Abstract
Nonsense-mediated mRNA decay (NMD) is a conserved proofreading mechanism that protects eukaryotic cells from the potentially deleterious effects of truncated proteins. Studies of Saccharomyces cerevisiae imply that NMD is a predominantly cytoplasmic decay pathway, while studies of mammalian systems suggest that decay of most substrate mRNAs may occur while they are still associated with the nucleus, possibly during a round of translation that occurs during their export to the cytoplasm. Complete entry of the latter mRNAs into the cytoplasm appears to render them immune to further NMD; i.e., they escape further susceptibility to this decay pathway. To determine if yeast cytoplasmic nonsense-containing mRNAs that evade decay are subsequently immune to NMD, we examined the consequences of placing each of the three UPF/NMD genes under the control of a galactose-inducible promoter. The decay kinetics of ADE2 and PGK1 nonsense-containing mRNAs were then analyzed when expression of UPF1, NMD2, or UPF3 was either repressed or subsequently induced. Results from these experiments demonstrated that activation of NMD caused rapid and immediate degradation of both substrate transcripts, with half-lives of both stable mRNA populations shortened to approximately 7 min. These findings make it unlikely that yeast nonsense-containing mRNAs can escape degradation by NMD and indicate that such mRNAs are available to this decay pathway at each round of translation.
Intricate mechanisms that safeguard against errors in gene expression have been identified in eukaryotes (10, 17, 19, 25, 37, 85). The phenomenon of nonsense-mediated mRNA decay (NMD) exemplifies one such mechanism, eliminating mRNAs containing premature nonsense codons within their protein coding regions and thus minimizing the synthesis of truncated polypeptides (25, 35, 36, 55, 66, 67). The process of NMD has been studied extensively in Saccharomyces cerevisiae, where rapid degradation of nonsense-containing mRNAs involves recognition of a premature translation termination codon, deadenylation-independent decapping, and subsequent 5′→3′ exonucleolytic digestion of the remainder of the mRNA (36, 60). In addition to the decapping enzyme Dcp1p (3, 43) and the exonuclease Xrn1p (32), three additional trans-acting factors are essential for NMD in yeast: Upf1p, Nmd2p/Upf2p, and Upf3p (12, 24, 27, 44-46). Consistent with their roles in the response to aberrant translation, all three of the latter UPF/NMD proteins have been shown to localize predominantly to the cytoplasm and to associate with polyribosomes (1, 2, 53, 64, 73). These observations indicated that yeast NMD occurred in the cytoplasm and was linked to translation, conclusions consistent with other results showing that (i) drugs or mutations that inhibit translation also eliminate NMD (49, 81, 90); (ii) nonsense-containing polysomal mRNAs stabilized in cycloheximide-treated cells reinitiate NMD as soon as the drug is withdrawn (90); and (iii) a dominant-negative form of Nmd2p/Upf2p inhibits decay only when localized to the cytoplasm (24).
In mammalian cells, NMD may not be limited to the cytoplasm. Nonsense-containing derivatives of mammalian β-globin, APRT, and HEXA mRNAs, as well as GPx1 mRNA, have been shown to decay in the cytoplasm (39, 54, 58, 59, 68). However, other nonsense-containing mRNAs, e.g., those encoded by the TPI, TCR-β, and Lκ genes (7-9, 15, 50), as well as globin mRNAs expressed in nonerythroid cells and a portion of APRT mRNA (39, 41, 87, 88), appear to be degraded while still associated with the nucleus. Of particular interest is the observation that the fraction of nonsense-containing TPI mRNAs that avoid nucleus-associated decay and are exported to the cytoplasm appear to be as stable as wild-type TPI mRNA (9). These findings suggest that recognition of premature nonsense codons in at least some mammalian mRNAs occurs solely in the nucleus, or during nuclear export, and that those mRNAs which escape to the cytoplasm can acquire immunity to further degradation by the NMD pathway (76).
