Abstract
The mammalian tooth forms by a series of reciprocal epithelial-mesenchymal interactions. Although several signaling pathways and transcription factors have been implicated in regulating molar crown development, relatively little is known about the regulation of root development. Four genes encoding nuclear factor I (NFI) transcription-replication proteins are present in the mouse genome: Nfia, Nfib, Nfic, and Nfix. In order to elucidate its physiological role(s), we disrupted the Nfic gene in mice. Heterozygous animals appear normal, whereas Nfic−/− mice have unique tooth pathologies: molars lacking roots, thin and brittle mandibular incisors, and weakened abnormal maxillary incisors. Feeding in Nfic−/− mice is impaired, resulting in severe runting and premature death of mice reared on standard laboratory chow. However, a soft-dough diet mitigates the feeding impairment and maintains viability. Although Nfic is expressed in many organ systems, including the developing tooth, the tooth root development defects were the prominent phenotype. Indeed, molar crown development is normal, and well-nourished Nfic−/− animals are fertile and can live as long as their wild-type littermates. The Nfic mutation is the first mutation described that affects primarily tooth root formation and should greatly aid our understanding of postnatal tooth development.
Tooth formation is a complex developmental process that is mediated through a series of epithelial-mesenchymal interactions (29). Several signaling pathways required for early molar tooth development have been identified, including the BMP (31), FGF (13, 27), SHH (5, 10), and WNT (25, 33) pathways. In addition, targeted disruption of a number of transcription factors, including Msx1 and -2 (1), Dlx1 and -2 (22), Pax-9 (21) and others, severely disrupts early tooth development. In contrast, only one pathway regulating late tooth developement has been identified. The Tabby (Ta), Downless (Dl), and Crinkled (Cr) mouse mutations each cause similar defects of postnatal development (32) by disrupting different components of the Ectodysplasin-A hormone signaling system.
The nuclear factor I (NFI) family of transcription-replication factors is encoded by four genes in mammals (NFI-A, -B, -C, and -X) and a single gene in Drosophila melanogaster and Caenorhabditis elegans (8, 14, 23). Prokaryotic homologues of the NFI genes have not been identified. NFI was discovered as a protein required for adenovirus DNA replication in vitro (19, 20), but it is now clear that NFI proteins play an important role in the expression of many cellular genes (reviewed in reference 8). NFI-C/CTF was the initial NFI gene cloned (24) and contains a prototypical proline-rich transcriptional activation domain, as well as a heptamer repeat homologous to the C-terminal domain of RNA polymerase II (18). Because products of the four NFI genes are often coexpressed (3) and bind to similar DNA sequences (15), it has been difficult to determine the roles of individual NFI family members in gene expression during development. We are analyzing the role of NFI genes in mouse development by generating mice deficient in each of the four NFI genes.
We disrupted the Nfic gene in mice by removal of its second exon, which encodes the NFI-C/CTF DNA-binding and dimerization domain. The most striking defect in Nfic−/− mice is that their molar teeth form no roots. In addition, their mature mandibular incisors are thin and brittle, and their maxillary incisors are dysmorphic. Consistent with these tooth defects, Nfic−/− mice exhibit compromised feeding on standard lab chow, resulting in increased morbidity and mortality. Since loss of Nfic is the first mouse mutation to specifically affect root development, these mice should be an important tool for understanding the molecular pathways essential for this process.
MATERIALS AND METHODS
Gene targeting.
Mouse genomic clones containing Nfic exon 2 were isolated from a mouse 129/Sv phage library as described previously (7). Endonuclease restriction, Southern blotting, PCR, and DNA sequencing were used to characterize clones, including mapping restriction sites and mapping and sequencing exons 1 and 2. The targeting vector contains 9.5- and 3.5-kb regions of 5′ and 3′ homology, respectively, flanking a PGK-Neo-bGHpA gene cassette (26) in the reverse transcriptional orientation. The Neo cassette replaces ∼1.3 kb of Nfic from an NdeI site 98 bp 5′ of exon 2 to an NheI site ∼700 bp 3′ of exon 2. The negatively selectable gene cassette MCI-TK (herpes simplex virus thymidine kinase with a mutant polyomavirus enhancer [17]) was adjacent to the shorter region of Nfic homology in the opposite transcriptional orientation. A total of 5 × 106 E14-1 mouse embryonic stem (ES) cells (12) were electroporated with 5 μg of XhoI-linearized targeting vector and then selected in 0.2 mg/ml of G418 (Gibco-BRL) and 2 μM ganciclovir (Roche Laboratories) for 6 days. Colonies were isolated and expanded twice in 96-well plates. Two plates were cryopreserved, and two were used for DNA isolation and Southern blot analyses.
PCR genotyping.
Toe or 0.5-cm tail-tip biopsies were shaken at 55°C overnight in 0.5 ml of tail lysis buffer (0.05 M Tris-Cl, pH 8.0; 0.1 M EDTA; 0.5% sodium dodecyl sulfate), including 0.5 mg of proteinase K/ml. Then, 1 μl of the lysate diluted 1:25 in H2O was analyzed in 20-μl multiplex PCRs (2.5 U of platinum Taq DNA polymerase (Gibco-BRL), 1.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate, 1× Rediload (Research Genetics), 1× PCR buffer (supplied with the Taq), and five primers at 0.2 μM each). The following two Nfic-specific primers (a and c), Neo-specific primer (b), and two mouse Y chromosome-specific primers (SRY) were used: a (5′-CATCTGTGTGAAACAGTCTGG-3′), b (5′-CCGCTTCCTCGTGCTTTACGG-3′), c (5′-AGCAGCTCATCCTTCACCGCG-3′), SRY1 (5′-AACAACTGGGCTTTGCACATTG-3′), and SRY2 (5′-GTTTATCAGGGTTTCTCTCTAGC-3′). PCR products were resolved on 2.5% agarose gels and visualized with ethidium bromide.
RT-PCR.
Total RNA was isolated by using TRIzol reagent (Gibco-BRL) according to the manufacturer's instructions. RNA from mandible was purified over a G50 spin column (Roche) prior to reverse transcription (RT). cDNA was generated from 2 to 5 μg of RNA by using Superscript (Gibco-BRL) according to the manufacturer's random-prime protocol. A total of 1 μl of each RT product was analyzed by PCR with a sense primer from exon 1 (5′-GATCGGCCTCACGGGCCGATGTATTCCTCCCCGCTCTGC-3′) and an antisense primer from exon 3 (5′-TTGCTGTCCTCCTGGTCTGAGC-3′). Products were resolved on 2.5% agarose gels. Analysis of Nfia, Nfib, Nfix, amelogenin, ameloblastin, dentin sialophosphoprotein (DSPP), and β2-microglobulin transcript levels was done by real-time quantitative PCR with an ABI 7700 or Bio-Rad real-time thermocycler with SYBRgreen detection. The primer sequences and conditions are available upon request.
