Abstract
Having opposing enzymatic activities, histone acetylases (HATs) and deacetylases affect chromatin and regulate transcription. The activities of the two enzymes are thought to be balanced in the cell by an unknown mechanism that may involve their direct interaction. Using fluorescence resonance energy transfer analysis, we demonstrated that the acetylase PCAF and histone deacetylase 1 (HDAC1) are in close spatial proximity in living cells, compatible with their physical interaction. In agreement, coimmunoprecipitation assays demonstrated that endogenous HDACs are associated with PCAF and another acetylase, GCN5, in HeLa cells. We found by glycerol gradient sedimentation analysis that HATs are integrated into a large multiprotein HDAC complex that is distinct from the previously described HDAC complexes containing mSin3A, Mi-2/NRD, or CoREST. This HDAC-HAT association is partly accounted for by a direct protein-protein interaction observed in vitro. The HDAC-HAT complex may play a role in establishing a dynamic equilibrium of the two enzymes in vivo.
Specific lysines on the core histones are acetylated by a series of histone acetylases (HATs). The status of acetylation constitutes one basis for the histone code, an important basis of chromatin-mediated regulatory processes, including transcription, replication, and chromosome dynamics (12, 23, 29). Acetylation of histone tails can be reversed by a diverse series of histone deacetylases (5, 15).
Both HATs and histone deacetylases are classified into several distinct groups. The GNAT family of HATs, one of the best-studied families, is conserved throughout eukaryotes (23, 28). In mammalian species there are two GNAT members, GCN5 and PCAF. They are structurally similar to each other and are expressed in overlapping sets of cells and tissues. They predominantly regulate acetylation of histone H3 and are generally involved in transcriptional activation (12, 25, 39). Both GCN5 and PCAF form large multiprotein complexes whose compositions are also conserved (9, 21, 23, 28). Among histone deacetylases, class I histone deacetylases (HDACs) were the first to be identified and are conserved from yeasts such as Saccharomyces cerevisiae to humans. Four HDACs are currently known in humans: HDAC1, HDAC2, HDAC3, and HDAC8 (15). These histone deacetylases are generally associated with transcriptional repression mediated by various DNA binding transcription factors (5, 15, 20, 24). HDAC1 and HDAC2 form at least three distinct complexes that contain representative factors, mSin3A, Mi-2/NRD, and CoREST/kiaa0071 (13, 32, 40, 44, 45, 46). HDAC3 also forms a complex that contains N-CoR and SMRT, among other components (10, 16, 36).
Although rapid progress has been made on understanding the structure and function of individual HATs and HDACs, a remaining question is how the activities of these two enzymes, which exert opposite functions, are mutually balanced in the cell (23). A series of genetic studies and promoter analyses suggest that the two enzymes may not act independently and that their activities in some cases may be linked to one another (22, 34, 37). Other lines of evidence indicate that some HATs and HDACs occupy the common space in the nucleus and coordinately regulate the same set of target genes. For example, transcriptional activation mediated by nuclear receptors such as retinoic acid receptor and retinoid X receptor involves ligand-dependent association and dissociation of HAT and HDAC, respectively, on a given promoter (2, 38). Coordinated activity of the two enzymes may also be inferred for cell growth-regulated genes controlled by E2F, whose promoter activity is repressed by the HDAC-associated retinoblastoma protein but is activated by subsequent HAT recruitment (5). Furthermore, YY1 and Sp1 interact with both HATs and HDACs, thereby acquiring an activator or repressor function depending on promoter context and other factors (5). A close interrelationship between the two enzymes may also be presumed, based on the earlier observation that histones are rapidly acetylated and deacetylated with a half-life of less than 10 min in some regions of a nucleus while in other regions histone acetylation is turned over more slowly (4, 6). More-recent studies demonstrate that HATs and HDACs are engaged in a rapid cycle of global histone acetylation and deacetylation that affects the whole yeast genome (1, 14, 34). The global, untargeted alteration of histone acetylation is likely to be critical for rapid reversal of targeted chromatin modification in a specific promoter associated with transcriptional activation and/or repression (14, 34).
In this work we wished to address the mechanisms that may balance the activities of the two enzymes. We surmised that among the mechanisms that help coordinate their activities, one might involve a physical interaction between the two enzymes. To search for evidence indicating the presence of HDAC-HAT interaction in vivo, we first employed a novel flow cytometry technique based on fluorescence resonance energy transfer (FRET) (26, 30). This technique allows an assessment of molecular interactions between two proteins in the living cell. Although not heretofore applied extensively to the analysis of nuclear events, this approach provides a powerful new tool to investigate the molecular behavior of transcription factors and chromatin modifiers in the nucleus. By introducing PCAF and HDAC1 labeled with distinct fluorochromes into HeLa cells, we observed clear FRET signals ascribable to their physical proximity. Using coimmunoprecipitation assays, it was shown that HDAC1, HDAC2, and HDAC3 are all associated with GCN5 and PCAF in HeLa cells. Glycerol gradient sedimentation analysis of HDAC1 complexes revealed that GCN5 is contained in a large multiprotein complex(es) distinct from the three HDAC complexes reported before. In vitro binding analysis indicated that the HATs are incorporated into the HDAC complex(es) at least in part by directly binding to HDACs. Finally, we present evidence suggesting that HDAC-HAT interactions occur in a dynamic fashion depending on the physiological state of cells. Taken together, these results point to a mechanism that internally maintains HDAC-HAT equilibrium in the cell.
MATERIALS AND METHODS
FRET analysis.
