Abstract
The growth hormone (GH)-GH receptor (GHR) axis modulates growth and metabolism and contributes to complications of diabetes mellitus. We analyzed the promoter region of the dominant transcript (L2) of the murine GHR to determine that a cis element, L2C1, interacts with transcription factors NF-Y, BTEB1, and HMG-Y/I. These proteins individually repress GHR expression and together form a repressosome complex in conjunction with mSin3b. The histone deacetylase inhibitor trichostatin A increases expression of the murine GHR gene, enhances association of acetyl-H3 at L2C1, inhibits formation of the repressosome complex, and decreases NF-Y's association with L2C1. Our studies reveal that murine models of experimental diabetes mellitus are characterized by reduced hepatic GHR expression, decreased acetyl-H3 associated with L2C1, and increased formation of the repressosome complex. In contrast, in the kidney diabetes mellitus is associated with enhanced GHR expression and lack of alteration in the assembly of the repressosome complex, thus permitting exposure of kidneys to the effects of elevated levels of GH in diabetes mellitus. Our findings define a higher-order repressosome complex whose formation correlates with the acetylation status of chromatin histone proteins. The delineation of the role of this repressosome complex in regulating tissue-specific expression of GHR in diabetes mellitus provides a molecular model for the role of GH in the genesis of certain microvascular complications of diabetes mellitus.
The growth hormone (GH)-insulin-like growth factor 1 (IGF-1) axis plays a critical permissive role in the pathogenesis of chronic microvascular complications of diabetes mellitus (DM). The absence of functional GH receptor (GHR) confers a protective effect against diabetic nephropathy and retinopathy in murine models of insulin-dependent DM (IDDM) (2, 29). For humans, acquired hypopituitarism ameliorated retinopathy and iatrogenic hypopituitarism was the standard care for proliferative retinopathy until the advent of laser coagulation therapy (24). In contrast there is resistance to GH's actions in promoting linear growth and IGF-1 generation in poorly controlled IDDM (30); animal and human studies indicate that decreased hepatic expression of GHR contributes to this resistance to GH's actions (1, 19, 21). However, the molecular mechanisms underlying the paradox of the liver becoming resistant but the kidney and retina retaining sensitivity to GH in DM is not known.
The GHR gene is characterized by sequence heterogeneity in the 5′ untranslated region (UTR) (8). For the mouse, three 5′ UTRs (L1, L2, and L5) have been characterized in some detail (20, 34, 36). The L2 transcript constitutes 50 to 80% of the hepatic GHR transcripts in the nonpregnant adult animal (8, 34). The 5′ flanking region of the L2 transcript exhibits promoter activity. A cis element, L2A, interacts with the Sp family of proteins and plays a role in the development-specific expression of the GHR gene (34). Recent studies have focused attention on the role of chromatin in modulating the transcriptional activity of genes (4). The present model for involvement of chromatin in gene activation or gene repression envisages the effects of a diverse array of posttranslational modifications that impinge on the histone amino terminus, thereby modulating access of transcription factors and other regulatory proteins to the underlying DNA. Whereas the role of chromatin structure and function in DNA template-dependent cellular processes such as transcription, DNA replication, DNA recombination, cell cycle progression, and epigenetic silencing is recognized, there is paucity of information regarding the significance of chromatin structure and function in specific disease states such as DM. Here we present evidence that a second cis element in the promoter of the murine GHR gene, which we designate L2C1, interacts with transcription factors NF-Y, HMG-Y/I, and BTEB1 and that these factors in conjunction with the corepressor mSin3b form a higher-order repressosome complex that decreases transcriptional activation of the mouse GHR. Our studies demonstrate novel in vivo associations between members of this quartet. We further demonstrate that alterations in the interaction of this repressosome complex with L2C1 play a role in the tissue-specific dysregulation of expression of the GHR gene in DM. To the best of our knowledge this is the first report demonstrating a direct correlation between the acetylation status of histone proteins and the transcriptional activity of a specific gene in DM.
MATERIALS AND METHODS
Oligonucleotides.
The following synthetic oligonucleotides (Invitrogen) were used in this study (residues altered in mutant oligonucleotides are indicated in lowercase): L2C1, 5′-TCTAGGAGGAGCCCCGCCGCCCAATTGAGAGCGACAC-3′; NF-Y consensus, v5′-ATTTTTCTGATTGGTTAAAAGT-3′; NF-Y mutant, 5′-GGCAGCCCCGCCGCaaccgTGAGAGCGACACGCACC-3′; BTEB1mutant, 5′-TTCCTCTAGGAGGAGCCatgataaCCAATTGAGAGCGACACGCACC-3′; NF-Y+BTEB1 mutant, 5′-TCCTCTAGGAGGAGCCatgataaaactgtGAGAGCGACACGCACC-3′; Tq-L2C1-forward, 5′-CCCGCAACTACCAATATTTTCC-3′; Tq-L2C1-reverse, 5′-GGTGCGTGTCGCTCTCAATT-3′; Tq-L2C1 probe, FAM5′-CTAGGAGGAGCCCCGCCGCC-3′TAMRA.
Where necessary, double-stranded oligonucleotides were generated by annealing the synthetic oligonucleotides with respective complementary sequences.
Plasmids.
Luciferase reporter constructs were generated as described previously (34). Deletion constructs were engineered by using a PCR-based strategy. Site-specific mutations in L2C1 were introduced by using QuikChange (Stratagene). pCDNA/BTEB1, dominant-negative (D/N) NF-Y, and high-mobility group of protein (HMG Y/I) constructs were kindly provided by H. Imataka (McGill University, Montreal, Canada), R. Mantovani (University of Milan, Milan, Italy), and A. Fusco (Universita degli Studi di Catanzaro, Catanzaro, Italy), respectively.
Cell culture, transient transfections, and luciferase assays.
BNL CL.2 cells (mouse liver; American Type Culture Collection, Manassas, Va.) were cultured and transiently transfected by using Lipofectamine as described previously (20). Three or four independent transfections were performed in triplicate, and transfection results are represented as means ± standard deviations (SD).