Mouse rearing.
The initial 17 F1 litters were fed a diet of autoclaved mouse chow (Purina 5010 and, for breeders, Purina 5021) and autoclaved distilled water. For studies requiring adults, several grams of the soft transgenic dough diet (S3472; Bio-Serv) were added to cages twice per week beginning 3 days prior to the P21 wean.
Histology and in situ hybridization.
Postnatal mice were anesthetized with isoflurane and cardiac perfused with 4% paraformaldehyde-phosphate-buffered saline (PBS). Heads were removed and depelted, the craniums were evacuated, and the skulls were decalcified by soaking them in formaldehyde-formic acid at 4°C overnight. After paraffin embedding, 4-μm sections were cut and stained with hematoxylin and eosin (H&E). For embryos, fixing, embedding, sectioning, and in situ hybridization with 35S-labeled Nfic cRNA sense and antisense probes were performed as described previously (3, 16). The control sense probe shows a low background level of silver grains (not shown). For postnatal tissue in situ hybridizations, digoxigenin (DIG)-labeled Nfic sense and antisense cRNA probes were prepared by using the T7/Sp6 DIG RNA labeling kit (Roche). Sections were deparaffinized and hybridized according to the manufacturer's protocol. Briefly, sections were hydrated, treated for 20 min with proteinase K (20 μg/ml), washed with PBS, fixed in 4% paraformaldehyde, washed again in PBS, incubated with 0.2 N HCl for 10 min to inhibit endogenous alkaline phosphatase, and acetylated by incubation for 10 min in 0.1 M triethanolamine containing 0.25% acetic anhydride. After a washing step, hybridization was performed overnight at 50°C in hybridization solution (50% deionized formide, 10% dextran sulfate, 1× Denhardt solution, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10 mM dithiothreitol, 1 mg of yeast tRNA/ml, 1 mg of salmon sperm DNA/ml) containing the DIG-UTP-labeled Nfic cRNA sense or antisense probes. After being washed for 30 min with 2× SSC-50% formamide and 10 min each in 2× SSC and 0.2× SSC at 50°C, sections were incubated for 30 min with blocking solution, followed by incubation for 1 h with blocking solution containing 1:500 sheep anti-DIG-alkaline phosphatase Fab fragments. After being washed, DIG probes were detected by incubating the sections with alkaline phosphatase substrates (368 μg of nitroblue tetrazolium salt and 184 μg of BCIP (5-bromo-4-chloro-3-indolylphosphate) in 1 ml of 100 mM Tris-HCl, 100 mM NaCl, and 50 mM MgCl2) until the color was developed. Sections were then used directly or counterstained for 2 to 3 min with methyl green, air dried, and mounted by using VectaMount (Vector Laboratories).
Skull bones and molar teeth isolation.
Mouse heads were depelted, the muscles and fascia were clipped away, and the brains and eyes were removed before soaking the heads in 1% KOH for 3 days. After three rinses with H2O, the heads were individually incubated in 50 mM Tris-Cl (pH 8.0), 0.5% SDS, and 0.2 mg of proteinase K/ml at 55°C for 20 min and then washed with H2O. Molars of Nfic−/− mice separated from the jaw during proteolysis, whereas those of the other Nfic genotypes were isolated by crown extraction or fracturing the mandible or maxilla.
RESULTS
Nfic gene targeting and confirmation of Nfic-null allele.
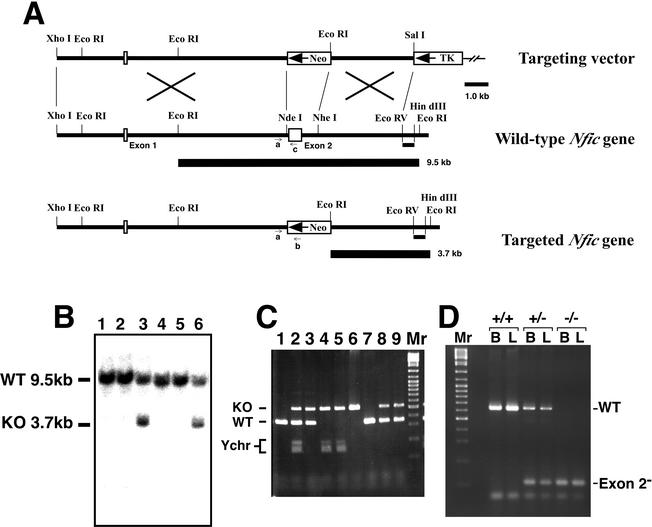
We constructed a replacement-type targeting vector substituting Nfic exon 2 with a neomycin resistance gene that is constitutively transcribed in the reverse direction (Neo in Fig. 1A). Since both the DNA-binding and the dimerization activities of NFI-C reside in exon 2, homologous recombination of the native gene and the targeting vector should yield a Nfic-null allele. On Southern blots an Nfic probe external to the targeting vector detects a 3.7-kb band from the targeted allele and a 9.5-kb band from the native allele on EcoRI digests of genomic DNA (the two thickest bars in Fig. 1A). A screen of 90 G418+ganciclovir-resistant ES clones identified four that harbored a targeted mutation and a composite of the results of the screen is shown (Fig. 1B).
FIG. 1.
Targeted mutation of the mouse Nfic gene. (A) Targeting vector (top), the Nfic region encompassing exons 1 and 2 (middle), and their homologous recombination product (bottom). The termini of homologous sequences and relevant restriction sites are noted. The positive and negative selection gene expression cassettes are boxes labeled Neo and TK, respectively, and their transcription orientations are indicated by arrows. Exons are indicated by open boxes labeled Exon. The 3′-flanking probe (EcoRV-HindIII; short bar) and the wild-type and mutant EcoRI restriction fragments it identifies in Southern analyses (thick bars) are shown. Plasmid backbone sequences are denoted by a thin line. Locations of the three primers (a, b, and c) used for PCR genotyping are indicated by small arrows. (B) Data from Southern blot screen of G418- and ganciclovir-resistant ES clones. Each of six clones is numbered above, and the hybridizing wild-type (WT 9.5kb) and mutant (KO 3.7kb) EcoRI fragments are indicated on the left. Clones 3 and 6 have a targeted mutation, whereas clones 1, 2, 4, and 5 do not. (C) PCR genotyping of one litter from an Nfic heterozygous cross. PCRs employing the three primers indicated in panel A plus two Y-chromosome-specific primers were used to screen tail biopsies of a litter of nine (lanes 1 to 9). Labels on the left indicate the 545-bp targeted-gene product (KO), the 358-bp wild-type product (WT), and the male-specific products (Ychr). Lanes (lane/progeny, Nfic genotype, sex): 1, +/+, female; 2, +/−, male; 3, +/−, female; 4, −/−, male; 5, −/−, male; 6, −/−, female; 7, +/+, female; 8, +/−, female; 9, +/−, female. Lane Mr is a 100-bp DNA ladder. (D) RT-PCR analysis of Nfic transcripts in tissues of each Nfic genotype. The total RNA isolated from brains (lanes B) or livers (lanes L) of Nfic wild-type (+/+), heterozygote (+/−), and homozygote (−/−) male P193 littermates was reverse transcribed, and the cDNA was then analyzed by PCR with exon 1- and exon 3-specific primers. The products corresponding to wild-type (WT) and aberrant (Exon 2−) transcripts are indicated. Lane Mr is a 100-bp DNA ladder.