Full-length hPCAF (42), hHDAC1 (31), and ICSBP (17) were cloned into cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) expression vectors from Clontech, creating ECFP-C1-PCAF, EYFP-N1-HDAC1, and EYFP-C1-ICSBP. Using Polyfect (Qiagen), HeLa cells (8 × 105) were transfected with a total of 3 μg of plasmid DNA encoding ECFP and YFP fusion proteins. At 24 h following transfection, cells were harvested in phosphate-buffered saline and FRET analysis was performed on a FACS Vantage cytometer (Becton Dickinson) as previously described (26). EYFP was excited with an air-cooled argon laser tuned to 514 nm, and signals were detected with a 546-to-10-nm bandpass filter. ECFP was separately excited with a krypton laser tuned to 413 nm, and signals derived from ECFP and EYFP due to FRET were simultaneously detected with 470-to-20-nm and 546-to-10-nm bandpass filters, respectively. Data were analyzed by FlowJo software. Microscopic images of live cells were obtained with a Leica TCS SP2 confocal microscope.
Immunoprecipitation and sedimentation analysis of HDAC complexes.
HeLa cells were infected with a retrovirus vector expressing Flag-tagged HDAC1, HDAC2, or HDAC3 as previously described (13). Nuclear extracts were prepared from ∼107 to 109 cells as previously described (7) and were dialyzed against a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 10 mM β-mercaptoethanol and readjusted to 300 mM NaCl supplemented with 0.1% Tween 20 before use. Extracts (10 to 30 mg) were absorbed to anti-Flag M2 affinity gel (Sigma), washed, and eluted with Flag peptides (200 μg/ml). For immunoprecipitation of endogenous HDAC1, extracts from uninfected HeLa cells prepared in the same buffer as described above and containing 150 mM NaCl were precipitated with anti-HDAC1 antibody conjugated to agarose beads. Bound proteins were eluted with 100 mM glycine (pH 2.5)-0.1% Tween 20 (13). Eluted samples were resolved by sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis (SDS-4 to 20% PAGE) and immunoblotted with rabbit antibodies to GCN5 and PCAF (21), HDAC1, Mta-L1, MBP2, Kiaa0601 and Kiaa0071 (13), Mi-2 (Upstate Biotechnology), RbAp48 (GeneTex), mSin3A (Santa Cruz), or Flag peptide (Sigma). The same procedure was used for analysis of U937 cell extracts. Sedimentation analysis of Flag-HDAC1 complexes was performed with 200 μl of anti-Flag antibody eluates obtained from HeLa cells expressing Flag-tagged HDAC1 in a 10 to 35% glycerol gradient in 20 mM Tris-HCl (pH 8.0)-100 mM NaCl as previously described (13). A total of 26 fractions, containing 170 μl each, were collected and analyzed by silver staining and immunoblotting. Chromatographic fractionation of Flag-HDAC1 immunoprecipitates was performed as follows. Nuclear extracts from HeLa cells (3 × 109 to 6 × 109 cells) were affinity purified on anti-Flag M2 agarose. Bound materials were eluted in buffer A (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol) containing Flag peptides (200 μg/ml). They were diluted in 20 mM NaCl and fractionated on a DEAE-Sepharose column (bed volume, 100 μl) (Amersham Pharmacia Biotech) with a stepwise elution in buffer A (containing 100 mM, 300 mM, 500 mM, and 1 M NaCl). Each fraction was dialyzed against 20 mM Tris-HCl (pH 8.0), concentrated with a SpeedVac Concentrator (Sovant), and analyzed by Western blotting.
Recombinant HDACs and PCAF deletions.
Full-length HDAC1 tagged with the Flag epitope at the C terminus and PCAF tagged with the Flag epitope at the N terminus and their deletions were cloned by a BaculoGold (Pharmingen) or a Bac-to-Bac (GIBCO-BRL) system into baculovirus vectors. Recombinant proteins were affinity purified from extracts prepared from infected Sf9 cells (multiplicity of infection > 10). Briefly, cells suspended in a buffer containing 50 mM Tris-HCl (pH 8.0), 20% glycerol, 0.2 mM EDTA, 600 mM NaCl, and 0.1% Triton X-100 were subjected to freezing and thawing. Supernatants were applied on M2 anti-Flag antibody beads and eluted with buffer supplemented with 100 mM NaCl containing 100 μg of Flag peptide/ml. Purification of recombinant histidine-tagged HDAC2 was as previously described (24).
In vitro binding assays.
Sf9 extracts containing recombinant PCAF (100 ng) were incubated with in vitro transcribed and translated 35S-labeled HDAC1 at 4°C for 1 h and precipitated with anti-PCAF antibody conjugated to protein G Sepharose beads (2). Bound materials were resolved on SDS-10% PAGE and visualized by fluorography. To test binding of full-length and truncated PCAF, recombinant HDAC2 (24) was conjugated to Ni2+-nitrilotriacetic acid (NTA) agarose beads (Qiagen) in NS-1-10 buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 1 mM EDTA, 0.01% Triton X-100, 10 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride, 10 mM β-mercaptoethanol) containing 2% bovine serum albumin and incubated with 50 ng or 500 ng of full-length or truncated recombinant PCAF at 4°C for 1 h. Proteins were eluted with NS-1-10 buffer with 200 mM imidazole and were evaluated by immunoblot analysis using anti-PCAF antibody.
HAT and deacetylase enzymatic activities.
HAT assays were performed as previously described (2, 25). [3H]acetyl coenzyme A (Amersham) (5 nmol) was incubated with 2 μg of core histones (Sigma) in 30 μl of each reaction mixture at 30°C for 10 to 30 min, and the reaction mixtures were separated on SDS-4 to 20% PAGE. Deacetylase assays were performed as previously described (13, 24).
RESULTS
FRET analysis reveals spatial proximity of PCAF and HDAC1 in living cells.
The previous observations that HATs and HDACs generate a dynamic equilibrium of chromatin acetylation across the genome (14, 34) suggest the possibility that HATs and HDACs are closely associated with each other in at least some regions of the nucleus. To investigate the molecular proximity of HAT and HDAC in real time in living cells, we have employed a novel, flow cytometry-based FRET technique (19, 30). This type of FRET analysis utilizes two proteins fused to green fluorescent protein variants CFP and YFP that are transfected into appropriate cells. When a CFP protein and a YFP are in proximity with each other, excitation of the CFP triggers a fluorescent emission of the YFP. CFP and YFP interact with maximal energy transfer at approximately 50 to 60 Å (26) (for reference, the diameter of a nucleosome is 110 Å). Given this fact and the assumption that a globular form of HDAC1 and PCAF would have a diameter of ∼50 to 60 Å, the presence of FRET would indicate a direct interaction of the two proteins. We have used the flow cytometry-based FRET procedure for this analysis, rather than a single-cell-based microscopic procedure, since it enables the detection of FRET in a large, heterogeneous population of cells (26).
