Abstract
Cyclin-dependent kinase inhibitors (CDKIs) have been shown to block human immunodeficiency virus and herpes simplex virus. It is hypothesized that CDKIs block viral replication by inhibiting transcription of specific cellular genes. Here we find that three CDKIs, flavopiridol, purvalanol A, and methoxy-roscovitine, block Moloney murine leukemia virus (MLV) transcription events. Using gene expression microarray technology to examine the inhibitory effects of CDKIs, we observed a cellular gene, the pre-B-cell leukemia transcription factor 1 (Pbx1) gene, down-regulated by CDKI treatment. The PBX consensus element (PCE), TGATTGAC, is conserved in the long terminal repeats of several murine retroviruses, including Moloney MLV. Mutations in the PCE completely inhibited viral transcription whereas overexpression of PBX1 and a PBX1-associated protein, PREP1, enhanced viral transcription. The interaction between the PCE and PBX1-PREP1 proteins was confirmed by gel shift experiments. Blocking PBX1 protein synthesis resulted in a significant decrease in viral transcription. Collectively, our results represent the first work demonstrating that the homeodomain proteins PBX1 and PREP1 are cellular factors involved in Moloney MLV transcription regulation.
Cyclin-dependent kinases (CDKs) are key regulators of cell cycle control and transcription. Nine CDKs have been identified thus far. CDK1 to CDK7 are required for cell cycle regulation, and CDK7 to CDK9 are involved in RNA polymerase II-dependent transcription (33). In fact, CDK2, CDK4, and CDK6 also regulate transcription by activating E2F via specific phosphorylation of the retinoblastoma gene product (Rb protein) in complex with E2F (13, 49). It has been shown elsewhere that approximately 90% of all neoplasias are associated with CDK hyperactivation and subsequent Rb pathway inactivation (21). Therefore, CDKs represent attractive targets for cancer therapy, and several CDK inhibitors (CDKIs) have been developed (4, 16, 18, 48). Two CDKIs, flavopiridol (Flavo) and UCN-1, are already in clinical trials as potential anticancer therapeutics (44, 49).
The antiviral activities of CDKIs were first observed in studies describing their interaction with positive transcription elongation factor b (P-TEFb), a factor required for human immunodeficiency virus (HIV) replication (32, 54). P-TEFb, composed of CDK9 and a cyclin subunit derived from one of three different genes (cyclin T1, T2, or K), controls RNA polymerase II-dependent transcription elongation by phosphorylating the carboxyl-terminal domain of the large subunit of RNA polymerase II (41). Most importantly, P-TEFb is a cellular cofactor required for Tat-dependent transactivation of the HIV genome (54). One study used a combination of in vitro and in vivo assays to screen more than 100,000 compounds for inhibitors of HIV Tat transactivation (32). All the compounds identified, including 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), were found to inhibit P-TEFb (32). These results suggest that CDK9 represents a potential target for the development of novel HIV therapeutics. Flavo has been recently identified as the most potent P-TEFb inhibitor (6, 7). Infection of cultured cells by HIV has been shown to be potently inhibited by Flavo (50% inhibitory concentration [IC50] = 8 nM) without significant toxic effects (6). Surprisingly, at 30 and 100 nM Flavo (the conditions which completely block HIV Tat transactivation and replication), negligible effects on cellular transcription were observed (7). It is possible that specific sets of cellular genes are inhibited by low intracellular concentrations of Flavo and that some of these genes might be involved in HIV replication. If such hypersensitive genes do exist, they could represent ideal targets for development of novel anti-HIV therapeutics. Thus far, no such cellular genes have been identified. Other CDKIs, including purvalanol A (Purv), roscovitine (Ros), and olomoucine (Olo), have also been shown elsewhere to block HIV basal and Tat-activated transcription, but much less potently than Flavo (53).
Ros and Olo were also shown previously to block herpes simplex virus (HSV) replication and transcription (45, 46). It has been shown previously that Ros and Olo inhibit human CDK7 and CDK9 effectively (IC50 = 0.6 μM) (20, 53). These results again raise the possibility that Ros and Olo block HSV by blocking transcription of specific cellular genes. It is important to determine if such CDKI-hypersensitive genes indeed exist and to examine the possible involvement of these cellular targets in viral transcription and replication. Furthermore, it should be very interesting to determine if CDKIs could be utilized as broad-spectrum antiviral reagents by examining their effects on other viruses.
In this paper, we examine the effects of CDKIs on murine leukemia virus (MLV), which is implicated in oncogenesis and used in the development of gene therapy strategies (19, 52). We found that three CDKIs, including Flavo, Purv, and methoxy-roscovitine (MeO-Ros), were able to block Moloney MLV transcription. We observed that both viral and cellular transcription were inhibited by CDKIs. Cellular genes whose expression is dramatically inhibited by CDKIs were identified by microarray technology. Since our goal was to identify cellular genes affecting viral transcription, we further investigated the subset of CDKI-affected cellular genes involved in transcription regulation. One cellular homeodomain gene, the pre-B-cell leukemia transcription factor 1 (Pbx1) gene, was identified as an important regulator of MLV transcription. We found a PBX1 binding sequence, PBX consensus element (PCE), TGATTGAC, that was perfectly conserved in the U5 region of the Moloney MLV long terminal repeat (LTR) and 14 other murine retroviruses. The PCE was first identified by Knoepfler and Kamps (25). Previous work has demonstrated that PBX1 binds the PCE as a heterodimer with other homeodomain factors such as MEIS1 or PREP1 (5, 25). Selection studies with degenerate oligonucleotides have shown that PBX1 binds to the first half of the PCE (i.e., TGAT) and that MEIS1 or PREP1 contacts the second half (i.e., TGAC) (24). Gel shift assays demonstrated the interaction between the PCE and PBX1-PREP1 proteins, whereas mutations in the PCE resulted in complete inhibition of viral transcription. In addition, overexpression of Pbx1 and Prep1 increased MLV transcription and partially eliminated the inhibitory effects of CDKIs. Furthermore, we demonstrated that transfection of antisense oligonucleotides and short interfering RNAs (siRNAs) directed against Pbx1 resulted in a significant inhibition of MLV transcription. We conclude that the homeodomain proteins PBX1 and PREP1 are involved in the Moloney MLV replication cycle through regulation of viral transcription.
MATERIALS AND METHODS
Compounds and cell lines.
MeO-Ros (a derivative of Ros) was synthesized as described previously (17). Purv (Calbiochem), Ros (Calbiochem), MeO-Ros, DRB (Sigma), and Flavo (obtained from the National Cancer Institute) were dissolved in dimethyl sulfoxide (DMSO) to 10 mM. NIH 3T3 and human embryonic kidney 293 cells were obtained from the American Type Culture Collection.
Plasmids.