We injected three Nfic heterozygous (+/−) ES clones into C57BL/6 blastocysts in order to generate mouse chimeras. Male chimeras were crossed to Black Swiss, 129S6, and C57BL/6 strains and genomic DNA from tail biopsies of progeny was screened by PCR for the presence of the mutated allele (data not shown). All three ES clones transmitted the mutated allele into the germ line. Nfic+/− mice were crossed and the progenies' genotypes were identified by PCR (Fig. 1C). The single PCR distinguishes both genotype and sex. Primers a and c amplify 358 bp of the native gene, whereas primers a and b amplify 545 bp of the targeted allele. Primers SRY1 and 2 amplify ∼150 bp of the Y chromosome. Southern blot analysis of these same DNAs confirmed the PCR results (data not shown).
To show that the targeted Nfic allele was null, we analyzed Nfic transcripts from brain and liver RNA of adult littermates of each Nfic genotype. Total RNA was isolated, reverse transcribed, and the cDNA was used as a template in PCRs with Nfic exon 1-specific (forward) and exon 3-specific (reverse) primers (Fig. 1D). As predicted, the PCR produced a single 638-bp product from brain and liver cDNA of the wild-type (+/+) mouse. This product is from a transcript that includes exon 2 (532 bp) and the termini of exons 1 and 3 (Fig. 1D, lanes +/+). The PCR generated two products from brain and liver cDNA of the Nfic+/− littermate (Fig. 1D, lanes +/−). The larger product corresponds to the wild-type transcript, whereas the smaller product corresponds to a 106-bp product resulting from a direct splice of exon 1 to 3. Only the smaller product was amplified from brain and liver cDNA of the Nfic−/− littermate (Fig. 1D, lanes −/−). Sequencing of the smaller product confirmed that it is generated from a direct splice from exon 1 to exon 3 (data not shown). Because the exon 1-exon 3 splice junction is out of frame, these aberrantly spliced Nfic transcripts are predicted to encode a 31-amino-acid missense polypeptide that terminates in exon 3.
Viability, growth, and fertility of Nfic−/− mice.
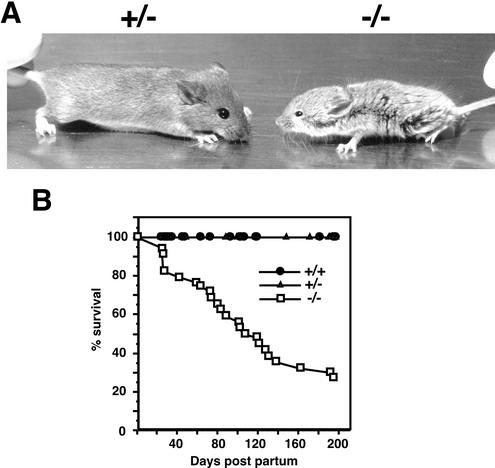
Our initial 17 heterozygote crosses produced 154 F1 progeny distributed in a Mendelian ratio: 33 wild-type (21.4%), 87 Nfic+/− (56.5%), and 34 Nfic−/− mice (22.1%). We noted in early litters that postweaning Nfic−/− mice became severely runted compared to their wild-type or Nfic+/− littermates (Fig. 2A). Continued rearing of Nfic−/− mice on standard lab chow resulted in continued runting and premature death (Fig. 2B and data not shown). Upon discovery of tooth defects in the Nfic−/− mice (see below, Fig. 3 to 6), we added a soft-dough dietary supplement 3 days prior to weaning and continued the diet supplementation of Nfic−/− mice postweaning. The dough diet supplementation allowed all (n = 18) Nfic−/− mice to survive longer than 3 months and into adulthood. Of three male and four female healthy adult Nfic−/− mice bred with Nfic+/− partners, all but one female mated successfully. The resulting litters were of normal size, the Nfic genotype ratio was the expected 1:1 (Nfic+/−:Nfic−/−), and the litters were reared successfully. These data indicate that Nfic−/− mice have greatly increased morbidity and mortality when fed standard lab chow and that a soft diet supplement rescues the mortality, permitting survival and fertility.
FIG. 2.
Growth retardation and increased mortality of Nfic−/− mice. (A) Female P54 Nfic+/− and Nfic−/− littermates. Note the severe runting of the −/− animal. A mirror image is shown in order to present the −/− sibling on the right. (B) Mortality of Nfic mice reared on standard chow. The percent survival of Nfic−/− (n = 34), Nfic+/− (n = 84), and Nfic+/+ (n = 33) mice are plotted versus time. The points of the +/− and +/+ curves indicate the time of sacrifice of littermates of Nfic−/− animals that had died. These studies were performed prior to observing tooth defects in Nfic−/− animals. Also, the increased mortality of Nfic−/− animals was not observed if they are reared on a soft-dough diet (see Results).
FIG. 3.