To determine if yeast cytoplasmic nonsense-containing mRNAs can become immune to rapid turnover, we examined the decay kinetics of two NMD substrate mRNAs in response to repressing or activating the NMD pathway. Both the ade2-1 and the pgk1-UAG-2 nonsense-containing mRNAs were stabilized by repressing the pathway, and activation of NMD caused rapid and immediate degradation of each transcript. These findings demonstrate that nonsense-containing mRNAs residing in the cytoplasm of yeast cells are potentially susceptible to NMD at each round of translation.
MATERIALS AND METHODS
Strains, plasmids, and general methods.
The yeast strains used in this study are listed in Table 1. Preparation of standard yeast media and methods of cell culture were conducted as described by Rose et al. (69). Transformation of yeast was done by the rapid method described by Soni et al. (75). DNA manipulations were performed according to standard techniques (71). All PCR amplifications were performed with Taq DNA polymerase (84) and confirmed, where appropriate, by DNA sequencing using the method described by Sanger et al. (72). Plasmid DNAs were prepared from Escherichia coli DH5α.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| HFY1200 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 | 24 |
| HFY870 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 NMD2 UPF3 | 27 |
| HFY1300 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 nmd2::HIS3 UPF3 | 24 |
| HFY861 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 upf3::HIS3 | 27 |
| HFY1067 | MATaade2-1 HIS3 his4-38 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 dcp1::URA3 | F. He and A. Jacobson, unpublished data |
| HFY1081 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 xrn::ADE2 | F. He and A. Jacobson, unpublished data |
| HFY456 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 rpb1-1 UPF1 nmd2::HIS3 UPF3 [pRS315] | 24 |
RNA extraction, Northern blotting, and mRNA decay rate measurements.
For the galactose induction experiments, RNA was isolated from yeast using the hot phenol method (29). Aliquots (20 μg) of each RNA sample were analyzed by Northern blotting. For isolation of RNA from polyribosome fractions, the method described by Benard et al. (5) was used. DNA probes were prepared by either random priming with [α-32P]dCTP (16) or by 5′ end-labeling of single-stranded oligodeoxyribonucleotides with [γ-32P]ATP (71). Relative mRNA levels were determined by quantitating Northern blots with a Bio-Rad Molecular Imager. The DNA probes used to detect specific transcripts included ADE2 (a 2-kb BglII fragment from an xrn1::ADE2 disruption plasmid), PGK1 (oligonucleotide 1 from reference 66), CYH2 (a 600-bp EcoRI-HindIII fragment from pGEM4Z-CYH2 which hybridizes to both the pre-mRNA and the mRNA [29]), and SCR1 (a 400-bp fragment amplified from yeast genomic DNA using oligonucleotides SCR1-1 and SCR1-2 [52]). mRNA decay rates were measured in wild-type and nmd2 cells grown in either rich medium (YEPD) or SC-galactose minimal medium, using thermal inactivation of RNA polymerase II to inhibit ongoing transcription (24, 29, 69).
Protein gels, Western blotting, and antibodies.
Whole-cell lysates were prepared by collecting culture aliquots equivalent to 0.2 ml at an optical density at 600 nm (OD600) of 1. The appropriate volume was centrifuged for 10 min, resuspended in 10 μl of 1× sample buffer (71), and boiled for 5 min just before loading onto the respective polyacrylamide gel. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described by Laemmli (42). Gels were electroblotted to Immobilon-P membranes (Millipore) under conditions recommended by the manufacturer. The binding conditions used for antibodies were as described by Harlow and Lane (23). Detection was enhanced by chemiluminescence with either the ECL or ECL(+) kit from Amersham Corp. Antibodies used included polyclonal affinity-purified anti-Upf1p antibody (53), polyclonal affinity-purified anti-Nmd2p antibody (26), and the monoclonal antihemagglutinin (anti-HA) antibody, 12CA5 (from Boehringer Mannheim Biochemicals), for detection of Upf1p, Nmd2p, and HA epitope-tagged Upf3p, respectively.
