Abstract
NF-κB1 p105 functions both as a precursor of NF-κB1 p50 and as a cytoplasmic inhibitor of NF-κB. Following the stimulation of cells with tumor necrosis factor alpha (TNF-α), the IκB kinase (IKK) complex rapidly phosphorylates NF-κB1 p105 on serine 927 in the PEST region. This phosphorylation is essential for TNF-α to trigger p105 degradation, which releases the associated Rel/NF-κB subunits to translocate into the nucleus and regulate target gene transcription. Serine 927 resides in a conserved motif (Asp-Ser927-Gly-Val-Glu-Thr-Ser932) homologous to the IKK target sequence in IκBα. In this study, TNF-α-induced p105 proteolysis was revealed to additionally require the phosphorylation of serine 932. Experiments with IKK1−/− and IKK2−/− double knockout embryonic fibroblasts demonstrate that the IKK complex is essential for TNF-α to stimulate phosphorylation on p105 serines 927 and 932. Furthermore, purified IKK1 and IKK2 can each phosphorylate a glutathione S-transferase-p105758-967 fusion protein on both regulatory serines in vitro. IKK-mediated p105 phosphorylation generates a binding site for βTrCP, the receptor subunit of an SCF-type ubiquitin E3 ligase, and depletion of βTrCP by RNA interference blocks TNF-α-induced p105 ubiquitination and proteolysis. Phosphopeptide competition experiments indicate that βTrCP binds p105 more effectively when both serines 927 and 932 are phosphorylated. Interestingly, however, βTrCP affinity for the IKK-phosphorylated sequence on p105 is substantially lower than that on IκBα. Thus, it appears that reduced p105 recruitment of βTrCP and subsequent ubiquitination may contribute to delayed p105 proteolysis after TNF-α stimulation relative to that for IκBα.
NF-κB transcription factors play an important role in the regulation of genes involved in immune and inflammatory responses, apoptosis, and development (7, 11). Mammals express five NF-κB proteins that bind DNA as homo- and heterodimers: RelA, RelB, c-Rel, NF-κB1 p50, and NF-κB2 p52 (22). NF-κB1 and NF-κB2 are synthesized as large precursors of 105 kDa (p105) and 100 kDa (p100), respectively, that require proteolytic processing by the proteasome to produce their respective p50 and p52 NF-κB subunits (1, 22).
Mature NF-κB dimers are retained in the cytoplasm of unstimulated cells through their interaction with a family of inhibitory proteins, termed IκBs (7). A number of agonists activate NF-κB, including proinflammatory cytokines, lipopolysaccharide, double-stranded RNA, and the viral transactivator Tax. These stimuli induce IκB phosphorylation via the IκB kinase (IKK) complex, which contains two related kinases, IKK1 (IKKα) and IKK2 (IKKβ), and a structural subunit, NEMO (IKKγ). This triggers IκB ubiquitination and subsequent proteolysis by the proteasome (11). NF-κB dimers are thereby released to translocate into the nucleus, where their transcriptional activity is also regulated by phosphorylation (7).
The NF-κB1 precursor p105 functions as an IκB due to the presence of ankyrin repeats in its C terminus and retains associated p50, c-Rel, and RelA in the cytoplasm (16, 20). Two proteolytic pathways control cellular levels of p105: limited (processing to p50) and complete (degradation) (11). Following the stimulation of cells with proinflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) or interleukin-1 (IL-1), p105 is phosphorylated and then more rapidly proteolysed by the proteasome (14-16). This predominantly results in accelerated p105 degradation, rather than increased processing to p50, which is largely a constitutive process (9, 24). Freed Rel subunits translocate into the nucleus to modulate target gene transcription. Analysis of knockout mice which lack the C-terminal (IκB-like) half of p105 while still expressing p50 has revealed an essential role for p105 in the proper regulation of p50 homodimers, which is required for the correct control of inflammatory responses (10).
The C-terminal PEST domain of p105 contains a conserved motif (Asp-Ser927-Gly-Val-Glu-Thr-Ser932) which is homologous to the IKK target sequence in IκBα (Fig. 1A) (8, 21). Our laboratory has recently demonstrated that phosphorylation of serine 927 is essential for the signal-induced degradation of p105 triggered by either IKK2 overexpression or stimulation of cells with TNF-α (21). Mutation of serine 932 to alanine was also found to reduce the rate of proteolysis triggered by IKK2 overexpression. In the present study, the role of serine 932 in signal-induced degradation was investigated in more detail. These experiments reveal that the phosphorylation of both serine 927 and serine 932 by the IKK complex is essential for TNF-α to trigger p105 ubiquitination and proteolysis.
FIG. 1.
IKK target sequences, p105 point mutants, and synthetic phosphopeptides. (A) Schematic representation of mammalian NF-κB1 p105. The relative positions of the Rel homology domain (RHD), nuclear localizing signal (NLS), glycine-rich region (GRR), ankyrin repeats (vertical rectangles), death domain (DD), and PEST domain are shown. An alignment of the IKK target sequence in the p105 PEST region with those in IκBα, IκBβ, and IκBɛ is also shown (all human proteins). (B) Serine-to-alanine point mutants of HA-p105. WT, wild type. (C) Sequences of synthetic p105 and IκBα peptides. P, phosphorylated residue.
MATERIALS AND METHODS
cDNA constructs and antibodies.
cDNAs encoding hemagglutinin (HA) epitope-tagged p105 and serine-to-alanine point mutant HA-p105 derivatives were described in an earlier study (21). For transient transfection experiments, these were subcloned into the pcDNA3 expression vector (Invitrogen Corp.), whereas the pMX-1 expression vector (Ingenius) was used to generate stably transfected HeLa cell lines. Expression vectors encoding Flag-epitope-tagged IKK2 (FL-IKK2), βTrCP1 (FL-βTrCP1), and βTrCP2 (FL-βTrCP2) have been described previously (17, 23).
HA-p105 was immunoprecipitated using 12CA5 monoclonal antibody (MAb) and detected in Western blots with a commercial anti-HA MAb (3F10; Roche Molecular Biochemicals). Endogenous p105 was immunoprecipitated and detected in Western blots by using the anti-p105C antibody, which recognizes the C terminus of human p105 (21). The anti-phospho-S927-p105 antibody has been described in previous work (21). To generate the anti-phospho-S932-p105 antibody, a peptide was synthesized corresponding to residues 922 through 935 of human p105 in which serine 932 was phosphorylated. After high-pressure liquid chromatography purification, this phosphopeptide was coupled to keyhole limpet hemocyanin (Pierce) and injected into rabbits. For use in Western blots, immunoglobulin G was isolated from 4 ml of antiserum by using protein G-Sepharose (Amersham Bioscience) and then affinity purified on a column of Affi-Gel 10 (Bio-Rad) coupled to 10 mg of phosphopeptide after first passing through an Affi-Gel 10 column coupled to 10 mg of unphosphorylated peptide from residues 922 to 935. Antibodies recognizing IκBα (sc-371; Santa Cruz), βTrCP1 and βTrCP2 (sc-8863; Santa Cruz), and the Flag epitope (M2 MAb; Sigma) were purchased from the indicated suppliers. Anti-glutathione S-transferase (GST) MAb was kindly provided by Steve Dillworth (Hammersmith Hospital, London, United Kingdom).
Cell culture and stable transfections.
HeLa cells (Ohio subline from the European Collection of Cell Cultures) and mouse embryonic fibroblasts (EFs) were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Corp.) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (50 U/ml). For NIH 3T3 fibroblasts, 10% newborn calf serum was substituted for FCS. IKK1 and IKK2 double knockout (IKK1/2−/−) EFs were generously provided by Mark Schmitt and Inder Verma (The Salk Institute, San Diego, Calif.) (13).