Two similar MLV-based retroviral vectors were used to measure MLV transcription, MLV-LTR-Luc and WZL-Luc. The first, MLV-LTR-Luc, used exclusively in retroviral infection assays, was derived from pBabe-neo (34), wherein (i) the simian virus 40 promoter was replaced with the internal ribosome entry site (IRES) of hepatitis C virus, (ii) the neomycin resistance gene was changed to the luciferase gene from the pGL2 control plasmid (Promega), and (iii) the U3 region of the 5′ LTR was replaced with the strong cytomegalovirus (CMV) promoter for higher-titer virus production. MLV-LTR-Luc was used in the Moloney MLV infection assay to generate viral supernatants (see below). During retroviral replication, the wild-type U3 region is recovered before integration so that measurements of MLV transcription are dependent on wild-type LTR (U3-R-U5)-driven luciferase expression. The second vector, WZL-Luc, used exclusively in transient-transfection assays, was also derived from pBabe-neo (34), wherein (i) the simian virus 40 promoter was replaced with the IRES of encephalomyocarditis virus and (ii) the firefly luciferase gene from the pGL2 control plasmid (Promega) was cloned upstream of the IRES. In the resulting configuration, WZL-Luc, the MLV LTR drives luciferase expression, and the IRES maintains expression of the neomycin resistance gene. Therefore, in both MLV-LTR-Luc, which measures LTR-driven transcription from integrated proviral sequences, and the WZL-Luc vector, which measures LTR-driven transcription from unintegrated, transfected plasmid, luciferase expression is driven by identical MLV LTRs. The mammalian expression vector for human NFAT1 has been described previously (37). CMV-PBX1a and CMV-PBX1b were obtained from Michael L. Cleary. pSV-SPORT-MEIS1 and pSV-SPORT-PREP1, containing the cDNAs for mouse Meis1 and human Prep1, respectively, were provided by F. Bradley Hillgartner. The coding sequences of Meis1 and Prep1 were subcloned into pcDNA6 (Invitrogen) to generate CMV-MEIS1 and CMV-PREP1. The pCITE-PBX1a, pCITE-MEIS1, and pCITE-PREP1 constructs were generated by cloning the coding sequences of Pbx1a, Meis1, and Prep1 into pCITE plasmids (Novagen). The pCITE constructs were used to synthesize translated PBX1a-MEIS1-PREP1 protein in vitro.
Moloney MLV infection assay.
The production of MLV supernatants was conducted as previously described (39) with a retroviral producer cell line transfected with MLV-LTR-Luc. 3T3 or 293 cells were grown to 50 to 80% confluence in 96-well plates and incubated with individual CDKIs for 30 min. MLV-LTR-Luc supernatants were then added and incubated for 24 h. Luciferase activity was measured with the Bright-Glo assay system (Promega), and the activity was determined with an Acquest Ultra-HTS system (LJL Biosystems, Inc.). For the overexpression assays, 293 cells were transfected with the indicated expression vectors (i.e., CMV-PBX1a, -PBX1b, -MEIS1, -PREP1, or -NFAT) and a CMV-β-galactosidase (β-Gal) internal control plasmid for 24 h with Fugene 6 reagent as described in the manufacturer's manual (Roche). MLV-LTR-Luc supernatants were then added and incubated with cells for another 48 h. Luciferase and β-Gal activities were measured with the dual-light system (Applied Biosystems). β-Gal activity was used to normalize luciferase activity to account for differences in transfection efficiency. The IC50s were determined with the GraphPad Prism program (GraphPad Software, Inc.).
Cell proliferation assay.
3T3 cells were incubated with CDKIs at indicated concentrations for 24 h. Effects of the CDKIs on proliferation were determined with the cell proliferation enzyme-linked immunosorbent assay system, version 2, based on the measurement of 5-bromo-2′-deoxyuridine (BrdU) incorporation during DNA synthesis in proliferating cells (Amersham Pharmacia Biotech, Inc.).
Moloney MLV transcription assay.
Two different approaches were utilized to examine the effects of CDKIs on MLV transcription. 3T3 or 293 cells were grown to 50 to 80% confluence in 96-well plates and transfected with WZL-Luc vector DNA with Fugene 6 or Lipofectamine Plus reagent as described in the manufacturer's manual (Invitrogen). Three hours posttransfection, CDKIs were added at the indicated concentrations for another 18 to 24 h. Effects of CDKIs on unintegrated MLV LTR-driven transcription were measured by assaying luciferase activity. Another approach with cells stably expressing integrated MLV LTR proviral sequences was used to measure viral transcription. 293 cells were incubated with MLV supernatants (MLV-LTR-Luc, as described previously) for 18 h to allow viral entry and integration. The medium was removed, and the cells were washed with phosphate-buffered saline. The MLV-LTR-Luc-integrated 293 cells were treated with CDKIs for 24 h followed by luciferase activity measurements. For the overexpression assays, 293 cells were cotransfected with the WZL-Luc retroviral vector, the indicated expression vectors (i.e., CMV-PBX1a, -PBX1b, -MEIS1, -PREP1, or -NFAT), and a CMV-β-Gal internal control plasmid for 24 h. Detection of β-Gal and luciferase activity was performed as described above.
Microarray procedure.
Microarray analysis was performed essentially as previously described (31). Briefly, murine NIH 3T3 cells were treated with 10 μM Purv, 10 μM MeO-Ros, 0.3 μM Flavo, or vehicle (DMSO) for 6 h (final concentration of DMSO in the medium, 0.1%) followed by total cellular RNA isolation with the RNeasy Mini kit (Qiagen). Five micrograms of total RNA was used to synthesize cDNA that was then used as a template to generate biotinylated cRNA. cRNA was fragmented and added to Affymetrix (Santa Clara, Calif.) MGU74Av2 chips (containing probes for over 6,000 mouse genes) as described in the standard protocol outlined in the gene chip expression analysis technical manual (Affymetrix). After sample hybridization, microarrays were washed and scanned with a laser scanner (Agilent, Palo Alto, Calif.). Each treatment condition was represented with duplicate arrays as total RNA preparation and cDNA and cRNA synthesis, and array hybridizations were performed in parallel from duplicate drug treatments.
Microarray data analysis.
Regulated probe sets were chosen from Affymetrix MAS 4.0 output files by filtering for differences in the average difference values for each CDKI and vehicle (DMSO). Filtering was done by two methods. The first method involved using Student's t test for each probe set with the average difference values of the duplicates. Probe sets with differences between drug and vehicle were chosen if P values were less than 0.01, the fold change in expression was at least an absolute value of 2.0, and the average difference values were at least a value of 200 from at least two of the four chips. The second filter also selects for probe sets with a fold change of at least 2.0 and average difference values of at least 200 from at least two of the four chips but also requires that the standard deviation of the duplicate values divided by the mean of the duplicate values be no more than 10% for either condition. Results from both filtering methods were combined, and comparisons between the different CDKIs were made.
Quantitative RT-PCR.
Oligonucleotide probe and primers for Pbx1 (the probe, forward, and reverse primers corresponded to regions 565 to 589, 532 to 552, and 604 to 623, respectively, after the start codon of Pbx1; accession no. NM_002585) were purchased from Applied Biosystems. Reverse transcription-PCR (RT-PCR) was done with the TaqMan One-Step RT-PCR Master Mix reagents kit as described by the manufacturer (Applied Biosystems). Amplification and detection of specific products were performed with the ABI Prism 7700 sequence detection system (Applied Biosystems).
Western blots.
3T3 cells were treated with 10 μM Purv, 10 μM MeO-Ros, 0.3 μM Flavo, or vehicle (DMSO) for 6 h. Cells were lysed in a solution containing 10 mM Tris (pH 7.6), 150 mM NaCl, 2 mM MgCl2, 1% Triton X-100, 15% glycerol, 0.1% of a saturated solution of phenylmethanesulfonyl fluoride in isopropanol, and protease inhibitor cocktail tablets (Roche). The cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immobilized on nitrocellulose filters. PBX1 proteins were detected by Western blot analysis and quantitated by the NIH Java-based image processing program (NIH Image/J). The polyclonal antibodies against PBX1a-PBX1b and against γ-tubulin were obtained from Santa Cruz Biotechnology, Inc. A monoclonal antibody against the PBX1b isoform was kindly provided by M. L. Cleary (47).
Gel mobility shift assay.