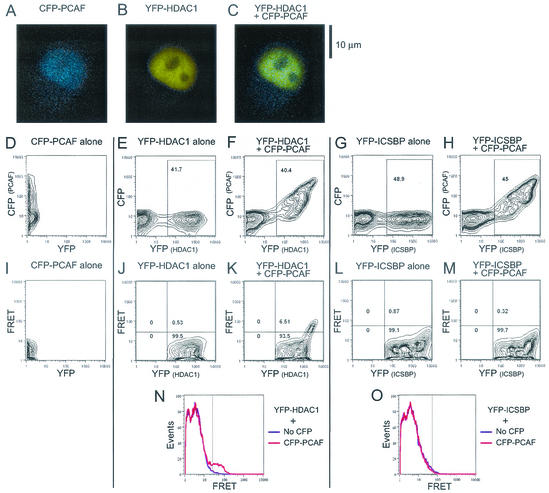
HeLa cells were cotransfected with CFP-PCAF along with YFP-HDAC1. As a control, CFP-PCAF was also transfected along with YFP-ICSBP. The latter protein is a nuclear transcription factor that forms a multiprotein complex but does not interact with PCAF (18). Fluorescence microscopy analysis of transfected cells (Fig. 1A) showed that CFP-CAF and YFP-HDAC1 localized to the nucleus as fine speckles, with the CFP and YFP signals apparently colocalizing in most areas of the nucleus except for those of the nucleoli. Figure 1D to H show the results of flow cytometry analysis of CFP and YFP signals detected by the emission spectrum characteristic of each fluorochrome. YFP-HDAC1 and YFP-ICSBP were detected in ∼40 to 50% of transfected cells, irrespective of whether cells were cotransfected with CFP-PCAF or transfected alone. Figure 1C and E show that CFP-PCAF and YFP-HDAC1 (or YFP-ICSBP) were coexpressed in most of the transfected cells (the transfection efficiency of CFP-PCAF was also ∼40 to 45%). FRET signals were monitored, as shown in the lower middle panels (I to M) of Fig. 1. As expected, when CFP-PCAF or YFP-HDAC1 was expressed alone, no FRET signals were detected (Fig. 1I and J). In contrast, when CFP-PCAF was coexpressed with YFP-HDAC1, 6.5% of YFP-positive cells exhibited FRET signals (panel K), indicating that PCAF and HDAC1 were spatially close to each other in these cells. The relatively low percentage of cells that showed FRET signals may be due to the fact that CFP-PCAF was expressed at a somewhat low level and/or to the possible interaction of CFP-PCAF with endogenous HDACs abundantly expressed in HeLa cells (see below). To ascertain whether the FRET signals detected by PCAF and HDAC1 were attributable to their physical proximity rather than mere coexpression in the nucleus, a similar analysis was performed with CFP-PCAF and YFP-ICSBP (Fig. 1L and M). No FRET signals were observed with this combination. The absence of FRET signals from ICSBP was not due to the amounts of protein expressed, since YFP-ICSBP was expressed at levels higher than those of YFP-HDAC1. Likewise, YFP-ICSBP did not exhibit FRET signals with CFP-HDAC1 (data not shown). An overlay of the corresponding histograms of the FRET signals elicited by PCAF and HDAC1 is shown in panels N and O in Fig. 1, in which a prominent FRET-positive population is evident in the cells coexpressing CFP-PCAF and YFP-HDAC1 but not in the cells coexpressing CFP-PCAF and YFP-ICSBP. We also found FRET signals by analysis of PCAF and HDAC1 labeled with the alternate fluorochrome (i.e., YFP-PCAF and CFP-HDAC1) (data not shown). These results led us to conclude that a significant fraction of transfected PCAF and HDAC1 are in close proximity to each other in living cells.
FIG. 1.
FRET detection of PCAF-HDAC1 interaction in living cells. HeLa cells were transfected with CFP-PCAF and YFP-HDAC1 or YFP-ICSBP (control). Microscopic images of live cells are shown in panels A to C. In panels D to H, relative fluorescence levels of CFP and YFP in individual transfected cells are presented as contour plots reflecting the number of cells exhibiting either one or both fluorochrome signals. YFP-positive populations (shown in rectangles within panels) were subjected to YFP-FRET plotting (I to M). Each number represents the percentage of cells in the compartment. The FRET signals detected are presented in panel K. Panels N and O show overlays of the FRET histograms obtained with HDAC1- or ICSBP-positive cells.
Endogenous HDAC1 forms a complex with GCN5 and PCAF in HeLa cells.