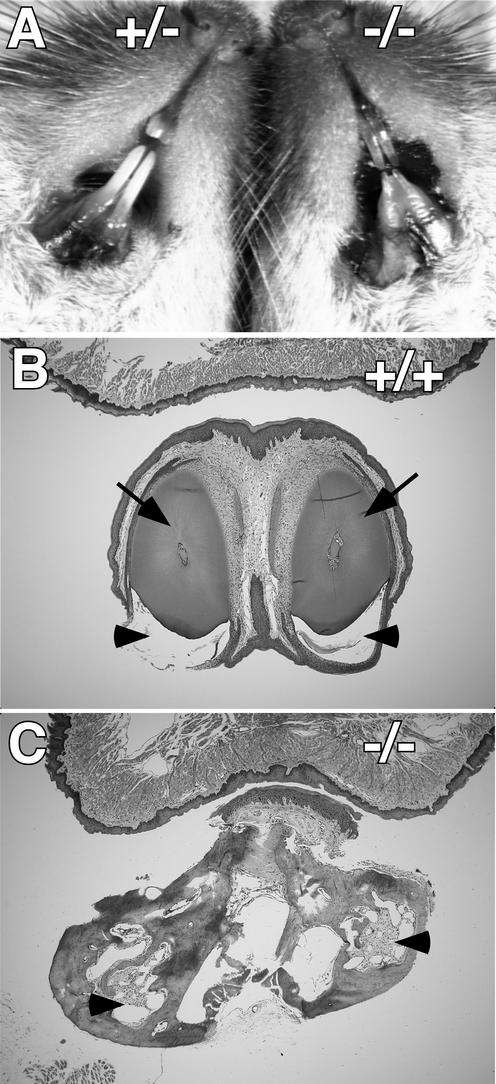
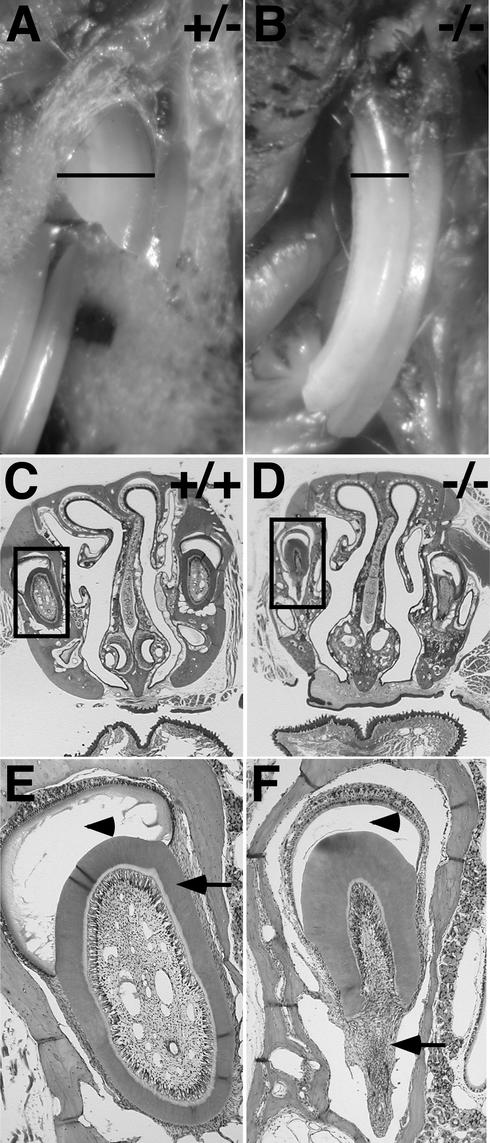
Abnormal mandibular incisors of Nfic−/− mice. (A) View of mouths of two P40 female littermates (Nfic+/− and Nfic−/−) reared on a soft-dough diet. Lower labia and tongues were excised to help expose incisors. Note the abnormally thin mandibular incisors and the extended gingiva of the −/− animal. (B) Mandible portion of an H&E-stained coronal section of the jaw of an Nfic+/+ P135 male. The tongue is at the top. An arrowhead and an arrow indicate the enamel and dentin, respectively. The majority of the enamel was lost during decalcification. (C) Same as panel B except the individual is a Nfic−/− littermate. Arrowheads indicate disorganized tissue where incisor structures are expected.
FIG. 6.
Abnormal alveolar bone formation in Nfic−/− mice. (A) View of maxillae of skulls prepared from P97 male +/− and −/− littermates. (B) Closeup of molar root areas in center of panel A. Note that the +/− mouse maxilla's left molars were extracted for the closeup, whereas the −/− mouse molars dislodged during skull preparation. (C) Lingual view of left mandible of the same +/− animal as in panel A. (D) Lingual view of left mandible of the same −/− animal as in panel A. (E) Lingual view of left mandibles isolated from P15 male +/+ and −/− littermates. Bar, 1 mm. (F) Closeup view of molar root sockets of mandibles shown in panel E. Both the +/+ and −/− mouse mandibles are forming bone between the three molars (arrowheads). However, the −/− mouse mandible is not forming bone at the regions between the missing roots of the first and second molars (arrows). The third molar of the +/+ mouse mandible did not dislodge during skull preparation (bottom).
Abnormal incisor development in Nfic−/− mice.
Gross observations of the mouth and jaw of Nfic−/− mice revealed virtually no mandibular (lower) incisors and overgrown, thin maxillary (upper) incisors (Fig. 3A). The structurally compromised incisors are most likely responsible for the morbidity and mortality of Nfic−/− mice reared on standard lab chow that was described above. H&E staining of coronal sections of adult mandibles indicated grossly abnormal mandibular incisor development (Fig. 3B versus C). The wild-type mandibular incisors had a normal structure consisting of an oval core of dentin with enamel deposition mainly on one surface (Fig. 3B). However, mandibular incisors in Nfic−/− mice were highly disorganized, frequently showing scattered pockets of differentiated tissue (Fig. 3C, arrowheads).
A similar analysis of coronal sections of maxillae revealed a less pronounced but clearly abnormal maxillary incisor development in Nfic−/− mice (Fig. 4). The maxillary incisors of wild-type (not shown) and Nfic+/− animals were substantially thicker than those of Nfic−/− mice (Fig. 4A versus B). The maxillary incisors of wild-type and heterozygous animals have dentin around the entire circumference and enamel on one surface (Fig. 4C [expanded in panel E]). In contrast, the maxillary incisors of Nfic−/− animals have near-normal labial dentin and enamel deposition but nearly absent lingual dentin deposition, leading to a grossly nonsymmetric structure (Fig. 4D [expanded in panel F]).
FIG. 4.
Abnormal maxillary incisors of Nfic−/− mice. (A) Close view of maxillary incisors in situ of a P135 Nfic+/− male reared on standard chow. The black bar indicates thickness of the right maxillary incisor. (B) Same as panel A except the individual is an Nfic−/− littermate. Note that the incisor is thin compared to panel A. (C) Upper portion of an H&E-stained coronal section of the head of a P135 Nfic+/+ male. The tongue is at the bottom. The box locates the right maxillary incisor. (D) Same as panel C except the individual is an Nfic−/− littermate. (E) Magnified view of boxed area in panel C. An arrowhead and an arrow indicate the enamel and dentin, respectively. Note the uniform layer of dentin around the circumference of the incisor. (F) Magnified view of boxed area in panel D. An arrowhead indicates the enamel deposition at the labial side, whereas the arrow indicates the absence of dentin at the lingual side of the incisor.
Abnormal molar root development in Nfic−/− mice.