The U5 region oligonucleotides, which contained wild-type PCE (5′-GGGAGGGTCTCCTCTGAGTGATTGACTACCCGTCAGCGGG [sequences of PCE are underlined]), mutant PCE 1mt (5′-GGGAGGGTCTCCTCTGAGCCCCTGACTACCCGTCAGCGGG [sequences of PCE mutant are underlined]), or mutant PCE 2mt (5′-GGGTCTCCTCTGAGTGATCCCCTACCCGTCAGCGGGGGTC), were used as the DNA probes. Double-stranded oligonucleotides were prepared by combining equal amounts of complementary single-stranded DNAs in a solution containing 10 mM Tris (pH 8.0) and 1 mM EDTA followed by heating to 95°C for 2 min and then cooling to room temperature. DNA binding reactions and 32P labeling of oligonucleotides were performed with gel shift assay systems (Promega). Anti-PBX1, anti-MEIS1 (Santa Cruz Biotechnology, Inc.), anti-PREP1 (Santa Cruz Biotechnology, Inc.), or anti-γ-tubulin antibodies were incubated with nuclear extracts of 293 cells for 10 min before the 32P-labeled probe was included. PBX1a, MEIS1, and PREP1 proteins were produced in vitro from T7 expression plasmids (i.e., pCITE plasmids) with a coupled reticulocyte lysate system (TNT Quick Coupled transcription-translation systems; Promega). DNA binding reactions were performed at 4°C for 30 min when in vitro-translated proteins were used. DNA and DNA-protein complexes were resolved on 6% nondenaturing polyacrylamide gels at room temperature in 0.3× TBE (27 mM Tris-borate [pH 8.3], 0.6 mM EDTA). Following electrophoresis, the gels were dried and exposed to X-ray film.
PCE mutants.
To generate mutations in the PCE of the Moloney MLV LTR, site-directed mutagenesis was performed according to the Stratagene manual (QuikChange site-directed mutagenesis kit) with a WZL plasmid as the template. The primers used to create the PCE mutants are as follows: PCE 1mt (5′-GGGAGGGTCTCCTCTGAGCCCCTGACTACCCGTCAGCGGG and 5′-CCCGCTGACGGGTAGTCAGGGGCTCAGAGGAGACCCTCCC), PCE 2mt (5′-GGGTCTCCTCTGAGTGATCCCCTACCCGTCAGCGGGGGTC and 5′-GACCCCCGCTGACGGGTAGGGGATCACTCAGAGGAGACCC), and PCE 12mt (5′-GGGAGGGTCTCCTCTGAGCCCCCCCCTACCCGTCAGCGGGGGTC and 5′-GACCCCCGCTGACGGGTAGGGGGGGGCTCAGAGGAGACCCTCCC). Luciferase sequences (obtained from the BamHI and SalI digestions of WZL-Luc) were cloned into the PCE-mutated WZL plasmids (i.e., PCE 1mt, PCE 2mt, and PCE 12mt). Mutations were confirmed by DNA sequencing. 293 cells were transiently transfected with wild-type or mutant WZL-Luc, and luciferase activity was determined 48 h posttransfection.
Antisense oligonucleotides and siRNAs.
Two sets of sense and antisense oligonucleotides were utilized to inhibit PBX1 protein synthesis: 61Pbx (sense) and as61Pbx (antisense) corresponded to region 61 to 80 after the start codon (accession number NM_002585) and 421Pbx and as421Pbx targeted positions 421 to 440 from the start codon. The sense oligonucleotides served as controls. In addition, a pcDNA3.1 reverse primer (5′-TAGAAGGCACAGTCGAGG) was used as another control. 293 cells were cotransfected with 0.035 μg of WZL-Luc and 250 nM oligonucleotides. Luciferase activity was measured 24 h after transfection. An siRNA targeting positions 220 to 243 after the start codon of Pbx1 was also generated (Dharmacon Research, Inc.). The siRNA of Renilla luciferase was used as a control (14). 293 cells were transfected with 0.02 μg of siRNAs for 24 h. Ten microliters of MLV-LTR-Luc viral supernatants was added to the cells and incubated for another 24 h. Luciferase activity was measured as described previously.
RESULTS
CDKIs block Moloney MLV infection.
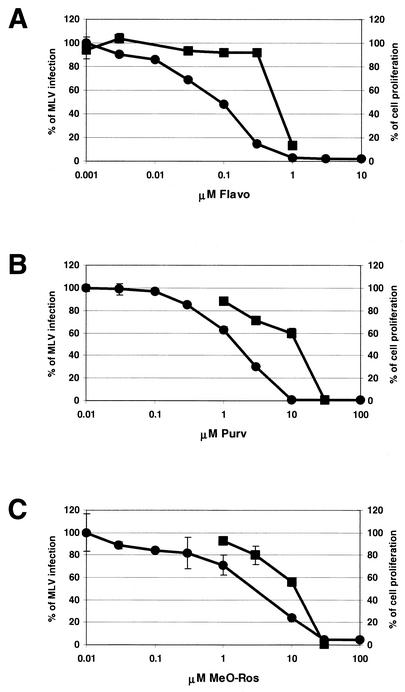
Besides the potential use of CDKIs in cancer therapy, previous studies have demonstrated that CDKIs, including Flavo, DRB, Purv, and Ros, also inhibit HIV and HSV infection (6, 32, 46, 53). We first examined the effects of Flavo, Purv, and MeO-Ros in the Moloney MLV infection assay (Fig. 1). MeO-Ros is a derivative of Ros and a more potent CDK1 inhibitor than Ros (N. S. Gray, unpublished data). Using a genetically modified Moloney MLV, whose viral genome has been replaced by firefly luciferase under the control of the MLV LTR (i.e., the viral promoter), we measured the effects of the CDKIs on MLV retroviral infection. In these experiments, luciferase would have been expressed only upon successful integration of the provirus, thus allowing luciferase activity to be used as a surrogate reporter for infection. All three CDKIs blocked MLV infection but with different efficiencies. Flavo demonstrated the most potent inhibition (IC50 = 0.083 μM) whereas Purv (IC50 = 1.6 μM) and MeO-Ros (IC50 = 6.3 μM) had similar inhibitory effects (Fig. 2, solid circles). We also examined the effects of Ros and DRB on MLV infection. Both compounds were able to inhibit MLV infection but less effectively than Flavo (Ros IC50 = 13.7 μM, DRB IC50 = 15.4 μM; data not shown). Since CDKIs induce apoptosis in numerous cell types, we next examined the CDKIs' effect on cell viability by performing Alamar blue proliferation assays (36). No significant toxic effects were detected when concentrations of Flavo were ≤300 nM and when concentrations of Purv and MeO-Ros were ≤10 μM (data not shown).
FIG. 1.
Structures of Flavo, Purv, and MeO-Ros.
FIG. 2.
Effects of the CDKIs on Moloney MLV replication and cell proliferation. MLV assays were performed with 3T3 cells as described in Materials and Methods. MLV infection was blocked by treatment with Flavo (A), Purv (B), and MeO-Ros (C) (solid circles). The IC50s were determined for Flavo (IC50 = 0.083 μM), Purv (IC50 = 1.6 μM), and MeO-Ros (IC50 = 6.3 μM). The effects of Flavo, Purv, and MeO-Ros on 3T3 cell proliferation were determined by BrdU assays (solid squares). Results were reproducible, and data from one experiment are presented. Bars indicate the errors between duplicates.
It has been shown previously that Moloney MLV infects replicating but not growth-arrested cells (42). Since CDKIs target CDK-dependent cell cycle regulation, it is possible that these CDKIs block Moloney MLV by causing cell cycle arrest. To clarify the inhibitory mechanisms of the CDKIs on MLV, we performed a BrdU incorporation assay to determine the effects of CDKIs on cell proliferation. Increasing concentrations of all three CDKIs blocked proliferation (Fig. 2, solid squares); however, the IC50 of each CDKI for proliferation was consistently higher than the IC50 for MLV infection. For example, 0.3 μM Flavo resulted in an 85% decrease in MLV infection but had very little effect on cell proliferation (only 8% inhibition [Fig. 2]). Therefore, a window between inhibition of MLV infection and induction of cell cycle arrest exists wherein the CDKIs block the MLV infection readout independently of their cell cycle effects (Fig. 2). The data indicate that Flavo, Purv, and MeO-Ros function as novel Moloney MLV infection inhibitors and that induction of cell cycle arrest might not be the only mechanism utilized by the CDKIs in blocking MLV infection.