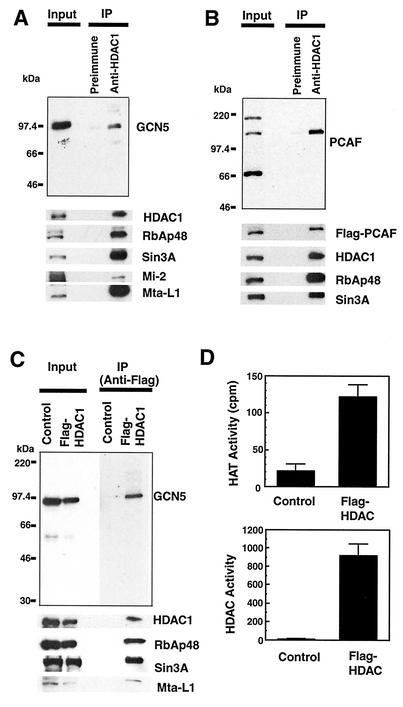
Because the FRET results relied on ectopically expressed HAT and HDAC, it was of importance to assess whether endogenous HDACs and HATs interact with each other. To investigate biochemical evidence for HDAC-HAT interaction, coimmunoprecipitation analysis was performed with antibody specific for HDAC1. For this purpose, HeLa cell nuclear extracts adjusted in a buffer with 150 mM to 300 mM NaCl were tested. To preserve complexes of weak associations, extracts were not processed by a urea-based chromatography method previously employed for purification of stable HDAC complexes (13). Precipitated materials were immunoblotted with antibody for the endogenous GCN5. As shown in Fig. 2A, HDAC1 antibody coprecipitated GCN5 along with HDAC1, RbAp48, Sin3A, and other components known to be contained in the stable HDAC1 complexes (13, 32, 44-46). Preimmune sera did not precipitate HDAC1 or GCN5, indicating that GCN5 was specifically associated with HDAC1. We assessed whether the association of HDAC1 with GCN5 described above is mediated by DNA by including ethidium bromide in immunoprecipitation experiments (43). A comparable level of GCN5 was coprecipitated in the presence of 1.2 μg of ethidium bromide/ml (data not shown). In addition, immunoblot analysis of precipitated materials did not reveal an appreciable amount of histone H3 or H4, indicating that this association is not dependent on the presence of chromatin.
FIG. 2.
Endogenous HDAC1 and Flag-tagged HDAC1 associate with GCN5 and PCAF in HeLa cells. (A) Nuclear extracts were precipitated with preimmune sera or anti-HDAC1 antibody conjugated to agarose beads. Eluted materials were immunoblotted with antibody for GCN5 (top panel) or the indicated proteins (bottom panels). The input represents immunoblot analysis of 5% of total extracts. (B) Extracts from HeLa cells expressing Flag-tagged PCAF were immunoprecipitated with anti-HDAC1 antibody and subjected to blotting for PCAF (top) or other proteins as described for panel A. (C) Extracts from HeLa cells infected with Flag-tagged HDAC1 (Flag-HDAC1) or control virus (Control) were immunoprecipitated with M2 anti-Flag antibody, eluted with Flag peptide, and analyzed by immunoblotting as described for panel A. (D) Enzymatic activity of HDAC1 immunoprecipitates. HAT (top panel) and HDAC (bottom panel) activities were measured with 30 μl and 100 μl, respectively, of eluates obtained from the experiments described for panel C. Increasing the amounts of control eluates revealed neither HAT nor HDAC activity.
Another GNAT member, PCAF, was not detected in the HDAC1 immunoprecipitates at an appreciable level, most likely because PCAF is expressed at a very low level in HeLa cells (27, 39). Because PCAF and GCN5 are 75% identical to each other in their amino acid sequences (39), it was of interest to test whether ectopically expressed PCAF associates with the HDAC1 complexes. As shown in Fig. 2B, extracts from HeLa cells transduced with a retrovirus vector with Flag-tagged PCAF (21) were immunoprecipitated with antibody for HDAC1 as described above and precipitated materials were tested for Flag-PCAF. Flag-PCAF was clearly coprecipitated along with HDAC1, as detected by both anti-PCAF antibody and anti-Flag antibody. As expected, Flag-PCAF was not precipitated when tested with preimmune sera.
To further establish the association of GCN5 with HDAC1, coimmunoprecipitation analysis was performed with HeLa cells expressing Flag-tagged HDAC1 (13). In Fig. 2C, extracts were immunoprecipitated with M2 anti-Flag antibody and blotted with antibody for GCN5. GCN5 was coprecipitated by anti-Flag antibody, along with Flag-HDAC1 and its components RbAp48, Sin3A, and Mta-L1. However, cells transduced with control vector did not precipitate HDAC1 or GCN5. Coomassie blue staining of the precipitated materials indicated that HDAC1 was in large excess over GCN5 in the precipitates. In addition, a relatively small proportion (<5%) of total GCN5 in the cells associated with HDAC1 complex (data not shown).
Given that HATs are associated with HDAC1, it was of interest to examine whether HAT activity is associated with the HDAC1 immunoprecipitates. As shown in Fig. 2D, extracts from cells expressing Flag-HDAC1 were precipitated with anti-Flag antibody and the precipitated materials were tested for HAT and deacetylase enzymatic activities. Interestingly, a significant HAT activity was detected with precipitates from cells expressing Flag-HDAC1; the levels of activity detected in 30 μl of eluates in Fig. 2D corresponded to ∼3 ng of recombinant PCAF. In contrast, no acetylase activities were detected in eluates from control cells (Fig. 2D, top panel). As expected, deacetylase activity was also detected in the same Flag-HDAC1 samples but not in control samples (Fig. 2D, lower panel). These results indicate that HDAC1 complexes possess both HAT and deacetylase activities, further supporting the association of HDAC1 and HAT.
PCAF and GCN5 interact with members of another acetylase family, p300 and CBP (28, 41). Since p300 and CBP display a slight structural similarity to GNAT family members (23), we examined whether these proteins are also coprecipitated with HDAC1. Tests with various available antibodies against p300 and CBP did not reveal either protein in the HDAC1 immunoprecipitates (data not shown). PCAF forms a large multiprotein complex that contains a set of conserved proteins, such as ADA2 and ADA3, and GCN5 forms a similar complex (9, 21, 33). To assess whether these GCN5- and PCAF-containing complexes associate with HDAC1, we searched for ADA2 and ADA3, as well as for PAF65α and hSPT3, components of the HAT complex (33), in the HDAC1 and Flag-HDAC1 immunoprecipitates. These proteins were not detected in either precipitate (data not shown), indicating that GCN5 and PCAF incorporated into the HDAC1 complex are independent of the previously described stable HAT complexes. It should also be noted that reciprocal immunoprecipitation in which Flag-PCAF was immunoprecipitated first, followed by immunoblotting for HDAC1, did not reveal a detectable level of HDAC1 in the eluates (data not shown). Although the basis for the absence of HDAC1 in Flag-PCAF immunoprecipitates is not entirely clear, it is possible that PCAF incorporated into HDAC1 complexes is sequestered and not readily accessible to the antibody.