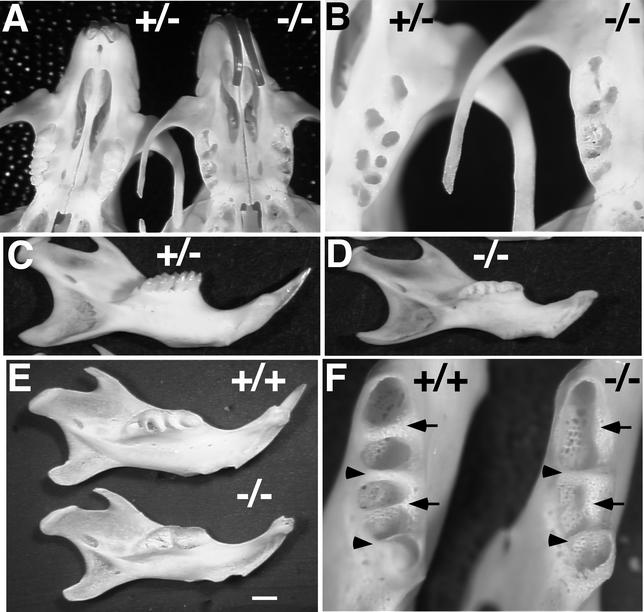
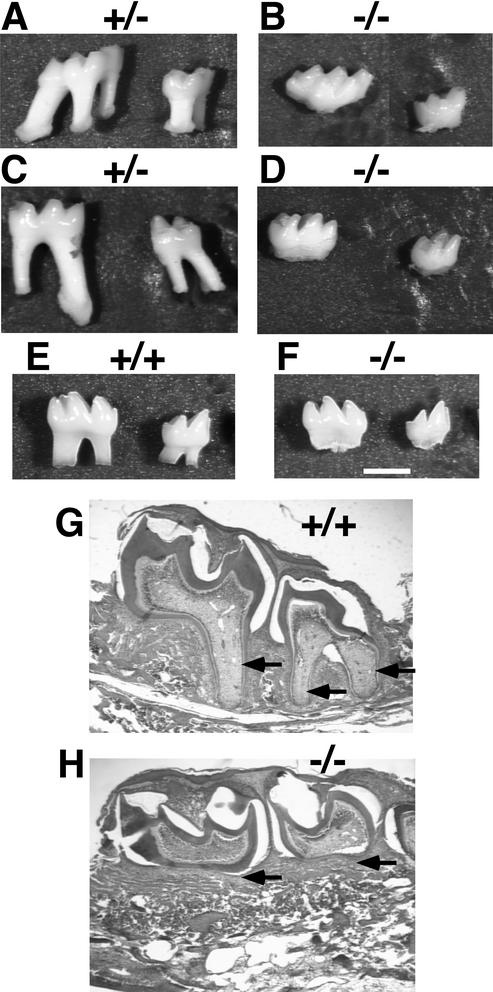
To examine the structure and morphology of the molar teeth, we isolated first and second mandibular and maxillary molars from adult littermates. The molars of wild-type (not shown) and Nfic+/− mice had normal crowns and roots (Fig. 5A and C). The mandibular and maxillary molars had the appropriate two and three roots, respectively. In contrast, molars from Nfic−/− mice had normal crowns but no roots (Fig. 5B and D). The lack of roots was observed both bilaterally and in all Nfic−/− molars examined. In order to determine whether the molar roots of the adult Nfic−/− mice failed to form or were resorbed following development, we isolated molars from P15 littermates, an age at which roots are still forming. The molars of both wild-type and Nfic−/− P15 mice have normal-appearing crowns (Fig. 5E and F). In contrast, although the wild-type molars show immature roots extending from the base of the crown (Fig. 5E), Nfic−/− molars show no developing roots (Fig. 5F). To further confirm the failure of molar root formation, serial sagittal sections were taken through the mandibles of P14 mice. Again, wild-type molars show normal root development (Fig. 5G), whereas Nfic−/− molars show no root development (Fig. 5H). The histology reveals that the only apparent difference between wild-type and Nfic−/− mouse molar development is root formation. There appear to be no structural changes in the jaw that might impede root outgrowth. These data indicate that molar root formation in the Nfic−/− mouse fails at either the initial outgrowth of the epithelial sheath or during early extension of the root.
FIG. 5.
Failure of molar root development in Nfic−/− mice. The panels compare the molars of P190 male (A to D), P15 male (E and F), and P14 male (G and H) littermates of the indicated Nfic genotype. (A and C) Lateral views of first (left) and second maxillary (A) and mandibular (C) molars of a +/− mouse. (B and D) Lateral views of first (left) and second maxillary (B) and mandibular (D) molars of a −/− mouse. (E and F) Lateral views of first (left) and second (right) mandibular molars of +/+ (E) and −/− (F) littermates. Bar, 1 mm. (G and H) H&E-stained sagittal sections of the developing left mandibles of +/+ (G) and −/− (H) littermates. The perimeter of the immature first and second molars in each is demarcated by dark-staining dentin. Arrows denote the immature roots present in panel G but absent in panel H.
Decreased alveolar (tooth socket) bone formation in Nfic−/− mice.
Since molar roots fail to form in Nfic−/− mice, we examined the jaw and the alveolar bone that normally surrounds the molar roots. Mandibles and maxillae of adult wild-type, Nfic+/− and Nfic−/− mice were prepared and examined by light microscopy. In the maxillae of both wild-type (not shown) and Nfic+/− mice, the incisors and molars remained embedded in bone during skull preparation (Fig. 6A). In contrast, the molars fell out during the preparation of Nfic−/− maxillae (Fig. 6A) and mandibles (Fig. 6D). Extraction of the molars from the maxillae of Nfic+/− animals revealed the deep root sockets of alveolar bone present in normal jaw (Fig. 6B). In contrast, the molar tooth sockets of Nfic−/− maxillae were shallow and contained an unorganized mesh of bony spicules (Fig. 6B). The mandible skeletons of Nfic−/− mice showed similar defects, both the incisors and molars detaching during preparation and the molar tooth sockets being filled with a coarse meshwork of alveolar bone (Fig. 6C and D and data not shown). In addition, the mandibles of Nfic−/− mice appear ∼10% smaller than those of their wild-type and heterozygous littermates (Fig. 6D versus C). No such size difference was noted in maxillae (Fig. 6A).
To examine earlier stages of alveolar bone formation, we isolated skeletonized mandibles from P15 littermates. In contrast to what was seen with adult skulls, molars fell out of both +/+ and −/− mouse mandibles (Fig. 6E and F). At this age alveolar bone has begun to surround the emerging molar roots in +/+ mice, whereas no such bone formation is seen in the tooth sockets of −/− mice (Fig. 6F, +/+ versus −/−). In contrast, bone formation between the teeth appears similar in both +/+ and −/− animals (Fig. 6F, +/+ versus −/−).
Nfic expression during tooth development.