CDKIs inhibit transcription of Moloney MLV.
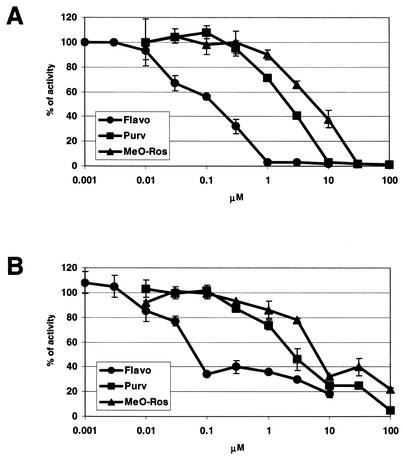
CDK7 and CDK9 are essential transcription factors involved in transcription initiation and elongation, respectively. Flavo was previously identified as the most potent CDK9 inhibitor, which associates with CDK9 at a 1:1 stoichiometric ratio even at enzyme concentrations in the low-nanomolar range (6, 7). In addition, it has been shown previously that Ros blocks CDK7 and CDK9 effectively (32, 53). These observations led us to examine if viral transcription driven by the Moloney MLV LTR could be blocked by the CDKIs. The results of the MLV infection assay (Fig. 2) show that the CDKIs inhibit MLV infection, resulting in decreased levels of luciferase activity. However, the MLV infection assay cannot distinguish between inhibition of events leading up to proviral integration into host chromatin and inhibition of retroviral LTR-driven transcription. To determine if the CDKIs might inhibit the latter, two different experimental approaches were utilized. First, murine NIH 3T3 cells were transiently transfected with a WZL-Luc retroviral construct that contains luciferase coding sequences under the control of the Moloney MLV LTR. A decrease in activity of the reporter gene was detected in response to treatment with all three CDKIs (Fig. 3A). The IC50s for inhibition of viral transcription by the CDKIs (Flavo IC50 = 0.11 μM, Purv IC50 = 2.1 μM, and MeO-Ros IC50 = 6.8 μM) closely approximated the IC50s affecting viral infection (Fig. 2 and 3A). Identical experiments were performed with 293 cells, and similar IC50s were obtained (data not shown). Another set of experiments was performed to determine if the CDKIs could inhibit MLV LTR-driven transcription from an integrated provirus. Cells containing stably integrated MLV-LTR-Luc proviruses were utilized. As shown in Fig. 3B, all three CDKIs blocked transcription with similar inhibitory efficiencies as those determined for Fig. 3A. The results suggest that the CDKIs specifically block transcription of Moloney MLV, and not necessarily the processes upstream of viral integration.
FIG. 3.
Effects of the CDKIs on transcription of Moloney MLV. (A) 3T3 cells were transiently transfected with WZL-Luc (i.e., MLV-LTR-Luc) in the presence of the CDKIs. Luciferase activity was measured 24 h after transfection, and the IC50s were determined for Flavo (IC50 = 0.11 μM), Purv (IC50 = 2.1 μM), and MeO-Ros (IC50 = 6.8 μM). (B) An MLV-LTR-Luc integrated cell line (MLV-LTR-Luc 293 cell line) was incubated with the CDKIs for 24 h followed by luciferase activity measurements. Assays were repeated twice in duplicate. Shown are representative experiments where bars indicate the errors between duplicates.
Identification of cellular genes involved in Moloney MLV transcription.
By targeting CDK7 and CDK9, factors required for transcription, CDKIs block not only viral but also cellular transcription. It is possible that the CDKIs could inhibit MLV transcription by blocking expression of critical cellular genes that regulate viral transcription. Previous studies of HIV inhibition by Flavo suggest that Flavo-hypersensitive cellular genes might be involved in HIV replication (6, 7). To determine if such CDKI-hypersensitive cellular genes could be involved in MLV transcription, microarray experiments were performed to identify genes whose transcripts were affected by CDKIs. Since the CDKIs presented in this study are all transcription inhibitors, we expected to find on the order of hundreds of genes influenced by a CDKI, while only a few would be involved in regulating MLV transcription. To overcome this specificity problem, we selected only those genes whose expression was affected by at least two of the three CDKIs. Since all three CDKIs target CDK proteins and all three can block MLV transcription, it follows that the CDKIs might affect expression of the same cellular genes to exert their inhibitory mechanism. Therefore, we reasoned that genes affected by more than one CDKI would represent more likely targets of the CDKIs and thus required cellular factors for MLV transcription. Through this simple selection process, we could eliminate nonspecific genes and single out candidate genes that regulate MLV transcription.
Utilizing microarray analysis, we attempted to identify cellular genes selectively deregulated by CDKIs. Briefly, 3T3 cells were treated with 10 μM Purv, 10 μM MeO-Ros, and 0.3 μM Flavo and then total RNAs were isolated for analysis. Under these conditions, 76 to 99% of MLV transcription was blocked whereas 56 to 92% of the cells were still replicating without significant cellular toxicity (Fig. 2 and data not shown). The microarray data were analyzed as described in Materials and Methods. In agreement with these observations, the results showed that the majority of cellular transcripts were not significantly affected (85 to 89% of cellular RNAs had less than twofold changes; data not shown). We found that 261 cellular genes were significantly affected by Purv, MeO-Ros, and Flavo. Fifty-seven out of 261 genes were affected by two or three CDKIs and were classified as putative MLV regulatory genes. These candidates include 25 known genes (Table 1) and 32 expression sequences-RIKEN cDNAs (data not shown). Since our goal in this study was to identify cellular genes affecting viral transcription, we focused only on the candidate genes involved in regulation of transcription.
TABLE 1.
CDKI-regulated cellular genes—putative regulators of MLV
| Gene product | Avg difference value for conditiona:
|
|||
|---|---|---|---|---|
| DMSO | Flavo | Purv | MeO-Ros | |
| PBX1 | 1,572 | 377 | 769 | 1,307 |
| High-mobility group protein HMG I-C | 1,959 | 1,452 | 485 | 427 |
| CCAAT-box-binding transcription factor | 431 | 103 | 374 | 110 |
| Putative myelin regulatory factor 1 | 1,402 | 519 | 697 | 389 |
| Forkhead box protein G1B | 301 | −36 | 350 | 50 |
| DEAD-box protein 3 | 690 | 147 | 395 | 189 |
| DNA-directed RNA polymerase II, largest subunit | 415 | 24 | 380 | −34 |
| Histone H1 | 1,747 | 151 | 1,506 | 625 |
| Histone H3 | 2,526 | 871 | 2,260 | 1,185 |
| Serine/threonine protein phosphatase PP1-γ catalytic subunit | 1,117 | 412 | 609 | 303 |
| Serine/threonine-protein kinase PLK | 431 | 67 | 232 | 101 |
| Prohibitin | 294 | 213 | 85 | 126 |
| Protein translation factor SUI1 homolog | 960 | 403 | 2,586 | 1,259 |
| Casein kinase II, alpha chain | 906 | 397 | 1,015 | 206 |
| Myristoylated alanine-rich C-kinase substrate | 403 | 193 | 336 | 121 |
| Mus musculus, similar to RAS p21 protein activator | 270 | 74 | 223 | 98 |
| Quaking | 748 | 204 | 635 | 251 |
| Nucleolar protein GU2 | 273 | 121 | 262 | 94 |
| Cell division cycle 20 homolog (Saccharomyces cerevisiae) | 1,757 | 864 | 1,297 | 869 |
| Secretory leukocyte protease inhibitor | 280 | 703 | 699 | 735 |
| Ubiquitin-conjugating enzyme E2, 23 kDa | 133 | 310 | 266 | 268 |
| Growth arrest and DNA damage-inducible protein GADD45 β | 131 | 222 | 272 | 473 |
| Cd27 binding protein | 413 | 843 | 610 | 969 |
| Phosphodiesterase 4B, cyclic AMP specific | 11 | 233 | 255 | 162 |
| Wild-type p53-induced gene 1 | 307 | 485 | 617 | 672 |
Affymetrix average difference values for means of duplicate chips.