Other class I histone deacetylases also form a complex with GCN5.
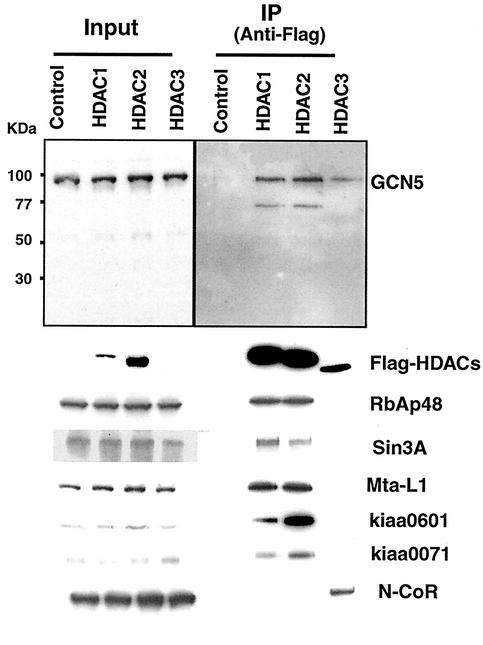
Class I deacetylases share a common structural fold (5, 15). HDAC1 and HDAC2 form stable multiprotein complexes that are similar to each other; in addition, HDAC1 and HDAC2 complexes can form a dimer (13). HDAC3, although it also forms stable complexes, appears to associate with unique components SMRT and N-CoR (16). To determine whether association with HATs is a general property among class I deacetylases, coimmunoprecipitation analysis was performed with Flag-tagged HDAC2 and HDAC3 expressed in HeLa cells. As shown in Fig. 3, GCN5 was coprecipitated along with Flag-HDAC2 and Flag-HDAC3. These data show that all class I deacetylases can associate with GCN5.
FIG. 3.
Flag-tagged HDAC2 and HDAC3 associate with GCN5 in HeLa cells. Extracts from HeLa cells infected with control virus or with recombinant virus with Flag-tagged HDAC1, HDAC2, or HDAC3 were immunoprecipitated with M2 anti-Flag antibody as described for Fig. 2 and subjected to blotting analysis for GCN5 and other indicated proteins.
Sedimentation analysis of the HDAC1-GCN5 complex.
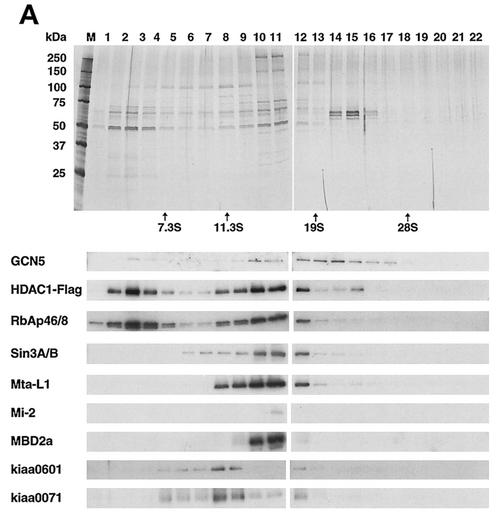
HDAC1 and HDAC2 form at least three distinct complexes containing, respectively, (i) mSin3A (11, 20, 45), (ii) Mi-2/NRD (32, 35, 40, 46), and (iii) CoREST/Kiaa0071 (13, 44). These complexes are each at minimum 200 to 400 kDa and are thought to assume distinct functions, e.g., transcriptional repression by the mSin3 complex and chromatin remodeling by the NRD complex. To assess whether HDAC-associated GCN5 is contained in any of the above complexes or whether it exists in a separate complex, immunoprecipitated Flag-HDAC1 complexes were subjected to velocity sedimentation analysis in glycerol gradient (13). Sediments were separated into 26 fractions corresponding to sedimentation coefficients of 4 to 28S. Figure 4 illustrates the results of silver staining (Fig. 4A, top two panels) and immunoblot analysis (4B and remaining panels of 4A) of each of the fractions. The majority of HDAC1 was found in fractions 8 through 12, containing RbAp48, mSin3A, Mta-L1 and Mi-2, and CoREST/kiaa0071. Importantly, HDAC1 was also found in the fractions exhibiting more-rapid sedimentation (factions 13 through 17), with sedimentation coefficients ranging from 15 to 26S. The bulk of the GCN5 detected was found in this most rapidly sedimenting fraction(s), peaking roughly at 21S. The GCN5-containing fractions may represent a previously unidentified large HDAC1 complex, because they have little RbAp48, mSin3A, MtaL1, and Kiaa0071 and because silver staining reveals several protein bands not present in fractions below 13S. The predicted average molecular mass of the GCN5-containing fractions is ∼1 MDa. Considering the broad spread over multiple fractions, the GCN5-containing fractions may represent several heterogeneous complexes. These results indicate that HDAC1 can form a large complex(es) distinct from the previously described complexes that contain GCN5. Although reproducibly detected in glycerol gradient analyses, the GCN5-containing complex(es) appears to represent a relatively small component of the total HDAC1 (<5%). We also noted that a certain amount of HDAC1 was detected in the fractions representing a smaller molecular mass corresponding to ∼5S (Fig. 4). Some of these components likely represent the HDAC core recruitment complex described by Zhang et al. (46).
FIG. 4.
