Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) exerts potent cytotoxic activity against transformed keratinocytes, whereas primary keratinocytes are relatively resistant. In several cell types, inhibition of the proteasome sensitizes for TRAIL-induced apoptosis by interference with NF-κB activation. Here we describe a novel intracellular mechanism of TRAIL resistance in primary cells and how this resistance is removed by proteasome inhibitors independent of NF-κB in primary human keratinocytes. This sensitization was not mediated at the receptor-proximal level of TRAIL DISC formation or caspase 8 activation but further downstream. Activation of caspase 3 was critical, as it only occurred when mitochondrial apoptotic pathways were activated, as reflected by Smac/DIABLO, HtrA2, and cytochrome c release. Smac/DIABLO and HtrA2 are needed to release the X-linked inhibitor-of-apoptosis protein (XIAP)-mediated block of full caspase 3 maturation. XIAP can effectively block caspase 3 maturation and, intriguingly, is highly expressed in primary but not in transformed keratinocytes. Ectopic XIAP expression in transformed keratinocytes resulted in increased resistance to TRAIL. Our data suggest that breaking of this resistance via proteasome inhibitors, which are potential anticancer drugs, may sensitize certain primary cells to TRAIL-induced apoptosis and could thereby complicate the clinical applicability of a combination of TRAIL receptor agonists with proteasome inhibitors.
Apoptotic cell death is an important biological process that is required to maintain the integrity and homeostasis of multicellular organisms. Inappropriate or impaired apoptosis has been implicated in the development of many human diseases, including cancer (71). The death-inducing members of the tumor necrosis factor (TNF) family, TNF, CD95/APO-1/Fas ligand (CD95L), and TNF-related apoptosis-inducing ligand (TRAIL/APO-2L), have been studied most intensively. These studies have resulted in elucidation of their role in activation-induced cell death, autoimmune disorders, immune privilege, and tumor evasion from the immune system (reviewed in references 79, 82, and 85).
TRAIL has attracted attention for its ability to preferentially kill tumor cells while most normal cells were resistant both in vitro (56, 86) and in vivo (1, 20, 80). The functional analysis of the TRAIL receptor-ligand system has been complicated by the fact that a total of five different receptors for this cytokine has been identified (reviewed in reference 46). Although the major role of TRAIL is the induction of apoptosis, it has also been shown in earlier overexpression studies to activate other signaling pathways, such as the transcription factor NF-κB (11, 62). NF-κB is known to induce genes involved in apoptosis resistance. Inhibition of NF-κB can sensitize cells for TNFα- or TRAIL-induced apoptosis, depending on the cell type, suggesting that distinct signaling pathways modulate the effect of TRAIL in a cell type-specific manner (2, 27, 76).
The early biochemical events resulting in apoptosis induction by ligand-induced death receptor cross-linking have been studied by the analysis of the so-called death-inducing signaling complex (DISC) (33, 81). Cross-linking of CD95 or the two apoptosis-inducing TRAIL receptors (TRAIL-R1 and TRAIL-R2) results in the recruitment of Fas-associated death domain (FADD; also called MORT1) and caspase 8 to the DISC (3, 34, 68). In a homotypic interaction, the death domain of FADD binds to the death domain of CD95. The death effector domain of FADD in turn interacts with the death effector domain of procaspase 8 and thereby recruits this proenzyme to the DISC (51). Procaspase 8 is proteolytically cleaved and thereby activated at the DISC. Activated caspase 8 then initiates the apoptosis-executing caspase cascade (81). This cascade is further controlled by “cross talk” between the intrinsic (mitochondrial) and extrinsic (death receptor) cell death pathways, thereby modulating the outcome of death receptor triggering (58). In that respect it has been shown for CD95L and recently also for TRAIL (24, 41) that its proapoptotic signaling can be blocked by Bcl-2 or Bcl-XL overexpression in some cell types, whereas other cell types cannot be protected by overexpression of these molecules, leading to the concept of two different cell types utilizing distinct signaling pathways with or without the necessity for mitochondrial contribution (60).
Further complexity is added to the regulatory pathways involved in death receptor sensitivity by proteins that are capable of inhibiting active caspases. These proteins are called inhibitor-of-apoptosis proteins (IAPs) (14, 25). IAPs are a family of proteins defined by baculovirus repeat (BIR) domains and, in some cases, a zinc ring finger domain (14, 25). IAPs like X-linked IAP (XIAP), Livin/MLIAP, cIAP1, and cIAP2 block apoptosis by directly inhibiting caspases. For some IAPs, an involvement in caspase-independent pathways of apoptosis was postulated (14, 65). XIAP is the most potent inhibitor of caspases among the above-mentioned IAPs (14).
The ubiquitin-proteasome pathway plays a central role in the regulation of essential cellular processes such as cell cycle control, transcription, signal transduction, and apoptosis (28, 36). Many key regulatory proteins are controlled by ubiquitination, which targets them for degradation by the 26S proteasome (29, 39). A recent report suggested that proteasome inhibitors are able to induce apoptosis in transformed but not in normal lymphocytes (50). Well-known proteasomal targets include the NF-κB/IκB system, p53, and IAPs (28). Earlier studies showed that NF-κB activation is efficiently blocked by proteasome inhibitors (27). These data support the view that sensitization to TNFα- or TRAIL-induced apoptosis by proteasome inhibitors is explained by the inhibition of NF-κB transcriptional activity (27, 30).
The mechanisms responsible for the resistance of primary cells to TRAIL are not fully understood. Although initial reports suggested that differential expression of TRAIL-R3 and TRAIL-R4 might determine resistance to TRAIL in primary cells, later studies demonstrated the important contribution of intracellular regulatory pathways in primary cells (42). To further investigate the resistance to TRAIL in primary human keratinocytes, we analyzed proteasome-mediated resistance to TRAIL-induced apoptosis and its neutralization by proteasome inhibitors. We show that inhibition of proteasomal function effectively sensitizes primary keratinocytes to TRAIL independent of NF-κB and downstream of the TRAIL DISC. This sensitization required the activation of mitochondrial signaling pathways necessary for XIAP inhibition. Our data suggest that one important step to overcome the intracellular resistance of primary keratinocytes to TRAIL is the inhibition of XIAP by activation of mitochondrial apoptosis signaling pathways.
MATERIALS AND METHODS
Materials.
The protease inhibitor z-Val-Ala-Asp-fluoromethyl ketone (zVAD-FMK) was obtained from Bachem (Heidelberg, Germany). If not indicated otherwise, all other reagents were of reagent grade and obtained from Serva (Heidelberg, Germany). The following antibodies were used: poly(ADP-ribose) polymerase (G. Poirier, CHUL Research Center, Quebec, Canada), caspase 3 (MF 393 and MF 397), and XIAP (MF 478; gifts from D. Nicholson, Merck Frosst Corp, Quebec, Canada); Smac (kindly provided by M. MacFarlane, Leicester, United Kingdom); and HtrA2/Omi (generously provided by R. Takahashi, RIKEN Brain Science Institute, Saitama, Japan). The FLICE (C-15) (51) and cFLIP (NF-6) (61) monoclonal antibodies were kindly provided by P. H. Krammer and are available from Alexis (Gruenberg, Germany). Monoclonal antibodies against FADD and XIAP were purchased from Transduction Laboratories (San Diego, Calif.), caspase 10 monoclonal antibodies (clone 4C1) were from MBL International (Watertown, Mass.), cytochrome c monoclonal antibodies were from Pharmingen (San Diego, Calif.), antibodies to IκBα (C-21) were from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.), and antibodies to IKK2 (antibody 2684) were from New England Biolabs, Boston, Mass.
The monoclonal antibodies specific for the different TRAIL receptors were generated by immunizing mice with TRAIL receptor-Fc fusion proteins. The specificity of the monoclonal antibodies was confirmed by staining cells transfected with expression plasmids for TRAIL-R1 to TRAIL-R4 (H. Walczak, unpublished observation). Anti-TRAIL-R1 (clone HS101) and anti-TRAIL-R2 (clone HS201), both of the murine immunoglobulin G1 (IgG1) isotype, were used for fluorescence-activated cell sorting staining. The monoclonal antibodies are also available from Alexis (Gruenberg, Germany). Recombinant leucine zipper (LZ)-TRAIL was generated as described before (78), and Flag-TRAIL was expressed as previously described (32). Horseradish peroxidase-tagged donkey anti-rabbit immunoglobulin antibodies and horseradish peroxidase-tagged goat anti-mouse IgG were from Pharmingen (Hamburg, Germany), and horseradish peroxidase-conjugated goat anti-mouse IgG1 and IgG2b were obtained from Southern Biotechnology Associates (Birmingham, Ala.).
Tissue culture.
Primary keratinocytes were prepared from neonatal foreskins as previously described (42). Cells were kept in serum-free KGM medium (Cellsystems, St. Katharinen, Germany). Cells were only used up to passage 3 for all experiments. The spontaneously transformed keratinocyte line HaCaT was kindly provided by N. Fusenig (German Cancer Research Center, Heidelberg, Germany) and cultured as described before (4).
Subfractionation of cytoplasmic proteins.
Cytosolic extracts were generated with a modified digitonin lysis protocol (66). Following trypsinization, cells were washed with phosphate-buffered saline, permeabilized with digitonin (Fluka, Taufkirchen, Germany) at a concentration of 150 μg/ml for 10 min, and centrifuged at 5,000 × g for 5 min to remove cellular debris. The resulting supernatants were spun again at 13,000 × g for 30 min at 4°C. Equal amounts of supernatant were analyzed by Western blot. Blots were subsequently rehybridized with monoclonal antibodies to cytochrome c oxidase subunit IV (Molecular Probes Inc., Eugene, Oreg.) to control for mitochondrial lysis (data not shown).
Western blot analysis.
Total cellular proteins were collected as described before (44) with the exception that complete protease inhibitor cocktail (Roche Molecular Diagnostics, Mannheim, Germany) was used. From 20 to 75 μg of protein was electrophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes. Blocking of membranes and incubation with the indicated primary and appropriate secondary antibodies were performed essentially as described elsewhere (43). Bands were visualized with the ECL detection kit (Amersham).
Analysis of death-inducing signaling complex.