The generation of stably transfected HeLa cells was carried out as described previously (21). Expression of HA-p105 was assessed by Western blotting.
Protein analyses.
To analyze HA-p105 proteolysis in stably transfected HeLa clones, cells were plated at 6 × 105 per well of a six-well plate (Nunc). After 18 h in culture, cells were washed with phosphate-buffered saline and cultured in methionine- and cysteine-free minimal essential Eagle's medium (Sigma) for 90 min. Cells were pulse-labeled with 2.65 MBq of [35S]methionine-[35S]cysteine (Pro-Mix; Amersham Bioscience) for 45 min and chased for the indicated times in complete medium (DMEM plus 2% FCS) alone (control) or complete medium supplemented with TNF-α (20 ng/ml; Amersham Bioscience). Cells were lysed in buffer A (1% NP-40, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 2 mM Na4P2O7, plus a cocktail of protease inhibitors [Roche]) supplemented with 0.1% sodium dodecyl sulfate (SDS) and 0.5% deoxycholate (RIPA buffer), and HA-p105 was immunoprecipitated as described previously (3). Labeled bands were quantified by laser densitometry (calibrated imaging densitometer; Bio-Rad). Pulse-chase experiments were carried out at least three times with similar results.
For ubiquitination experiments, HeLa cells were seeded at 2 × 106 cells per 90-mm-diameter dish and left in culture until confluent (1 to 2 days). Cells were then treated with 50 μM N-acetyl-l-leucyl-l-leucyl-l-norleucinal (LLnL) proteasome inhibitor (Sigma) for 1 h and incubated for a further 15 min in the presence of TFN-α or control medium. Cells were extracted in 1.2 ml of buffer A per dish. After centrifugation, HA-p105 was immunoprecipitated from lysates of four dishes, resolved on a SDS-6% polyacrylamide gel electrophoresis (PAGE) gel, and Western blotted.
Analysis of p105 phosphorylation.
To analyze p105 phosphorylation on serines 927 and 932 in NIH 3T3 cells, HA-p105 was transiently expressed alone or together with FL-IKK2 by using Lipofectamine (Invitrogen Corp.). Cells were treated with 20 μM MG132 proteasome inhibitor (Biomol Research Labs) for the last 4 h of a 48-h culture and then lysed in buffer A. Phosphorylation was determined by immunoblotting of anti-HA immunoprecipitates with the indicated phosphopeptide-specific antibodies. Analysis of the phosphorylation of serines 927 and 932 on endogenous p105 in HeLa cells was carried out as described previously (21).
In vitro kinase assays.
His6-IKK1 and His6-IKK2 were expressed in Sf9 insect cells by baculovirus infection and purified to near homogeneity as described previously (21). One hundred nanograms of recombinant IKK protein was incubated at room temperature in 50 μl of kinase buffer (25 mM Tris [pH 7.5], 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM sodium vanadate, 10 mM MgCl2) plus 20 μM ATP and 5 μg of purified GST-p105758-967, GST-p105758-967(S927A), or GST-p105758-967(S932A) protein. The reaction was stopped by the addition of 50 μl of 2× Laemmli sample buffer (12), and proteins were Western blotted with anti-phospho-Ser927 or anti-phospho-Ser932 antibody. Equal protein loading was confirmed by reprobing blots with an anti-GST MAb.
For in vitro phosphorylation experiments with endogenous IKK complex, control or TNF-α-stimulated HeLa cells (5 × 106 per 90-mm dish) were lysed in buffer A and immunoprecipitated by using anti-NEMO antibody (sc-8330; Santa Cruz). Immunoprecipitates were washed four times in buffer A and twice in kinase buffer. Kinase reactions were carried out as described above.
RNA interference.
Depletion of endogenous βTrCP expression was achieved by RNA interference (6). Small interfering RNAs (siRNAs) for human βTrCP1 and βTrCP2 were synthesized by Xeragon Inc. The sequences of βTrCP siRNAs were GUGGAAUUUGUGGAACAUCTT (sense) and GAUGUUCCACAAAUUCCACTT (antisense). At 12 to 16 h prior to transfection, 105 HeLa cells were seeded in six-well plates (Costar) in DMEM supplemented with 10% FCS. Cells were transfected with βTrCP siRNAs (0.4 nmol per well) by using Oligofectamine (Invitrogen Corp.) according to the manufacturer's instructions. After 72-h culture, cells were stimulated with TNF-α and then lysed in buffer A. Experiments with other synthetic siRNAs demonstrated that the effects of βTrCP siRNA treatment were specific by using IκBα degradation as a readout (data not shown).
Endogenous βTrCP1 and βTrCP2 were not detected on the Western blot of total cell lysates. Therefore, to determine the effectiveness of siRNAs in decreasing βTrCP1 and βTrCP2 levels, siRNAs or control transfected cells were lysed in buffer A (1 ml) and centrifuged lysates were incubated overnight with phospho-S32/S36 ΙκBα peptide (Fig. 1C) coupled to Affi-Gel 10 beads (Bio-Rad). Beads were then washed four times in buffer A and isolated endogenous βTrCP1 and βTrCP2 quantified by Western blotting. The same methodology was used to determine whether endogenous βTrCP1 and βTrCP2 could directly bind to p105-derived phosphopeptides (Fig. 1C).
Peptides.
Synthetic p105 peptides (doubly, singly, and non-phosphorylated) were purchased from the University of Bristol Biochemistry Department, and IκBα peptides were purchased from SynPep (Dublin, Calif.). All peptides were purified by high-pressure liquid chromatography (>90%) and verified by mass spectrometry. The sequences of the peptides used are shown in Fig. 1C.
βTrCP binding assays.
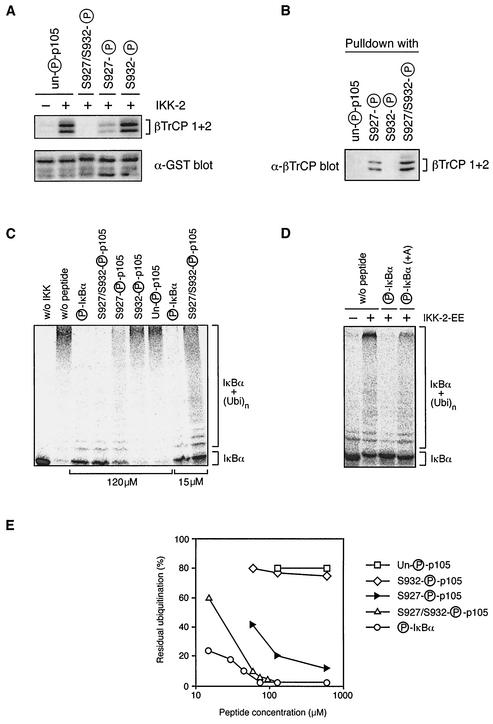
FL-βTrCP1 and FL-βTrCP2 were synthesized together from their respective expression vectors and labeled with [35S]methionine (Amersham Bioscience) by cell-free translation (25-μl reaction volume; Promega TNT-coupled rabbit reticulocyte system). GST-p105758-967 fusion protein was phosphorylated with His6-IKK2 as described above, bound to glutathione-Sepharose beads (10 μl; Amersham Bioscience), and washed three times in buffer A. Translated proteins, diluted to 200 μl in buffer A, were then incubated with the phospho-GST-p105758-967 beads overnight at 4°C with the indicated phosphopeptides or with no addition. Beads were washed a further four times in buffer A, and 35S-labeled protein was visualized by autoradiography after SDS-8% PAGE.