TSA treatment.
BNL CL.2 cells (106 cells/10 cm-diameter plate) were plated 16 h prior to exposure to trichostatin A (TSA) (100 ng/ml) or vehicle (alcohol) for 24 h.
Protein studies.
Nuclear extracts were prepared from cells by using the NE-PER kit (Pierce) or from mouse tissues as described previously (10). Coimmunoprecipitations and immunoblotting experiments were performed by using standard protocols.
Fluorescent 5′-nuclease RT-PCR.
Total RNA was extracted using TRI-reagent (Molecular Research Center). Real-time quantitative reverse-transcription (RT)-PCR using the ABI Prism 7700 sequence detection system (PE Biosystems) was performed and analyzed following protocols described previously (20).
DNase I footprinting.
The PCR-derived radiolabeled probe was incubated with nuclear extracts from adult mouse liver and processed for DNase I treatment, electrophoresis, and analysis as previously described (22). The exact locations of the DNase I footprints were determined by comparison to a concurrently electrophoresed sequence ladder.
EMSAs.
Electromobility shift assays (EMSAs) were performed with 2 μg of nuclear proteins by using standard protocols and samples resolved on a 4% nondenaturing polyacrylamide gel. For supershift experiments, anti-BTEB1 (kindly provided by H. Imataka, McGill University), anti-NFY-A, anti-Sin3b, or anti-HMG Y/I (Santa Cruz Biotechnology) antibodies were used. Recombinant HMG Y/I protein was a gift from D. Thanos (Columbia University, New York, N.Y.).
ChIP assay.
BNL CL.2 cells exposed to vehicle or TSA were subjected to chromatin immunoprecipitation assay (ChIP) using acetyl histone-H3 immunoprecipitation assay kit (Upstate Biotechnology) with minor modifications. Following un-cross-linking, DNA was extracted (QIAquick; Qiagen) and 1 μl of the sample was used in PCR with [α-32P]ATP in the reaction mixture to amplify L2C1. The PCR products were resolved on a 10% nondenaturing polyacrylamide gel.
For ChIP assay using intact organs, tissue samples from normal and diabetic mice were suspended in ice-cold phosphate-buffered saline, minced, washed, and homogenized. The homogenate was centrifuged, and the pellet was resuspended in cell dissociation buffer (Invitrogen) and incubated for 15 min at room temperature. The cells were resuspended in culture medium, and proteins were cross-linked for 10 min at room temperature and processed for ChIP.
Murine models of DM.
Both spontaneous (NOD) and chemically induced (streptozocin [STZ]) murine models of experimental diabetes were used for these studies. All experiments were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Children's Hospital of Pittsburgh. The animals were allowed free access to food (Purina rodent diet) and water, and their glycemic status was monitored as previously described (20). Hyperglycemia was defined as glucose values of >300 mg/dl on three consecutive occasions. Female NOD mice (Jackson Laboratory) were maintained in a pathogen-free environment. The blood sugar concentrations of the nondiabetic NOD (n = 4) and diabetic NOD (n = 6) mice at the time of sacrifice were 103 ± 16 and 482 ± 65 mg/dl (mean ± SD), respectively (P < 0.001). Diabetic mice were sacrificed 2.6 ± 0.5 weeks (mean ± SD) following the detection of hyperglycemia. For the STZ model, 6- to 8-week-old female C56/Black mice were injected intraperitoneally with 300 mg of STZ (Sigma-Aldrich)/kg of body weight. The blood sugar concentrations of the nondiabetic (n = 4) and diabetic (n = 6) mice at the time of sacrifice were 94 ± 15 and 473± 28 mg/dl (mean ± SD), respectively (P < 0.001). The diabetic mice were sacrificed 1.3 ± 0.3 weeks (mean ± SD) following the detection of hyperglycemia.
RESULTS
Proteins regulating expression of the L2 transcript.
Deletional analysis of the 5′-flanking region of the L2 transcript of the murine GHR identified a regulatory region located 344 to 210 bp 5′ of the transcription start site. Exclusion of this region enhanced the activity of the L2 promoter (Fig. 1a), indicating that the proteins interacting with a cis element(s) within this region repress transcription of GHR. Extending the analysis via the DNase footprint assay identified a 72-bp stretch within this region that is protected by adult mouse liver extract (Fig. 1b). Computer-based analysis for consensus binding sites revealed the presence of CCAAT and GC box motifs within this putative cis element. A 37-bp oligonucleotide encompassing these CCAAT and GC boxes, which we designated L2C1, was tested for protein binding activity by EMSA. Addition of nuclear extracts from adult mouse liver to an aliquot of 32P-labeled L2C1 resulted in the formation of two distinct sequence-specific protein-DNA complexes, CI and CII (Fig. 2a). Supershift assays demonstrated the presence of the canonical CCAAT box binding protein NF-Y in the CI DNA-protein complex (Fig. 2a). Upon excluding the canonical members of the GC box/Kruppel-like family of proteins, Sp1, Sp2, Sp3, and Sp4, as candidates for protein(s) binding to the GC box (data not shown), we established another member of this family, BTEB1, to be a component of the CII protein-DNA complex (Fig. 2a). The presence of NF-Y and BTEB in the CI and CII complexes, respectively, was also investigated by analyzing the effect of mutations within the L2C1 sequence on the ability to compete for protein binding. Hence, altering the NF-Y binding site in the L2C1 sequence abolished competition for binding to the CI complex (Fig. 2b) and a mutation in the putative BTEB1 binding site resulted in a failure of the mutant oligonucleotide to compete for binding at the CII complex (Fig. 2b). These results demonstrate that NF-Y is a component of the CI complex and BTEB1 participates in the formation of the CII protein-DNA complex. Overlapping the NF-Y binding site on L2C1 is a recognition site for HMG. The presence of HMG in the CI complex was revealed by a concentration-dependent inhibition of the formation of the CI complex following preincubation with graded concentrations of either poly(dI-dC) · poly(dI-dC) or HMG-Y/I antibody (Fig. 2c) and by direct binding of recombinant HMG-Y/I to L2C1 (Fig. 2d). Inhibition of NF-Y binding following depletion of HMG-Y/I from the CI complex (Fig. 2c) indicates that HMG-Y/I is necessary for the participation of NF-Y in the assembly of the CI complex at L2C1.