For precipitation of the native TRAIL DISC, 5 × 106 human primary keratinocytes were used for each condition. Cells were washed once with RPMI medium at 37°C and subsequently incubated for the indicated time periods at 37°C in the presence of 1 μg of Flag-TRAIL per ml precomplexed with 2 μg of anti-Flag antibody M2 (Sigma, Taufkirchen, Germany) per ml for 15 min or, for the unstimulated control, in the absence of Flag-TRAIL. DISC formation was stopped by washing the monolayer twice with ice-cold phosphate-buffered saline. Cells were lysed on ice by addition of 1 ml of lysis buffer (30 mM Tris-HCl [pH 7.5] at 21°C, 120 mM NaCl, 10% glycerol, 1% Triton X-100, complete protease inhibitor cocktail [Roche Molecular Diagnostics, Mannheim, Germany]). After 15 min of lysis, the lysates were centrifuged at 50,000 × g for 15 min to pellet cellular debris. DISC complexes were precipitated from the lysates by coincubation with 20 μl of protein G beads (Roche, Mannheim, Germany) for 12 h on an end-over-end shaker at 4°C.
For precipitation of the nonstimulated receptors, 200 ng of Flag-TRAIL and 400 ng of anti-Flag M2 were added to the lysates prepared from nonstimulated cells at 200 ng to control for protein association with a nonstimulated receptor(s). Ligand affinity precipitates were washed five times with lysis buffer before the protein complexes were eluted from the beads by addition of 15 μl of 2× standard reducing sample buffer. Subsequently, proteins were separated by SDS-PAGE on 4 to 12% NuPage Bis-Tris gradient gels (Invitrogen, Karlsruhe, Germany) before detection of DISC components by Western blot analysis.
Determination of DEVDase activity.
Cells were lysed following treatment in buffer containing 50 mM HEPES-KOH (pH 7.0), 2 mM EDTA, 10% sucrose, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 5 mM dithiothreitol. Equal amounts of protein were incubated with 2,5 μM biotinylated DEVD-acyloxymethyl ketone (biotin-DEVD-AOMK) (kindly provided by D. Nicholson, Merck Frosst Corp, Quebec, Canada) for 30 min at 37°C (57). Biotin-DEVD-AOMK is an irreversible caspase inhibitor that covalently binds to DEVD-cleaving caspases, resulting in the biotin labeling of active caspase fragments (57, 72). Samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. Following overnight incubation with peroxidase-conjugated streptavidin (Dako Diagnostika, Hamburg, Germany), biotin-labeled fragments were visualized with the ECL detection kit (Amersham, Arlington Heights, Ill.). Membranes were subsequently stripped and reprobed with antibodies to caspase 3 (MF 393) to detect caspase 3-specific fragments.
Coimmunoprecipitation of XIAP.
Primary and transformed keratinocytes (2 × 106 cells) were treated with 250 ng of TRAIL per ml for 3 h and lysed in TENT buffer (50 mM Tris [pH 8.0], 2 mM EDTA, 150 mM NaCl, and 1% Triton X-100) on ice. Equal amounts of cellular proteins were precipitated with 1 μg of either rabbit IgG, rabbit caspase 3 (MF 397), or XIAP (MF 478) antiserum (kindly provided by D. Nicholson, Merck Frosst Corp, Quebec, Canada) in the presence of 20 μl of protein A-Sepharose beads overnight. Precipitates were washed four times with lysis buffer before the protein complexes were eluted from the beads by addition of 2× standard reducing sample buffer. Subsequently, proteins were separated by SDS-PAGE before detection of XIAP by Western blot analysis with monoclonal antibodies to XIAP (Transduction Laboratories, San Diego, Calif.).
FACScan analysis.
For analysis of the mitochondrial transmembrane potential (ΔΨm), 5 × 105 cells were resuspended in Dulbecco's modified Eagle's medium containing 40 nM tetramethylrhodamine ethyl ester (59) and incubated at 37°C for 15 min as described before (21). After being washed once with ice-cold phosphate-buffered saline, cells were immediately analyzed. For surface staining of TRAIL-R1 and TRAIL-R2, 2 × 105 cells were detached by incubation with phosphate-buffered saline containing 5 mM EDTA, followed by brief trypsinization, and incubated with monoclonal antibodies against TRAIL-R1 (HS101), TRAIL-R2 (HS201), or control murine IgG1 at 10 μg/ml, followed by biotinylated secondary goat anti-mouse immunoglobulin antibodies and phycoerythrin-conjugated streptavidin (Pharmingen, Hamburg, Germany). For all experiments, 104 cells were analyzed by FACScan (Becton Dickinson & Co, San Jose, Calif.).
Apoptosis and cytotoxicity assays.
Crystal violet staining of surviving attached cells was performed 16 to 24 h after addition of LZ-TRAIL in 24-well plates (1 × 105 cells) or 96-well plates (2 × 104 cells) as described before (42). Parallel plates were examined by propidium iodide staining and flow cytometric analysis of subdiploid DNA content as described before (42). For annexin-propidium iodide staining, 2 × 105 to 5 × 105 cells were stained with 10 μl of annexin V-fluorescein isothiocyanate conjugate (Becton Dickinson & Co, San Jose, Calif.) and 5 μl of propidium iodide (50 ng/ml) according to the manufacturer's instructions, incubated for 15 min at room temperature in the dark, and immediately analyzed.
Electrophoretic mobility shift assays.
Nuclear and cytoplasmic extracts of primary keratinocytes were prepared according to Schreiber et al. (63) with slight modifications as previously described (53). The oligonucleotide probe used was a consensus NF-κB binding site derived from the κB element of the murineinterleukin-2 promoter (TCEdA>C) (6): 3′-GACCAAGAGGGATTTCACCCCTAAATC-5′. The following antibodies were used for the supershift electrophoretic mobility shift assay: anti-p65-NF-κB (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.; A, sc-109), anti-c-Rel (Santa Cruz; N, sc-070), anti-RelB (Santa Cruz; C-19, sc-226), anti-p50-NF-κB (Santa Cruz; NLS, sc-114), and anti-p52-NF-κB (Upstate Biotechnology Inc., Lake Placid, N.Y.; 06-413).
Retroviral infection.
The pCFG5-IEGZ retroviral vector containing inserts of the transdominant ΙκBα (ΙκBα-TD) mutant or kinase-dead IKK2 (IKK2-KD) mutant was used for infection of primary human keratinocytes as described earlier (13) with the exception that viral supernatants were generated in KGM. Infection efficiency of keratinocytes ranged from 30% to 80%, as determined by green fluorescent protein fluorescence 72 h after infection. At this time point, keratinocytes were used for cytotoxicity assays and biochemical characterization.
Transient-transfection assay for XIAP.
Cells were transfected with the Nucleofector technology (Amaxa, Köln, Germany) according to the recommended protocols. Briefly, 2 × 106 transformed keratinocytes were resuspended in 100 μl of Cell Line R solution. Then 3 μg of pEBB-XIAP vector or pEBB control vector (kindly provided by Colin Duckett, University of Michigan Medical School) was mixed with the suspension, added to an Amaxa cuvette, and nucleofected with program U27. Cells were then diluted with 500 μl of warm RPMI and added to six-well plates containing 1 ml of warm medium. After a 10-h incubation period at 37°C, biochemical and functional studies were performed as indicated in the figure legends. Transfection efficiency, as determined by green fluorescent protein expression in parallel plates, varied between 50% and 70%.
RESULTS
Primary keratinocytes are sensitized to TRAIL-induced apoptosis by proteasome inhibition.
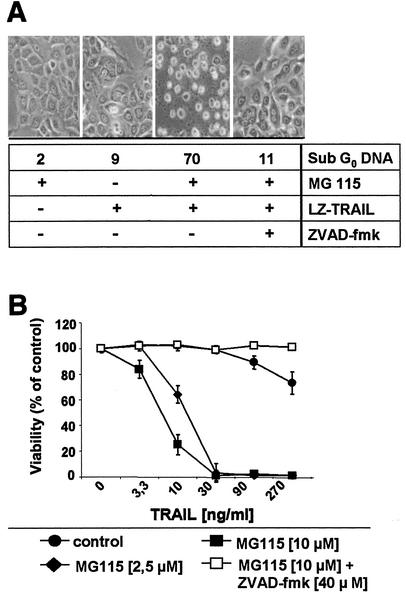
We have previously shown that primary keratinocytes are relatively resistant to TRAIL-induced apoptosis compared to transformed keratinocytes (42). Inhibition of proteasome function modulates apoptotic signaling pathways in a variety of tumor cells (16, 28). We thus explored the potential role of proteasomal function for the resistance of primary keratinocytes to TRAIL-mediated apoptosis. To this end, we used MG115 as a proteasomal inhibitor in our studies. MG115 treatment resulted in a dramatic sensitization of primary keratinocytes for TRAIL-induced apoptosis in a concentration-dependent manner (Fig. 1) (55). Moreover, two other known proteasome inhibitors, lactacystin (19) and epoxomicin (52), similarly sensitized primary keratinocytes for TRAIL-induced apoptosis (data not shown). The broad-spectrum caspase inhibitor zVAD-FMK completely abolished TRAIL-induced apoptosis under these conditions, confirming the critical role of caspases in this process (Fig. 1). Our data suggest that peptide aldehyde inhibitors such as MG115 interfere with proteasome-regulated signaling pathways, resulting in increased sensitivity of primary keratinocytes to TRAIL-induced apoptosis.
FIG. 1.
Proteasome inhibitor MG115 sensitizes primary keratinocytes to TRAIL-induced apoptosis. (A) Primary keratinocytes were treated with 50 ng of TRAIL per ml for 16 h. Caspase-dependent cytotoxicity of TRAIL (sub-G0 DNA analysis) is only detectable in the presence of the proteasome inhibitor MG115 (10 μM). zVAD-FMK (40 μM) completely abolishes TRAIL-induced apoptosis. (B) Dose-dependent effect of MG115 on TRAIL-induced cell death in primary keratinocytes was determined by crystal violet staining 16 h after addition of the indicated concentrations of TRAIL.
TRAIL induces NF-κB activation in primary keratinocytes.