PBX1 and PREP1 are required for Moloney MLV transcription.
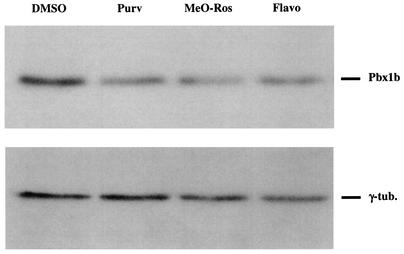
We further investigated the involvement of the transcription factor Pbx1 gene, one of the candidate cellular genes, in Moloney MLV transcription. We first performed quantitative RT-PCR to confirm the decreased expression of Pbx1 mRNA observed by microarray analysis. In agreement with the microarray data, the results showed that treatment with 10 μM Purv, 10 μM MeO-Ros, or 0.3 μM Flavo resulted in 2.1-, 4.9-, and 3.1-fold decreases in Pbx1 mRNA, respectively. In previous studies, alternative splicing of Pbx1 transcripts has been observed, resulting in generation of two isoforms, PBX1a and PBX1b (23). Western blot analysis performed with antibodies recognizing both PBX1a and PBX1b isoforms showed that only the PBX1b proteins were present in NIH 3T3 cells (data not shown). Cell lysates of NIH 3T3 cells treated with 10 μM Purv, 10 μM MeO-Ros, or 0.3 μM Flavo were analyzed by Western blotting. As shown in Fig. 4, treatment with any of the three CDKIs caused a decrease in PBX1b protein levels. These data indicate that treatments of cells with Purv, MeO-Ros, and Flavo all result in a significant decrease in PBX1 protein expression in treated cells.
FIG. 4.
Effects of the CDKIs on PBX1b protein levels. Cell lysates of NIH 3T3 cells treated with DMSO, 10 μM Purv, 10 μM MeO-Ros, or 0.3 μM Flavo (final concentration of DMSO, 0.1%) were analyzed by Western blotting with a monoclonal antibody against PBX1b isoforms and quantitated by NIH Image/J. γ-Tubulin was utilized as a loading control. After normalization of the quantitated data with the loading control, the effect of CDKI treatment resulted in a 34% (Purv), 38% (MeO-Ros), and 27% (Flavo) decrease in PBX1b protein levels. The results shown are representative of three assays with different anti-PBX1 antibodies.
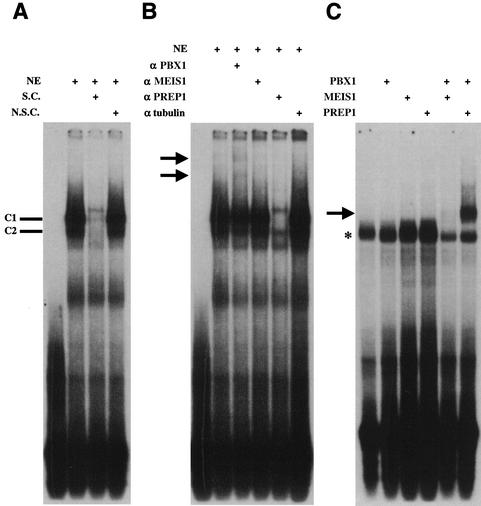
A direct connection between Moloney MLV and PBX1 was detected upon careful observation of the MLV LTR DNA sequence which contains an exact match of the PCE, TGATTGAC, in the U5 region of Moloney MLV LTR. The potential importance of this element is highlighted by its inclusion in 14 other murine retroviruses, including Abelson MLV, CAS-BR-E MLV, HoMuLV MLV, AKV MLV, DG-75 MLV, MLV SL3-3, Friend MLV, Rauscher MLV, MLV MCF1233, Moloney murine sarcoma virus, myeloproliferative sarcoma virus, Kirsten murine sarcoma virus, FBR murine osteosarcoma virus, and bl/ka(b) murine nonleukemogenic retrovirus. It has been shown previously that PBX1-MEIS1 or PBX1-PREP1 heterodimers bind the PCE (5, 24). To test the possibility that PBX and PBX1-associated proteins bind this element in the MLV LTR, we performed a gel mobility shift assay. A U5 region oligonucleotide containing the PCE was labeled with 32P and incubated with nuclear extracts of 293 cells, resulting in the formation of two major DNA-protein complexes (C1 and C2, Fig. 5A). The specificity of the C1 and C2 complexes for PCE binding was confirmed with unlabeled specific competitor oligonucleotides (i.e., unlabeled PCE, Fig. 5A). No effects on C1 and C2 complexes were observed in the presence of an unlabeled nonspecific competitor (i.e., SP1 consensus oligonucleotide, Fig. 5A). We next examined the interaction between the PCE and PCE-associated proteins by using antibodies specific to these factors. Incubation of nuclear extracts with antibodies raised against PBX1 proteins resulted in partial disruption of the C1 and C2 complexes and appearance of two supershifted complexes (Fig. 5B, arrows). Incubation of nuclear extracts with antibodies against MEIS1 protein did not have any detectable effects, whereas both C1 and C2 complexes were significantly disrupted when incubated with anti-PREP1 antiserum (Fig. 5B). Identical experiments were performed with nuclear extracts of 3T3 or HeLa cells, and the same effects of anti-PBX1 as well as anti-PREP1 antibodies on C1-C2 complexes were observed (data not shown). We also performed a gel shift assay with in vitro-translated PBX1a, MEIS1, and PREP1 proteins and 32P-labeled PCE oligonucleotides. The results show that PBX1a, MEIS1, PREP1 protein alone, or PBX1a-MEIS1 protein complexes are not able to bind to the PCE (Fig. 5C). However, a specific PCE-protein complex was observed when PBX1a-PREP1 protein mixtures were included in the reaction mixture (Fig. 5C). Collectively, the gel shift experiments indicate that PBX1 and PREP1 proteins are included in the PCE-protein complexes.
FIG. 5.
Characterization of proteins binding to the PCE. (A) A gel mobility shift assay was performed with nuclear extracts (NE) of 293 cells and a radiolabeled probe containing the PCE. While the unlabeled specific competitor (i.e., unlabeled PCE oligonucleotide [S.C.]) was included, two specific PCE-protein complexes were disrupted (C1 and C2). No effects on C1 or C2 were observed with unlabeled nonspecific competitor (i.e., unlabeled SP1 consensus oligonucleotide [N.S.C.]). (B) Nuclear extracts (NE) of 293 cells were incubated with antibodies against PBX1, MEIS1, PREP1, and γ-tubulin prior to addition of the PCE probe. Anti-γ-tubulin antibodies were used as the negative control and had no effect on the PCE-protein complexes. The position of each supershifted complex is indicated by arrows. Identical assays were performed with nuclear extracts of 3T3 or HeLa cells on different days with the same results (data not shown). (C) Gel shift assays were performed with the PCE probe and in vitro-translated PBX1a, MEIS1, and PREP1 proteins produced by a coupled reticulocyte lysate system. The arrow indicates the binding of PBX1a and PREP1 proteins. The asterisk indicates the binding of endogenous complexes in lysates. The assays were performed four times on different days with the same results.