FIG. 1.
Identification of the cis element, L2C1. (a) Transient expression analyses of the promoter-regulatory region of L2 transcript of the murine GHR. Luciferase expression plasmids containing various portions of the 5′-flanking sequence of L2 transcript were transiently transfected in BNL CL.2 cells and assayed for luciferase activity. The arrow indicates the orientation of GHR DNA relative to the direction of GHR gene transcription. Results represent means ± standard errors. Using analysis of variance, P < 0.05 in comparison with pGL3B (*) and pGL3B-L2[-344] (#). (b) DNA footprint analysis. 32P-single-end-labeled DNA fragment (generated by PCR) was incubated with 20 μg of nuclear extract from adult mouse liver (lanes 2 and 3) or no protein (lane 1) and tested for the presence of DNase I footprint by the addition of DNase I (0.5 ng [lane 2] and 2 ng [lane 3]). Arrowheads indicate the protected bands within the footprinted region. The sequence of the 72-bp footprinted region is indicated with the putative GC (double-underlined) and CCAAT (single-underlined) boxes.
FIG. 2.
Identification of the proteins interacting at L2C1. (a) Nuclear proteins from mouse liver interact with the L2C1 site in the promoter of the L2 transcript of the murine GHR gene. 32P-labeled L2C1 was incubated with (lanes 1 to 3) or without (lane P) nuclear extracts prepared from liver tissues of adult mice in the absence (lane 1) or presence of antibodies against the NF-YA (lane 2) or BTEB1 (lane 3), electrophoresed, and subjected to autoradiography. The bands representing specific DNA-protein complexes (CI, CII), supershifted complexes (SS), and the free probe (FP) are indicated. (b) NF-YA and BTEB1 interact with L2C1. 32P-labeled L2C1 was incubated with nuclear extract from adult mouse liver without (lane NC) or with excess unlabeled oligonucleotide (lanes a [25×], b [50×], and c [100×]). The unlabeled oligonucleotide used was either the wild-type sequence (L2C1) or oligonucleotides with mutations in the NF-Y (NF-Ym), BTEB1 (BTEBm), or both (L2C1m) binding sites. The bands representing specific DNA-protein complexes (CI, CII) and the free probe (FP) are indicated. (c) HMG Y/I is a component of the CI complex. 32P-labeled L2C1 was incubated with (lanes 1 to 6) or without (lane P) nuclear extracts prepared from liver tissues from adult mice in the presence of either poly(dI-dC) · poly(dI-dC) (0 μg [lane 1], 1 μg [lane 2], and 2 μg [lane 3]) or anti-HMG-Y/I antibody (20 ng [lane 4], 40 ng [lane 5], 100 ng [lane 6]). The bands representing specific DNA-protein complexes (CI, CII) and the free probe (FP) are indicated. (d) L2C1 binds HMG Y/I. 32P-labeled L2C1 was incubated without (lane P) or with recombinant HMG-Y/I protein (lane 1), electrophoresed, and subjected to autoradiography. The bands representing L2C1-HMG complex (◂) and the unbound 32P-labeled L2C1 (FP) are indicated.
Mutation of the composite L2C1 element comprising NF-Y, HMG-Y/I, and BTEB1 binding sites resulted in a significant increase in transcriptional activity (Fig. 3a). We next tested for the individual effects of these factors on GHR transcription. Inhibition of endogenous NF-Y by overexpression of a D/N NF-YA resulted in increased activity of the L2 promoter (Fig. 3b). Overexpression of the HMG-Y/I sense construct decreased L2 activity, and conversely overexpressing antisense HMG-Y/I increased L2 promoter activity (Fig. 3b). Since the effects of HMG-Y/I were absent when the D/N NF-YA was coexpressed, we conclude that NF-Y binding is obligatory for the effect of HMG-Y/I at the L2CI site (Fig. 3b). The specificity of the actions of the D/N NF-YA and HMG-Y/I was confirmed by the loss of these effects when the L2C1 element was mutated (Fig. 3c). Overexpression of BTEB1 resulted in decreased L2 promoter activity (Fig. 3d). Altering the BTEB1 binding site within L2C1 abolished the effect of overexpressing BTEB1 on L2 promoter activity, indicating that the action of BTEB1 was dependent on an intact L2CI-BTEB1 site (Fig. 3d). NF-Y and HMG-Y/I binding are a prerequisite for BTEB1 activity, since mutation of the NF-Y binding site that abolished both NF-Y and HMG-Y/I binding (data not shown) abolished the response to BTEB1 (Fig. 3d). These data indicate that collectively NF-Y, HMG-Y/I, and BTEB1 proteins repress L2 promoter activity via interaction with the L2CI element.
FIG. 3.
L2C1 represses the activity of the murine GHR gene promoter. (a) Transient transfection of luciferase reporter plasmids whose expression was controlled by wild-type GHR promoter (L2) or GHR promoter with a mutation designed to abolish the L2C1 binding sites for NF-Y, HMG-Y/I, and BTEB1 (L2C1 mutant) in BNL CL.2 cells. Results represent means ± SD of three or four independent transfections performed in triplicate. For luciferase activity, P < 0.05 compared to that of the empty vector (pGL3B) (*) or the wild-type GHR promoter (#), respectively. (b) NF-Y, BTEB1, and HMG-Y/I individually repress transcriptional activation. Transient cotransfection assays in BNL CL.2 cells with L2 luciferase reporter plasmid with constructs expressing the dominant negative form of NF-YA (DN/NF-Y, 250 ng), sense HMG-Y/I (HMG-S, 250 ng), or antisense HMG-Y/I (HMG-AS, 250 ng). Results represent means ± SD of three or four independent transfections performed in triplicate. *, P < 0.05 (luciferase activity compared to that of the wild-type GHR promoter); ns, statistically not significantly different from DN/NF-Y. (c) BNL CL.2 cells transiently cotransfected with the L2C1m-GHR reporter construct and expression plasmid for either D/N NF-Y, BTEB1, or sense HMG-Y/I. Results represent means ± SD of three or four independent transfections performed in triplicate. (d) Cotransfection of BTEB1 (500 ng) expression plasmid with luciferase constructs whose expression was controlled by wild-type promoter or GHR promoter with mutated NF-Y+HMG-Y/I or BTEB binding sites at L2C1. Results represent means ± SD of three or four independent transfections performed in triplicate. *, P < 0.05 in comparison to respective transfections carried out without BTEB1 (control).