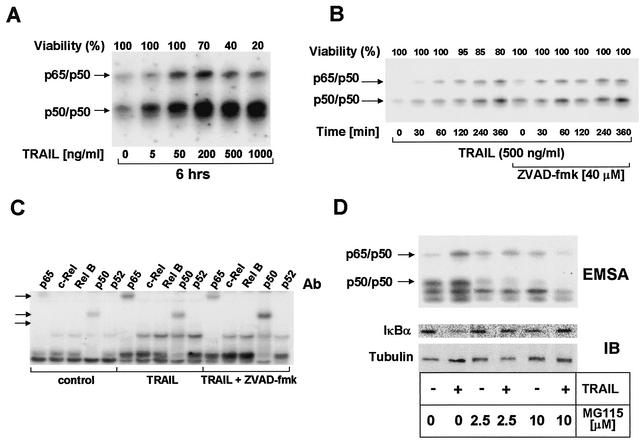
One important target of proteasomal degradation is the IκB/NF-κB system (29, 48). NF-κB has potent antiapoptotic properties, particularly in TNFα-induced apoptosis (30), whereas its role in TRAIL-induced apoptosis is more controversial (26, 27). To study whether the increased sensitivity to TRAIL-induced apoptosis may be mediated by inhibition of NF-κB function, we first analyzed TRAIL-induced NF-κB activation in primary keratinocytes and the impact of the proteasome inhibitor MG115. Employing electrophoretic mobility shift assays, we detected an increase in NF-κB DNA binding activity upon stimulation with TRAIL at concentrations of between 50 and 500 ng/ml (Fig. 2A). This induction was observed within 30 min and increased further up to 6 h after stimulation (Fig. 2B).
FIG. 2.
TRAIL dose-dependently enhances NF-κB binding in primary keratinocytes. (A) Primary keratinocytes were treated with 5 to 1,000 ng of TRAIL per ml for 6 h and analyzed for NF-κB DNA binding by electrophoretic mobility shift assay with nuclear extracts. Induction of p65/p50 heterodimers (upper arrow) or p50 homodimers (lower arrow) is shown. As a control for the specificity of DNA binding, control experiments with an excess of unlabeled probe were performed (not shown). Cell viability was determined in parallel experiments by crystal violet staining and is indicated as a percentage of the control value. (B) Primary keratinocytes were exposed to 250 ng of TRAIL per ml for 6 h in the presence or absence of 40 μM zVAD-FMK. Survival of cells was determined in parallel experiments as described for A. (C) Composition of TRAIL-induced nuclear NF-κB complexes in primary keratinocytes. Following incubation of primary keratinocytes with TRAIL (250 ng/ml) for 4 h, supershift electrophoretic mobility shift analysis of inducible NF-κB-specific complexes was performed with nuclear extracts. Each of the Rel-specific antibodies (1 μl of antibody [Ab]; see the text) was added to nuclear protein extracts. Control supershift electrophoretic mobility shift assays (EMSAs) were performed with normal rabbit serum (not shown). Arrows indicate supershifted complexes. (D) MG115 leads to dose-dependent inhibition of TRAIL-induced NF-κB activation (upper panel) and IκBα degradation (lower panel). Membranes were rehybridized with antitubulin monoclonal antibodies to confirm comparable loading of cytoplasmic proteins.
Although zVAD-FMK has been shown to completely block TRAIL-induced apoptosis (42), it did not negatively affect TRAIL-induced NF-κB activation (Fig. 2B), indicating that caspase activation is not required for TRAIL-induced NF-κB activation. The supershift electrophoretic mobility shift assay demonstrated that TRAIL-induced NF-κB binding activity consisted mainly of p50 homodimers as well as p65/p50 heterodimers. Nevertheless, weak supershift signals of p65/c-Rel complexes were also detectable (Fig. 2C). In line with reports for other cell types (27), TRAIL-induced NF-κB DNA binding activity and degradation of IκBα were dose-dependently inhibited by the proteasome inhibitor MG115 (Fig. 2D).
NF-κB activation does not modulate TRAIL sensitivity in keratinocytes.
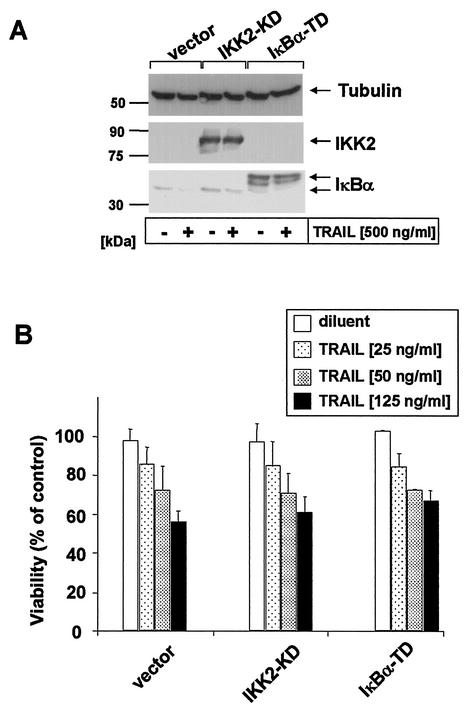
The proteasome has been shown to modulate multiple signaling pathways, including NF-κB, p53, and IAPs (9, 88). In order to inhibit NF-κB more specifically, we next studied whether ectopic expression of dominant negative mutants of IKK2 (IKK2-KD) or ΙκBα (ΙκBα-TD) influenced TRAIL sensitivity in keratinocytes. We thus examined primary keratinocytes following retroviral infection with either IKK2-KD, ΙκBα-TD, or a viral control construct. Similar to the data shown in Fig. 2D, overexpression of these proteins efficiently inhibited TRAIL-induced degradation of ΙκBα and NF-κB activation compared to vector-transfected control cells (Fig. 3A and data not shown). However, sensitivity to TRAIL-induced apoptosis was not significantly modulated by these mutants (Fig. 3B).
FIG. 3.
Overexpression of dominant negative mutant of IKK2 (IKK2-KD) or transdominant IκBα (IκBα-TD) inhibits TRAIL-induced degradation of IκBα without modulation of TRAIL-induced apoptosis. (A) Primary human keratinocytes were retrovirally transduced with IKK2-KD, IκBα-TD, or control vector and subsequently analyzed for expression of mutant proteins as well as degradation of endogenous IκBα following stimulation with 500 ng of TRAIL per ml for 1 h by Western blotting. Membranes were rehybridized with antitubulin monoclonal antibodies to confirm comparable loading of cellular proteins. (B) Primary keratinocytes overexpressing IKK2-KD or IκBα-TD as described for A were either left untreated or incubated with 25, 50, or 125 ng of LZ-TRAIL per ml for 6 h. Thereafter, the percentage of dead green fluorescent protein-positive cells was determined by tetramethylrhodamine ethyl ester staining. Data are shown as mean ± standard deviation of two independent experiments.
Consistent with these data, neither TRAIL nor the proteasome inhibitor MG115 modulated mRNA levels of antiapoptosis genes such as cIAP-1, cIAP-2, TRAF-1, or TRAF-2 in keratinocytes when assayed by an RNase protection assay (not shown). Taken together, our results indicate that neither TRAIL-induced NF-κB activation nor constitutive NF-κB activation significantly modulates resistance to TRAIL. More importantly, our data suggest that the peptide aldehyde inhibitor MG115 interferes with other proteasome-regulated signaling pathways which finally result in increased sensitivity of keratinocytes to TRAIL-induced apoptosis.
Sensitization to TRAIL-induced apoptosis by proteasome inhibition is mediated downstream of the death-inducing signaling complex.
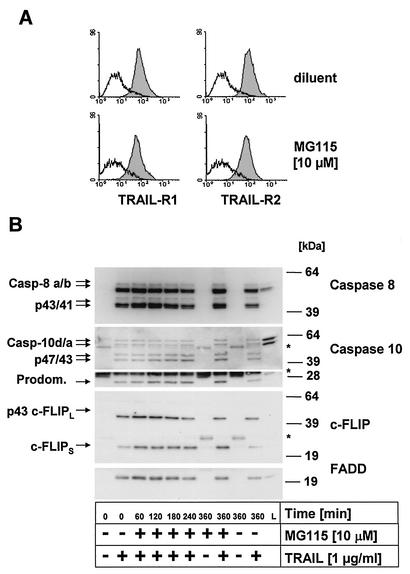
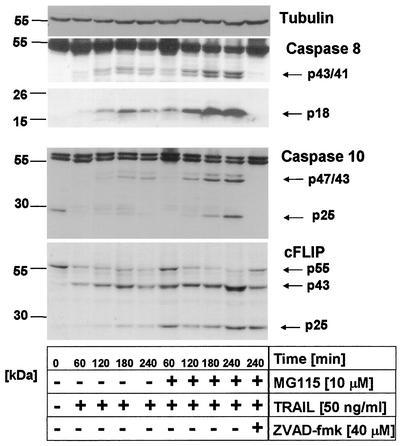
We next determined whether proteasome inhibitors modulate surface expression of the proapoptotic TRAIL-R1 and TRAIL-R2. As shown in Fig. 4A, exposure of primary keratinocytes to MG115 did not significantly modulate surface expression of either TRAIL-R1 or TRAIL-R2, which are both expressed in primary keratinocytes. TRAIL stimulation of these receptors leads to rapid activation of caspase 8 and, as recently shown, also of caspase 10 (35, 67, 84) at the death-inducing signaling complex (DISC) initiating the caspase cascade (3, 34, 68). Thus, increased caspase 8 or 10 recruitment to and/or subsequent activation at the DISC could be responsible for the observed sensitization to TRAIL-induced apoptosis following MG115 treatment.
FIG. 4.
Caspases 8 and 10 are recruited and cleaved at the native TRAIL DISC independent of MG115. (A) Primary keratinocytes were treated with 10 μM MG115 or diluent for 6 h and subsequently analyzed for surface expression of TRAIL-R1 and TRAIL-R2 by fluorescence-activated cell sorting. MG115 does not modulate TRAIL-R1 or TRAIL-R2 surface expression on primary keratinocytes. (B) Analysis of the native TRAIL DISC in primary keratinocytes. Cells were treated for the indicated times with MG115 or diluent alone. The stimulated TRAIL DISC (+) or unstimulated receptors (−) were precipitated with Flag-TRAIL precomplexed with 2 μg of anti-Flag antibodies per ml (clone M2). The resulting protein complexes were separated by SDS-PAGE and analyzed by Western blot for components of the TRAIL DISC. The adaptor protein FADD was detectable only when stimulated receptors were precipitated. Also, recruitment of the two isoforms of caspase 8, caspase 8a and 8b, was detected only following stimulation of TRAIL receptors. In addition, the intermediate cleavage products, p43/p41, of the two isoforms of caspase 8 were detectable in the DISC. For caspase 10, the full-length forms caspase 10d (p59) and caspase 10a (p55) as well as the cleavage products p47/43 and the p25 fragment, corresponding to the prodomain (Prodom.), were detectable in the immunoprecipitates. Analysis of cFLIP levels revealed that both isoforms, cFLIPL and cFLIPS, were present in the TRAIL DISC, with comparable levels in control cells and MG115-treated cells. For cFLIPL, only the p43 cleavage product was detectable in the DISC, indicative of proteolytic processing by active caspases in the DISC. The migration position of the IgG heavy and light chains of the precipitating anti-Flag M2 antibody as well as that of protein G cross-reacting with the antibody used for Western blot detection (caspase 10, lower part) are indicated by asterisks.