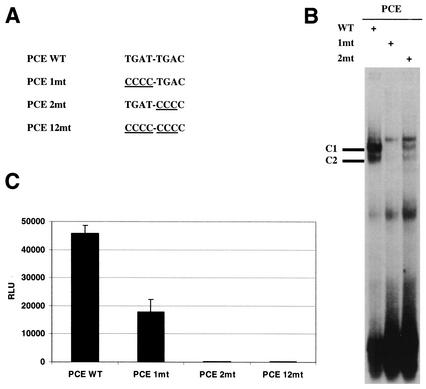
It has been shown previously that PBX1 binds to the first half of the PCE (i.e., TGAT) and that MEIS1 or PREP1 contacts the second half (i.e., TGAC) (24). To determine the importance of the PCE in DNA-protein interactions, probes with mutations in the first (PCE 1mt) and second (PCE 2mt) tetramers of the PCE were generated (Fig. 6A) and utilized in gel shift assays. Both the PCE 1mt and PCE 2mt probes caused significant or even complete disruption of two PCE-protein complexes (Fig. 6B). To determine the importance of the PCE in MLV transcription, WZL-Luc plasmids containing either PCE 1mt or PCE 2mt were generated and used to transiently transfect cells. The data show that PCE 1mt causes a 60% decrease in viral transcription whereas PCE 2mt causes complete inhibition (Fig. 6C). A WZL-Luc vector with mutations on both halves of the PCE (PCE 12mt, Fig. 6A) was also generated which completely blocked viral transcription as well (Fig. 6C). The data indicate that the PCE is an essential element in controlling MLV transcription.
FIG. 6.
Effects of mutations in the PCE on MLV transcription. (A) Nucleotide sequences of the wild-type (WT) and mutant PCEs. (B) A gel shift assay was performed with 293 cell nuclear extracts with different DNA probes including wild-type PCE (WT), PCE 1mt (1mt), or PCE 2mt (2mt). C1 and C2, PCE-protein complexes. (C) 293 cells were transiently transfected with the WZL-Luc vector containing wild-type or mutant PCEs. Luciferase activity was measured 48 h after transfection. Results were reproducible, and the assay was repeated more than three times in triplicate.
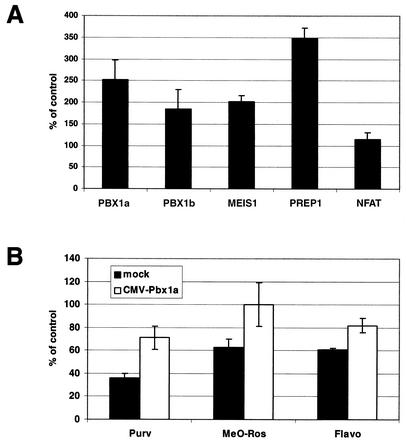
The involvement of PCE binding proteins, PBX1, MEIS1, and PREP1, in MLV transcription was examined next. Cells were transiently transfected with expression plasmids containing MEIS1, PREP1, or one of the PBX1 isoforms, PBX1a or PBX1b, followed by the addition of MLV viral supernatants (i.e., MLV-LTR-Luc) 24 h later. Both PBX1a and PBX1b enhanced MLV transcription with similar fold induction (2- to 2.5-fold, Fig. 7A). PBX1-associated proteins, MEIS1 and PREP1, stimulated 2- and 3.5-fold increases in MLV transcription, respectively (Fig. 7A). Although the gel shift assay did not demonstrate the involvement of MEIS1 proteins in the PCE-protein complexes, overexpression of MEIS1 enhanced levels of MLV transcription (Fig. 7A). On the other hand, overexpression of NFAT (nuclear factor of activated T cell), a protein involved in HIV transcriptional regulation, had no effects on MLV (Fig. 7A). To confirm that the effects of PBX1-MEIS1-PREP1 were on viral transcription alone and not mechanistically associated with other aspects of the MLV infection process, another assay was performed. Cells were cotransfected with WZL-Luc and an expression vector encoding PBX1a, MEIS1, or PREP1. Overexpression of PBX1a or MEIS1 resulted in a twofold increase in MLV transcription, and overexpression of PREP1 resulted in a threefold increase (data not shown). We next tested whether PBX1 was one of the CDKIs' targets by examining if PBX1 could complement the inhibitory effects of the CDKIs on MLV transcription. 293 cells were transiently transfected with a CMV-PBX1a expression plasmid for 24 h to overexpress PBX1a followed by CDKI treatment (i.e., 1 μM Purv, 1 μM MeO-Ros, and 0.03 μM Flavo) and addition of MLV viral supernatants. Western blotting of cell lysates subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-PBX1 antibodies indicated that PBX1a proteins were overexpressed within 24 h of transfection (data not shown). Even in the presence of any one of the three CDKIs, PBX1a-overexpressing cells recovered 20 to 40% of MLV viral transcription (Fig. 7B) compared with mock-transfected cells, suggesting that PBX1 might be a cellular target of the CDKIs. Full recovery was not seen since PBX1 might be just one of the cellular genes targeted by the CDKIs, as evidenced by the chip results, or the expression levels of PBX1 were not high enough to compensate for all the CDKI molecules present in the cells.
FIG. 7.
Involvement of PBX1 and PBX1-associated proteins in MLV transcription. (A) 293 cells were transiently transfected with an expression plasmid (i.e., CMV-PBX1a, -PBX1b, -MEIS1, -PREP1, or -NFAT) for 24 h before retroviral supernatants of MLV were added. Luciferase activity was determined 48 h after addition of viral supernatants. (B) 293 cells were transiently transfected with mock or CMV-PBX1a plasmids. MLV supernatants were added 24 h after transfection in the presence of the CDKIs (i.e., 1 μM Purv, 1 μM MeO-Ros, or 0.03 μM Flavo). Luciferase activity was measured 48 h after MLV was included. Results were reproducible, and data from one experiment are presented here.
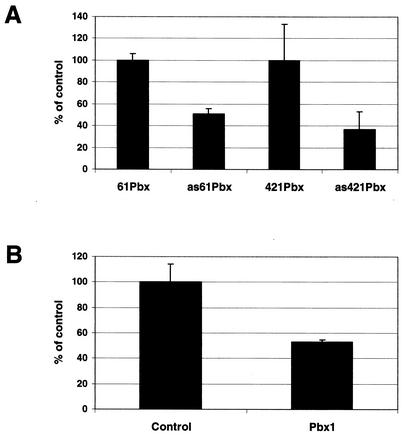
Overexpression studies were complemented with gene knockdown experiments to address the influence of PBX1 on MLV transcription events. Antisense oligonucleotides and siRNAs targeting Pbx1 were utilized to block PBX1 protein synthesis, and the effects on MLV transcription were examined. Two antisense Pbx1 oligonucleotides were generated, and each antisense oligonucleotide was paired with a sense oligonucleotide as control. Cells were transiently transfected with WZL-Luc in concert with a given oligonucleotide, and luciferase activities were measured 24 h afterwards. The presence of antisense Pbx1 oligonucleotides resulted in a significant inhibition, diminishing MLV transcription by as much as 63% (Fig. 8A). We also used Pbx1-targeted siRNA to inhibit PBX1 protein synthesis in experiments where siRNA targeting Renilla luciferase was used as control (all the reporter constructs used in this paper contained firefly luciferase). The inhibitory effects of Pbx1 siRNA on PBX1 protein synthesis were confirmed by Western blotting (data not shown). MLV-LTR-Luc viral supernatants were added to cells transfected with siRNAs and incubated for 24 h. As shown in Fig. 8B, viral infection was blocked by 47% in the presence of Pbx1-targeted siRNA. Furthermore, in agreement with the antisense oligonucleotide experiments, Pbx1-targeted siRNA also caused a 53% decrease in MLV transcription (i.e., transient transfection with WZL-Luc; data not shown). The results of the gene knockdown experiments confirm the involvement of PBX1 in MLV infection and transcription. Collectively, we conclude that PBX1 and PREP1 are involved in Moloney MLV infection through the regulation of viral transcription.
FIG. 8.