To determine the association of βTrCP1 and βTrCP2 with phospho-p105 in vivo, HA-p105 HeLa cells were transiently transfected with vectors encoding FL-βTrCP1 and FL-βTrCP2 (0.5 μg each) by using Lipofectamine (Invitrogen Corp.). After 48-h culture, cells were stimulated with TNF-α for 15 min or left unstimulated. HA-p105 was then isolated from cell lysates by immunoprecipitation with anti-HA MAb, and Western blots were probed with anti-FL MAb to detect HA-p105-associated FL-βTrCP1 or FL-βTrCP2.
In vitro IκBα ubiquitination.
[35S]Methionine-labeled IκBα was prepared as described previously (26) and then phosphorylated by recombinant, constitutively active IKK2 (IKK2-EE) (17). Immunopurified SCFβTrCP complex was preincubated with 5 to 650 μM concentrations of phosphorylated or unphosphorylated p105 or IκBα peptide (Fig. 1C) for 15 min and then mixed with phospho-IκBα, E1, and E2. Ubiquitination was performed as described previously (26), and ubiquitinated products were analyzed by SDS-PAGE (8% acrylamide) and subsequent autoradiography.
RESULTS
Serine 932 is essential for TNF-α-induced p105 degradation.
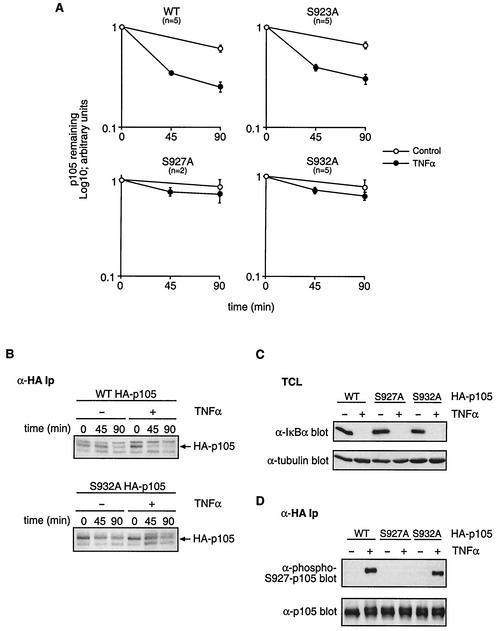
The Ley laboratory has previously demonstrated that a serine 932-to-alanine mutant of HA-p105 displays delayed proteolysis compared to wild-type HA-p105 when coexpressed with FL-IKK2 in NIH 3T3 cells. This suggested that the residue might play a regulatory role in signal-induced p105 proteolysis (21). Since the IKK complex is essential for cytokine-induced p105 degradation (21), it was important to determine whether serine 932 is important for p105 proteolysis triggered by cytokine stimulation of the endogenous IKK complex. To investigate this, HeLa cells were transfected with plasmids encoding HA-p105 or HA-p105(S932A) and stable clones were isolated after G418 selection (Fig. 1B). A pulse-chase metabolic labeling experiment was carried out with a representative clone that expressed levels of HA-p105(S932A) similar to that of a control clone expressing HA-p105. Stimulation with TNF-α increased the rate of turnover of HA-p105 as expected (Fig. 2A and B). In contrast, TNF-α stimulation consistently had no detectable effect on the rate of proteolysis of HA-p105(S932A). Analysis of the 90-min point confirmed that there was no significant difference in the amount of HA-p105(S932A) remaining ± TNF-α (P = 0.23; n = 5). However, the mobility of the HA-p105(S932A) band was retarded and became less distinct after TNF-α stimulation, probably due to phosphorylation on amino acids other than serine 932 (Fig. 2B). As reported previously (21), TNF-α stimulation did not increase the proteolysis of HA-p105(S927A) (Fig. 2A). Control experiments demonstrated that IκBα was rapidly and completely degraded following TNF-α stimulation in the HA-p105(S932A) clone similar to that in the wild-type HA-p105 clone, confirming that the TNF-α signaling pathway controlling the IKK complex was intact in the former cell line (Fig. 2C). Thus, serine 932 is essential for TNF-α to efficiently induce the proteolysis of HA-p105.
FIG. 2.
Serine 932 is essential for TNF-α to trigger proteolysis of p105. (A) Clones of HeLa cells stably transfected with HA-p105, HA-p105(S923A), HA-p105(S927A), or HA-p105(S932A) were metabolically pulse-labeled with [35S]methionine-[35S]cysteine (45 min) and then chased for the indicated times in complete medium (control) or complete medium supplemented with TNF-α. Anti-HA immunoprecipitates were resolved by SDS-7.5% PAGE and revealed by fluorography. Amounts of immunoprecipitated wild-type (WT) and point-mutated HA-p105 were quantified by laser densitometry (mean ± standard error of the mean; n = 5). (B) HeLa cells stably transfected with either HA-p105 or HA-p105(S932A) were analyzed by pulse-chase metabolic labeling as described for panel A. Anti-HA immunoprecipitates (Ip) were resolved by SDS-7.5% PAGE and revealed by fluorography. The position of HA-p105 is indicated with an arrow. (C) Cell lysates from the indicated HeLa clones with or without TNF-α stimulation (15 min) were Western blotted for endogenous IκBα. (D) Stably transfected HeLa cells were preincubated for 1 h with LLnL proteasome inhibitor and then stimulated for 15 min with TNF-α or control medium. Immunoprecipitates of wild-type and point-mutated HA-p105 were then sequentially Western blotted with the indicated antibodies.
It was possible that the inhibitory effect of the S932A mutation on TNF-α-induced proteolysis of HA-p105 was due to the indirect effects on the phosphorylation of serine 927, which has previously been shown to be essential for the signal-induced degradation of p105 (8, 21). To investigate this possibility, HA-p105 was immunoprecipitated from the HA-p105(S932A) clone with or without TNF-α stimulation and Western blotted with anti-phospho-S927-p105 antibody. TNF-α-induced serine 927 phosphorylation of HA-p105(S932A) was similar to that of wild-type HA-p105 (Fig. 2D). Thus, the S932A mutation did not mediate its inhibitory effects on HA-p105 proteolysis by blocking the phosphorylation of serine 927.
A recent study demonstrated that recombinant IKK2 phosphorylated a p105917-968 fusion protein on a residue equivalent to serine 923 in the full-length p105 protein (8). Furthermore, proteolysis of a serine 923-to-alanine point mutant of p105 was reduced when coexpressed in 293 cells with IKK2 and βTrCP compared to the wild-type protein. In contrast, previous experiments from this laboratory did not suggest an important role for serine 923 in degradation of HA-p105 triggered by FL-IKK2 overexpression in NIH 3T3 fibroblasts (21). To investigate whether serine 923 is required for the cytokine-induced degradation of p105, stable HeLa clones expressing HA-p105(S923A) were generated. Stimulation of a representative clone with TNF-α increased the rate of HA-p105(S923A) proteolysis to a degree similar to that for wild-type HA-p105 (Fig. 2A). Analysis of the 90-min-chase time point confirmed that there was no significant difference in the amount of HA-p105(S923A) remaining (65%) compared to that of wild-type HA-p105 (60%) in TNF-α-stimulated cells (P = 0.372; n = 5). Thus, consistent with previous data (21), serine 923 is not required for stimulus-induced p105 degradation.
In conclusion, these data indicate an essential role for both serines 927 and 932 in the proteolysis of p105 triggered by TNF-α stimulation, whereas serine 923 is not required for TNF-α-induced p105 proteolysis.
TNF-α induces the rapid phosphorylation of p105 on serine 932.