We therefore examined whether MG115 modifies the activation of caspase 8 or 10 at the DISC by immunoprecipitation of the native TRAIL DISC from primary keratinocytes (Fig. 4B). DISC analysis demonstrated rapid recruitment and cleavage of caspases 8 and 10 at the DISC. The structurally related protein cFLIP, which is also expressed in primary keratinocytes, acts as an inhibitor of CD95L- and TRAIL-induced apoptosis by inhibiting the activation of caspase 8 at the DISC (38, 42, 61). In line with our previous observation (42), large amounts of cFLIPL and cFLIPS (where L indicates long and S indicates short) were found in the DISC. Importantly, the DISC composition remained unchanged after treatment with the proteasome inhibitor MG115 (Fig. 4B). Thus, neither DISC activation nor surface expression of the TRAIL-R proteins explains the increased sensitivity to TRAIL of primary keratinocytes treated with MG115. However, it is currently not known whether recruitment and activation of caspases is the sole predictor of the activity of DISC constituents. Therefore, we next examined activation kinetics of the known DISC-associated proteins in cellular lysates of primary keratinocytes in the presence and absence of MG115.
Indeed, TRAIL induced rapid cleavage of caspase 8 to p43/41 as well as to the active p18 fragment, which were detectable within 60 to 120 min of stimulation. Furthermore, rapid cleavage of cFLIPL (p43) was seen within 60 to 120 min. Similarly, specific cleavage of caspase 10 to the p47/p43 intermediates as well as to a 25-kDa fragment representing the fully cleaved prodomain was detectable (Fig. 5). Slight differences in the amount of the active fragment of caspase 8 (p18) or the fully cleaved prodomain of caspase 10 (p25) were observed at later time points in the presence of the inhibitor MG115. Addition of zVAD-FMK fully inhibited TRAIL-induced cleavage of caspase 8, caspase 10, and, to a lesser extent, cFLIPL (Fig. 5). In the presence of MG115, levels of cFLIPS were increased compared to the barely detectable levels found under control conditions.
FIG. 5.
Proteasome inhibitor MG115 does not influence early TRAIL-induced caspase 8, caspase 10, and cFLIP cleavage. Primary keratinocytes were incubated for 60 min with 10 μM MG115 or diluent alone and subsequently treated with 50 ng of TRAIL per ml for the indicated times. TRAIL treatment led to cleavage of caspase 8 (p43/p41/p18), caspase 10 (p47/43/25), and cFLIP (p43) under both conditions, as determined by Western blot analysis. Membranes were rehybridized with antitubulin monoclonal antibodies to confirm comparable loading of proteins.
These data show that MG115-induced sensitization of primary keratinocytes to TRAIL is not regulated at the level of caspase 8 or 10 activation. Collectively, our findings indicate that the dramatic sensitization of primary keratinocytes for TRAIL-mediated apoptosis induced by proteasome inhibition is not due to differential activation of the apoptotic signaling pathway at the TRAIL DISC. Thus, this sensitization of primary keratinocytes for TRAIL-induced apoptosis must be regulated at a step downstream of the DISC.
Autocatalytic maturation of caspase 3 to active enzyme is blocked in primary keratinocytes.
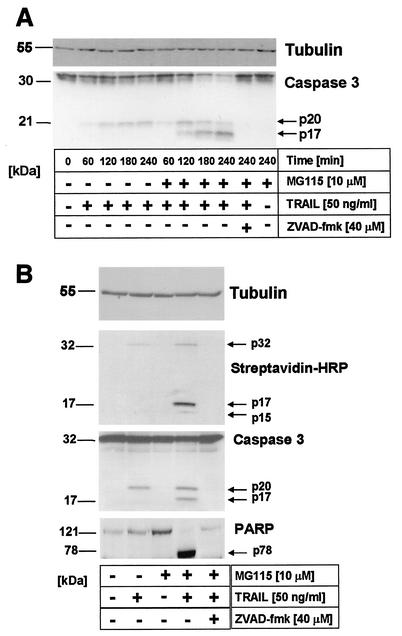
Caspase 3 is the major effector caspase in most cell types (54). The activation of caspase 3 is tightly regulated at several levels, beginning with a proteolytic cut between the large and small subunits, followed by removal of the prodomain. While the first step occurs mainly via active initiator caspases such as caspase 8, 9, or 10, the second step is mediated by autoproteolytic maturation of caspase 3 (54). In order to further examine the level at which MG115 acts to sensitize primary keratinocytes for TRAIL-induced apoptosis, we examined the activation pattern of caspase 3. Interestingly, TRAIL stimulation of primary keratinocytes in the absence of MG115 resulted in the cleavage of caspase 3 to a 20-kDa fragment within 60 to 120 min without detectable induction of apoptosis. However, full processing to the 17-kDa fragment was detectable when cells were treated with TRAIL in the presence of MG115 (Fig. 6A).
FIG. 6.
Full processing of caspase 3 to a 17-kDa fragment in TRAIL-treated primary keratinocytes upon proteasome inhibition. (A) Primary keratinocytes were incubated for 60 min with 10 μM MG115 (right part) or diluent alone (left part) and subsequently treated with 50 ng of TRAIL per ml for the indicated time intervals. TRAIL leads to substantial cleavage of caspase 3 to a 20-kDa fragment (p20), whereas only in the presence of the proteasome inhibitor is a 17-kDa fragment (p17) detectable. No cleavage fragments were detected in the presence of zVAD-FMK or after incubation with MG115 alone. Membranes were rehybridized with antitubulin monoclonal antibodies to confirm comparable loading of proteins. (B) TRAIL-induced effector caspase activity with affinity labeling with biotinylated DEVD-AOMK (see Materials and Methods). Activity of cleaved effector caspases by 50 ng of TRAIL per ml requires the presence of the proteasome inhibitor MG115. Primary keratinocytes were treated as above, and DEVD-specific caspase activity in cellular lysates was analyzed 3 h after TRAIL stimulation. DEVDase activity of cleaved fragments (p17/p15, upper panel) was only detectable in cells treated with TRAIL in the presence of MG115, whereas TRAIL or MG115 alone did not induce detectable caspase activity. Slight labeling of full-length caspase 3 was detected in cells treated with TRAIL alone. Subsequent rehybridization of the blot with caspase 3-specific antibody confirmed that the slower-migrating caspase 3 species (p20) did not have detectable enzymatic activity in primary keratinocytes (middle panel). In line, poly(ADP-ribose) polymerase (PARP) cleavage was only detected upon treatment with TRAIL in the presence of MG115 (lower panel). Membranes were rehybridized with antitubulin monoclonal antibodies to confirm comparable loading of proteins.
To further examine the activity of caspase 3 under these conditions, we used biotinylation of catalytically active caspase 3 in cellular lysates of primary keratinocytes (57). This technique allows the simultaneous detection of enzymatic activity and molecular weight, enabling the parallel monitoring of cleavage and activity of caspase 3. Biotin-labeled fragments of caspase 3 were clearly detectable in lysates of cells treated with TRAIL in the presence of MG115. Interestingly, in cells treated with TRAIL alone, full-length caspase 3 was marginally labeled, indicating some enzymatic activity of the full-length caspase 3, confirming the recent report by Roy et al. (57) (Fig. 6B, upper panel). Consistent with the data shown in Fig. 6A, reprobing of the same membrane with caspase 3-specific antibodies revealed the comparable presence of the cleaved p20 fragment of caspase 3 in lysates treated with TRAIL alone (Fig. 6B, middle panel). These data demonstrate that the initial cleavage of caspase 3 is not blocked in TRAIL-treated primary keratinocytes but that the resulting cleavage product was enzymatically inactive. In line with this observation, cleavage of poly(ADP-ribose) polymerase, a known substrate of caspase 3 (40), was only detected when caspase 3 was further processed to the p17 fragment after treatment with MG115 (Fig. 6B, lower panel). Taken together, our findings indicate that TRAIL-induced full processing of caspase 3 to the active 17-kDa fragment which is required for full enzymatic activity is blocked in primary human keratinocytes and that this block is released upon treatment of primary keratinocytes with the proteasome inhibitor MG115.
Differential expression of XIAP in primary versus transformed keratinocytes.
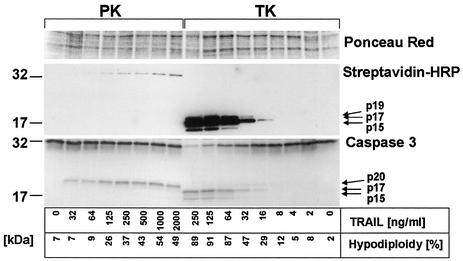
In order to further investigate the interplay between caspase 3 maturation and TRAIL receptor triggering, we compared two parameters of TRAIL-induced apoptosis in primary versus transformed keratinocytes: the dose-dependent cleavage pattern of caspase 3 and induction of apoptosis as determined by hypodiploidy analysis (Fig. 7). In line with our previous findings (42), low concentrations of TRAIL were sufficient to induce significant apoptosis in transformed keratinocytes, whereas primary keratinocytes were dose-dependently protected but nonetheless succumbed to high concentrations of TRAIL (Fig. 7). In contrast to the findings in primary keratinocytes (Fig. 7, lower panel, left lanes), the p20 fragment of caspase 3 was hardly detectable in transformed keratinocytes, whereas the p17 and a p15 fragment of caspase 3 were readily detected in these cells when stimulated with low concentrations of TRAIL (Fig. 7, lower panel, right lanes). Full cleavage correlated with detectable activity of effector caspases, as examined by affinity labeling (Fig. 7, upper panel). Interestingly, high concentrations of TRAIL sufficient to induce partial apoptosis led to labeling of full-length caspase 3 in primary keratinocytes (Fig. 7, upper panel).
FIG. 7.