Plasmid constructions.
The galactose-inducible UPF1 construct was made by ligating a 3.6-kb EcoRI-SalI fragment from pMA424-UPF1 (24) to pRS426 (11), containing the GAL1 promoter (664-bp fragment immediately upstream of the initiation codon, generated by PCR), cut with the same enzymes. The galactose-inducible NMD2 plasmid was constructed by ligating a 3.7-kb XbaI-SalI fragment cut from the pRS315-NMD2 plasmid (27) to the pMW29 vector (91) cut with the same enzymes. The galactose-inducible UPF3 plasmid was constructed by ligating a 1.7-kb NcoI-SalI fragment cut from the pRS314-HA-UPF3 plasmid (27) to pRS316 (74), containing the GAL1 promoter, cut with the same enzymes. The latter plasmid was obtained by restriction digestion of pRS314-GALp-HA-NMD3 [4]).
Preparation of polyribosome fractions.
Yeast cell extracts were prepared and fractionated on sucrose gradients as described previously (53).
Galactose induction.
Yeast strains containing galactose-inducible UPF/NMD gene constructs were grown in SC medium (69) without uracil and with raffinose to mid-log phase (OD600 = 0.5). Strains containing the inducible UPF/NMD gene constructs and either of the pgk1 nonsense alleles were grown in SC medium without uracil and leucine and with raffinose (to maintain selection for the GAL-UPF/NMD plasmid and the plasmid harboring the pgk1 allele, respectively) to mid-log phase(OD600 = 0.5). Galactose was then added to a final concentration of 2%. Culture aliquots for RNA and protein isolations were taken at 10-min intervals for 40 min.
RESULTS
The ade2-1 transcript is a substrate for NMD.
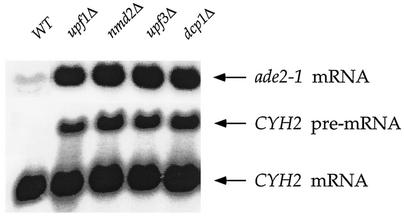
To address the stability of cytoplasmic nonsense-containing mRNAs, we took advantage of an allele of the ADE2 gene, ade2-1. Earlier studies showed that the ade2-1 mutation could be suppressed in yeast strains containing an ochre tRNA suppressor (86), suggesting that the ade2-1 allele was attributable to a nonsense (UAA) mutation and that the ade2-1 mRNA was likely to be a substrate for NMD. To test the latter possibility, single deletions of UPF1, NMD2, or UPF3 were constructed in yeast strains that harbored the ade2-1 allele, and the effects of these mutations on the abundance of the ade2-1 transcript were examined. Northern analyses of mRNA steady-state levels demonstrated that mutations in genes regulating the stability of nonsense-containing transcripts affected the ade2-1 transcript in precisely the same manner that they affected a well-characterized NMD substrate, the CYH2 pre-mRNA (Fig. 1) (25). The ade2-1 mRNA was approximately sevenfold more abundant in upf/nmd mutant cells than in the isogenic UPF/NMD (wild-type) strain (Fig. 1). Likewise, deletion of a gene encoding a general factor involved in mRNA decay (i.e., DCP1) (62) also promoted a sevenfold increase in ade2-1 transcript abundance (Fig. 1). These differences in mRNA abundance were consistent with the respective differences in the decay rates of the ade2-1 mRNA in wild-type and upf/nmd mutant cells. In YEPD medium at 37°C, the half-life (t1/2) of the ade2-1 mRNA was found to be less than 5 min in the UPF/NMD strain and approximately 35 min in upf/nmd cells (data not shown), suggesting that the wild-type gene, ADE2, encodes a relatively stable mRNA. The latter conclusion is supported by previous experiments, reporting t1/2s of 30 and 33 min, respectively, for the ADE2 mRNA (80; htt://web.wi.mit.edu/young/expression/halflife.html). Collectively, these results indicate that the ade2-1 mRNA requires Upf1p, Nmd2p, Upf3p, and Dcp1p for its degradation and is, therefore, a typical substrate for NMD.