The previous experiments indicate an important role for serine 932 in p105 proteolysis triggered by stimulation with TNF-α. However, it was important to obtain direct evidence that serine 932 was actually phosphorylated in vivo to rule out the inhibitory effects on p105 proteolysis resulting from putative structural alterations caused by the S932A mutation. To investigate this, rabbits were immunized with a synthetic peptide corresponding to residues 922 to 935 of human p105 in which serine 932 was phosphorylated and specific antibody was purified on a phosphopeptide affinity column.
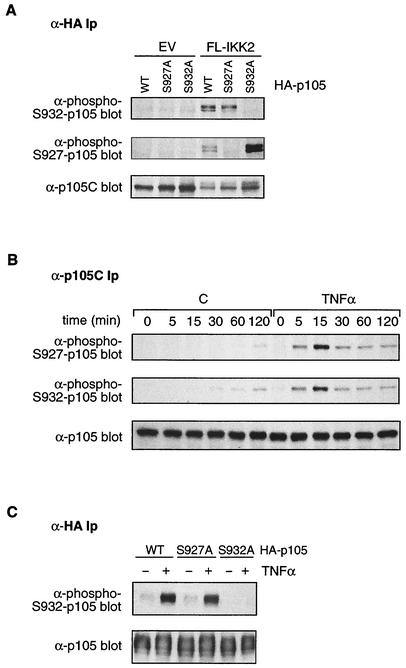
To initially investigate the possibility of the signal-induced phosphorylation of p105 on serine 932, NIH 3T3 cells were transfected with plasmids encoding wild-type HA-p105 or HA-p105(S932A) plus FL-IKK2 or control empty vector (EV). Cells were incubated with MG132 inhibitor for the last 4 h of a 48-h culture to block proteasome-mediated proteolysis. HA-p105 immunoprecipitated from cells coexpressing FL-IKK2 was clearly recognized by the anti-phospho-S932-p105 antibody but not from cells cotransfected with EV (Fig. 3A). However, no signal was obtained in cells cotransfected with HA-p105(S932A) and FL-IKK2, although HA-p105(S932A) protein was present in immunoprecipitates and phosphorylated on serine 927. In addition, incubation of blots with anti-phospho-S932-p105 antibody plus S932 phosphopeptide reduced the phospho-HA-p105 signal in FL-IKK2-cotransfected cells to background whereas the addition of unphosphorylated peptide had no effect (data not shown). Together, these data confirm the specificity of the anti-phospho-S932-p105 antibody and demonstrate that overexpression of FL-IKK2 induces the phosphorylation of HA-p105 on serine 932. Furthermore, these data indicate that the reduced proteolysis of HA-p105(S932A) in response to FL-IKK2 coexpression was not the consequence of decreased serine 927 phosphorylation (21). Conversely, the reduced IKK2-triggered proteolysis of HA-p105(S927A) did not result from the blockade of serine 932 phosphorylation (Fig. 3A).
FIG. 3.
TNF-α induces rapid phosphorylation of p105 on serine 932. (A) NIH 3T3 cells were transiently cotransfected with expression vectors encoding wild-type (WT) HA-p105 or the indicated mutants and IKK2 or EV and treated with MG132 proteasome inhibitor for the last 4 h of a 48-h culture. HA-p105 was immunoprecipitated, and isolated protein was sequentially Western blotted with anti-phospho-Ser932 (upper gel), anti-phospho-Ser927 (middle gel), and anti-p105C (lower gel) antibodies. Ip, immunoprecipitates. (B) HeLa cells were preincubated for 30 min with MG132 and then stimulated for the indicated times with TNF-α or control medium (C). Immunoprecipitates of endogenous p105 were then Western blotted sequentially with the indicated antibodies. (C) HeLa cells stably transfected with the indicated HA-p105 constructs were preincubated for 1 h with LLnL inhibitor and then stimulated for 15 min with TNF-α or control medium. Anti-HA immunoprecipitates were then sequentially Western blotted with the indicated antibodies.
To investigate whether proinflammatory cytokines induce the phosphorylation of serine 932 of p105, HeLa cells were preincubated with MG132 inhibitor for 30 min and then stimulated for the indicated times with TNF-α. Endogenous p105 was isolated from cell lysates by immunoprecipitation and then immunoblotted with anti-phospho-S932-p105 antibody. TNF-α stimulation induced rapid serine 932 phosphorylation that reached a maximum at 15 min and then declined (Fig. 3B, middle panel). The kinetics of p105 serine 927 phosphorylation after TNF-α stimulation were very similar (Fig. 3B, upper panel). Low levels of p105 phosphorylated on both serines 927 and 932 also accumulated in control unstimulated cells by 1 to 2 h, which were presumably due to basal IKK activity.
It was important to determine whether the inhibitory effect of the S927A mutation on the TNF-α-induced proteolysis of HA-p105 (21) was due to the indirect inhibition of serine 932 phosphorylation. To investigate this, HA-p105 was immunoprecipitated from the HeLa clone expressing HA-p105(S927A) and cultured for 1 h with LLnL proteasome inhibitor and for a further 15 min in the presence or absence of TNF-α. Western blotting with the anti-phospho-S932-p105 antibody indicated that HA-p105(S927A) was phosphorylated on serine 932 to a similar degree as wild-type HA-p105 phosphorylation after TNF-α stimulation (Fig. 3C). Thus, the S927A mutation does not inhibit the TNF-α induction of HA-p105 proteolysis by blocking serine 932 phosphorylation.
The data in this section confirm that serine 932 of p105 is phosphorylated under stimulatory conditions that promote p105 proteolysis and indicate that the inhibitory effect of the S932A mutation on TNF-α-induced p105 proteolysis is due to the removal of a regulatory phosphorylation site.
Serine 932 of p105 is phosphorylated by the IKK complex.
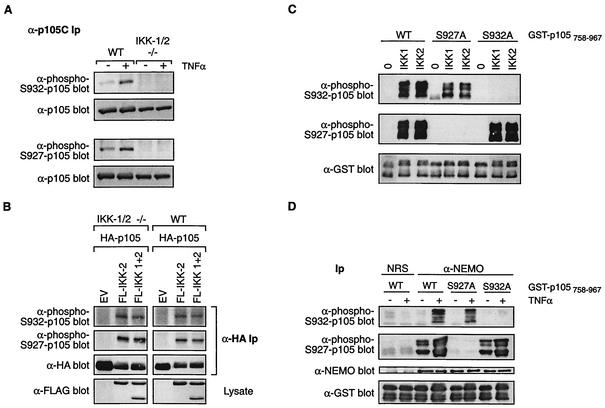
Since the IKK complex is essential for signal-induced p105 proteolysis, it was important to determine whether cytokine-induced phosphorylation of p105 on serine 932 required the IKK complex. To investigate this, wild-type mouse EFs and IKK1/2 double knockout EFs (12), which lack a functional IKK complex, were pretreated for 30 min with MG132 inhibitor and then stimulated with TNF-α for 15 min. Western blotting of immunoprecipitated endogenous p105 indicated that TNF-α induced serine 932 phosphorylation in wild-type EF cells but not in the IKK1/2-/- cells (Fig. 4A). TNF-α also failed to induce serine 927 phosphorylation in the IKK1/2-/- EF cells, whereas serine 927 phosphorylation was induced in wild-type cells. Similar experiments with IKK1 or IKK2 single-knockout EFs demonstrated that IKK1 and IKK2 are redundant for the TNF-α induction of p105 phosphorylation on serines 927 and 932 (data not shown), consistent with the ability of TNF-α to induce p105 proteolysis in such cells (21). However, TNF-α-induced phosphorylation of these residues was consistently detected to be higher in wild-type cells than in the single knockouts, suggesting that IKK complexes containing both IKK1 and IKK2 are required for optimal p105 phosphorylation. TNF-α induction of p38 phosphorylation in the IKK1/2-/- EFs was similar to that in the wild type, confirming that the TNF-α signaling was intact (data not shown). Furthermore, transient overexpression of FL-IKK2 or FL-IKK1 and FL-IKK2 in the IKK1/2-/- EFs strongly induced phosphorylation of p105 on both serines 927 and 932 (Fig. 4B), supporting the hypothesis that the defect in TNF-α-induced p105 phosphorylation in these cells is indeed due to the lack of a functional IKK complex.