Caspase 3 activation pattern in primary and transformed keratinocytes. (A) DEVD-specific caspase activity in cellular lysates was analyzed 3 h after TRAIL stimulation in primary keratinocytes (PK) and transformed HaCaT keratinocytes (TK). DEVDase activity of p19, p17, and p15 (upper panel) was detectable in transformed keratinocytes treated with TRAIL, whereas the same concentrations of TRAIL did not induce detectable DEVDase activity of cleaved fragments of caspase 3 in primary keratinocytes. Induction of apoptosis was monitored in parallel plates by hypodiploidy analysis. Higher concentrations of TRAIL sufficient to induce partial cell death in primary keratinocytes led to a dose-dependent increase in full-length caspase 3 labeling, indicative of some enzymatic activity of full-length caspase 3. Subsequent reprobing of blots with caspase 3-specific antibody confirmed that low concentrations of TRAIL were sufficient to induce the slower-migrating caspase 3 form (p20) in primary keratinocytes. In contrast, mainly the mature cleaved forms p17 and a smaller fragment (p15) were detected in transformed keratinocytes, even at low concentrations of TRAIL. Ponceau Red staining of the membrane confirmed comparable loading of proteins. HRP, horseradish peroxidase.
Taken together, these data pointed to a potential inhibitor of effector caspases blocking the full maturation of cleaved caspase 3 in primary keratinocytes. XIAP, the most potent caspase inhibitor of the IAP family, has been shown to selectively inhibit the cleaved forms of caspase 3 and 7 (14). We therefore examined the expression level of XIAP in primary and transformed keratinocytes. Intriguingly, Western blot analysis revealed that the 57-kDa XIAP protein was present in primary but not in transformed keratinocytes (Fig. 8A, lysate). Furthermore, a 29-kDa fragment of XIAP generated by caspase-mediated cleavage (15) was only detected in TRAIL-treated primary keratinocytes. These data suggest that XIAP may act as an inhibitor of TRAIL-induced apoptosis in primary keratinocytes by blocking the full cleavage and, thus, the activity of caspase 3.
FIG. 8.
XIAP confers resistance to TRAIL-induced apoptosis in primary keratinocytes. (A) XIAP is expressed in primary keratinocytes (PK) but not in transformed keratinocytes (TK) and can be coimmunoprecipitated under native conditions with caspase 3 following TRAIL treatment. Total cellular lysates from primary keratinocytes and transformed keratinocytes were prepared, and equal amounts of protein were either analyzed by Western blotting (25 μg) with XIAP monoclonal antibodies (lysate, left and right side) or subjected to immunoprecipitation (500 μg). A 57-kDa protein representing full-length XIAP was detected in primary (right) but not transformed (left) keratinocytes. An additional band of roughly 65 kDa (asterisks) detected in total lysates was shown to be a nonspecific band (immunoprecipitation: XIAP, right lanes). Following immunoprecipitation with caspase 3 antiserum, full-length but not cleaved XIAP was detected only in immunoprecipitates of TRAIL-treated primary keratinocytes PK (immunoprecipitation: caspase 3). (B) Transfection of transformed keratinocytes with XIAP leads to decreased cleavage of full-length caspase 3 and increased detection of caspase 3 p20. Transformed keratinocytes were transiently transfected with 3 μg of pEBB-XIAP vector or pEBB control vector as indicated in Materials and Methods. Then 125 ng of TRAIL per ml was added to the six-well plates, and cellular lysates were collected 3 h later. Primary keratinocytes were treated in parallel plates. The same membrane was first incubated with monoclonal antibodies to XIAP and subsequently reprobed with caspase 3 antibodies. Rehybridization with antitubulin monoclonal antibodies confirmed comparable loading of cellular proteins. Shown is a representative experiment of a total of three independent experiments. (C) XIAP overexpression leads to relative resistance of transformed keratinocytes to TRAIL-induced apoptosis. Cells were transfected as described for B and treated with the indicated concentrations of TRAIL, and viability was determined 6 h later with annexin/propidium iodide staining. Shown are means and standard deviations for two independent experiments. (D) Primary keratinocytes were incubated for 60 min with 10 μM MG115 (right part) or diluent alone (left part) and subsequently treated with 50 ng of TRAIL per ml for the indicated time periods. TRAIL treatment resulted in cleavage of XIAP to a 29-kDa fragment (p29), with similar cleavage at early time points in the presence or absence of the proteasome inhibitor. No cleavage fragments were detected in the presence of zVAD-FMK or after incubation with MG115 alone for 4 h, at which time XIAP levels were comparable to those found under control conditions.
In order to investigate further the interaction of caspase 3 with XIAP, immunoprecipitation studies were performed. While antibodies to XIAP readily precipitated XIAP in untreated as well as TRAIL-treated primary keratinocytes, coimmunoprecipitation of XIAP with caspase 3 was only detected following TRAIL treatment (Fig. 8A). These data demonstrate the interaction of caspase 3 with XIAP in TRAIL-treated primary keratinocytes under physiological conditions.
In order to address the question of whether the lack of XIAP in transformed keratinocytes contributes to the high TRAIL sensitivity of these cells, transient-transfection studies were performed. Following overexpression of XIAP in transformed keratinocytes (Fig. 8B, middle lanes) resistance to TRAIL-induced apoptosis was increased (Fig. 8C). Furthermore, increased levels of caspase 3 p32 and p20 were detected following TRAIL treatment in XIAP-overexpressing cells. Interestingly, high levels of overexpressed XIAP (Fig. 8B, upper panel) did not completely block TRAIL-induced caspase 3 maturation compared to primary keratinocytes (Fig. 8B, lower panel). Because the transfection efficiency in these experiments was between 50 and 70%, the remaining detectable fully cleaved caspase 3 (Fig. 8B) as well as the partial apoptosis protection in functional assays (Fig. 8C) might, at least in part, be explained by the percentage of cells without efficient XIAP expression. However, we cannot exclude that additional differences in the inhibition of effector caspase activation exist between primary and transformed keratinocytes. Future studies are required to delineate this point. Taken together, our data demonstrate that the lack of XIAP contributes to the higher sensitivity of transformed keratinocytes to TRAIL.
We next investigated whether the expression level or cleavage pattern of XIAP following TRAIL treatment of primary keratinocytes is modulated by MG115. We therefore examined XIAP levels during TRAIL treatment in the presence and absence of MG115. Limited cleavage of XIAP to a 29-kDa fragment was detected in the presence and absence of MG115. However, a slight increase in cleaved XIAP was detected at later time points in MG115-treated cells (Fig. 8D). This increase correlated with full caspase 3 processing (compare Fig. 6A), suggesting that XIAP cleavage is a secondary event mediated by active caspase 3. Importantly, MG115 treatment alone did not significantly change XIAP levels (Fig. 8D). These data provide evidence that the mechanism of MG115-induced increase of TRAIL sensitivity does not involve cleavage of XIAP or modulation of XIAP expression but rather direct inhibition of caspase 3 by XIAP.
MG115 sensitizes primary keratinocytes to TRAIL-induced apoptosis upstream of or at the mitochondrial level.
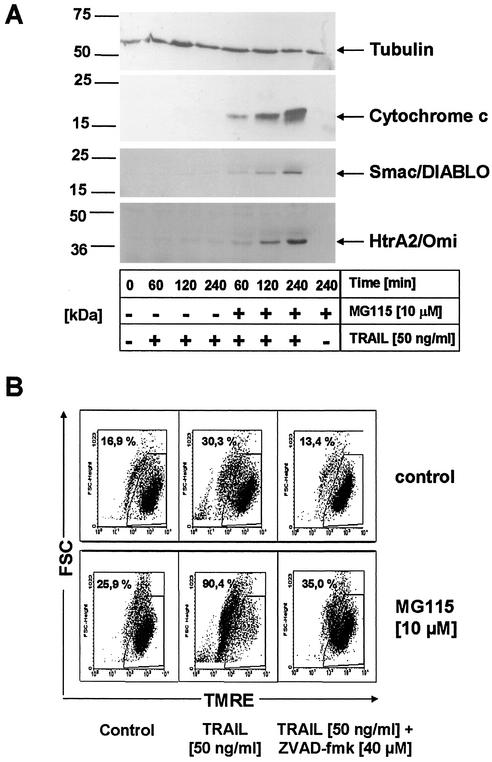
Apoptotic activation of the mitochondrion results in the release of proteins such as cytochrome c, Smac/DIABLO, and HtrA2/Omi from the intermitochondrial membrane space (49, 73). Smac/DIABLO and HtrA2/Omi release from mitochondria in turn leads to caspase activation by inhibiting the antiapoptotic function of several IAPs, particularly XIAP (8, 17, 18, 45, 74). Our finding that XIAP is highly expressed in primary keratinocytes prompted us to investigate whether proteasome inhibition may allow TRAIL-induced release of Smac/DIABLO or HtrA2/Omi from mitochondria, resulting in XIAP inhibition.
We therefore examined the kinetics of the release of mitochondrial proteins into the cytosol during MG115-induced sensitization of primary keratinocytes to TRAIL. In cells treated with TRAIL alone, release of cytochrome c, Smac/DIABLO, or HtrA2/Omi could not be detected (Fig. 9A). In contrast, in cells treated with TRAIL in the presence of MG115, cytochrome c, Smac/DIABLO, and HtrA2/Omi were rapidly released from the mitochondria, whereas exposure to MG115 alone had no effect (Fig. 9A). Consistently, the quantitative disruption of the inner mitochondrial membrane potential (Δψm) which represents an important “point of no return” of apoptosis (37), was detected in TRAIL-treated primary keratinocytes in the presence but not in the absence of MG115 (Fig. 9B). Taken together, these data suggest that proteasome inhibition sensitizes primary keratinocytes for TRAIL-induced apoptosis by disabling the XIAP-mediated inhibition of autoproteolytic maturation of caspase 3 by allowing Smac/DIABLO and HtrA2/Omi release from the mitochondria and, thus, inhibition of XIAP.
FIG. 9.
MG115 sensitizes primary keratinocytes upstream or at the mitochondrial level. (A) Cytochrome c, Smac/DIABLO, and HtrA2/Omi release after TRAIL treatment of primary keratinocytes. Cells were treated for the indicated time intervals with 50 ng of TRAIL per ml in the presence or absence of 10 μM MG115. Cytoplasmic lysates were subsequently prepared as described in Materials and Methods and analyzed for cytochrome c, Smac/DIABLO, and HtrA2/Omi by Western blot analysis. Equal loading of cytoplasmic proteins was determined by reprobing of blots with antitubulin antibodies. (B) TRAIL-induced loss of mitochondrial transmembrane potential ΔΨm requires the presence of the proteasome inhibitor MG115. Cells were incubated for 60 min with either diluent alone or 10 μM MG115, followed by treatment with 50 ng of TRAIL per ml. Cells were analyzed for ΔΨm by staining with 40 nM tetramethylrhodamine ethyl ester 4 h later. Quantitative disruption of ΔΨm was only seen in cells treated with TRAIL in the presence of the proteasome inhibitor MG115.