FIG. 1.
The ade2-1 transcript is a substrate for NMD. Total RNA isolated from yeast strains with the indicated UPF/NMD genotypes was analyzed by Northern blotting with DNA probes that detected the ade2-1 or CYH2 transcripts. WT, wild type. The yeast strains used for this experiment were grown in YEPD at 30°C and included HFY1200 (WT), HFY870 (upf1Δ), HFY1300 (nmd2Δ), HFY861 (upf3Δ), and HFY1067 (dcp1Δ).
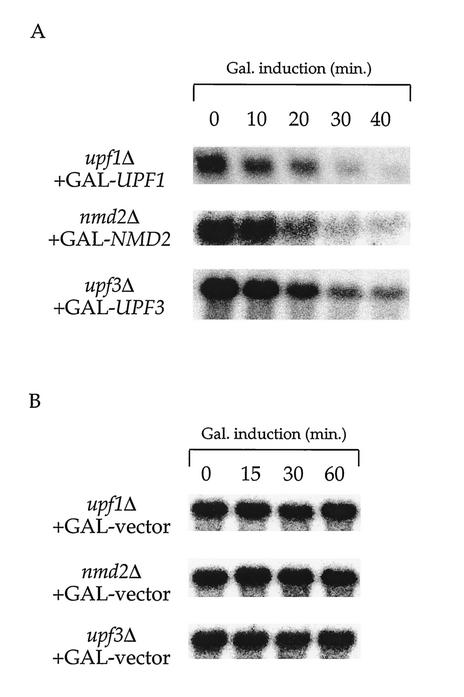
Galactose-inducible expression of UPF1, NMD2, and UPF3.
To assess the stability of ade2-1 transcripts that had avoided degradation by the NMD pathway, we sought a mechanism to regulate the activity of the pathway. To accomplish this, the UPF1, NMD2, and UPF3 genes were cloned into either single- or high-copy-number plasmids containing the inducible GAL1 promoter, and the resulting plasmids were transformed into the respective UPF/NMD deletion strains. Each of the resulting strains contained a galactose-regulated UPF/NMD gene. As shown in Fig. 2, Upf1p, Nmd2p, and Upf3p are not detectable in the respective regulated strains prior to galactose induction, but these proteins accumulate substantially postinduction. Quantitation of each of the Western blots shown in Fig. 2, and others, indicated that (i) Upf1p, Nmd2p, and Upf3p all begin to accumulate approximately 12 to 14 min after galactose addition and (ii) by 20 min after galactose addition, the cellular levels of each of the induced proteins are comparable to those present in the isogenic UPF/NMD strains (data not shown). From these data, we conclude that use of these constructs allows for inducible expression of UPF1, NMD2, and UPF3.
FIG. 2.
Galactose-inducible expression of UPF1, NMD2, and UPF3. Yeast strains harboring upf1Δ (HFY870), nmd2Δ (HFY1300), or upf3Δ (HFY861) mutations and the respective, complementary galactose-inducible NMD gene construct were grown in SC raffinose liquid medium without uracil to mid-log phase (OD600 = 0.5). Galactose was then added to a final concentration of 2%, culture aliquots were taken at 10-min intervals, and all samples were subsequently analyzed by Western blotting. Gal., galactose.
The ade2-1 transcript is rapidly degraded upon activation of NMD.