FIG. 4.
The IKK complex directly phosphorylates serine 932 of p105. (A) Wild-type (WT) and IKK1/2-/- (−/−) EFs were pretreated with MG132 for 30 min and then incubated for a further 30 min with TNF-α or control medium. p105 was then immunoprecipitated from cell lysates and Western blotted with the indicated antibodies. Ip, immunoprecipitates. (B) IKK1/2−/− or wild-type EFs were transiently cotransfected with the indicated expression vectors. Cells were treated with MG132 for the last 4 h of a 48-h culture. HA-p105 was isolated from cell lysates by immunoprecipitation and immunoblotted with the antibodies shown. (C) In vitro kinase assays were carried out with baculovirus-produced purified His6-IKK1 or His6-IKK2 (100 ng), using as substrates 5 μg of wild-type or point-mutated GST-p105758-967 protein, as indicated. Phosphorylation was assessed by sequentially immunoblotting with anti-phospho-Ser932 (upper gel) or anti-phospho-Ser927 (middle gel) antibodies. Equal loading of fusion proteins was confirmed by reprobing blots with anti-GST MAb (lower gel). (D) HeLa cells were stimulated for 15 min with TNF-α or left unstimulated. The endogenous IKK complex was isolated from cell lysates by immunoprecipitation with an anti-NEMO antibody. Control immunoprecipitations were carried out with nonimmune rabbit serum (NRS). Phosphorylation was assessed as described for panel C. Blots were probed with anti-NEMO antibody to confirm that equal amounts of IKK complex were immunoprecipitated.
These data raise the possibility that the IKK complex might directly phosphorylate p105 serine 932. To investigate this, GST fusion proteins were purified from Escherichia coli encoding the C terminus of p105 (residues 758 to 967; GST-p105758-967) and corresponding mutants in which the residues equivalent to p105 serine 927 or 932 were mutated to alanine [GST-p105758-967(S927A) and GST-p105758-967(S932A), respectively]. These fusion proteins were phosphorylated in vitro with baculovirus-expressed His6-IKK1 or His6-IKK2. Western blotting with anti-phospho-S932-p105 antibody confirmed that both kinases phosphorylated GST-p105758-967 and GST-p105758-967(S927A) on serine 932, whereas no signal was detected with GST-p105758-967(S932A), as expected (Fig. 4C). Similarly, both His6-IKK1 and His6-IKK2 also phosphorylated serine 927 of the wild type and the S932A mutants of GST-p105758-967 but not of the S927A mutant, as reported previously (21). Endogenous IKK complex, purified by immunoprecipitation from TNF-α-stimulated HeLa cells, also phosphorylated both serine 927 and serine 932 of p105 (Fig. 4D). Thus, p105 serine 932 is a direct target of the IKK complex in vitro. The combined biochemical and genetic data in this section therefore demonstrate that TNF-α induces the phosphorylation of both serine 927 and serine 932 of p105 via the IKK complex.
Signal-induced phosphorylation of serine 927 and serine 932 regulates p105 ubiquitination.
IKK2-induced phosphorylation of p105 triggers its ubiquitination, targeting it for proteolysis by the proteasome (8, 19). The role of the phosphorylation of serines 927 and 932 on TNF-α-induced p105 ubiquitination was therefore investigated. HeLa cells stably transfected with HA-p105, HA-p105(S927A), or HA-p105(S932A) were pretreated with the LLnL proteasome inhibitor for 1 h. The cells were then incubated for a further 15 min with TNF-α or left unstimulated. HA-p105, immunoprecipitated from cell lysates with anti-HA MAb, was Western blotted with anti-p105C antibody. TNF-α stimulation promoted the appearance of a high-molecular-weight ladder of more slowly migrating forms of HA-p105, characteristic of ubiquitinated p105 (8, 19), which were not visible if LLnL inhibitor was omitted (Fig. 5A). Direct immunoblotting of p105 immunoprecipitates with antiubiquitin antibody confirmed that TNF-α-induced laddering was due to HA-p105 polyubiquitination (data not shown). However, TNF-α stimulation failed to induce high-molecular-weight forms of either HA-p105(S927A) or HA-p105(S932A) (Fig. 5B). These data indicate that both the S927A and S932A mutations block the TNF-α-induced ubiquitination of p105, suggesting that the phosphorylation of both serines 927 and 932 is required for TNF-α to trigger p105 ubiquitination. This is consistent with the observation that serines 927 and 932 are both essential for TNF-α-induced p105 proteolysis (Fig. 2A).
FIG. 5.
Serines 927 and 932 are required for TNF-α to trigger p105 ubiquitination. (A) HeLa cells stably transfected with HA-p105 were pretreated for 1 h with LLnL proteasome inhibitor or vehicle control (dimethyl sulfoxide [DMSO]). Cells were then stimulated for 15 min with TNF-α or control medium. HA-p105 was isolated from cell lysates by immunoprecipitation, resolved by SDS-6% PAGE, and then Western blotted with anti-p105C antibody. Ip, immunoprecipitates; WT, wild type. (B) The indicated HeLa clones were pretreated with LLnL inhibitor and stimulated with TNF-α as described for that in panel A. Anti-HA immunoprecipitates were Western blotted with anti-p105C antibody.
βTrCP is essential for the TNF-α induction of p105 and IκBα proteolysis.
Biochemical experiments have demonstrated that the phosphorylation of p105 by IKK creates a binding site for βTrCP1 and/or βTrCP2, F-box WD40 repeat proteins that are the receptor subunits of an SCF-type E3 ubiquitin ligase (19). In vitro experiments indicate that this association promotes the polyubiquitination of p105 and subsequent proteolysis by the proteasome (8, 19). Consistent with this model, the transient expression of an F-box deletion mutant of βTrCP1, which can bind to IKK-phosphorylated p105 but cannot assemble with other SCF subunits, blocks p105 proteolysis induced by IKK2 overexpression (19).
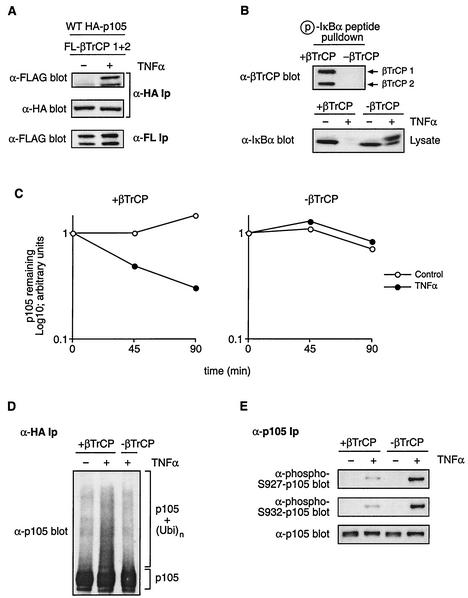
To determine whether TNF-α stimulation also induces binding of βTrCP (used as a collective term referring to both βTrCP1 and βTrCP2) to p105, HeLa cells stably expressing HA-p105 were transiently transfected with expression vectors encoding βTrCP1 and βTrCP2. HA-p105 was isolated by immunoprecipitation from lysates of cells stimulated for 15 min with TNF-α or left unstimulated. TNF-α stimulation induced a clear association between HA-p105 and both βTrCP1 and βTrCP2 (Fig. 6A).