DISCUSSION
Primary keratinocytes are resistant to TRAIL concentrations that induce apoptosis in transformed keratinocytes but nonetheless succumb to high levels of TRAIL (42). This observation led us to further explore intracellular signaling pathways in primary keratinocytes which might modulate sensitivity to death ligands such as TRAIL (26, 27, 76). One important step for the regulation of such signaling pathways is the programmed degradation of specific proteins via the proteasome (16). Our initial finding that proteasome inhibition dramatically sensitized primary cells to TRAIL led us to examine a prime candidate for the regulation of TRAIL sensitivity, the transcription factor NF-κB. Several reports have suggested that TNFα-mediated apoptosis in particular is regulated by a group of genes induced by NF-κB, including cIAP-1, cIAP-2, TRAF-1, and TRAF-2 (83, 87). Regarding TRAIL, conflicting data have been published (26, 27), suggesting that for this death ligand, cell type-specific differences may exist. Here we show that TRAIL induced a moderate but reproducible activation of NF-κB-specific DNA binding and Iκβα degradation under nonoverexpression conditions of TRAIL-R proteins. Our data are in line with recent reports utilizing 293 and HeLa cell lines (23, 27, 76). However, specific blockade of NF-κB by ectopically expressed inhibitors of NF-κB activation (i.e., IKK2 or Iκβα) failed to sensitize keratinocytes to TRAIL. These data clearly indicate that other signaling pathways modulated by proteasome function play a more important role for sensitivity to TRAIL, at least in keratinocytes.
TRAIL-induced signaling pathways have thus far been studied mainly in cell lines that are highly sensitive to TRAIL or in cell lines that are resistant to receptor ligation due to artificial overexpression of intracellular inhibitors such as cFLIP (31). In order to study the possible involvement of TRAIL DISC formation in the mechanism of TRAIL resistance of primary keratinocytes, we first had to elucidate the composition of the native TRAIL DISC in these cells. We determined that the TRAIL DISC in primary keratinocytes contains FADD, caspase 8, caspase 10, the second initiator caspase recently shown to be recruited to the native TRAIL DISC in several cell lines (35, 67, 84), cFLIPL, and cFLIPS. Earlier overexpression studies have demonstrated that cFLIPL is capable of inhibiting TRAIL-induced apoptosis in keratinocytes (42) and melanoma cells (22). Detailed comparative and kinetic studies of the proteolytic activity of the TRAIL DISC are required to determine the exact contribution of all DISC components in these cells. Nonetheless, our studies clearly showed that in primary keratinocytes, a cell type that expresses TRAIL-R1 and TRAIL-R2 at high levels, inhibition of the proteasome did not modulate the recruitment or cleavage of FADD, cFLIP, and caspase 8 or 10 at the DISC, suggesting that the proteasome inhibitor MG115 sensitizes primary keratinocytes downstream of the TRAIL DISC. Intriguingly, active initiator caspases can be detected in TRAIL-treated primary keratinocytes without significant apoptosis. Cleavage of caspase 3 to the 20-kDa fragment is found at similar levels in cells treated with TRAIL in the absence versus the presence of proteasome inhibitors. These data further support the notion that caspase 8 is sufficiently activated to achieve initial cleavage of caspase 3. Thus, our data indicate that besides the activity of the TRAIL DISC, additional intracellular regulatory mechanisms exist. These mechanisms control TRAIL-induced effector caspase activity before a “point of no return” is passed, at which apoptosis induction is inevitable. Importantly, in primary cells, as exemplified here for primary keratinocytes, low levels of initiator caspase activation might be tolerated without induction of apoptosis, thereby activating nonapoptotic signaling pathways.
To date, little is known about the signaling events associated with TRAIL receptor ligation downstream of the DISC. In particular, the role of mitochondrial signaling pathways remains to be elucidated for TRAIL-induced apoptosis. In some cell lines examined, TRAIL-induced apoptosis was shown to be largely independent of mitochondrial signaling pathways (32, 77). In other studies, Bcl-xL protected pancreatic carcinoma cells from TRAIL-induced apoptosis (24), and inactivation of Bax was recently shown to confer resistance to TRAIL-induced apoptosis in colon cancer lines (41). Taken together, these data support the view that cell type-specific differences downstream of active initiator caspases might be important for the cell fate decision following stimulation of TRAIL receptors.
Our studies show that proteasome inhibitors induce major and early differences in the TRAIL-dependent release of cytochrome c, Smac/DIABLO, and HtrA2 (64) despite similar early activation of initiator caspases. Importantly, the proteasome inhibitor alone had no effect on mitochondrial cytochrome c, Smac/DIABLO, or HtrA2 release which is in contrast to findings obtained in transformed cell lines (75). This difference might be due to a variable sensitivity of tumor cells to proteasome inhibitors compared to primary cells (50). Our data suggest that in primary keratinocytes, activation of the mitochondrial apoptotic pathway is required for the execution of TRAIL-induced apoptosis and that proteasome inhibitors enable the release of proapoptotic molecules such as Smac/DIABLO, cytochrome c, and HtrA2/Omi from mitochondria in response to TRAIL treatment.
Our data showing partial cleavage of caspase 3 in primary keratinocytes are consistent with recent reports demonstrating that TRAIL-induced activation of Smac/DIABLO release is dependent on Bax and Bak, promoted by Bid, and inhibited by Bcl-2 and Bcl-xL downstream of caspase 8 activation in different tumor cell lines (12, 69). In line with these reports, the release of Smac/DIABLO, HtrA2/Omi, and cytochrome c in the presence of the proteasome inhibitor correlated with full maturation of caspase 3 to the 17-kDa fragment in primary keratinocytes. Interestingly, analysis of several Bcl-2 family members, including Bcl-XL, Bax, and Bid, did not reveal changes in the expression levels or cleavage patterns of these molecules at time points when full caspase 3 maturation and release of mitochondrial proapoptotic proteins is already detected in primary keratinocytes (data not shown). Thus, it remains to be determined if modulation of the expression levels of Bcl-2 family members and their translocation to mitochondria or other potential targets are modulated by proteasome inhibitors.
Moreover, it is possible that cytochrome c-mediated activation of the apoptosome (5, 7) might also be involved in increased caspase 3 activation. However, our data clearly show that synergistic action of mitochondrion-derived proapoptotic molecules in concert with death receptor-mediated caspase 3 activation is ultimately necessary and sufficient to overcome inhibitory molecules such as XIAP in primary keratinocytes. In support of this concept, recent in vitro experiments demonstrated that an increase in Smac/DIABLO levels can overcome XIAP-mediated inhibition of caspase 3 maturation (69). Of interest, the removal of the prodomain of the effector caspase 6 is required for full enzymatic activity, further supporting our findings for caspase 3 (10).
When comparing the dose-dependent cleavage patterns of TRAIL-resistant primary keratinocytes and highly sensitive transformed keratinocytes, low concentrations of TRAIL were sufficient to induce fully active p17 fragments in transformed cells but not in primary cells, indicating that primary keratinocytes require an additional signal for full caspase 3 maturation. As shown in this study, one explanation for this difference is the differential expression of XIAP, which inhibits the autoproteolytic maturation of caspase 3 (14, 15). Primary keratinocytes might require the release of Smac/DIABLO and HtrA2/Omi (64) from the mitochondria for inactivation of XIAP. Thus, the stoichiometry of DISC-generated active caspase 8, cytoplasmic XIAP, and mitochondrion-released Smac/DIABLO or HtrA2/Omi might define resistance or sensitivity to TRAIL-induced apoptosis in primary keratinocytes and possibly also in other cells, as suggested recently (69).
Remarkably, there may be important cell type-specific differences in the action of proteasome inhibitors, as primary human hepatocytes are not sensitized to TRAIL by these agents (T. Ganten and H. Walczak, unpublished data). These differences might be explained by differential expression of TRAIL receptors, distinct DISC components, or downstream inhibitory molecules, as described in this report. Our data demonstrating interaction of XIAP with caspase 3 under physiological conditions as well as increased resistance to TRAIL-induced apoptosis following ectopic expression of XIAP in highly TRAIL-sensitive transformed keratinocytes further support the important role of XIAP in TRAIL resistance in primary keratinocytes.
Taken together, our data support a model in which proteasome inhibitors allow initiator caspase-mediated activation of mitochondrial pathways of apoptosis in primary cells. The activation of mitochondrial pathways is necessary for inactivation of XIAP and, consequently, of full caspase 3 activity in primary keratinocytes (Fig. 10). Alternatively, XIAP might exert its caspase-inhibitory role by its ubiquitin-protein ligase activity (70). Since polyubiquitination labels proteins for proteasomal degradation, proteasome inhibition may thereby lead to increased levels of active caspase 3 and subsequent apoptotic cell death in cells expressing high levels of XIAP. This mechanism might not be required in cells expressing low levels of XIAP, as supported by our overexpression studies. However, another possibility of the action of the proteasomal inhibitor may be decreased XIAP-mediated degradation of Smac/DIABLO, as suggested by a very recent report (47). Future studies are required to delineate these points.
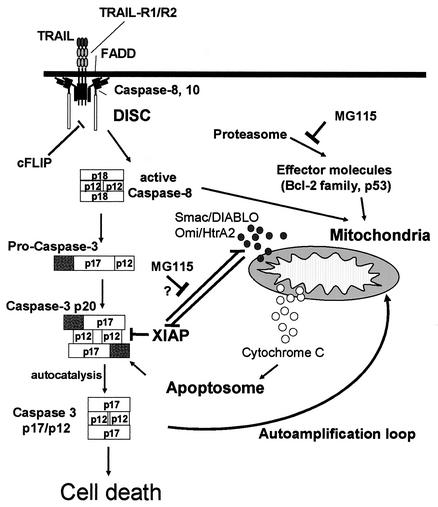
FIG. 10.
Model of action of proteasome inhibitors mediating sensitization of primary keratinocytes to TRAIL. TRAIL activates caspases 8 and 10 at the DISC. Initial processing of caspase 3 produces a p20/p12 intermediate of caspase 3 which is inhibited by XIAP. Proteasome inhibitors interfere with caspase 8-mediated activation of mitochondrial pathways of apoptosis, leading to the caspase 8-activated release of Smac/DIABLO and HtrA2/Omi and subsequent removal of the caspase 3 prodomain (hatched bars). This activation of mitochondrial pathways is necessary for inactivation of XIAP and subsequent full caspase 3 activity in primary keratinocytes.