The availability of the strains described above makes it possible to determine the stability of ade2-1 transcripts before and after activation of the NMD pathway. Under conditions where NMD is inactive, these nonsense-containing mRNAs accumulate in the cytoplasm and are relatively stable. Upon activation of the NMD pathway, the fate of these mRNAs can be monitored by simply measuring their relative abundance over time, leading to an approximation of the decay kinetics of the steady-state ade2-1 mRNA population. If the accumulated ade2-1 transcripts are susceptible to NMD, then activation of this decay pathway should result in their rapid degradation. If the ade2-1 transcripts are immune to NMD, then activation of the decay pathway should have no effect on the stability of these mRNAs. The overall ade2-1 mRNA population would then consist of newly synthesized mRNAs that are rapidly degraded and the stable cytoplasmic transcripts that had accumulated prior to activation of NMD. Under these circumstances, the expected decay rate of the entire ade2-1 mRNA population would initially be slow (approximating that of the stabilized ade2-1 transcripts), and then, after substantial dilution with newly synthesized mRNA, the population would begin to reflect a more rapid decay rate.
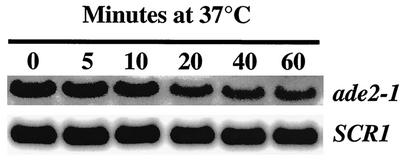
These possibilities were evaluated by Northern blot analyses of yeast strains expressing regulatable UPF1, NMD2, or UPF3. As a first step, the t1/2 of the ade2-1 mRNA was measured in nmd2 cells in SC-galactose medium. In experiments in which temperature-sensitive rpb1-1/nmd2Δ cells were shifted to 37°C (24), the ade2-1 mRNA had a t1/2 of 22 to 24 min (Fig. 3). Extrapolation from previous comparisons of mRNA decay rates at 30 and 37°C indicates that the t1/2 of this mRNA at 30°C must approximate 44 to 48 min (29, 65). The abundance of this stabilized ade2-1 mRNA was then assessed (at 30°C) as a function of UPF1, NMD2, or UPF3 expression. Figure 4A shows that, as expression of these factors increases (Fig. 2), the abundance of the ade2-1 mRNA decreases (Fig. 4A). Subsequent to the time at which the UPF/NMD proteins begin to accumulate (12 to 14 min postinduction; see above), the ade2-1 mRNA disappears, with a t1/2 of approximately 7 min in all three strains. By 30 min after galactose induction of any of the three UPF/NMD genes, approximately 20% of the ade2-1 mRNA population remains, and by 40 min, the abundance of the ade2-1 mRNA returns to the low levels characteristic of a UPF/NMD (wild-type) strain. These experiments show that induction of Upf1p, Nmd2p, or Upf3p restores NMD and results in immediate destabilization of the entire ade2-1 mRNA population, i.e., the ade2-1 mRNA molecules present in the cell prior to galactose induction are not immune to degradation by NMD.
FIG. 3.
t1/2 of the ade2-1 mRNA in SC-galactose medium. Yeast cells (HFY456) harboring nmd2Δ and rpb1-1 mutations were grown in SC-galactose medium at 24°C and then shifted to 37°C to inhibit further transcription. RNA was isolated at different times after the temperature shift and subjected to Northern blotting as in Fig. 1. Hybridization to the SCR1 probe served as a loading control (52).
FIG. 4.
The ade2-1 transcript is rapidly degraded upon activation of NMD. (A) Activation of NMD causes rapid degradation of ade2-1 mRNA. Total RNA was isolated from yeast strains (HFY870, HFY1300, and HFY861) harboring upf1Δ, nmd2Δ, or upf3Δ mutations and the respective, complementary galactose-inducible NMD gene constructs (GAL-UPF1, GAL-NMD2, or GAL-UPF3). Galactose induction was performed as described in Fig. 2, and RNA was analyzed by Northern blotting with a DNA probe that detected the ade2-1 transcript. (B) The addition of galactose does not destabilize ade2-1 mRNA. Total RNA isolated from yeast strains with the indicated UPF/NMD genotypes, and harboring only the vector plasmid (GAL-vector), was analyzed by Northern blotting as in Fig. 4A. Gal., galactose.