Role of chromatin in the transcriptional activation of the L2 promoter in BNL CL.2 cells.
NF-Y and HMG-Y/I can alter chromatin architecture via changes in the acetylation status of histone (3, 18, 31). To investigate the possible effects of chromatin structure and in particular histone acetylation on the expression of L2, we engineered BNL CL.2 (murine liver) cell lines stably expressing the L2-promoter construct pGL3B-L2, containing approximately 2.0 kb of the 5′-flanking region of L2. Incubation of these cells with TSA, a histone deacetylase-specific inhibitor, resulted in increased activity of the L2 promoter (Fig. 4a). To ascertain that this effect of histone acetylation is transduced by interaction(s) at the L2C1 site, BNL CL.2 cells stably transfected with a mutant L2-reporter construct (pGL3B-L2[mCCAAT]) that incorporated a mutation within the NF-Y binding site proven to obviate NF-Y binding were exposed to TSA. The effect of TSA on cells stably transfected with pGL3B-L2[mCCAAT] was significantly less than that on the wild-type construct (Fig. 4a). Thus, the effect of histone acetylation on the L2 transcript is dependent on the interaction(s) at the L2C1 site. To validate the biological significance of these results, RNAs from TSA-treated BNL CL.2 cells were analyzed by real-time RT-PCR. Compared with control cells, cells exposed to TSA exhibited increased levels of L2C1 mRNA (Fig. 4b). Furthermore, EMSA revealed that exposure to TSA decreased binding of CI and CII complexes at L2C1 (Fig. 4c). To correlate transcriptional activation of GHR with changes in chromatin at the L2C1 site, BNL CL.2 cells exposed to TSA were subjected to ChIP with anti-acetyl-H3, anti-NF-YA, and anti-HMG-Y/I antibodies. In parallel with EMSA results, TSA treatment resulted in decreased association of NF-Y to chromatin at the L2C1 region (Fig. 4d). In contrast, significantly higher levels of acetylated histone H3 were associated with L2C1 in TSA-treated cells than in control cells (Fig. 4d). We did not observe a significant change in the levels of HMG-Y/I associated with chromatin at the L2C1 site (data not shown). We conclude that transcriptional activation of the L2 promoter due to perturbation of the chromatin architecture results from decreased binding of NF-Y to the L2C1 region.
FIG. 4.
Role of chromatin in modulating the activity of L2C1. (a) Effect of TSA exposure on BNL CL.2 cells stably transfected with L2- and -mCCAAT reporter constructs. Results represent mean ± SD of three independent experiments. *, P < 0.05 in comparison to the effect of TSA on cells stably transfected with an intact L2C1 site. (b) Fluorescent 5′-nuclease (TaqMan) real time RT-PCR analysis of expression of endogenous L2 transcript after exposure of BNL CL.2 cells to vehicle (alcohol) or TSA. The results are depicted as mean and range. *, P < 0.05, in comparison to the cells exposed to vehicle only. (c) 32P-labeled L2C1 was incubated with or without nuclear extracts prepared from BNL CL.2 cells exposed to vehicle or TSA in the absence or presence of antibodies against the NF-YA or BTEB1, electrophoresed, and subjected to autoradiography. The inset shows total NF-Y content in these cells as ascertained by EMSA with 32P-labeled NF-Y consensus oligonucleotide. The bands representing specific DNA-protein complexes (CI, CII), supershifted complexes (SS), and the free probe (FP) are indicated. (d) ChIP assay on BNL CL.2 cells treated without (vehicle) and with TSA. Immunoprecipitation with anti-NF-YA (top panel) and anti-acetyl-H3 (middle panel) antibodies. The input DNA and the sample without addition of the antibody (unbound) are also shown. The linearity of the PCR assay was established by assaying serial concentrations of the input DNA (bottom panel).
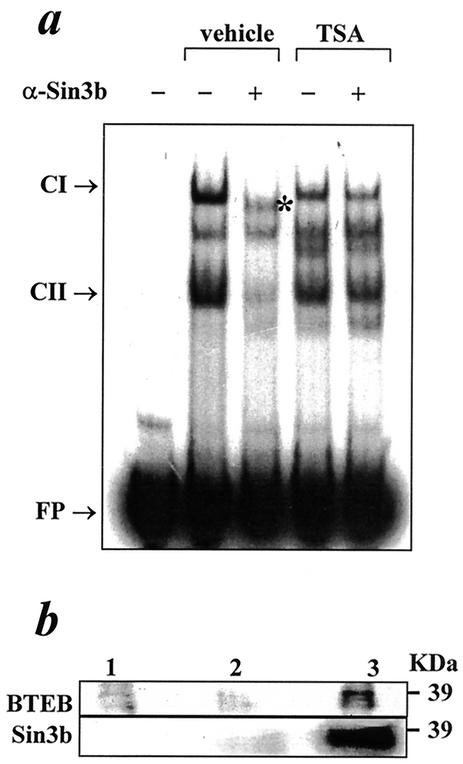
Establishment of a repressive role for the L2C1 cis element indicated a possible association of canonical corepressor proteins with the DNA-protein complex at L2C1. In EMSA, preincubation of nuclear proteins from BNL CL.2 cells with anti-mSin3b antibody resulted in the inhibition of the CI and CII complexes and the appearance of a new DNA-protein complex (Fig. 5a). This new complex could either represent a supershift of the CII complex or the faster migration of the CI complex devoid of mSin3b protein. In cells exposed to TSA, mSin3b antibody failed to alter the formation of either complex (Fig. 5a), suggesting that TSA prevents the association of mSin3b with CI and CII. Prior studies have demonstrated physical interaction between NF-Y and HMG-Y/I (5, 15) and between BTEB1 and Sin3a (35). In coimmunoprecipitation experiments we established an in vivo association of NF-Y with BTEB and of NF-Y with mSin3b (Fig. 5b). These experiments provide evidence for the combined roles of NF-Y, BTEB1, HMG-Y/I and Sin3b in the formation of a repressosome complex.