Inhibition of MLV transcription by blocking PBX1 protein synthesis. (A) 293 cells were cotransfected with WZL-Luc and a Pbx1-targeted antisense oligonucleotide. Each Pbx1 antisense nucleotide was paired with a sense nucleotide as control. Luciferase activity was measured 24 h after transfection. A pcDNA3.1 reverse primer, used for sequencing the insert of pcDNA plasmids (Invitrogen), served as another control. The pcDNA3.1 primer and two sense Pbx1 oligonucleotides had similar effects on viral transcription (data not shown). (B) 293 cells were transiently transfected with an siRNA of Pbx1 for 24 h and then incubated with MLV viral supernatants for another 24 h. An siRNA of Renilla luciferase was used as control. The assay was repeated twice in triplicate with the same results.
DISCUSSION
Here we identify three CDKIs, Flavo, Purv, and MeO-Ros, as novel MLV inhibitors. However, since CDK2, CDK4, CDK6, CDK7, and CDK9 are directly or indirectly involved in transcription, inhibition of these CDKs would be expected to lead to broad transcriptional repression regardless of how specific the CDKIs are. It has been shown previously that gene expression profiles of Flavo-treated cells closely resemble those of cells incubated with actinomycin D, a general transcription inhibitor (26). This result might help to explain the cytotoxicity of Flavo detected in numerous cell lines and the severe side effects observed in clinical trials against cancers when high doses of the drug are used, suggesting that the usage of CDKIs could be limited in treating cancers and viral infections (49).
However, in the treatment of retroviruses, a window does exist where the CDKIs block HIV-MLV very effectively with no significant effects on cellular proliferation or transcription (Fig. 2) (6, 7). It has been hypothesized that CDKIs block viral replication through the inhibition of specific cellular genes. In this paper, we test this idea by utilizing a microarray approach to identify cellular targets of Flavo, Purv, and MeO-Ros and to further investigate the possible involvement of these cellular genes in MLV transcription. Since CDKIs have broad effects on cellular transcription, it is likely that most cellular genes are nonspecifically inhibited by the CDKIs and not involved in MLV transcriptional regulation. To identify cellular cofactors required for MLV transcription, we focused only on genes that were dramatically affected by two or three CDKIs. Although each CDKI exhibits specificity in blocking specific CDKs, different CDKIs might utilize similar mechanisms to block MLV transcription. Based on this assumption, the genes commonly inhibited by CDKIs could represent candidate genes involved in MLV transcription. Using this simple strategy, we were able to reduce the number of putative cellular genes potentially targeted by CDKIs. Identification of PBX1 as a cellular cofactor for MLV transcription suggests that this approach indeed works. The strategy should be useful in identifying cellular targets involved in HIV, HSV, and cancers that are blocked by CDKIs, which could represent novel drug targets (6, 7, 45, 49).
There are at least three different mechanisms utilized by the CDKIs in the inhibition of MLV: (i) induction of cell arrest by blocking the CDKs controlling the cell cycle, (ii) inhibition of viral transcription through direct inhibition of CDK7-CDK9, and (iii) indirect down-regulation of the cellular gene PBX1 through the inhibition of CDK7-CDK9. Our results demonstrate that PBX1 binds to the PCE present in the U5 region of Moloney MLV LTR (Fig. 5B) and functions as a cellular regulator for MLV transcription, as supported by overexpression and gene knockdown experiments (Fig. 7 and 8). Three candidate genes (those for serine/threonine protein phosphatase PP1-γ catalytic subunit, serine/threonine-protein kinase PLK, and prohibitin) identified by microarray are required for cell division (8, 38, 43), which could help to explain how CDKIs induce cell cycle arrest. In addition to CDKs and PBX1, there might be more cellular genes involved in MLV transcription. HMG I-C, for example, is involved in HIV and MLV integration in vitro (28), and its gene is one of the candidate genes identified in this study. There are three members in the high-mobility group protein family: HMG I, HMG Y, and HMG I-C (3). It has been shown previously that HMG I modulates the binding of transcription factors to the LTR of HIV (22). It is possible that HMG I-C also functions as a regulator of HIV-MLV transcription. Furthermore, it is likely that both murine and human retroviruses require the same cellular proteins during viral replication. It should be interesting to determine the possible involvement of the other MLV candidate genes in HIV infection.
The LTR of MLV is divided into three regions termed U3, R, and U5 regions. The U3 region contains the viral promoter and transcriptional enhancers. It has been shown previously that both the pathogenic potential and disease specificity of MLV are dependent on the enhancer sequences in the U3 regions (11, 12, 27, 29). Several binding sites for cellular proteins and transcription factors have been identified, including a binding site for leukemia virus factor b, a viral core-like element, the consensus motif for nuclear factor 1, and the glucocorticoid response element (15). The R region is used by reverse transcriptase to mediate translocation of a DNA intermediate during viral DNA synthesis (30). In addition, the transcriptional initiation and postinitiation of MLV are also regulated by the R region (9, 10). On the other hand, our understanding of the functions mapping to the U5 region remains incomplete. Previous studies suggest that the U5 region might be involved in RNA packaging and reverse transcription (35). In addition, it has been reported previously that no enhancer is identified in the U5 region by computer-assisted sequence analysis (40).
The discovery of the involvement of homeodomain proteins PBX1 and PREP1 in MLV transcription is novel. The PBX1 binding motif is the first enhancer identified in the U5 region of MLV. The strong sequence conservation of U5 region compared with the U3 region suggests that the PCE and PCE-associated proteins might be key regulators in retroviral transcription. Gel shift assays confirmed the association between the PCE and PBX1-PREP1 proteins (Fig. 5B and C). No association between the PCE and MEIS1 (or PBX1-MEIS1) proteins was detected in gel shift assays (Fig. 5B and C). However, overexpression of MEIS1 proteins indeed resulted in increased MLV transcription (Fig. 7A). It has been shown previously that the Drosophila Meis homolog, Hth, induces nuclear translocation of PBX1 proteins (1). It is possible that overexpression of MEIS1 causes more PBX1 proteins to be translocated into the nucleus and therefore induces MLV transcription. Mutations in the PCE indicate that the MEIS1-PREP1 tetramer (TGAC) is more important than the PBX1 tetramer (TGAT) in regulating viral transcription (Fig. 6C, PCE 1mt versus PCE 2mt). It has been shown previously that PBX1 interacts and binds DNA with HOX proteins to form trimeric protein complexes (PBX1-MEIS1-HOX or PBX1-PREP1-HOX) (2, 51). One study showed that PBX1 could function as a non-DNA-binding partner in the trimeric complexes (50). Future work will be required to investigate the association between PBX1-PREP1 complexes and the U5 region of the LTR in detail and the possible requirement of HOX proteins in MLV transcription.
Acknowledgments
We thank Michael L. Cleary for providing Pbx1 expression vectors and antibodies; F. Bradley Hillgartner for Meis1 and Prep1 plasmids; and Pedro Aza-Blanc, Jing Li, Dehua Yu, Cecilia Jiang, Jason Chyba, and Rick Soden for expert technical assistance.