To ensure that addition of galactose, by itself, does not result in destabilization of the ade2-1 mRNA, the galactose induction experiment was repeated in upf1Δ, nmd2Δ, and upf3Δ strains transformed with an empty GAL1 vector. Northern analyses of RNA isolated from these strains demonstrate that the ade2-1 transcript remains stable throughout the course of this control experiment (Fig. 4B). Side-by-side Northern analyses of ade2-1 mRNA in these control cells with those obtained in uninduced cells harboring GAL1-UPF1, GAL1-NMD2, or GAL1-UPF3 plasmids yielded virtually identical levels (data not shown), a result indicating that the observed induction is a bona fide switch from inactive to active states of NMD.
Degradation of the ade2-1 mRNA population occurs on polyribosomes.
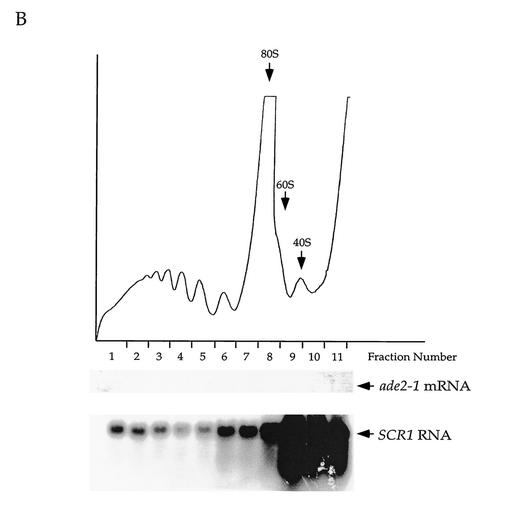
To confirm that the ade2-1 mRNA accumulated in upf/nmd cells is cytoplasmic and that its eventual decay occurs on polyribosomes (90), the association of the ade2-1 mRNA population with ribosomes was investigated under conditions where NMD was either inactive or active. Cytoplasmic extracts were prepared from a strain containing galactose-inducible UPF1, both prior to galactose induction and 30 min postinduction, and then resolved on sucrose gradients. Fractions collected from these gradients were analyzed by Northern blotting. Under circumstances when NMD is inactive, the ade2-1 mRNA was found to cosediment predominantly with the polyribosome fractions (Fig. 5A, fractions 1 to 7), suggesting that these transcripts are associated with actively translating ribosomes. The association of these transcripts with an average of four to five ribosomes is consistent with premature translational termination within a large mRNA (2.2 kb [77]). Upon restoration of NMD, the ade2-1 mRNA is rapidly degraded (Fig. 4) and is no longer detected in the polyribosome fractions (Fig. 5B). As a control for these experiments, the Northern blots of Fig. 5A and B were also probed for the SCR1 RNA. The latter blots demonstrate that the qualities and quantities of RNA isolated from the two sets of gradients (0 and 30 min post-galactose induction) were similar (Fig. 5A and B).
FIG. 5.
Degradation of the ade2-1 mRNA occurs on polysomes. (A) The ade2-1 mRNA is detected in the polysome fractions before galactose induction. Total RNA isolated from polysome fractions collected before the addition of galactose was analyzed by Northern blotting with DNA probes that detected the ade2-1 mRNA and the SCR1 RNA (the latter to serve as a control to ensure that RNA was isolated from the polysome fractions). (B) The ade2-1 mRNA is no longer detected in the polysome fractions upon activation of NMD. Total RNA isolated from polysome fractions collected 30 min after the addition of galactose was analyzed by Northern blotting as described for panel A. The results depicted in this figure were obtained from the upf1Δ yeast strain (HFY870) harboring the galactose-inducible UPF1 construct.