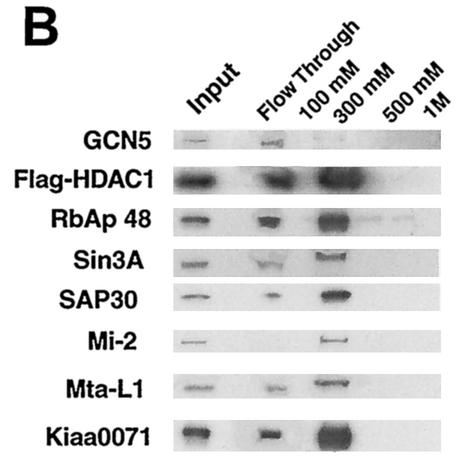
Sedimentation analysis of HDAC1-GCN5 complex(es). (A) Using M2 anti-Flag antibody agarose beads, Flag-HDAC1 complexes in HeLa cells were isolated and subjected to 10 to 35% glycerol gradient centrifugation. Each fraction was analyzed on SDS-10% PAGE. Upper two large panels: silver-staining patterns. Markers used were aldolase (7.3S), catalase (11.3S), thyroglobulin (19S), and 28S rRNA, as indicated at the bottom of the two large panels. Lower panels: immunoblot detection of indicated proteins. (B) Flag-HDAC1 complexes were fractionated on a DEAE-Sepharose column and eluted stepwise with buffer containing the indicated NaCl concentrations, and proteins were identified by immunoblotting analysis.
To obtain additional evidence of an HDAC1 complex(es) containing GCN5, we have fractionated the Flag-HDAC1 immunoprecipitates by DEAE-Sepharose chromatography. Figure 4B shows the results of immunoblot analysis of materials eluted stepwise with increasing concentrations of NaCl. HDAC1 was mostly found in two fractions, namely, the flow- through and the 300 mM NaCl eluates, but not in the 100 mM, 500 mM, or 1 M NaCl eluates. The HDAC-associated components, Sin3A, Mi-2, Mta-L1, and Kiaa0071, were predominantly eluted with 300 mM NaCl, indicating that this fraction corresponds to the previously described stable HDAC complexes (13, 32, 40, 44-46). In contrast, GCN5 was found almost totally in the flow-through fraction with a trace amount in the 300 mM NaCl eluates. A significant amount of RbAp48 appeared to be present in the flow through, as well as in 300 mM NaCl eluates. These results support the presence of a GCN5-containing HDAC1 complex that is distinct from the previously described HDAC complexes.
HDACs interact with PCAF in vitro.
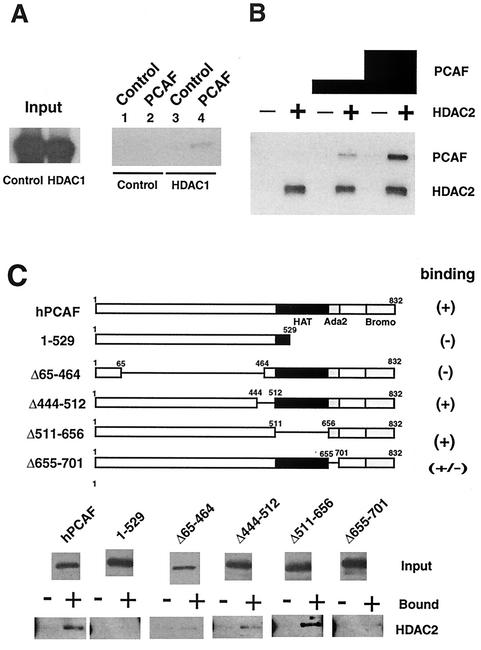
It was of importance to determine whether the association of HAT with HDAC is attributable to a direct protein-protein interaction or to an indirect association involving a third protein. As seen in the results shown in Fig. 5A, we tested whether HDAC1 binds to PCAF. 35S-labeled, in vitro-translated HDAC1 was incubated with recombinant PCAF and immunoprecipitated with anti-PCAF antibody. Radiolabeled HDAC1, but not radiolabeled luciferase, run as a control was coimmunoprecipitated with PCAF. Anti-PCAF antibody did not precipitate HDAC1 when incubated with an unrelated recombinant protein, ICSBP, verifying specific immunoprecipitation. A direct interaction between PCAF and HDAC2 was also observed (Fig. 5B). Histidine-tagged, recombinant HDAC2 conjugated to Ni2+ agarose beads (24) was incubated with increasing amounts of recombinant PCAF in these experiments, and bound PCAF was detected in immunoblot analysis. PCAF bound to HDAC2 in a dose-dependent manner but not to control beads conjugated to control protein, ICSBP.
FIG. 5.
HDAC-PCAF interactions in vitro. (A) 35S-labeled HDAC1 or 35S-labeled luciferase was incubated with Flag-tagged recombinant PCAF or a control protein (His-ICSBP) and coimmunoprecipitated with anti-PCAF antibody. Precipitated materials were detected by fluorography. The input represented 10% of the total reaction mixture. (B) Sf9 extracts containing recombinant His-HDAC2 (+) or the control (His-ICSBP) (−) were conjugated to Ni2+-NTA agarose beads and incubated with 50 ng or 500 ng of recombinant PCAF. Bound materials were analyzed by immunoblotting with anti-PCAF antibody. (C) PCAF domain analysis. HDAC2 (+) or control protein (ICSBP) (−) immobilized to Ni2+-NTA agarose was incubated with Flag-tagged PCAF deletions. Bound materials were detected by immunoblotting analysis with anti-FCAF antibody.
Using deletion analysis, we assessed domains of PCAF involved in the interaction with HDAC2. As shown in Fig. 5C, recombinant PCAF deletion mutants were incubated with full-length HDAC2 conjugated to Ni2+ beads. While constructs Δ444-512 and Δ511-656 bound to HDAC2, constructs 1-529 and Δ65-464 did not, indicating that an N-terminal region encompassing amino acids 65 to 464 and the C-terminal region encompassing amino acids 656 to 832 are required for interaction with HDAC2. These results indicate that HDACs and PCAF can interact with each other through protein-protein interaction.
Alteration of the HDAC1-HAT complex during U937 cell differentiation.