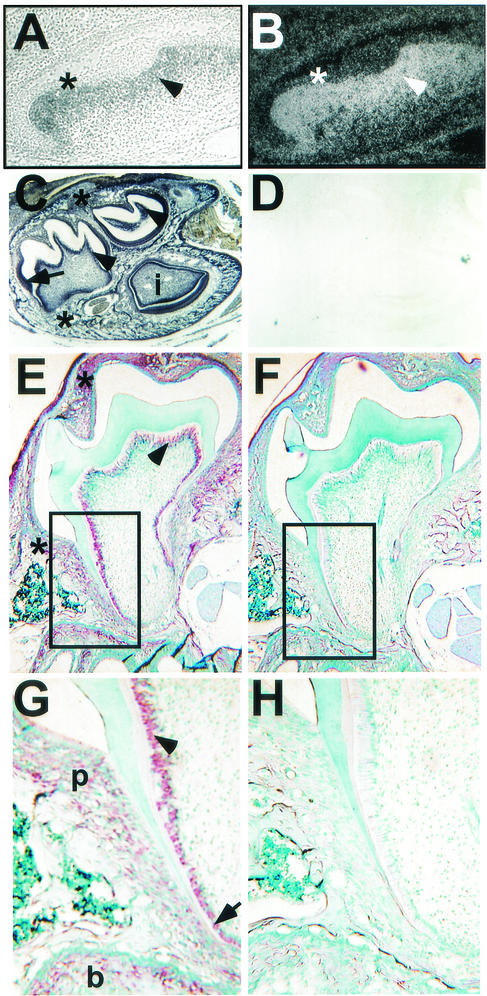
To assess when and where Nfic is expressed during tooth development, we performed in situ hybridization with the Nfic-specific probe we described previously (3). During tooth bud development at E15.5, Nfic is expressed strongly in the mesenchymal cells of the dental papilla and weakly in the epithelial components (Fig. 7A and B ). When root formation is beginning at P8 (Fig. 7C), Nfic is expressed strongly in most epithelial tissues of the tooth, including ameloblasts; in odontoblasts; in surrounding mesenchymal tissues, including the stellate reticulum; and in the dental papilla of the molars and incisor. No staining is seen with the control sense probe (Fig. 7D, F, and H). At P14, when molar root formation is in progress, Nfic expression appears more restricted being present within the odontoblasts and preodontoblasts of the molars (Fig. 7E and G) and within the periodontal ligament and developing bone (Fig. 7G). Figure 7G and H are magnifications of the boxed regions of Fig. 7E and F, respectively. Thus, Nfic is expressed at multiple stages of tooth development and strongly in odontoblasts and preodontoblasts during root formation.
FIG. 7.
Nfic expression during normal tooth development. Nfic expression was detected in wild-type animals by in situ hybridization using 35S-labeled (A and B) and DIG-labeled (C to H) antisense (B, C, E, and G) and control sense (D, F, and H) Nfic probes. Panels A and B show phase-contrast and dark-field images, respectively, of an E15.5 mouse first maxillary molar hybridized with a 35S-labeled Nfic antisense probe. Note the high Nfic expression in the molar mesenchymal cells (arrowhead), with weaker expression in the epithelium (asterisk). Panels C and D show staining by Nfic antisense and sense probes, respectively, of the mandible of a P8 mouse. Note the expression of Nfic in ameloblasts (arrow), odontoblasts (arrowheads), and surrounding connective tissue (top asterisk, stellate reticulum; bottom asterisk, mesenchymal tissues) and in the pulp of the molars and incisor (i) of panel C. No staining is seen with the control sense probe (D). Panels E and F show staining by Nfic antisense and control sense probes, respectively, in mouse P14 first maxillary molars. Sections were counterstained lightly with methyl green to identify cell types. Nfic is expressed in crown odontoblasts (arrow) and connective tissue (asterisks). Panels G and H are higher magnifications of panels E and F, respectively, showing the expression of Nfic in root odontoblasts (arrowhead), preodontoblasts (arrow), periodontal ligament (p), and bone (b) in panel G. Similar expression of Nfic was seen in mandibular molars at this stage (not shown).
Decreased tooth-specific gene expression in Nfic−/− mice.
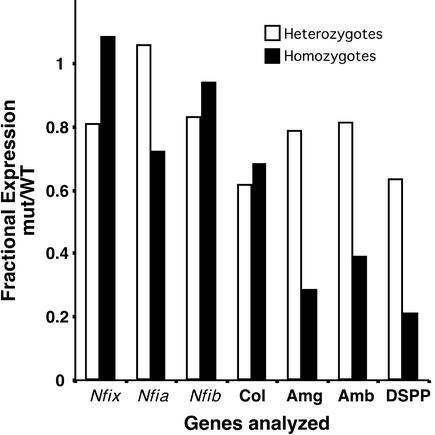
To determine whether the abnormal incisor and molar root development was reflected in changes in tooth-specific gene expression, we examined the levels of amelogenin, ameloblastin, and DSPP transcripts in RNA isolated from dissected P15 mandibles. Amelogenin (Amg) and ameloblastin (Amb) are expressed in ameloblasts, whereas DSPP is expressed in odontoblasts. Using real-time quantitative PCR, we found that levels of Amb, Amg, and DSPP were each reduced by >50% in mandibular RNA of Nfic−/− mice compared to their levels in heterozygous or wild-type mice (Fig. 8, lanes Amb, Amg, and DSPP). There were no consistent changes in transcript levels for α1 type I collagen in the Nfic−/− mice (Fig. 8, lane Col). These data are consistent with the histology of P15 Nfic−/− animals that show abnormal mandibular incisor and molar root development.
FIG. 8.
Reduced tooth-specific gene expression in the mandibles of Nfic−/− mice. Nfix, Nfia, Nfib, α1 type I collagen (col), amelogenin (Amg), ameloblastin (Amb), and DSPP transcript levels were quantified by real-time quantitative PCR and normalized to β2-microglobulin levels in the same samples. The bars represent the average of three animals of each genotype. Values are expressed as the transcript levels found in heterozygous (white bars, +/−) and Nfic−/− animals (black bars, −/−) relative to the levels seen in wild-type littermates.
To assess whether the loss of Nfic affected the expression of the other NFI genes, we measured Nfia, Nfib, and Nfix transcript levels in mandibular RNA. No changes were detected in the transcript levels for the other three NFI genes in Nfic−/− mice (Fig. 8, lanes Nfix, Nfib, and Nfia). The levels of Nfix, Nfib, and Nfia transcripts were also similar in liver RNA from wild-type and Nfic−/− mice (not shown). These data indicate that loss of Nfic does not appear to affect the expression of the other NFI genes in the tissues examined.