FIG. 5.
Protein-protein interaction at L2C1. (a) EMSA to demonstrate association of Sin3b with proteins interacting at L2C1. 32P-labeled L2C1 was incubated without and with nuclear extracts from vehicle or TSA-treated BNL CL.2 cells, in the absence or presence of antibody directed against Sin3b. The bands representing specific DNA-protein complexes (CI, CII), the band with altered electromobility (*), and the free probe (FP) are indicated. (b) Immunoblots to study in vivo interaction between NF-Y and mSin3b and NF-Y and BTEB. (Upper panel) Proteins from untransfected BNL CL.2 cells (lane 1) or BNL CL.2 cells transfected with BTEB1 expression plasmid (lanes 2 and 3) were immunoprecipitated with immunoglobulin G (IgG) (lane 2) or anti-BTEB antibody (lanes 1 and 3) and immunoblotted with anti-NF-YA antibody. The position of the molecular weight standard is indicated. (Lower panel) Total protein from BNL CL.2 cells not subjected to immunoprecipitation (lane 1) or immunoprecipitated with IgG (lane 2) or anti-Sin3b antibody (lane 3) and immunoblotted with anti-NF-YA antibody. The position of the molecular weight standard is indicated.
Interaction of the repressosome with the minimal promoter machinery.
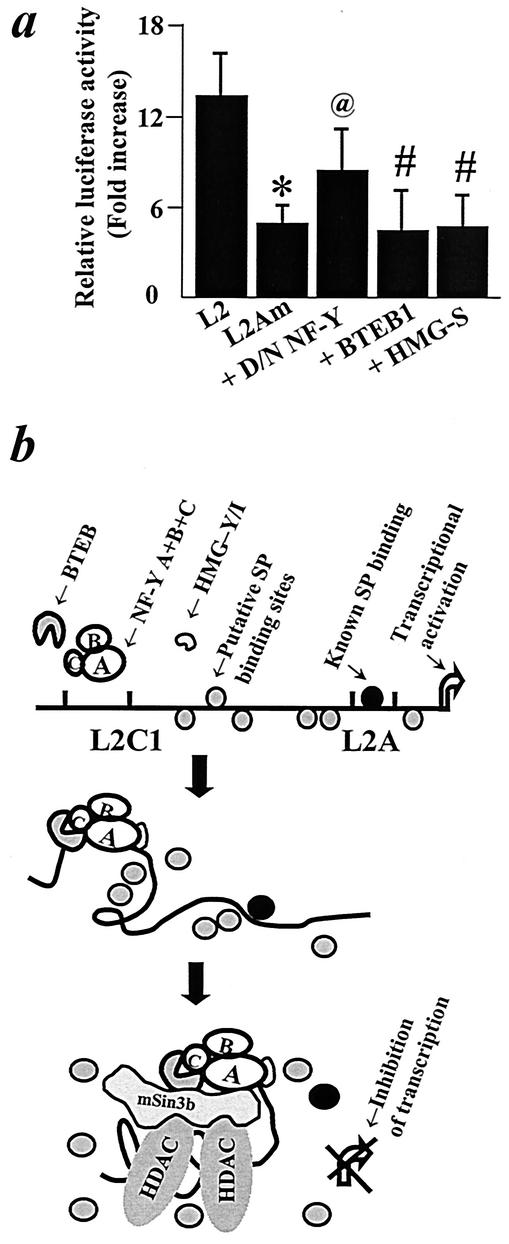
Our previous studies had identified a regulatory element, L2A, in the minimal L2 promoter located 261 bp downstream from the L2C1 site (34). We analyzed the effects of mutations in L2A on the activity of the L2C1 repressosome complex. The effects of BTEB1 and HMG-Y/I on L2 promoter activity were completely abolished in the L2A mutant (Fig. 6a). Whereas the effect of D/N NF-Y on the activity of the L2 promoter was statistically significant, the magnitude of response was substantially less than that observed with the wild-type promoter (Fig. 6a). These results support a model wherein the effects of the repressosome on the activity of the L2 promoter are transduced via interference with positive regulators of GHR transcription such as Sp factors at the basal promoter (Fig. 6b).
FIG. 6.
Mode of action of the repressosome. (a) Interaction of the repressosome complex with the minimal promoter. BNL CL.2 cells were transiently cotransfected with a wild-type GHR promoter construct containing an intact (L2) or mutated L2A (L2Am) site, and the latter was cotransfected with expression plasmids for D/N NF-Y (250 ng), HMG-Y/I (250 ng), or BTEB1(500 ng). Results represent means ± SD of three or four independent transfections performed in triplicate. *, P < 0.05 in comparison to luciferase activity of L2; P < 0.05 (@) or difference not significant (#) for luciferase activity compared to L2Am. (b) Model depicting the role of factors binding to the L2C1 site in the modulation of activity of the L2 promoter. Sp factors activate transcription of the GHR gene by interacting with GC boxes (such as the L2A site) in the minimal promoter of the L2 transcript. Binding of NF-Y/HMG-Y/I and BTEB1 at L2C1 results in DNA bending. Sin3b associates with BTEB and NF-Y and recruits histone deacetylases (HDACs) to the DNA, compacting the DNA. Changes in chromatin architecture brought upon by the repressosome result in loss of effect of the Sp factors at the L2A site and consequent inhibition of GHR transcription.
Role of the repressosome in modulation of GHR expression in DM.