It is possible that the resistance mechanism of primary human keratinocytes outlined in this study is also relevant for other types of primary cells and could thus be a more general mechanism of resistance of primary cells to TRAIL. In that respect, our findings suggest that the use of proteasome inhibitors as anticancer agents might lead to unwanted sensitization of primary cells to death receptor-mediated apoptosis. Thus, the clinical application of proteasome inhibitors, either alone or in combination with death receptor agonists such as TRAIL, must be closely monitored for unwanted toxicity that could be due to the apoptosis-sensitizing effect exerted by these agents on primary cells as identified in this study.
Acknowledgments
We thank P. H. Krammer for monoclonal antibodies to caspase 8 (C-15) and cFLIP (NF-6), D. Nicholson for caspase 3 and XIAP antisera as well as biotin-DEVD-aomk, M. MacFarlane for rabbit serum to Smac/DIABLO, X. Wang for rabbit serum to Bid, R. Takahashi for rabbit serum to HtrA2/Omi, S. Roy for the protocol to perform affinity labeling experiments, and C. S. Duckett for the XIAP expression vector. We are also grateful to Evi Horn for excellent technical assistance and Heiko Stahl for generating monoclonal antibodies to TRAIL-R1 to -R4.
Part of this study was funded by grants from the IZKF Würzburg (01 K5 9603), Sander-Stiftung (2000.092.1), and Deutsche Krebshilfe (10-1951-Le1) to Martin Leverkus. Henning Walczak is supported by a BioFuture grant from the Bundesministerium für Bildung und Forschung (BMBF). Manfred Neumann and Bernd Baumann were supported by grants from the Deutsche Forschungsgemeinschaft (NE 608/1-3 and/2-2 to M.N. and SFB 497/B1 to B.B.).
The first two authors contributed equally to this paper.
The last two authors share senior authorship of this paper.
Footnotes
This article is dedicated to Harald zur Hansen on the occasion of his retirement as head of the German Cancer Research Center (Deutsches Krebs forschungszentrum) in Heidelberg with gratitude and appreciation for 20 years of leadership.
REFERENCES
- 1.Ashkenazi, A., R. C. Pai, S. Fong, S. Leung, D. A. Lawrence, S. A. Marsters, C. Blackie, L. Chang, A. E. McMurtrey, A. Hebert, L. DeForge, I. L. Koumenis, D. Lewis, L. Harris, J. Bussiere, H. Koeppen, Z. Shahrokh, and R. H. Schwall. 1999. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 104:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer, J. L., N. Holler, S. Reynard, P. Vinciguerra, P. Schneider, P. Juo, J. Blenis, and J. Tschopp. 2000. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2:241-243. [DOI] [PubMed] [Google Scholar]
- 4.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratton, S. B., G. Walker, S. M. Srinivasula, X. M. Sun, M. Butterworth, E. S. Alnemri, and G. M. Cohen. 2001. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 20:998-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briegel, K., B. Hentsch, I. Pfeuffer, and E. Serfling. 1991. One base pair change abolishes the T cell-restricted activity of a κB-like proto-enhancer element from the interleukin 2 promoter. Nucleic Acids Res. 19:5929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cain, K., D. G. Brown, C. Langlais, and G. M. Cohen. 1999. Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J. Biol. Chem. 274:22686-22692. [DOI] [PubMed] [Google Scholar]
- 8.Chai, J., C. Du, J. W. Wu, S. Kyin, X. Wang, and Y. Shi. 2000. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406:855-862. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F., D. Chang, M. Goh, S. A. Klibanov, and M. Ljungman. 2000. Role of p53 in cell cycle regulation and apoptosis following exposure to proteasome inhibitors. Cell Growth Differ. 11:239-246. [PubMed] [Google Scholar]
- 10.Cowling, V., and J. Downward. 2002. Caspase 6 is the direct activator of caspase 8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase 6 prodomain. Cell Death Differ. 9:1046-1056. [DOI] [PubMed] [Google Scholar]
- 11.Degli-Esposti, M. A., W. C. Dougall, P. J. Smolak, J. Y. Waugh, C. A. Smith, and R. G. Goodwin. 1997. The novel receptor TRAIL-R4 induces NF-κB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 7:813-820. [DOI] [PubMed] [Google Scholar]
- 12.Deng, Y., Y. Lin, and X. Wu. 2002. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 16:33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denk, A., M. Goebeler, S. Schmid, I. Berberich, O. Ritz, D. Lindemann, S. Ludwig, and T. Wirth. 2001. Activation of NF-κB via the IκB kinase complex is both essential and sufficient for proinflammatory gene expression in primary endothelial cells. J. Biol. Chem. 276:28451-28458. [DOI] [PubMed] [Google Scholar]
- 14.Deveraux, Q. L., and J. C. Reed. 1999. IAP family proteins-suppressors of apoptosis. Genes Dev. 13:239-252. [DOI] [PubMed] [Google Scholar]
- 15.Deveraux, Q. L., R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388:300-304. [DOI] [PubMed] [Google Scholar]
- 16.Drexler, H. C. 1997. Activation of the cell death program by inhibition of proteasome function. Proc. Natl. Acad. Sci. USA 94:855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 18.Ekert, P. G., J. Silke, C. J. Hawkins, A. M. Verhagen, and D. L. Vaux. 2001. DIABLO promotes apoptosis by removing MIHA/XIAP from processed caspase 9. J. Cell Biol. 152:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 20.Gliniak, B., and T. Le. 1999. Tumor necrosis factor-related apoptosis-inducing ligand's antitumor activity in vivo is enhanced by the chemotherapeutic agent CPT-11. Cancer Res. 59:6153-6158. [PubMed] [Google Scholar]
- 21.Goldstein, J. C., N. J. Waterhouse, P. Juin, G. I. Evan, and D. R. Green. 2000. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2:156-162. [DOI] [PubMed] [Google Scholar]
- 22.Griffith, T. S., W. A. Chin, G. C. Jackson, D. H. Lynch, and M. Z. Kubin. 1998. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J. Immunol. 161:2833-2840. [PubMed] [Google Scholar]
- 23.Harper, N., S. N. Farrow, A. Kaptein, G. M. Cohen, and M. MacFarlane. 2001. Modulation of TRAIL-induced NF-κB activation by inhibition of apical caspases. J. Biol. Chem. 276:34743-34752. [DOI] [PubMed] [Google Scholar]
- 24.Hinz, S., A. Trauzold, L. Boenicke, C. Sandberg, S. Beckmann, E. Bayer, H. Walczak, H. Kalthoff, and H. Ungefroren. 2000. Bcl-XL protects pancreatic adenocarcinoma cells against CD95- and TRAIL-receptor-mediated apoptosis. Oncogene 19:5477-5486. [DOI] [PubMed] [Google Scholar]
- 25.Holcik, M., and R. G. Korneluk. 2001. XIAP, the guardian angel. Nat. Rev. Mol. Cell. Biol. 2:550-556. [DOI] [PubMed] [Google Scholar]
- 26.Hu, W. H., H. Johnson, and H. B. Shu. 1999. Tumor necrosis factor-related apoptosis-inducing ligand receptors signal NF-κB and JNK activation and apoptosis through distinct pathways. J. Biol. Chem. 274:30603-30610. [DOI] [PubMed] [Google Scholar]
- 27.Jeremias, I., C. Kupatt, B. Baumann, I. Herr, T. Wirth, and K. M. Debatin. 1998. Inhibition of nuclear factor κB activation attenuates apoptosis resistance in lymphoid cells. Blood 91:4624-4631. [PubMed] [Google Scholar]
- 28.Jesenberger, V., and S. Jentsch. 2002. Deadly encounter: ubiquitin meets apoptosis. Nat. Rev. Mol. Cell. Biol. 3:112-121. [DOI] [PubMed] [Google Scholar]
- 29.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Ann. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 30.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 31.Kataoka, T., R. C. Budd, N. Holler, M. Thome, F. Martinon, M. Irmler, K. Burns, M. Hahne, N. Kennedy, M. Kovacsovics, and J. Tschopp. 2000. The caspase-8 inhibitor FLIP promotes activation of NF-κB and Erk signaling pathways. Curr. Biol. 10:640-648. [DOI] [PubMed] [Google Scholar]
- 32.Keogh, S. A., H. Walczak, L. Bouchier-Hayes, and S. J. Martin. 2000. Failure of Bcl-2 to block cytochrome c redistribution during TRAIL-induced apoptosis. FEBS Lett. 471:93-98. [DOI] [PubMed] [Google Scholar]
- 33.Kischkel, F. C., S. Hellbardt, I. Behrmann, M. Germer, M. Pawlita, P. H. Krammer, and M. E. Peter. 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kischkel, F. C., D. A. Lawrence, A. Chuntharapai, P. Schow, K. J. Kim, and A. Ashkenazi. 2000. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 12:611-620. [DOI] [PubMed] [Google Scholar]
- 35.Kischkel, F. C., D. A. Lawrence, A. Tinel, H. LeBlanc, A. Virmani, P. Schow, A. Gazdar, J. Blenis, D. Arnott, and A. Ashkenazi. 2001. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J. Biol. Chem. 276:46639-46646. [DOI] [PubMed] [Google Scholar]
- 36.Koepp, D. M., J. W. Harper, and S. J. Elledge. 1999. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97:431-434. [DOI] [PubMed] [Google Scholar]
- 37.Kroemer, G., and J. C. Reed. 2000. Mitochondrial control of cell death. Nat. Med. 6:513-951. [DOI] [PubMed] [Google Scholar]
- 38.Krueger, A., I. Schmitz, S. Baumann, P. H. Krammer, and S. Kirchhoff. 2001. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 276:20633-20640. [DOI] [PubMed] [Google Scholar]
- 39.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 40.Lazebnik, Y. A., S. H. Kaufmann, S. Desnoyers, G. G. Poirier, and W. C. Earnshaw. 1994. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371:346-347. [DOI] [PubMed] [Google Scholar]
- 41.LeBlanc, H., D. A. Lawrence, E. Varfolomeev, K. Totpal, J. Morlan, P. Schow, S. Fong, R. Schwall, D. Sinicropi, and A. Ashkenazi. 2002. Tumor-cell resistance to death receptor-induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat. Med. 8:274-281. [DOI] [PubMed] [Google Scholar]