DISCUSSION
We show here that disruption of Nfic causes major defects in postnatal murine tooth development, the most striking defect being loss of molar root formation (Fig. 5). In addition, there are clear defects in mandibular and maxillary incisor formation and in alveolar bone formation in molar tooth sockets (Fig. 3, 4, and 6). These tooth and bone defects cause runting and lethality unless the Nfic−/− animals are reared on a soft- dough diet (Fig. 2). Since Nfic−/− animals can survive and be fertile if maintained on nutrient dough, it appears that the tooth defects cause the lethal phenotype seen with loss of Nfic. Whether additional nonlethal developmental defects are present in the Nfic−/− animals awaits further analyses.
The specific combination of defects seen in Nfic−/− mice (loss of molar roots with apparently normal crown formation and severe mandibular incisor disruption with milder maxillary incisor defects) is unique to this mouse and indicates a relatively late role for Nfic in tooth development. In contrast, loss of either Msx-1 (1) or Lef1 (33) causes the arrest of tooth development at the bud stage, which precludes analysis of late events in tooth formation. Loss of both Dlx-1 and Dlx-2 results in complete agenesis of maxillary molars, whereas mandibular molars and incisors appear normal (22, 30). This differential effect of loss of the Dlx genes emphasizes the apparently unique molecular requirements of mandibular versus maxillary molar development. In addition, the lack of a dental phenotype in individual Dlx knockouts demonstrates the apparent redundancy of some transcription factors in maxillary molar development. Redundant functions in tooth formation have also been observed for the Gli family of transcription factors. The loss of Gli3 had no observable dental phenotype, whereas the loss of Gli2 resulted in relatively mild abnormalities in maxillary incisor development (10). However, in Gli2/Gli3 double mutants there was no normal tooth development with apparent arrest at the early bud (incisors) or prebud stages (molars). Given this redundancy in Dlx and Gli family members, it will be of interest to determine whether the Nfic−/− phenotype is affected by loss of other NFI genes.
Compared to the study of early tooth development described above, few molecules are known to be required for late events in tooth morphogenesis, including root formation. For example, although Msx-1 is essential for early tooth bud formation, transplantation and rescue studies have shown that Msx-1 is not essential for later cap and crown formation but appears to again play a role in the long-term survival of odontoblasts and dental pulp (1). Thus, it is difficult to predict which, if any, of the genes identified as essential for early tooth development function in molar root formation. Two mutations, Tabby (Ta) and Downless (Dl), are known to affect late tooth development (32). However, these mutations affect both molar crown and root formation, whereas the Nfic−/− mutation affects predominantly root formation. Since Nfic is expressed during both molar crown and root formation (Fig. 7), while the major phenotype we see is restricted to root formation (Fig. 3 to 6), it will be of interest to determine whether one of the other NFI genes compensates for the loss of Nfic during crown formation.
Molar root development begins at ∼P9 by outgrowth of a collar of cells (Hertwig's epithelial root sheath) from the molar crown. These epithelial cells then induce the differentiation of dental papillary mesenchymal cells into odontoblasts which, through migration, dentin deposition, and mineralization form the hard structure of the root. Our data suggest that this collar of cells either fails to grow out of the crown and/or fails to induce odontoblast differentiation (Fig. 5). Whether this molar root defect is related mechanistically to the abnormal incisor development seen in the Nfic−/− mice is unclear. However, it is intriguing that the maxillary incisor defects seen in Nfic−/− mice appear to be due to a failure of differentiation and/or dentin formation in lingual regions of the incisor (Fig. 4F, arrow), much as root formation requires appropriate differentiation and dentin formation at the base of the molar (Fig. 5). Unfortunately, there are no genes or signaling pathways known to be expressed solely in the epithelial root sheath, and the incisors' cellular homologue of the molar root sheath has not been well characterized. In addition, since incisors continue to grow throughout adult life, whereas molar root and crown development and growth terminate, it is possible that there are incisor-specific mechanisms that allow for continuous growth. It will be important to define gene expression patterns in the affected regions of both the molars and incisors in the Nfic−/− mice to determine whether these defects are due to changes in similar regulatory pathways. The Nfic−/− mouse should provide a unique and important resource to determine the transcriptional and cell-signaling pathways essential for molar root formation.
Given the previously noted widespread expression of Nfic during development (3), it is perhaps surprising that the phenotype of the Nfic−/− mouse is so restricted. Indeed, most other mouse mutations affecting tooth development also affect other developmental programs, including hair follicle and skin formation (Ta and Dl), bone formation (Msx1 and -2), craniofacial development (Msx1 and -2, Pax9), cardiac development (Msx1), and others. However, some of these genes are expressed in migrating neural crest cells that populate many organ systems, which most likely explains their pleiotropic phenotypes (2, 6). Detailed genetic analysis of Nfic interactions with other genes will be important in determining whether Nfic functions in other organ systems. Also, since a number of human dental and craniofacial defects appear to be due to mutations in human homologues of murine tooth development genes (11, 28, 34), it will be important to determine whether mutations in NFIC underlie any of the uncharacterized odontogenic syndromes.
The defects seen here are in striking contrast to the neuroanatomical defects present in Nfia−/− animals (agenesis of the corpus callosum, hydrocephalus, etc. [4]) and the lung defects seen in Nfib−/− animals (9) and indicate that these three NFI family members each have unique and essential roles in mouse development. The apparent lack of compensatory changes in the other NFI family members in both Nfic (Fig. 8) and Nfia−/− mice suggest that there is little if any “feedback” between the family members. This is surprising because there is frequently overlapping expression of the different family members during development (3). However, although the three phenotypes are very different, it is possible that similar changes in transcriptional pathways underlie the anatomical defects seen in all three mutants. We are currently taking two approaches to finding out whether Nfic and Nfia interact in similar transcriptional and developmental pathways in mice: (i) we are generating Nfia/Nfic double-knockout mice to assess their phenotype and (ii) we are assessing the earliest changes in gene expression patterns in the affected tissues of Nfic−/− and Nfia−/− mice to determine the pathways disrupted in each animal. These approaches should help us to understand the pathways underlying the anatomical defects seen in these animals and allow us to determine whether these two NFI genes share common regulatory mechanisms in mice.
Acknowledgments
We acknowledge the University of Missouri Research Animal Diagnostic and Investigative Laboratory (Columbia, Mo.), the SUNY Buffalo Histopathology Laboratory, and the Lerner Research Institute (LRI) histology core for histological analyses. We thank Clemencia Colmenares (LRI) for providing advice and assistance in ES cell culture and Valerie Stewart of the LRI transgenic and/or knockout core for blastocyst injections and chimeric mouse production. We also thank Christine E. Campbell (SUNY—Buffalo) for reading the manuscript and helpful discussions.