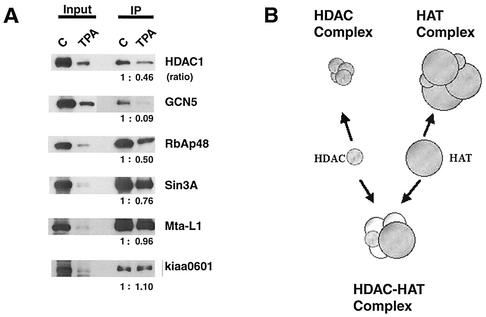
In an effort to understand the potential role of the HDAC-HAT complex(es), we sought to examine whether the formation of the HDAC-GCN5 complex changes during differentiation. For this purpose, a macrophage differentiation model using U937 cells was tested in which the myelomonocytic cells undergo differentiation in response to phorbol ester tetradecanoyl phorbol acetate (TPA) (3, 17). TPA treatment induced differentiation within 3 to 4 days, coinciding with increased adherence of cells to the petri dish and growth arrest. As shown in Fig. 6, nuclear extracts were prepared from cells treated with TPA for 4 days and tested for expression of HDAC1 and GCN5. As seen in the input lane, expression levels of HDAC1 and GCN5 were markedly reduced following TPA treatment in U937 cells, as measured on a protein content basis. Additionally, other proteins, including RbAp48, mSin3A, Mta-L1, and Kiaa0601, were also proportionally reduced in their levels, indicating that relative levels of these components were unchanged after TPA treatment. As shown in Fig. 6A, extracts were immunoprecipitated with antibody for the endogenous HDAC1 and tested for GCN5 and other proteins by immunoblotting. Endogenous GCN5 was coprecipitated along with HDAC1 as well as other HDAC1 components from untreated U937 cells, establishing that the association of HAT with HDAC1 is not restricted to HeLa cells. Surprisingly, HDAC1 precipitates from TPA-treated cells contained little GCN5, with levels almost undetectable by immunoblot assays (see quantification in Fig. 6A). The paucity of GCN5 was striking, since other HDAC1 components such as mSin3A and Mta-L1 as well as Kiaa0601 were coprecipitated from TPA-treated cells at levels comparable to or exceeding that in untreated cells. Quantification of the coprecipitated proteins (shown as the ratios in Fig. 6A) verified a selective reduction of GCN5, suggesting that the association of GCN5 with HDAC1 is modulated upon differentiation in U937 cells. PCAF appeared to be expressed at low levels in U937 cells and was not coprecipitated with HDAC1 at a detectable level before or after TPA treatment, suggesting that the reduction of GCN5 in the HDAC1 complex(es) was not due to the replacement of GCN5 by PCAF. These results were reproducibly observed with samples treated with TPA for 3 and 5 days. To assess whether the reduction in the HDAC1-GCN5 complex is a result of general growth arrest rather than differentiation, quiescent and exponentially growing WI38 fibroblasts were tested by a similar coimmunoprecipitation analysis. No discernible difference was detected for the relative amounts of GCN5 in the HDAC1 precipitates in these materials (data not shown), indicating that the selective reduction of GCN5-HDAC1 association is not a mere result of growth inhibition. These results suggest that interaction of HDAC1 with GCN5 is dynamic and is influenced by physiological processes such as differentiation.
FIG. 6.
Modulation of HDAC1-GCN5 interaction in differentiating U937 cells. (A) U937 cells were treated with TPA (20 ng/ml) for 4 days. Nuclear extracts were immunoprecipitated with anti-HDAC1 antibody and evaluated by immunoblotting analysis for the indicated proteins as described for Fig. 2A. The numbers indicate the ratios of precipitated proteins in untreated cells (assigned a value of 1) versus TPA-treated cells. (B) Model of an HDAC-HAT complex(es). A majority of HDACs and HATs form their own multiprotein complexes (top). A fraction of the two enzymes interact with each other and form an independent complex(es) (bottom), which may have a distinct function.
DISCUSSION
The aim of this study was to explore how HATs and deacetylases might mutually balance their activities in vivo. Our study began with the use of the novel flow cytometry-based FRET technique to search for an interaction of fluorescent protein-tagged PCAF and HDAC1 in living cells. Results demonstrated that in some cells, the acetylase and deacetylase reside within 50 to 60 Å of each other, a distance close enough to form a direct protein-protein interaction. The physical proximity of tagged PCAF and HDAC1 under these conditions is unambiguous, since another nuclear protein ICSBP displayed no FRET signals with either PCAF or HDAC1. To our knowledge this is the first demonstration of an in situ nuclear protein-protein interaction detected by FRET on a cell population basis. These observations provided a strong impetus to further study the interaction of the endogenous enzymes and its biochemical characteristics.
Coimmunoprecipitation experiments (Fig. 2 and 3) furnished a biochemical confirmation that endogenous HDAC1 is associated with endogenous GCN5. Furthermore, GCN5 was coprecipitated with Flag-tagged HDAC1, as well as HDAC2 and HDAC3, the latter having been shown to associate uniquely with N-CoR and SMRT (16). In addition, PCAF, another GNAT family member, was coprecipitated with HDAC1 when ectopically expressed in HeLa cells, indicating that the HDAC-HAT association is a general feature of the two enzymes. Supporting the presence of HAT in the complexes, Flag-tagged HDAC1 immunoprecipitates exhibited both HAT and deacetylase enzymatic activities.
Glycerol gradient sedimentation analysis revealed that GCN5 occurs in the fractions comprising a large protein complex(es) of an average size minimally estimated to be ∼1 MDa. This complex(es) appeared distinct from the previously reported stable HDAC complexes of 200 to 400 kDa that contain representative factors, mSin3, Mi-2/NRD, and Co-REST/Kiaa0071 (13, 32, 44-46). The presence of a distinct GCN5-HDAC complex(es) was further supported by the cofractionation of GCN5 and HDAC1 on an ion-exchange column (Fig. 4B). The GCN5-containing complex(es) may be heterogeneous in size and may contain a number of additional factors not included in the previously described HDAC complexes. GCN5 and PCAF contained in the HDAC complex(es) were associated neither with P300/CBP nor with other components of the previously characterized, stable HAT complexes (21, 33). Thus, GCN5 in the HDAC complex most likely represents an entity separate from the known HAT complexes and may have a distinct function (Fig. 6B). The HDAC-HAT association is in part attributed to a direct interaction of the two proteins, since recombinant HDACs and PCAF bound to each other in vitro. However, considering that the two proteins are in a large complex(es) and are capable of complexing with various other proteins, interaction of the two enzymes is most likely stabilized by other proteins in vivo.