Resistance to GH's action in stimulating IGF-1 in DM is in part due to decreased expression of GHR in the liver. In murine models of DM the decrease in hepatic GHR expression is due to a decrease in expression of the L2 transcript (20). To elucidate the molecular basis for the effect of DM on GHR expression, we analyzed the effect of DM on the formation of the repressosome complex at L2C1. In the liver, the levels of the NF-Y complex (CI) were more elevated in diabetic than in nondiabetic NOD mice (Fig. 7a, top panel). The specificity of this finding was established by demonstrating nonparallel changes in the binding of an unrelated DNA-binding protein, OCT-1 (Fig. 7a, middle panel) and BTEB1 at the L2C1 site (data not shown). The increase in the binding of NF-Y at the L2CI site was not due to an increase in total cellular content of NF-Y in DM as demonstrated by EMSA with an NF-Y consensus binding sequence (Fig. 7a, bottom panel). ChIP assay revealed that the amount of acetylated H3 associated with L2C1 was significantly lower in the livers of diabetic mice than in those of nondiabetic mice (Fig. 7b). In contrast, diabetes resulted in an increase in the level of NF-Y associated with L2C1 in the liver (Fig. 7b). DM did not alter HMG-Y/I binding at the L2C1 site (data not shown).
FIG. 7.
Increased binding activity of NF-Y to the L2C1 cis element in DM. (a) EMSA with liver nuclear extracts from nondiabetic (lanes 1 to 4) or diabetic (lanes 5 to 10) NOD mice. The probes used were L2C1 (upper panel), OCT-1 (middle panel), or NF-Y consensus binding site (lower panel). The densitometry of the blots is represented with the data normalized to the nondiabetic group. (b) ChIP assays were conducted with liver from nondiabetic and diabetic (STZ induced) mice and L2C1 detected by PCR. Immunoprecipitation with anti-acetyl-H3 (top panel) and anti-NF-YA (bottom panel). The input DNA and the sample without addition of the antibody (unbound) are also shown.
Previous studies have suggested tissue-specific differences in GHR expression in DM (21). Using the TaqMan assay, we observed an increase in L2 expression in kidneys of diabetic (STZ model) compared to that in nondiabetic mice (Fig. 8a). In contrast, in the liver DM resulted in a significant reduction in the expression of the L2 transcript. In the kidney, EMSA established that the binding of NF-Y to the L2C1 cis element was weak, with a relative abundance of BTEB1 binding (Fig. 8b), and that in contrast to what was observed for the liver, DM did not affect the formation of the CI DNA-protein complex at the L2C1 site (Fig. 8c). ChIP assays revealed that in the kidney DM is characterized by increased acetyl-H3 associated with L2C1. An association of NF-Y with chromatin at L2C1 in either the nondiabetic or the diabetic state was not demonstrable in the kidney (Fig. 8d), and DM did not influence levels of chromatin-associated HMG at the L2C1 site (Fig. 8d). These results support a model wherein lack of assembly of a NF-Y-dependent repressosome complex at the L2C1 site on the GHR promoter permits the kidney to avoid a DM-induced decrease in GHR expression and hence exposes the kidney to the effects of elevated levels of GH in DM.
FIG. 8.
Role of L2C1 in regulation of GHR expression in kidney in IDDM. (a) Fluorescent 5′-nuclease (TaqMan) real-time RT-PCR analysis of expression of L2 transcript in liver and kidney in murine experimental DM. The results (mean and range) are depicted relative to nondiabetic (control) mice. *, P < 0.05 in comparison to control. (b) 32P-labeled L2C1 was incubated with (lanes 1 to 3) or without (lane P) nuclear extracts prepared from kidney of adult male mice in the absence (lane 1) or presence of antibodies against the NF-YA (lane 2) or BTEB1 (lane 3), electrophoresed, and subjected to autoradiography. (Inset) Overexposure of the autoradiograph to illustrate the supershift of C1 complex by NF-YA antibody. The bands representing specific DNA-protein complexes (CI, CII), supershifted complexes (SS), and the free probe (FP) are indicated. (c) Lack of alteration in binding of proteins at the L2C1 complex in IDDM. EMSA was carried out with kidney nuclear extracts from nondiabetic or diabetic mice. The probes used were L2C1 (upper panel) or NF-Y consensus binding site (lower panel). (d) ChIP assays with immunoprecipitation with anti-acetyl H3, anti-HMG, and anti-NF-YA were conducted on kidney tissues from nondiabetic and diabetic (STZ induced) mice and L2C1 detected by PCR. The input DNA and the sample without addition of antibody (unbound) are also shown.
DISCUSSION
The two major findings of this study are that expression of the dominant transcript of the GHR in postnatal life is regulated by a higher-order repressosome complex consisting of NF-Y, HMG-Y/I, BTEB1, and mSin3b proteins and that the effect of insulinopenic DM on hepatic GHR gene transcription is mediated by the mechanisms involving chromatin remodeling at a defined site on the GHR promoter. The differential effects of DM on NF-Y's interactions at the GHR promoter in the liver and kidney results in tissue-specific regulation of expression of GHR in the liver and kidney in DM and thus plays a role in the genesis of diabetic nephropathy.
Our studies indicate that NF-Y is a critical member of the DNA-binding protein complex at the L2C1 site of the GHR promoter. NF-Y is a ubiquitous heterotrimeric complex (18), and its binding to DNA brings about structural changes resembling nucleosome organization (14). Whereas NF-Y usually plays a role as an activator of gene transcription (13, 16), the present report provides evidence that NF-Y participates in the assembly and action of a repressosome complex and thus adds to the select examples of NF-Y inhibiting gene transcription (9, 12, 17, 28). NF-Y forms partners with a disparate group of proteins (3, 5, 6, 15, 18). The present report establishes that NF-Y also associates in vivo with the transcription factor BTEB1 and corepressor Sin3b. Like NF-Y, BTEB is a bimodal transcription factor and can function either as an activator or as a repressor of transcription (27), and a previous report has demonstrated that BTEB can interact with mSin3a to repress transcription (35). Physical interactions between NF-Y and HMG-Y/I modulate NF-Y activity through stabilization of NF-Y binding (5). It is noteworthy that previous studies have demonstrated that acetylation of HMG results in the disruption of an enhanceosome complex (25). Thus, posttranslational modifications of HMG in response to alterations in the cellular milieu, such as in DM, could modulate NF-Y binding at the L2C1 site. In the context of the L2 transcript of the GHR gene, the ability of NF-Y to act as a repressor possibly reflects the association with other members of the repressosome complex and also the spatial relationship with cis elements in the minimal promoter.