FIG. 6.
βTrCP is required for TNF-α-induced p105 ubiquitination and proteolysis. (A) HeLa cells stably transfected with HA-p105 were transiently transfected with vectors encoding FL-βTrCP1 and FL-βTrCP2. After 48-h culture, cells were stimulated for 15 min with TNF-α or left unstimulated. Anti-HA immunoprecipitates (Ip) were Western blotted with the indicated antibodies. FL-βTrCP expression in cell lysates was confirmed by anti-FL MAb immunoprecipitation and Western blotting. WT, wild type. (B) HeLa cells were pretreated with βTrCP siRNAs complementary to both βTrCP1 and βTrCP2 (−βTrCP) or with control buffer (+βTrCP). After 72 h, cells were stimulated with TNF-α (15 min) or left unstimulated and then lysed. In the upper gel, βTrCP expression of unstimulated cells was determined by Western blotting of phospho-IκBα peptide pulldowns. In the lower gel, cell lysates were Western blotted for IκBα. (C) HeLa cells stably expressing HA-p105 were pretreated as described for panel B. Turnover of HA-p105 in TNF-α-stimulated and control unstimulated cells was determined by pulse-chase metabolic labeling as done for that shown in Fig. 2. Similar results were obtained on two other occasions. (D) βTrCP expression in HA-p105 HeLa cells was suppressed by siRNA pretreatment as described for panel B. HA-p105 ubiquitination induced by 15-min TNF-α stimulation was determined as described in the legend to Fig. 5. (E) βTrCP expression was decreased by siRNA treatment as described for panel B. Phosphorylation of endogenous p105 after TNF-α treatment (15 min) was determined by immunoprecipitation and Western blotting with the indicated antibodies.
Functional experiments on the role of βTrCP on p105 ubiquitination have relied on purified components for in vitro experiments or protein overexpression in cells (8, 19). It was therefore important to confirm genetically that βTrCP was indeed required for TNF-α-induced p105 proteolysis. siRNA-mediated gene suppression (6), based on a 21-bp double-stranded RNA complementary to both βTrCP1 and βTrCP2, was used to codeplete βTrCP1 and βTrCP2 from HeLa cells stably transfected with HA-p105. Western blotting confirmed that siRNA depleted βTrCP expression to undetectable levels (Fig. 6B, upper panel). Pulse-chase metabolic labeling revealed that βTrCP depletion blocked TNF-α-induced HA-p105 proteolysis (Fig. 6C). TNF-α-induced p105 ubiquitination, as revealed by the presence of high-molecular-weight p105 laddering, was also substantially reduced by βTrCP depletion (Fig. 6D). In addition, TNF-α-induced IκBα proteolysis was blocked in HeLa cells depleted of βTrCP (Fig. 6B, lower panel) whereas TNF-α-induced p38 phosphorylation was unaffected (data not shown). Thus, βTrCP is required for TNF-α to promote the ubiquitination and proteolysis of p105, supporting earlier models for βTrCP in regulating p105 function (8, 19). Furthermore, these experiments confirm the essential role of βTrCP in TNF-α-induced IκBα degradation (4).
The effect of βTrCP depletion on TNF-α-induced p105 phosphorylation was also investigated. TNF-α-induced phosphorylation of p105 on serines 927 and 932 was substantially increased in βTrCP-depleted cells compared to that in the control (Fig. 6E). This suggests that p105 phosphorylated on serines 927 and 932 is specifically targeted for ubiquitination by βTrCP, consistent with the essential role of these residues in TNF-α-induced p105 ubiquitination and proteolysis (Fig. 2A and 5B).
Efficient binding of βTrCP to p105 requires phosphorylation of both serines 927 and 932.
The siRNA experiments raise the possibility that the recruitment of βTrCP to p105 requires phosphorylation on both serines 927 and 932. A competition assay using synthetic phosphopeptides (Fig. 1C) corresponding to residues 922 to 935 of p105 was used to directly determine the role of p105 serine 927 and 932 phosphorylation in βTrCP binding.
For this, GST-p105758-967 fusion protein, phosphorylated in vitro by recombinant His6-IKK2 or left untreated, was coupled to glutathione-Sepharose beads and used as an affinity matrix to isolate [35S]methionine-labeled βTrCP1 and βTrCP2 which had been translated in vitro. As expected, the amount of βTrCP isolated by GST-p105758-967 was significantly increased following His6-IKK2 phosphorylation compared with that for the control unphosphorylated protein (Fig. 7A). The addition of phosphoserine 932 p105922-935 peptide had no effect on βTrCP binding compared to no peptide (data not shown) or to the addition of unphosphorylated p105922-935 peptide. Some inhibition of binding was detected upon addition of p105922-935 peptide phosphorylated on serine 927. However, addition of p105922-935 peptide phosphorylated on both serines 927 and 932 completely blocked binding to βTrCP.
FIG. 7.
Efficient binding of βTrCP to p105 requires phosphorylation of p105 serines 927 and 932. (A) GST-p105758-967 fusion protein, phosphorylated by His6-IKK2 or left unphosphorylated, was coupled to glutathione-Sepharose beads and used to affinity purify βTrCP translated and labeled with [35S]methionine in vitro. The indicated peptides were added to a final concentration of 100 μg/ml during the pulldown. Isolated βTrCP was detected by autoradiography of SDS-8% PAGE gels. (B) The indicated peptides were coupled to Affi-Gel 10 beads and used as affinity matrices to isolate from HeLa cell lysates endogenous βTrCP, which was detected by Western blotting. (C and D) IκBα, translated and labeled in vitro with [35S]methionine, was first phosphorylated with recombinant IKK2-EE. Labeled protein was then ubiquitinated in vitro (26) in the presence of the indicated peptides or with no added peptide (w/o peptide). Ubiquitination of unphosphorylated IκBα (w/o IKK) is shown as a negative control. IκBα ubiquitination was revealed by autoradiography of SDS-8% PAGE gels. (E) The extent of IκBα ubiquitination in vitro (carried out as described for panel C) was quantified on a phosphorimager over a range of competing peptide concentrations.
These data suggest that p105 requires phosphorylation on both serines 927 and 932 to most efficiently bind βTrCP but that weak binding can be achieved when only serine 927 is phosphorylated. To investigate this possibility directly, each of the phosphopeptides was individually coupled to Affi-Gel 10 beads and then used as affinity matrices to purify endogenous βTrCP from HeLa cell lysates. Western blotting revealed no βTrCP binding with unphosphorylated p105922-935 peptide or phosphoserine 932 p105922-935 peptide (Fig. 7B), consistent with the peptide competition experiment (Fig. 7A). However, βTrCP binding was detected to p105922-935 peptide phosphorylated on only serine 927, but to a lesser extent than that to the doubly phosphorylated p105922-935 peptide (Fig. 7B). These data confirm that βTrCP binds more effectively to p105 phosphorylated on both serines 927 and 932.
βTrCP has a higher affinity for phospho-IκBα than phospho-p105.
Previous experiments have indicated that p105 and IκBα are phosphorylated with very similar kinetics by IKK after TNF-α stimulation of HeLa cells (21). However, TNF-α stimulation only results in the slow and partial degradation of p105, in contrast to IκBα, whose degradation is very rapid and complete (Fig. 2C) (21). A possible explanation for this difference is that the two proteins recruit SCFβTrCP with different efficiencies after IKK-mediated phosphorylation and, as a result, are ubiquitinated at different rates.