- 42.Leverkus, M., M. Neumann, T. Mengling, C. T. Rauch, E. B. Bröcker, P. H. Krammer, and H. Walczak. 2000. Regulation of TRAIL sensitivity in primary and transformed human keratinocytes. Cancer Res. 60:553-559. [PubMed] [Google Scholar]
- 43.Leverkus, M., H. Walczak, H.-W. Fries, G. Terbeck, A. McLellan, E.-B. Bröcker, and E. Kämpgen. 2000. Maturation of dendritic cells leads to upregulation of cellular FLICE-inhibitory protein (cFLIP) and concomitant downregulation of death receptor mediated apoptosis. Blood 110:353-357. [PubMed] [Google Scholar]
- 44.Leverkus, M., M. Yaar, and B. A. Gilchrest. 1997. Fas/Fas ligand interaction contributes to UV-induced apoptosis in human keratinocytes. Exp. Cell Res. 232:255-262. [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z., C. Sun, E. T. Olejniczak, R. P. Meadows, S. F. Betz, T. Oost, J. Herrmann, J. C. Wu, and S. W. Fesik. 2000. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 408:1004-1008. [DOI] [PubMed] [Google Scholar]
- 46.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The tumor necrosis factor and tumor necrosis factor receptor superfamilies: integrating mammalian biology. Cell 104:487-501. [DOI] [PubMed] [Google Scholar]
- 47.MacFarlane, M., W. Merrison, S. B. Bratton, and G. M. Cohen. 2002. Proteasome-mediated degradation of Smac during apoptosis:XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 277:36611-36616. [DOI] [PubMed] [Google Scholar]
- 48.Marienfeld, R., F. Berberich-Siebelt, I. Berberich, A. Denk, E. Serfling, and M. Neumann. 2001. Signal-specific and phosphorylation-dependent RelB degradation: a potential mechanism of NF-κB control. Oncogene 20:8142-8147. [DOI] [PubMed] [Google Scholar]
- 49.Martins, L. M. 2002. The serine protease Omi/HtrA2: a second mammalian protein with a Reaper-like function. Cell Death Differ. 9:699-701. [DOI] [PubMed] [Google Scholar]
- 50.Masdehors, P., H. Merle-Beral, H. Magdelenat, and J. Delic. 2000. Ubiquitin-proteasome system and increased sensitivity of B-CLL lymphocytes to apoptotic death activation. Leukemia Lymphoma 38:499-504. [DOI] [PubMed] [Google Scholar]
- 51.Medema, J. P., C. Scaffidi, P. H. Krammer, and M. E. Peter. 1998. Bcl-xL acts downstream of caspase-8 activation by the CD95 death-inducing signaling complex. J. Biol. Chem. 273:3388-3393. [DOI] [PubMed] [Google Scholar]
- 52.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neumann, M., T. Grieshammer, S. Chuvpilo, B. Kneitz, M. Lohoff, A. Schimpl, B. R. Franza, Jr., and E. Serfling. 1995. RelA/p65 is a molecular target for the immunosuppressive action of protein kinase A. EMBO J. 14:1991-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicholson, D. W. 1999. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6:1028-1042. [DOI] [PubMed] [Google Scholar]
- 55.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 56.Pitti, R. M., S. A. Marsters, S. Ruppert, C. J. Donahue, A. Moore, and A. Ashkenazi. 1996. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 271:12687-12690. [DOI] [PubMed] [Google Scholar]
- 57.Roy, S., C. I. Bayly, Y. Gareau, V. M. Houtzager, S. Kargman, S. L. C. Keen, K. Rowland, I. M. Seiden, N. A. Thornberry, and D. W. Nicholson. 2001. Maintenance of caspase-3 proenzyme dormancy by an intrinsic “safety catch” regulatory tripeptide. Proc. Natl. Acad. Sci. USA 98:6132-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, S., and D. W. Nicholson. 2000. Cross-talk in cell death signalling. J. Exp. Med. 192:F21-F25. [PMC free article] [PubMed] [Google Scholar]
- 59.Scaduto, R. C., Jr., and L. W. Grotyohann. 1999. Measurement of mitochondrial membrane potential with fluorescent rhodamine derivatives. Biophys. J. 76:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, K. M. Debatin, P. H. Krammer, and M. E. Peter. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scaffidi, C., I. Schmitz, P. H. Krammer, and M. E. Peter. 1999. The role of c-FLIP in modulation of CD95-induced apoptosis. J. Biol. Chem. 274:1541-1548. [DOI] [PubMed] [Google Scholar]
- 62.Schneider, P., M. Thome, K. Burns, J. L. Bodmer, K. Hofmann, T. Kataoka, N. Holler, and J. Tschopp. 1997. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-κB. Immunity 7:831-836. [DOI] [PubMed] [Google Scholar]
- 63.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ′mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 65.Silke, J., P. G. Ekert, C. L. Day, C. J. Hawkins, M. Baca, J. Chew, M. Pakusch, A. M. Verhagen, and D. L. Vaux. 2001. Direct inhibition of caspase 3 is dispensable for the anti-apoptotic activity of XIAP. EMBO J. 20:3114-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Single, B., M. Leist, and P. Nicotera. 1998. Simultaneous release of adenylate kinase and cytochrome c in cell death. Cell Death Differ. 5:1001-1003. [DOI] [PubMed] [Google Scholar]
- 67.Sprick, M. R., E. Rieser, H. Stahl, A. Grosse-Wilde, M. A. Weigand, and H. Walczak. 2002. Caspase 10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner. EMBO J. 21:4520-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sprick, M. R., M. A. Weigand, E. Rieser, C. T. Rauch, P. Juo, J. Blenis, P. H. Krammer, and H. Walczak. 2000. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12:599-609. [DOI] [PubMed] [Google Scholar]
- 69.Sun, X. M., S. B. Bratton, M. Butterworth, M. MacFarlane, and G. M. Cohen. 2002. Bcl-2 and Bcl-xL inhibit CD95-mediated apoptosis by preventing mitochondrial release of Smac/DIABLO and subsequent inactivation of X-linked inhibitor-of-apoptosis protein. J. Biol. Chem. 277:11345-11351. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki, Y., Y. Nakabayashi, and R. Takahashi. 2001. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3. Proc. Natl. Acad. Sci. USA 98:8662-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson, C. B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267:1456-1562. [DOI] [PubMed] [Google Scholar]
- 72.Thornberry, N. A., E. P. Peterson, J. J. Zhao, A. D. Howard, P. R. Griffin, and K. T. Chapman. 1994. Inactivation of interleukin-1 beta converting enzyme by peptide (acyloxy)methyl ketones. Biochemistry 33:3934-3940. [DOI] [PubMed] [Google Scholar]
- 73.van Loo, G., X. Saelens, M. van Gurp, M. MacFarlane, S. J. Martin, and P. Vandenabeele. 2002. The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ. 9:1031-1042. [DOI] [PubMed] [Google Scholar]
- 74.Verhagen, A. M., P. G. Ekert, M. Pakusch, J. Silke, L. M. Connolly, G. E. Reid, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43-53. [DOI] [PubMed] [Google Scholar]
- 75.Wagenknecht, B., M. Hermisson, P. Groscurth, P. Liston, P. H. Krammer, and M. Weller. 2000. Proteasome inhibitor-induced apoptosis of glioma cells involves the processing of multiple caspases and cytochrome c release. J. Neurochem. 75:2288-2297. [DOI] [PubMed] [Google Scholar]
- 76.Wajant, H., E. Haas, R. Schwenzer, F. Muhlenbeck, S. Kreuz, G. Schubert, M. Grell, C. Smith, and P. Scheurich. 2000. Inhibition of death receptor-mediated gene induction by a cycloheximide-sensitive factor occurs at the level of or upstream of Fas-associated death domain protein (FADD). J. Biol. Chem. 275:24357-24366. [DOI] [PubMed] [Google Scholar]
- 77.Walczak, H., A. Bouchon, H. Stahl, and P. H. Krammer. 2000. Tumor necrosis factor-related apoptosis-inducing ligand retains its apoptosis-inducing capacity on Bcl-2- or Bcl-xL-overexpressing chemotherapy-resistant tumor cells. Cancer Res. 60:3051-3057. [PubMed] [Google Scholar]
- 78.Walczak, H., M. A. Degli-Esposti, R. S. Johnson, P. J. Smolak, J. Y. Waugh, N. Boiani, M. S. Timour, M. J. Gerhart, K. A. Schooley, C. A. Smith, R. G. Goodwin, and C. T. Rauch. 1997. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 16:5386-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walczak, H., and P. H. Krammer. 2000. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp. Cell Res. 256:58-66. [DOI] [PubMed] [Google Scholar]
- 80.Walczak, H., R. E. Miller, K. Ariail, B. Gliniak, T. S. Griffith, M. Kubin, W. Chin, J. Jones, A. Woodward, T. Le, C. Smith, P. Smolak, R. G. Goodwin, C. T. Rauch, J. C. Schuh, and D. H. Lynch. 1999. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5:157-163. [DOI] [PubMed] [Google Scholar]
- 81.Walczak, H., and M. R. Sprick. 2001. Biochemistry and function of the DISC. Trends Biochem. Sci. 26:452-453. [DOI] [PubMed] [Google Scholar]
- 82.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 83.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. Baldwin, Jr. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680-1683. [DOI] [PubMed] [Google Scholar]
- 84.Wang, J., H. J. Chun, W. Wong, D. M. Spencer, and M. J. Lenardo. 2001. Caspase-10 is an initiator caspase in death receptor signaling. Proc. Natl. Acad. Sci. USA 98:13884-13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wehrli, P., I. Viard, R. Bullani, J. Tschopp, and L. E. French. 2000. Death receptors in cutaneous biology and disease. J. Investig. Dermatol. 115:141-148. [DOI] [PubMed] [Google Scholar]
- 86.Wiley, S. R., K. Schooley, P. J. Smolak, W. S. Din, C. P. Huang, J. K. Nicholl, G. R. Sutherland, T. D. Smith, C. Rauch, C. A. Smith, and R. G. Goodwin. 1995. Identification and characterization of a new member of the tumor necrosis factor family that induces apoptosis. Immunity 3:673-682. [DOI] [PubMed] [Google Scholar]
- 87.Wu, M. X., Z. Ao, K. V. Prasad, R. Wu, and S. F. Schlossman. 1998. IEX-1L, an apoptosis inhibitor involved in NF-κB-mediated cell survival. Science 281:998-1001. [DOI] [PubMed] [Google Scholar]
- 88.Zhao, J., T. Tenev, L. M. Martins, J. Downward, and N. R. Lemoine. 2000. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J. Cell Sci. 113:4363-4371. [DOI] [PubMed] [Google Scholar]