Results virtually identical to those of Fig. 5A and B were obtained using the galactose-regulated NMD2 and UPF3 constructs (data not shown). Taken together, these findings indicate that the ade2-1 mRNA that accumulates when NMD is inactive associates with cytoplasmic ribosomes and that this mRNA disappears from the polyribosomal fraction when its degradation by the NMD pathway is activated.
Activation of NMD triggers rapid decay of PGK1 transcripts with early but not late nonsense codons.
To substantiate our findings with the ade2-1 mRNA, we investigated the effect that restoration of NMD had on the decay kinetics of another nonsense-containing transcript. The PGK1 mRNA is normally very stable, having a t1/2 in rpb1-1 temperature-shift experiments of 60 to 70 min (66, 80), but a derivative with a nonsense mutation at codon 22 (pgk1-UAG-2) is extremely unstable (t1/2 = 6 min [66, 81]). Inactivation of the NMD pathway (by mutations in UPF1, NMD2, or UPF3) restores the stability of this nonsense-containing mRNA (t1/2 >35 min [66]), confirming that it is a substrate for NMD. The large differences in the t1/2s of this transcript in the active and inactive states of NMD make it ideal for an investigation of the possible existence of mRNA immunity to rapid decay.
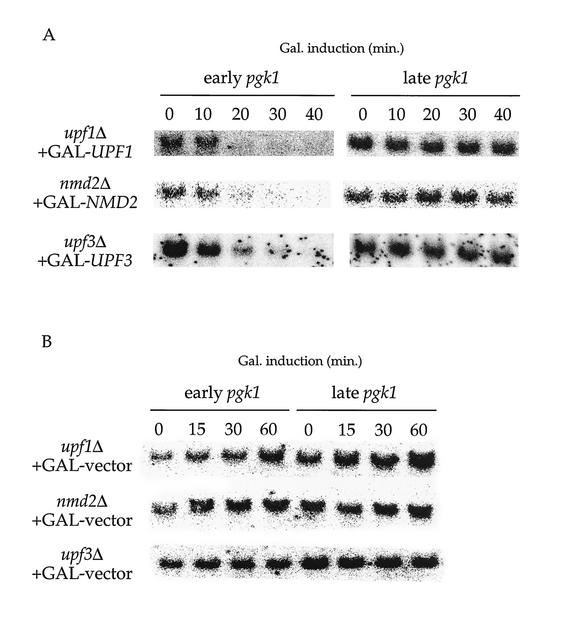
Figure 6A demonstrates that induction of UPF1, NMD2, or UPF3 (in the respective deletion strains) resulted in rapid disappearance of the pgk1-UAG-2 mRNA (see “early pgk1” panel). The decay kinetics for the steady-state population of this mRNA were comparable to those of the ade2-1 mRNA, such that (i) it disappeared with a t1/2 of approximately 7 min, after a lag for induction of the pathway, and (ii) by 30 min postinduction, most (85%) of the mRNA was degraded (Fig. 6A and 7). These results support previous findings that this transcript is a substrate for NMD and indicate that restoration of the NMD pathway causes its rapid and immediate degradation.
FIG. 6.
pgk1 mRNA harboring an early nonsense codon degrades rapidly upon activation of NMD. (A) Activation of NMD results in degradation of a pgk1 transcript containing an early nonsense codon but does not destabilize a pgk1 transcript with a late nonsense codon. Total RNA isolated from yeast strains (HFY870, HFY1300, and HFY861) with the indicated UPF/NMD genotypes harboring complementing galactose-inducible NMD gene constructs (GAL-UPF1, GAL-NMD2, or GAL-UPF3) was analyzed by Northern blotting with a DNA probe that detected nonsense-containing pgk1 transcripts. (B) The addition of galactose does not destabilize pgk1 mRNA harboring an early nonsense codon. Total RNA isolated from yeast strains with the indicated UPF/NMD genotypes harboring only the vector plasmid (GAL-vector) was analyzed by Northern blotting as described for panel A. Gal., galactose.