A previously developed in vitro IκBα ubiquitination assay (26) was used to compare the relative affinity of βTrCP for the IKK target sites on p105 and IκBα. In vitro-translated and [35S]methionine-labeled IκBα was allowed to associate with immunopurified NF-κB (from HeLa cells), phosphorylated with recombinant IKK2-EE, and subjected to βTrCP-mediated ubiquitination in a cell-free system which does not support IκBα phosphorylation or proteasome-mediated degradation. Under control conditions, phosphorylated IκBα, but not unphosphorylated IκBα, was efficiently converted to a polyubiquitinated species (Fig. 7C, compare lane without IKK to lane without peptide). The addition of IκBα28-37 peptide phosphorylated on serines 32 and 36 (Fig. 1C) completely inhibited IκBα ubiquitination with a 50% inhibitory concentration (IC50) of ≤5 μM (Fig. 7C and E). The addition of p105922-935 peptide phosphorylated on serines 927 and 932 also inhibited IκBα ubiquitination but was much less efficient (IC50 ≈ 25 μM) than the phospho-IκBα28-37 peptide. The phosphoserine 927 p105922-935 peptide was even less effective (IC50 ≈ 60 μM) in inhibiting IκBα ubiquitination, and phosphoserine 932 p105922-935 peptide had no detectable effect (Fig. 7C and E). These data confirm that βTrCP preferentially binds to p105 phosphorylated on both serines 927 and 932 and also indicate that βTrCP has a higher affinity for phospho-IκBα than for phospho-p105.
The Asp-Ser927(P)-Gly-Val-Glu-Thr-Ser932(P) motif in the PEST region of p105 is closely related to the N-terminal IKK target sequence of IκBα, Asp-Ser32(P)-Gly-Leu-Asp-Ser36(P), except for the introduction of an extra residue between the two phosphorylated serines in p105 (Fig. 1A). To determine whether this spacing might explain the differential affinity of βTrCP for the phospho-IκBα28-37 and phospho-p105922-935 peptides, an IκBα28-37 phosphopeptide was synthesized in which the spacer between phosphoserines 32 and 36 was increased by the introduction of an alanine residue (Fig. 1C). This peptide was less effective in blocking IκBα ubiquitination than the wild-type phospho-IκBα28-37 peptide (Fig. 7D) (IC50 ≈ 50 μM estimated from titration experiments). This suggests that the high affinity binding of βTrCP to phosphorylated targets requires a 3-amino-acid spacing between phosphorylated serines and that target proteins with a 4-amino-acid spacing, such as p105, bind with lower affinity.
DISCUSSION
The experiments in this study demonstrate that phosphorylation of serines 927 and 932 within the PEST region of NF-κB1 p105 is essential for its degradation triggered by TNF-α stimulation (Fig. 2A). This reflects a requirement for the phosphorylation of both serines to promote efficient recruitment of the E3 ubiquitin ligase SCFβTrCP to phospho-p105 and subsequent p105 ubiquitination (Fig. 6). Furthermore, in vitro experiments demonstrate that serine 927 and serine 932 are directly phosphorylated by the IKK complex, whose expression is necessary for the TNF-α-induced phosphorylation of both serines in EF cells (Fig. 4). IL-1 stimulation also triggers both serine 927 and 932 phosphorylation of p105, and IL-1-induced p105 proteolysis is blocked by the mutation of either serine 927 or serine 932 to alanine (21) (data not shown). Thus, although the signaling pathways triggered by TNF-α and IL-1 are distinct (18, 25), both cytokines appear to trigger p105 proteolysis via a common phosphorylation-based mechanism. It is likely that p105 proteolysis induced by other agonists, such as lipopolysaccharide and phorbol ester (5, 14), will also involve the phosphorylation of serines 927 and 932 by the IKK complex. Consistent with this hypothesis, p105 proteolysis triggered by overexpressed TPL2/Cot (3) also requires the phosphorylation of both serines 927 and 932 (data not shown).
Heissmeyer et al. have previously suggested a regulatory role for serine 923 (Asp-Ser923-Val-Cys-Asp-Ser927) in IKK2-induced HA-p105 proteolysis in 293 cells transiently cotransfected with βTrCP (8). However, p105 degradation triggered by phosphorylation by the endogenous IKK complex after TNF-α stimulation does not require serine 923 (Fig. 2A) and an earlier study by our laboratory also failed to reveal a role for this residue in IKK2-induced p105 proteolysis in NIH 3T3 cells (21). Serine 923, therefore, is unlikely to be an important physiological target of the IKK complex, and it has not yet been demonstrated that serine 923 is actually phosphorylated after cytokine stimulation. To investigate this directly, an antibody was raised to a synthetic peptide corresponding to residues 921 through 933 of human p105 in which serine 923 is phosphorylated. Although this antibody clearly recognized specifically the immunizing phosphopeptide on dot blots, it did not bind to p105 phophorylated either in vivo after TNF-α stimulation or in vitro by recombinant IKK2 (data not shown). These latter results contrast with those of Heissmeyer et al. (8), who reported that purified IKK2 phosphorylated the recombinant His6-p105917-968 fusion protein on serine 923 in vitro. The Ley laboratory has recently demonstrated that the efficient phosphorylation of p105 on both serines 927 and 932 in vitro requires the p105 death domain (p105 residues 802 to 893), which acts as a docking site for IKK2 (2) (data not shown). It is possible that serine 923 of the recombinant His6-p105917-968 fusion protein is artefactually phosphorylated by IKK2 due to the absence of p105 death domain-IKK2 interaction, which may be important both to ensure the efficiency of phosphorylation and for correct target amino acid specificity (2).
The Asp-Ser927-Gly-Val-Glu-Thr-Ser932 motif in the PEST domain of p105 is distinct from the N-terminal IKK target sequence of IκBα (Asp-Ser32-Gly-Leu-Asp-Ser36) due to the introduction of an extra residue between the two phosphorylated serines in p105 (Fig. 1A). In vitro binding experiments suggest that one functional consequence of this extra amino acid in p105 is to significantly decrease the affinity of βTrCP for the IKK phosphorylated sequence (Fig. 7). Consistent with this possibility, introduction of an extra amino acid between the IKK-phosphorylated serines in IκBα reduces the ability of IκBα phosphopeptides to block βTrCP-dependent IκBα ubiquitination (Fig. 7D). Interestingly, the IKK target sequence in p105 is completely conserved between human, mouse, rat, and chicken p105 (21), suggesting that the lower affinity of phospho-p105 for βTrCP confers some evolutionary advantage. Presumably one consequence of this decreased binding is to reduce the efficiency of βTrCP-mediated ubiquitination of phospho-p105 compared with that of phospho-IκBα. This may explain in part the slow and partial proteolysis of p105 in TNF-α-stimulated cells compared with that of IκBα, which is rapidly and completely degraded (21). However, although IKK phosphorylates p105 and IκBα with similar kinetics after TNF-α stimulation (21), it cannot be excluded that the IKK complex phosphorylates IκBα at a higher stoichiometry than p105, and this may also contribute to the different efficiency of their proteolysis in TNF-α-stimulated cells.