Activation of GHR transcription by TSA is associated with an increase in acetylated histone H3 at the L2C1 site. Conversely, inhibition of GHR transcription in DM is associated with decreased levels of acetylated histone H3 at the L2C1 site. These results are in agreement with the contemporary model wherein acetylation of histone H3 and/or H4 correlates with the opening of the nucleosome and activation of transcription (13). Based on the ability of TSA to alter the activity of the GHR promoter, our results indicate that histone acetyltransferases and deacetylases influence GHR promoter activity and that this effect is mediated in part due to interaction(s) at the L2C1 site. Both NF-Y and HMG-Y/I are known targets for acetylation by proteins possessing histone acetyltransferase activity (18, 31). Although TSA treatment results in activation and/or derepression of the GHR promoter, the effect of acetylation on the function of NF-Y is unclear. Thus, acetylation of NF-Y could alter its DNA-binding affinity, influence its interactions with other transcription factors or with general factors of the holoenzyme machinery, or modulate its association with nucleosomes, resulting in modification of nearby chromatin structures. Alternatively, the activity of nonhistone proteins like HMG-Y/I or NF-Y could be influenced by patterned display of modifications on one or more histone tails (“histone code” [32]). The inverse relationship between levels of NF-Y and acetylated histone H3 at L2C1 supports such a model, wherein modifications of histone at the L2C1 site initiate alterations in the binding activity of cognate DNA-binding proteins resulting in the dissolution of the repressosome complex with consequent activation of GHR transcription.
One paradox of the role of GH/IGF-1 in the genesis of microvascular complication of DM is that the liver is resistant to GH while GH action is required for the genesis of retinopathy and nephropathy (1, 19, 21, 23, 30). Tissue-specific differences in the expression of the GHR may play a role in this differential sensitivity of organs to the effects of DM (21). Our studies indicate that lack of recruitment of the repressosome complex to the L2C1 site of the GHR promoter plays a permissive role in the sensitivity of kidney to GH in DM. The increase in NF-Y binding to L2C1 in diabetic liver is specific to DM, since under other circumstances that lead to decreased expression of the L2 transcript of the GHR gene such as fasting, altered binding of NF-Y at the L2C1 site is not observed (data not shown). Our findings are also consistent across both the NOD and STZ models of insulinopenic DM. One of the questions raised by our study is the mechanism(s) involved in recruitment of the repressosome complex to the GHR promoter in the diabetic state. The regulation of gene transcription via effects of cellular redox on DNA-binding activity of NF-Y is known (26). Experimental and clinical DM are associated with altered cellular redox (7, 33), and the potential role of these metabolic changes in transducing the effects of DM on NF-Y activity at L2C1 remains to be defined. It will be of interest to investigate if drugs that either alter cellular redox (7) or modulate the activity of NF-Y (18) are potential targets for drug discovery in the prevention and/or therapy of diabetic nephropathy. Another potential target for DM-induced changes in chromatin is histone proteins. Thus, in addition to the various posttranslational modifications that histones are exposed to in the nondiabetic state (4), histones could also be the target of modification by advanced glycation end products in DM (11). The roles of these myriad mechanisms in modulating expression of GHR in DM remain to be clarified.
In summary, we have defined the role of a repressor complex in the control of transcription of the murine GHR gene. The differential effects of DM on the assembly of this higher-order repressor complex in liver and kidney provide a molecular basis for the tissue-specific regulation of expression of GHR in DM. The understanding of these mechanisms may permit the identification of pharmacophores designed to alter expression of the GHR in a tissue-specific manner and thus potentially abolish the development of certain complications of DM such as nephropathy.
Acknowledgments
This study was supported by grants from the NIH (DK49845 [to R.K.M.] and T32DK07729), Children's Hospital of Pittsburgh, and the Vira I. Heinz Foundation.
We thank M. Trucco and S. Bertera for the murine models of IDDM and M. K. Bennett (University of California, Irvine) for advice regarding ChIP assays. We also thank Clayton Mathews and Lee Denson for critically reviewing the manuscript.