The existence of a HDAC-HAT complex(es) appears compatible with the long-held notion that there is a mechanism for establishing and maintaining an equilibrium between HAT and deacetylase activities (23). It has been known that in vivo core histones are rapidly acetylated and deacetylated with a half-life of less than 10 min in ∼15% of chromatin (4). The chromatin regions that support rapid histone acetylation turnover are thought to coincide with the regions of high transcriptional activity where HATs and deacetylases can coexist (6). Thus, the HDAC-HAT complex(es) found in the present study may reside in this compartment of chromatin and participates in transcription. For example, the complex may play a role in ligand-dependent transcription by nuclear receptors, providing timely activation and repression of hormone-dependent gene expression. Consistent with this idea, HDACs and HATs both interact with a number of nuclear receptors (2, 38). In a similar context, signal-induced transcription is often followed by a rapid reversal of transcription leading to the restoration of basal gene expression. In these situations the HDAC-HAT complexes may help finely coordinate acetylation and deacetylation of local promoters. In support of this idea, there are other transcription factors such as YY1 and Sp1 (5) which interact with both HDAC and HAT and which may play a part in coordinated activation and repression of specific promoters.
On the other hand, the HDAC-HAT complexes may act in a more global manner, affecting a genome-wide state of histone acetylation (1, 14, 34). It has been shown that yeast core histones are rapidly and globally acetylated and deacetylated, creating a constant, dynamic flux in histone acetylation status. This rapid cycle of histone acetylation and deacetylation is dependent on the activity of gcn5 and rpd3, a yeast HDAC (34). It is possible that the HDAC-HAT complex(es) found in the present study contributes to this untargeted, globally acting enzymatic activity. This global acetylation and deacetylation is shown to confer a rapid return of transcription to a ground state, following activation or repression, occurring within a matter of minutes (14). Although this rapid, untargeted acetylation and deacetylation is likely to be important for transcriptional regulation, it may also be involved in a wider range of biological activities such as DNA replication and differentiation (1).
Lastly, we have shown that the GCN5-HDAC1 association is markedly reduced in U937 cells following TPA-induced differentiation. This result is interesting, since global alterations of chromatin structures and changes in transcriptional patterns are the main features of cellular differentiation (8). Our data indicate that assembly of HAT into the HDAC complex(es) is a dynamic process and is closely tied to the state of chromatin and cellular activities.
In conclusion, this study demonstrates that through a physical interaction, a certain fraction of HAT is integrated into a large HDAC complex(es) distinct from those described previously. The HDAC-HAT complex(es) may have a role in coordinating acetylation and deacetylation of chromatin affecting many cellular activities.
Acknowledgments
We thank S. Schreiber and E. Seto for HDAC plasmids, T. Howard for viral transduction of HeLa cells, and R. Swofford and K. Holmes for flow cytometry analysis.
REFERENCES
- 1.Berger, S. L. 2000. Local or global? Nature 408:412-415. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, J. C., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, Y. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, D. H., C. Angelin-Duclos, and K. Calame. 2000. BLIMP-1: trigger for differentiation of myeloid lineage. Nat. Immunol. 1:169-176. [DOI] [PubMed] [Google Scholar]
- 4.Covault, J., and R. Chalkley. 1980. The identification of distinct populations of acetylated histone. J. Biol. Chem. 255:9110-9116. [PubMed] [Google Scholar]
- 5.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Davie, J. R. 1998. Covalent modifications of histones: expression from chromatin templates. Curr. Opin. Genet. Dev. 8:173-178. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francastel, C., D. Schubeler, D. I. Martin, and M. Groudine. 2000. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell Biol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 9.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 10.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 12.Howe, L., C. E. Brown, T. Lechner, and J. L. Workman. 1999. Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr. 9:231-243. [DOI] [PubMed] [Google Scholar]
- 13.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 14.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 16.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masumi, A., and K. Ozato. 2001. Coactivator p300 acetylates the interferon regulatory factor-2 in U937 cells following phorbol ester treatment. J. Biol. Chem. 276:20973-20980. [DOI] [PubMed] [Google Scholar]
- 18.Masumi, A., I. M. Wang, B. Lefebvre, X. J. Yang, Y. Nakatani, and K. Ozato. 1999. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol. Cell. Biol. 19:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyawaki, A., and R. Y. Tsien. 2000. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 327:472-500. [DOI] [PubMed] [Google Scholar]
- 20.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 21.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Martin, J., and A. D. Johnson. 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki, S., L. A. Lesoon-Wood, A. Dey, T. Kuwata, B. D. Weintraub, G. Humphrey, W. M. Yang, E. Seto, P. M. Yen, B. H. Howard, and K. Ozato. 1999. Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin beta gene. EMBO J. 18:5389-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 26.Siegel, R. M., J. K. Frederiksen, D. A. Zacharias, F. K. Chan, M. Johnson, D. Lynch, R. Y. Tsien, and M. J. Lenardo. 2000. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288:2354-2357. [DOI] [PubMed] [Google Scholar]
- 27.Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 30.Stryer, L. 1978. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 47:819-846. [DOI] [PubMed] [Google Scholar]
- 31.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 32.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 33.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 34.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 35.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 36.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe, A. P. 1997. Transcriptional control. Sinful repression. Nature 387:16-17. [DOI] [PubMed] [Google Scholar]
- 39.Xu, W., D. G. Edmondson, and S. Y. Roth. 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 18:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 41.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 43.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]