metamorphic development of the adult epidermis, p. 843-897. In M. Bate and A. Martinez Arias (ed.), The development of Drosophila melanogaster, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Geng, F., Y. Cao, and B. C. Laurent. 2001. Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21:4311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gindhart, J., Jr., and T. C. Kaufman. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grand, F., S. Kulkarni, A. Chase, J. M. Goldman, M. Gordon, and N. C. Cross. 1999. Frequent deletion of hSNF5/INI1, a component of the SWI/SNF complex, in chronic myeloid leukemia. Cancer Res. 59:3870-3874. [PubMed] [Google Scholar]
- 30.Hartenstein, V., and J. W. Posakony. 1990. A dual function of the Notch gene in Drosophila sensillum development. Dev. Biol. 142:13-30. [DOI] [PubMed] [Google Scholar]
- 31.Kennison, J. A., and J. W. Tamkun. 1988. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 85:8136-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingston, R. E., C. A. Bunker, and A. N. Imbalzano. 1996. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 10:905-920. [DOI] [PubMed] [Google Scholar]
- 33.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolonin, M. G., and R. Finley, Jr. 1998. Targeting cyclin-dependent kinases in Drosophila with peptide aptamers. Proc. Natl. Acad. Sci. USA 95:14266-14271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 36.Landis, G., D. Bhole, L. Lu, and J. Tower. 2001. High-frequency generation of conditional mutations affecting Drosophila melanogaster development and life span. Genetics 158:1167-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 38.Laurent, B. C., and M. Carlson. 1992. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 6:1707-1715. [DOI] [PubMed] [Google Scholar]
- 39.Laurent, B. C., M. A. Treitel, and M. Carlson. 1991. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. USA 88:2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, D., H. Sohn, G. V. Kalpana, and J. Choe. 1999. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature 399:487-491. [DOI] [PubMed] [Google Scholar]
- 41.Lee, J. R., S. Urban, C. F. Garvey, and M. Freeman. 2001. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107:161-171. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, C. A., H. W. Jacobs, S. A. Datar, W. Du, B. A. Edgar, and C. F. Lehner. 2000. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19:4533-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morozov, A., E. Yung, and G. V. Kalpana. 1998. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc. Natl. Acad. Sci. USA 95:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muchardt, C., B. Bourachot, J. C. Reyes, and M. Yaniv. 1998. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J. 17:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller, C., and A. Leutz. 2001. Chromatin remodeling in development and differentiation. Curr. Opin. Genet. Dev. 11:167-174. [DOI] [PubMed] [Google Scholar]
- 46.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Östlund Farrants, A. K., P. Blomquist, H. Kwon, and Ö. Wrange. 1997. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol. 17:895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 49.Papoulas, O., G. Daubresse, J. A. Armstrong, J. Jin, M. P. Scott, and J. W. Tamkun. 2001. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl. Acad. Sci. USA 98:5728-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pazin, M. J., and J. T. Kadonaga. 1997. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88:737-740. [DOI] [PubMed] [Google Scholar]
- 51.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson, C. L., and I. Herskowitz. 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68:573-583. [DOI] [PubMed] [Google Scholar]
- 53.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 54.Petruk, S., Y. Sedkov, S. Smith, S. Tillib, V. Kraevski, T. Nakamura, E. Canaani, C. M. Croce, and A. Mazo. 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294:1331-1334. [DOI] [PubMed] [Google Scholar]
- 55.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 56.Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib, J. Blechman, T. Nakamura, C. M. Croce, A. Mazo, and E. Canaani. 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rozovskaia, T., S. Tillib, S. Smith, Y. Sedkov, O. Rozenblatt-Rosen, S. Petruk, T. Yano, T. Nakamura, L. Ben-Simchon, J. Gildea, C. M. Croce, A. Shearn, E. Canaani, and A. Mazo. 1999. Trithorax and ASH1 interact directly and associate with the Trithorax group-responsive bxd region of the Ultrabithorax promoter. Mol. Cell. Biol. 19:6441-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sevenet, N., A. Lellouch-Tubiana, D. Schofield, K. Hoang-Xuan, M. Gessler, D. Birnbaum, C. Jeanpierre, A. Jouvet, and O. Delattre. 1999. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum. Mol. Genet. 8:2359-2368. [DOI] [PubMed] [Google Scholar]
- 59.Sevenet, N., E. Sheridan, D. Amram, P. Schneider, R. Handgretinger, and O. Delattre. 1999. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am. J. Hum. Genet. 65:1342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shanahan, F., W. Seghezzi, D. Parry, D. Mahony, and E. Lees. 1999. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol. Cell. Biol. 19:1460-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staehling-Hampton, K., P. J. Ciampa, A. Brook, and N. Dyson. 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sturtevant, M. A., and E. Bier. 1995. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development 121:785-801. [DOI] [PubMed] [Google Scholar]
- 63.Sturtevant, M. A., M. Roark, and E. Bier. 1993. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 7:961-973. [DOI] [PubMed] [Google Scholar]
- 64.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takayama, M. A., T. Taira, K. Tamai, S. M. Iguchi-Ariga, and H. Ariga. 2000. ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells 5:481-490. [DOI] [PubMed] [Google Scholar]
- 66.Tamkun, J. W. 1995. The role of brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev. 5:473-477. [DOI] [PubMed] [Google Scholar]
- 67.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 68.Tripoulas, N. A., E. Hersperger, D. La Jeunesse, and A. Shearn. 1994. Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1). Genetics 137:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trouche, D., C. Le Chalony, C. Muchardt, M. Yaniv, and T. Kouzarides. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 94:11268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vazquez, M., L. Moore, and J. A. Kennison. 1999. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development 126:733-742. [DOI] [PubMed] [Google Scholar]
- 71.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 72.Vidal, M. 1997. The reverse two-hybrid system, p. 109-147. In P. L. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 73.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 74.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 76.Zraly, C. B., Y. Feng, and A. K. Dingwall. 2002. Genetic and molecular analysis of region 88E9;88F2 in Drosophila melanogaster, including the ear gene related to human factors involved in lineage-specific leukemias. Genetics 160:1051-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zraly, C. B., D. R. Marenda, R. Nanchal, G. Cavalli, C. Muchardt, and A. K. Dingwall. SNR1 is an essential subunit in a subset of Drosophila Brm complexes, targeting specific functions during development. Dev. Biol., in press. [DOI] [PubMed]