REFERENCES
- 1.Baxter, R. C., J. M. Bryson, and J. R. Turtle. 1980. Somatogenic receptors of rat liver: regulation by insulin. Endocrinology 107:1176-1181. [DOI] [PubMed] [Google Scholar]
- 2.Bellush, L. L., S. Doublier, A. N. Holland, L. J. Striker, G. E. Striker, and J. J. Kopchick. 2000. Protection against diabetes-induced nephropathy in growth hormone receptor/binding protein gene-disrupted mice. Endocrinology 141:163-168. [DOI] [PubMed] [Google Scholar]
- 3.Caretti, G., M. C. Motta, and R. Mantovani. 1999. NF-Y associates with H3-H4 tetramers and octamers by multiple mechanisms. Mol. Cell. Biol. 19:8591-8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 5.Currie, R. A. 1997. Functional interaction between the DNA binding subunit trimerization domain of NF-Y and the high mobility group protein HMG-I(Y). J. Biol. Chem. 272:30880-30888. [DOI] [PubMed] [Google Scholar]
- 6.Currie, R. A. 1998. NF-Y is associated with the histone acetlytransferase GCN5 and P/CAF. J. Biol. Chem. 273:1430-1434. [DOI] [PubMed] [Google Scholar]
- 7.de Cavanagh, E. M. V., F. Iinserra, J. Toblli, I. Stella, C. Fraga, and L. Ferder. 2001. Enalapril attenuates oxidative stress in diabetic rats. Hypertension 38:1130-1136. [DOI] [PubMed] [Google Scholar]
- 8.Edens, A., and F. Talamantes. 1998. Alternative processing of growth hormone receptor transcripts. Endocr. Rev. 19:559-582. [DOI] [PubMed] [Google Scholar]
- 9.Farsetti, A., M. Narducci, F. Moretti, S. Nanni, R. Mantovani, A. Sacchi, and A. Pontecorvi. 2001. Inhibition of ER alpha-mediated trans-activation of human coagulation factor XII gene by heteromeric transcription factor NF-Y. Endocrinology 142:3380-3388. [DOI] [PubMed] [Google Scholar]
- 10.Gorski, K., M. Carneiro, and U. Schibler. 1986. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell 47:767-776. [DOI] [PubMed] [Google Scholar]
- 11.Gugliucci, A., and M. Bendayan. 1995. Histones from diabetic rats contain increased levels of advanced glycation end products. Biochem. Biophys. Res. Commun. 212:56-62. [DOI] [PubMed] [Google Scholar]
- 12.Izumi, H., C. Molander, L. Z. Penn, A. Ishisaki, K. Kohno, and K. Funa. 2001. Mechanism for the transcriptional repression by c-Myc on PDGF B-receptor. J. Cell. Sci. 114:1533-1544. [DOI] [PubMed] [Google Scholar]
- 13.Jin, S., and K. W. Scotto. 1998. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol. Cell. Biol. 18:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liberati, C., A. Ronchi, P. Lievens, S. Ottolenghi, and R. Mantovani. 1998. NF-Y organizes the Y-Globin CCAAT boxes region. J. Biol. Chem. 273:16880-16889. [DOI] [PubMed] [Google Scholar]
- 15.Liberati, C., R. Sgarra, G. Manifioletti, and R. Mantovani. 1998. DNA binding of NF-Y: the effect of HMG-I proteins depends upon the CCAAT box. FEBS Lett. 433:174-178. [DOI] [PubMed] [Google Scholar]
- 16.Maity, S. N., and B. de Crombugghe. 1998. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends. Biochem. Sci. 23:174-178. [DOI] [PubMed] [Google Scholar]
- 17.Manni, I., G. Mazzaro, A. Gurtner, R. Mantovani, U. Haugwitz, K. Krause, K. Engeland, A. Sacchi, S. Soddu, and G. Piaggo. 2001. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 276:5570-5576. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 19.Menon, R. K., S. Arslanian, B. May, W. S. Cutfield, and M. A. Sperling. 1992. Diminished growth hormone-binding protein in children with insulin-dependent diabetes mellitus. J. Clin. Endocrinol. Metab. 74:934-938. [DOI] [PubMed] [Google Scholar]
- 20.Menon, R. K., A. Shaufl, J. H. Yu, D. A. Stephan, and R. P. Friday. 2001. Identification and characterization of a novel transcript of the murine growth hormone receptor gene exhibiting development- and tissue-specific expression. Mol. Cell. Endocrinol. 172:135-146. [DOI] [PubMed] [Google Scholar]
- 21.Menon, R. K., D. A. Stephan, R. H. Rao, Z. Shen-orr, Jr., L. S. Down, C. T. Roberts, D. LeRoith, and M. A. Sperling. 1994. Tissue-specific regulation of the growth hormone receptor gene in streptozotocin-induced diabetes in the rat. J. Endocrinol. 142:453-462. [DOI] [PubMed] [Google Scholar]
- 22.Menon, R. K., D. A. Stephan, M. Singh, S. M. Morris, Jr., and L. Zou. 1995. Cloning of the promoter-regulatory region of the murine growth hormone receptor gene: identification of a developmentally regulated enhancer element. J. Biol. Chem. 270:8851-8859. [DOI] [PubMed] [Google Scholar]
- 23.Mercado, M., M. E. Molitch, and G. Baumann. 1992. Low plasma growth hormone binding protein in IDDM. Diabetes 41:605-609. [DOI] [PubMed] [Google Scholar]
- 24.Munichoodappa, C. S., S. B. Rees, R. F. Bradley, M. C. Balodimos, and G. Boden. 1971. Bragg peak proton beam irradiation of the pituitary gland for proliferative diabetic retinopathy. Ann. Intern. Med. 74:491-498. [DOI] [PubMed] [Google Scholar]
- 25.Munshi, N., M. Y. Merika, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG-I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 26.Nakshatri, H., P. Bhat-Nakshatri, and R. A. Currie. 1996. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J. Biol. Chem. 271:28784-28791. [DOI] [PubMed] [Google Scholar]
- 27.Philipsen, S., and G. Suske. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, Q., K. W. Gross, and C. D. Sigmund. 2001. NF-Y antagonizes renin enhancer function by blocking stimulatory transcription factors. Hypertension 38:332-336. [DOI] [PubMed] [Google Scholar]
- 29.Smith, L. E., J. J. Kopchick, W. W. Chen, J. Knapp, F. Kinose, D. Daley, E. Foley, R. G. Smith, and J. M. Schaeffer. 1997. Essential role of growth hormone in ischemia-induced retinal neovascularization. Science 276:1706-1709. [DOI] [PubMed] [Google Scholar]
- 30.Sonksen, P. H., D. Russell-Jones, and R. H. Jones. 1993. Growth hormone and diabetes mellitus. Horm. Res. 40:68-79. [DOI] [PubMed] [Google Scholar]
- 31.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 33.West, I. C. 2000. Radicals and oxidative stress in diabetes. Diabet. Med. 17:171-180. [DOI] [PubMed] [Google Scholar]
- 34.Yu, J. H., G. Schwartzbauer, A. Kazlman, and R. K. Menon. 1999. Role of the Sp family of transcription factors in the ontogeny of growth hormone receptor gene expression. J. Biol. Chem. 274:34327-34336. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, J.-S., M. C. Moncrieffe, J. Kaczynski, V. Ellenrieder, F. G. Prendergast, and R. Urrutia. 2001. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 21:5041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou, L., and R. K. Menon. 1995. A member of the CTF/NF-1 transcription factor family regulates murine growth hormone receptor gene promoter activity. Endocrinology 136:5236-5239. [DOI] [PubMed] [Google Scholar]