REFERENCES
- 1.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthelsen, J., V. Zappavigna, E. Ferretti, F. Mavilio, and F. Blasi. 1998. The novel homeoprotein PREP1 modulates Pbx-Hox protein cooperativity. EMBO J. 17:1434-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, B. A., M. M. Dubay, E. A. Sausville, L. Brizuela, and P. J. Worland. 1996. Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK) 2 and CDK4 in human breast carcinoma cells. Cancer Res. 56:2973-2978. [PubMed] [Google Scholar]
- 5.Chang, C. P., Y. Jacobs, T. Nakamura, N. A. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. MEIS proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 7.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 8.Clay, F. J., S. J. McEwen, I. Bertoncello, A. F. Wilks, and A. R. Dunn. 1993. Identification and cloning of a protein kinase-encoding mouse gene, Plk, related to the polo gene of Drosophila. Proc. Natl. Acad. Sci. USA 90:4882-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cupelli, L., S. A. Okenquist, A. Trubetskoy, and J. Lenz. 1998. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J. Virol. 72:7807-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cupelli, L. A., and J. Lenz. 1991. Transcriptional initiation and postinitiation effects of murine leukemia virus long terminal repeat R-region sequences. J. Virol. 65:6961-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DesGroseillers, L., and P. Jolicoeur. 1984. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J. Virol. 52:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiFronzo, N. L., and C. A. Holland. 1999. Sequence-specific and/or stereospecific constraints of the U3 enhancer elements of MCF 247-W are important for pathogenicity. J. Virol. 73:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Golemis, E. A., N. A. Speck, and N. Hopkins. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J. Virol. 64:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, N., L. Detivaud, C. Doerig, and L. Meijer. 1999. ATP-site directed inhibitors of cyclin-dependent kinases. Curr. Med. Chem. 6:859-875. [PubMed] [Google Scholar]
- 17.Gray, N. S. 1995. Synthesis and biological application of 2,6,9-trisubstituted purine libraries. Ph.D. thesis. University of California, Berkeley.
- 18.Gray, N. S., L. Wodicka, A. M. Thunnissen, T. C. Norman, S. Kwon, F. H. Espinoza, D. O. Morgan, G. Barnes, S. Leclerc, L. Meijer, S. H. Kim, D. J. Lockhart, and P. G. Schultz. 1998. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science 281:533-538. [DOI] [PubMed] [Google Scholar]
- 19.Gunzburg, W. H., A. Fleuchaus, R. Saller, and B. Salmons. 1996. Retroviral vector targeting for gene therapy. Cytokines Mol. Ther. 2:177-184. [PubMed] [Google Scholar]
- 20.Hajduch, M., L. Havlieek, J. Vesely, R. Novotny, V. Mihal, and M. Strnad. 1999. Synthetic cyclin dependent kinase inhibitors. New generation of potent anti-cancer drugs. Adv. Exp. Med. Biol. 457:341-353. [PubMed] [Google Scholar]
- 21.Hatakeyama, M., and R. A. Weinberg. 1995. The role of RB in cell cycle control. Prog. Cell Cycle Res. 1:9-19. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, A., M. Bunce, N. Siddon, R. Reeves, and D. J. Tremethick. 2000. High-mobility-group protein I can modulate binding of transcription factors to the U5 region of the human immunodeficiency virus type 1 proviral promoter. J. Virol. 74:10523-10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagawa, N., A. Ogo, Y. Takahashi, A. Iwamatsu, and M. R. Waterman. 1994. A cAMP-regulatory sequence (CRS1) of CYP17 is a cellular target for the homeodomain protein PBX1. J. Biol. Chem. 269:18716-18719. [PubMed] [Google Scholar]
- 24.Knoepfler, P. S., K. R. Calvo, H. Chen, S. E. Antonarakis, and M. P. Kamps. 1997. MEIS1 and pKnox1 bind DNA cooperatively with PBX1 utilizing an interaction surface disrupted in oncoprotein E2a-PBX1. Proc. Natl. Acad. Sci. USA 94:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoepfler, P. S., and M. P. Kamps. 1997. The highest affinity DNA element bound by Pbx complexes in t(1;19) leukemic cells fails to mediate cooperative DNA-binding or cooperative transactivation by E2a-PBX1 and class I Hox proteins—evidence for selective targetting of E2a-PBX1 to a subset of Pbx-recognition elements. Oncogene 14:2521-2531. [DOI] [PubMed] [Google Scholar]
- 26.Lam, L. T., O. K. Pickeral, A. C. Peng, A. Rosenwald, E. M. Hurt, J. M. Giltnane, L. M. Averett, H. Zhao, R. E. Davis, M. Sathyamoorthy, L. M. Wahl, E. D. Harris, J. A. Mikovits, A. P. Monks, M. G. Hollingshead, E. A. Sausville, and L. M. Staudt. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2:research0041.1-0041.11. [Online.] http://genomebiology.com/2001/2/10/research/0041. [DOI] [PMC free article] [PubMed]
- 27.Lenz, J., D. Celander, R. L. Crowther, R. Patarca, D. W. Perkins, and W. A. Haseltine. 1984. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature 308:467-470. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., K. Yoder, M. S. Hansen, J. Olvera, M. D. Miller, and F. D. Bushman. 2000. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 74:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., E. Golemis, J. W. Hartley, and N. Hopkins. 1987. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J. Virol. 61:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobel, L. I., and S. P. Goff. 1985. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J. Virol. 53:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 32.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 34.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, J. E., and S. P. Goff. 1989. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J. Virol. 63:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolaychik, V. V., M. M. Samet, and P. I. Lelkes. 1996. A new method for continual quantitation of viable cells on endothelialized polyurethanes. J. Biomater. Sci. Polym. Ed. 7:881-891. [DOI] [PubMed] [Google Scholar]
- 37.Northrop, J. P., S. N. Ho, L. Chen, D. J. Thomas, L. A. Timmerman, G. P. Nolan, A. Admon, and G. R. Crabtree. 1994. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature 369:497-502. [DOI] [PubMed] [Google Scholar]
- 38.Nuell, M. J., D. A. Stewart, L. Walker, V. Friedman, C. M. Wood, G. A. Owens, J. R. Smith, E. L. Schneider, R. Dell'Orco, C. K. Lumpkin, D. B. Danner, and J. K. McClung. 1991. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 11:1372-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portis, J. L., S. Perryman, and F. J. McAtee. 1991. The R-U5-5′ leader sequence of neurovirulent wild mouse retrovirus contains an element controlling the incubation period of neurodegenerative disease. J. Virol. 65:1877-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki, K., H. Shima, Y. Kitagawa, S. Irino, T. Sugimura, and M. Nagao. 1990. Identification of members of the protein phosphatase 1 gene family in the rat and enhanced expression of protein phosphatase 1 alpha gene in rat hepatocellular carcinomas. Jpn. J. Cancer Res. 81:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sausville, E. A., J. Johnson, M. Alley, D. Zaharevitz, and A. M. Senderowicz. 2000. Inhibition of CDKs as a therapeutic modality. Ann. N. Y. Acad. Sci. 910:207-221. [DOI] [PubMed] [Google Scholar]
- 45.Schang, L. M., J. Phillips, and P. A. Schaffer. 1998. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J. Virol. 72:5626-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schang, L. M., A. Rosenberg, and P. A. Schaffer. 1999. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J. Virol. 73:2161-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. Cheah, J. L. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for PBX1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128:3543-3557. [DOI] [PubMed] [Google Scholar]
- 48.Senderowicz, A. M. 1999. Flavopiridol: the first cyclin-dependent kinase inhibitor in human clinical trials. Investig. New Drugs 17:313-320. [DOI] [PubMed] [Google Scholar]
- 49.Senderowicz, A. M., and E. A. Sausville. 2000. Preclinical and clinical development of cyclin-dependent kinase modulators. J. Natl. Cancer Inst. 92:376-387. [DOI] [PubMed] [Google Scholar]
- 50.Shanmugam, K., N. C. Green, I. Rambaldi, H. U. Saragovi, and M. S. Featherstone. 1999. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 19:7577-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, W. F., S. Rozenfeld, A. Kwong, L. G. Kömüves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sourvinos, G., C. Tsatsanis, and D. A. Spandidos. 2000. Mechanisms of retrovirus-induced oncogenesis. Folia Biol. (Prague) 46:226-232. [DOI] [PubMed] [Google Scholar]
- 53.Wang, D., C. de la Fuente, L. Deng, L. Wang, I. Zilberman, C. Eadie, M. Healey, D. Stein, T. Denny, L. E. Harrison, L. Meijer, and F. Kashanchi. 2001. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J. Virol. 75:7266-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]