Both phosphopeptide competition and pull-down experiments (Fig. 7) indicate that βTrCP can bind weakly when p105 serine 927 alone is phosphorylated, whereas no binding is detected to a phosphoserine 932 p105 peptide. However, phosphorylation of both serines 927 and 932 generates a higher affinity binding site for βTrCP. Interestingly binding of βTrCP1 to phospho-IκBα peptides shows a similar phosphorylation requirement. Thus, an IκBα peptide phosphorylated on both serines 32 and 36 binds to recombinant βTrCP1 with high affinity (Kd = 70 nM) whereas a peptide phosphorylated on only serine 32 binds much more weakly (Kd = 16 μM) (R. Hay, unpublished data). In contrast, binding to a phosphoserine 36 IκBα peptide is unmeasurable. Together these data suggest that βTrCP has a bipartite phosphopeptide binding site in which the N-terminal phosphoserine of either p105 or IκBα interacts more strongly than the C-terminal phosphoserine.
Heissmeyer et al. have reported that HA-p105 degradation triggered by coexpression with FL-IKK2 and βTrCP1 in 293 cells is unaffected by the introduction of a serine 932-to-alanine mutation but blocked by a serine 927-to-alanine mutation (8). Thus, HA-p105 proteolysis promoted by overexpressed FL-IKK2 and βTrCP1 can be triggered by serine 927 phosphorylation alone. In contrast, a serine 932-to-alanine mutation delays HA-p105 proteolysis promoted by FL-IKK2 expression in NIH 3T3 fibroblasts (21) and completely blocks p105 proteolysis triggered by TNF-α stimulation (Fig. 2A). Thus, in these latter experimental models, HA-p105 must be phosphorylated on both serines 927 and 932 for optimal signal-induced proteolysis. It is likely that βTrCP1 overexpression masks the need for serine 932 phosphorylation by artefactually promoting sufficient βTrCP1 binding to HA-p105 that is phosphorylated only on serine 927 (Fig. 7B). However, when ubiquitination is mediated by endogenous SCFβTrCP, phosphorylation of both serines is required for optimal βTrCP binding. The present study therefore highlights the importance of determining the function of phosphorylation sites on a protein where the participating regulatory proteins (i.e., IKK and SCFβTrCP) are expressed at physiological levels, rather than relying solely on transient overexpression experiments.
In conclusion, this study demonstrates that the TNF-α-induced proteolysis of NF-κB1 p105 involves the IKK complex-mediated phosphorylation of serines 927 and 932 in the PEST region, within a motif similar to that in the IKK target sequence of IκBα. Phosphorylation of both residues is shown to be required for the efficient recruitment of βTrCP, a subunit of the SCF-type E3 ligase which promotes the ubiquitination of phospho-p105 and triggers its subsequent proteolysis.
Acknowledgments
V.L. and J.J. contributed equally to this work.
We thank Nancy Bump, Alain Israel, Frank Mercurio, Keiji Tanaka, and Inder Verma for the reagents used in this study. The help of Lee Johnston, Steve Smerdon, and Steve Gamblin (National Institute for Medical Research) for suggestions on the manuscript, Sandra Watton (National Institute for Medical Research) for advice on affinity purification of antiphosphopeptide antibodies, and Lesley Thompson (University of St. Andrews) and Ada Hatzubai (Hebrew University) for helping with in vitro ubiquitination experiments is gratefully acknowledged. We are also indebted to the PhotoGraphics department (National Institute for Medical Research) for making the figures.
This work was supported by the U.K. Medical Research Council, the Arthritis Research Campaign (project grant L0536 to V.L.), and the AINP consortium, EC—5th framework.
REFERENCES
- 1.Baldwin, A. S. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-681. [DOI] [PubMed] [Google Scholar]
- 2.Beinke, S., M. P. Belich, and S. C. Ley. 2002. The death domain of NF-κB1 p105 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 277:24162-24168. [DOI] [PubMed] [Google Scholar]
- 3.Belich, M. P., A. Salmeron, L. H. Johnston, and S. C. Ley. 1999. TPL-2 kinase regulates the proteolysis of the NF-κB inhibitory protein NF-κB1 p105. Nature 397:363-368. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Neriah, Y. 2002. Regulatory functions of ubiquitination in the immune system. Nature Immunol. 3:20-26. [DOI] [PubMed] [Google Scholar]
- 5.Donald, R., D. W. Ballard, and J. Hawiger. 1995. Proteolytic processing of NF-κB/IκB in human monocytes. J. Biol. Chem. 270:9-12. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 8.Heissmeyer, V., D. Krappmann, E. N. Hatada, and C. Scheidereit. 2001. Shared pathways of IκB kinase-induced SCFβTrCP-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBa. Mol. Cell. Biol. 21:1024-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heissmeyer, V., D. Krappmann, F. G. Wulczyn, and C. Scheidereit. 1999. NF-κB p105 is a target on IκB kinases and controls signal induction of BCL-3-p50 complexes. EMBO J. 18:4766-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa, H., E. Claudio, D. Dambach, C. Raventos-Suarez, C. Ryan, and R. Bravo. 1998. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p) 105 precursor (NF-κB1) but expressing p50. J. Exp. Med. 187:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Li, Q., G. Estepa, S. Memet, A. Israel, and I. M. Verma. 2000. Complete lack of NF-κB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 14:1729-1733. [PMC free article] [PubMed] [Google Scholar]
- 14.MacKichan, M. L., F. Logeat, and A. Israel. 1996. Phosphorylation of p105 PEST sequences via a redox-insensitive pathway up-regulates processing to p50 NF-κB. J. Biol. Chem. 271:6084-6091. [DOI] [PubMed] [Google Scholar]
- 15.Mellits, K. H., R. T. Hay, and S. Goodbourn. 1993. Proteolytic degradation of MAD3 (IκBα) and enhanced processing of the NF-κB precursor p105 are obligatory steps in the activation of NF-κB. Nucleic Acids Res. 21:5059-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercurio, F., J. A. DiDonato, C. Rosette, and M. Karin. 1993. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 7:705-718. [DOI] [PubMed] [Google Scholar]
- 17.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. W. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science 278:860-866. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill, L. A. J., and C. A. Dinarello. 2000. The IL-1 receptor/toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol. Today 21:206-209. [DOI] [PubMed] [Google Scholar]
- 19.Orian, A., H. Gonen, B. Bercovich, I. Fajerman, E. Eytan, A. Israel, F. Mercurio, K. Iwai, A. L. Schwartz, and A. Ciechanover. 2000. SCFβTrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 19:2580-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice, N. R., M. L. MacKichan, and A. Israel. 1992. The precursor of NF-κB p50 has IκB-like functions. Cell 71:243-253. [DOI] [PubMed] [Google Scholar]
- 21.Salmeron, A., J. Janzen, Y. Soneji, N. Bump, J. Kamens, H. Allen, and S. C. Ley. 2001. Direct phosphorylation of NF-κB p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276:22215-22222. [DOI] [PubMed] [Google Scholar]
- 22.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10:405-455. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, H., T. Chiba, T. Suzuki, T. Fujita, T. Ikenoue, M. Omata, K. Furuichi, H. Shikama, and K. Tanaka. 2000. Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds IκBα for signal-dependent ubiquitination. J. Biol. Chem. 275:2877-2884. [DOI] [PubMed] [Google Scholar]
- 24.Syrovets, T., M. Jendrach, A. Rohwedder, A. Schule, and T. Simmet. 2001. Plasmin-induced expression of cytokines and tissue factor in human monocytes involves AP-1 and IKK-β-mediated NF-κB activation. Blood 97:3941-3950. [DOI] [PubMed] [Google Scholar]
- 25.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 26.Yaron, A., A. Hatzubai, M. Davis, I. Lavon, S. Amit, A. M. Manning, J. S. Andersen, M. Mann, F. Mercurio, and Y. Ben-Neriah. 1998. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396:590-594. [DOI] [PubMed] [Google Scholar]