This work was supported in part by Public Health Service grants HD34901 from the National Institute of Child Health and Development and DK58401 from the National Institute of Diabetes and Digestive and Kidney Diseases to R.M.G.
REFERENCES
- 1.Bei, M., K. Kratochwil, and R. L. Maas. 2000. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development 127:4711-4718. [DOI] [PubMed] [Google Scholar]
- 2.Chai, Y., X. Jiang, Y. Ito, P. Bringas, Jr., J. Han, D. H. Rowitch, P. Soriano, A. P. McMahon, and H. M. Sucov. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127:1671-1679. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry, A. Z., C. E. Lyons, and R. M. Gronostajski. 1997. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 208:313-325. [DOI] [PubMed] [Google Scholar]
- 4.das Neves, L., C. Duchala, F. Godinho, M. Haxhiu, C. Colmenares, W. Macklin, C. Campbell, K. Butz, and R. Gronostajski. 1999. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus and agenesis of the corpus callosum. Proc. Natl. Acad. Sci. USA 96:11946-11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dassule, H. R., P. Lewis, M. Bei, R. Maas, and A. P. McMahon. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127:4775-4785. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson, C. A., A. S. Tucker, and P. T. Sharpe. 2000. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development 127:403-412. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher, C. F., N. A. Jenkins, N. G. Copeland, A. Z. Chaudhry, and R. M. Gronostajski. 1999. Exon structure of the nuclear factor I DNA-binding domain from Caenorhabditis elegans to mammals. Mamm. Genome 10:390-396. [DOI] [PubMed] [Google Scholar]
- 8.Gronostajski, R. M. 2000. Role of NFI/CTF gene family in transcription and development. Gene 249:31-45. [DOI] [PubMed] [Google Scholar]
- 9.Grunder, A., T. T. Ebel, M. Mallo, G. Schwarzkopf, T. Shimizu, A. E. Sippel, and H. Schrewe. 2002. Nuclear factor I-B (Nfib)-deficient mice have severe lung hypoplasia. Mech. Dev. 112:69-77. [DOI] [PubMed] [Google Scholar]
- 10.Hardcastle, Z., R. Mo, C. C. Hui, and P. T. Sharpe. 1998. The Shh signaling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development 125:2803-2811. [DOI] [PubMed] [Google Scholar]
- 11.Headon, D. J., S. A. Emmal, B. M. Ferguson, A. S. Tucker, M. J. Justice, P. T. Sharpe, J. Zonana, and P. A. Overbeek. 2001. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature 414:913-916. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, M., K. Hardy, A. Handyside, S. Hunter, and M. Monk. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326:292-295. [DOI] [PubMed] [Google Scholar]
- 13.Kettunen, P., J. Laurikkala, P. Itaranta, S. Vainio, N. Itoh, and I. Thesleff. 2000. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 219:322-332. [DOI] [PubMed] [Google Scholar]
- 14.Kruse, U., F. Qian, and A. E. Sippel. 1991. Identification of a fourth nuclear factor I gene in chicken by cDNA cloning: NFIX. Nucleic Acids Res. 19:6641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruse, U., and A. E. Sippel. 1994. Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett. 348:46-50. [DOI] [PubMed] [Google Scholar]
- 16.Lyons, G., M. Ontell, R. Cox, D. Sassoon, and M. Buckingham. 1990. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell Biol. 111:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour, S. L., K. R. Thomas, and M. R. Capecchi. 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336:348-357. [DOI] [PubMed] [Google Scholar]
- 18.Meisterernst, M., L. Rogge, C. Donath, I. Gander, F. Lottspeich, R. Mertz, T. Dobner, and R. Fockler. 1988. Isolation and characterization of the porcine nuclear factor I (NFI) gene. FEBS Lett. 236:27-32. [DOI] [PubMed] [Google Scholar]
- 19.Nagata, K., R. Guggenheimer, T. Enomoto, J. Lichy, and J. Hurwitz. 1982. Adenovirus DNA replication in vitro identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc. Natl. Acad. Sci. USA 79:6438-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagata, K., R. A. Guggenheimer, and J. Hurwitz. 1983. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl. Acad. Sci. USA 80:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters, H., A. Neubuser, K. Kratochwil, and R. Balling. 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12:2735-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu, M., A. Bulfone, I. Ghattas, J. J. Meneses, L. Christensen, P. T. Sharpe, R. Presley, R. A. Pedersen, and J. L. Rubenstein. 1997. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 185:165-184. [DOI] [PubMed] [Google Scholar]
- 23.Rupp, R., U. Kruse, G. Multhaup, U. Gobel, K. Beyreuther, and A. Sippel. 1990. Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 18:2607-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoro, C., N. Mermod, P. Andrews, and R. Tjian. 1988. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature 334:218-224. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar, L., M. Cobourne, S. Naylor, M. Smalley, T. Dale, and P. T. Sharpe. 2000. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc. Natl. Acad. Sci. USA 97:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 27.St Amand, T. R., Y. Zhang, E. V. Semina, X. Zhao, Y. Hu, L. Nguyen, J. C. Murray, and Y. Chen. 2000. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 217:323-332. [DOI] [PubMed] [Google Scholar]
- 28.Stockton, D. W., P. Das, M. Goldenberg, R. N. D'Souza, and P. I. Patel. 2000. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24:18-19. [DOI] [PubMed] [Google Scholar]
- 29.Thesleff, I., and P. Sharpe. 1997. Signalling networks regulating dental development. Mech. Dev. 67:111-123. [DOI] [PubMed] [Google Scholar]
- 30.Thomas, B. L., A. S. Tucker, M. Qui, C. A. Ferguson, Z. Hardcastle, J. L. Rubenstein, and P. T. Sharpe. 1997. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development 124:4811-4818. [DOI] [PubMed] [Google Scholar]
- 31.Tucker, A. S., A. Al Khamis, and P. T. Sharpe. 1998. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 212:533-539. [DOI] [PubMed] [Google Scholar]
- 32.Tucker, A. S., D. J. Headon, P. Schneider, B. M. Ferguson, P. Overbeek, J. Tschopp, and P. T. Sharpe. 2000. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development 127:4691-4700. [DOI] [PubMed]
- 33.van Genderen, C., R. M. Okamura, I. Farinas, R. G. Quo, T. G. Parslow, L. Bruhn, and R. Grosschedl. 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 8:2691-2703. [DOI] [PubMed] [Google Scholar]
- 34.Wilkie, A. O., Z. Tang, N. Elanko, S. Walsh, S. R. Twigg, J. A. Hurst, S. A. Wall, K. H. Chrzanowska, and R. E. Maxson, Jr. 2000. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat. Genet. 24:387-390. [DOI] [PubMed] [Google Scholar]