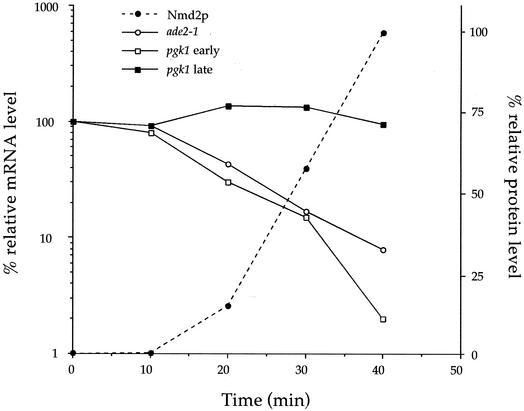
FIG. 7.
Activation of NMD results in the rapid decay of substrate transcripts. Quantitation of the relative levels of Nmd2p (•) and nonsense-containing mRNAs before and after the activation of NMD (by the addition of galactose): ade2-1 mRNA (○), pgk1 mRNA harboring an early nonsense codon (▪), and pgk1 mRNA harboring a late nonsense codon (□). The data for this graph were derived from the Northern blots of Fig. 4A (ade2-1) and 6A (early and late pgk1) and from the Western blot of Fig. 2 (Nmd2p).
Destabilization of mRNAs by premature nonsense codons is a position-dependent phenomenon wherein mRNAs with nonsense codons occurring in the last 20 to 30% of the coding region retain their wild-type decay rates (21, 35, 36, 66). As an additional means to determining whether the postinduction disappearance of the pgk1-UAG-2 and ade2-1 mRNAs was a direct consequence of restoration of NMD, we repeated the NMD induction experiments in cells harboring a pgk1 allele with a nonsense mutation at codon 385 (pgk1-UAG-7). This mutation does not affect the stability of the encoded mRNA (t1/2 > 35 min) and does not render it a substrate for NMD (66). As such, the pgk1-UAG-7 transcript serves as an ideal control to test whether galactose induction of the UPF/NMD genes results in selective degradation of bona fide NMD substrates. Figure 6A shows that galactose induction of UPF1, NMD2, or UPF3 does not affect the abundance of the pgk1-UAG-7 mRNA (see “late pgk1”). This result demonstrates that the decay pathway activated by induction of the UPF/NMD genes remains specific for proper substrate mRNAs and reiterates the finding that the pgk1-UAG-7 mRNA is not a substrate for NMD.
Control experiments were also conducted to ensure that destabilization of the pgk1-UAG-2 mRNA subsequent to restoration of NMD was not due to an effect of galactose addition. Figure 6B shows that upf1Δ, nmd2Δ, and upf3Δ strains containing either the early or late pgk1 nonsense alleles and an empty GAL1 vector do not alter the stability of either the early or late pgk1 nonsense mRNAs in response to galactose addition to the growth medium. Therefore, it is activation of the NMD pathway and not simply the addition of galactose that causes destabilization of the pgk1-UAG-2 mRNA.
DISCUSSION
Inducible NMD in yeast.
In the absence of a functional NMD pathway, yeast nonsense-containing transcripts exhibit t1/2s characteristic of their stable, wild-type counterparts (24, 46, 81). To determine whether nonsense-containing mRNAs that accumulate in NMD-deficient cells retain their ability to be recognized and degraded by the NMD pathway, we created a series of yeast strains in which NMD was galactose inducible. We then analyzed the cellular levels of specific nonsense-containing mRNAs when expression of UPF1, NMD2, or UPF3 was repressed, or subsequently induced. Induction of any of the UPF/NMD genes in their respective deletion strains resulted in a 12- to 14-min lag before any of the factors could be detected, followed by a rapid and immediate reduction of the levels of ade2-1 and pgk1-UAG-2 substrate transcripts. By 40 min postinduction, the levels of both substrate mRNAs in all induced strains were decreased to levels observed in wild-type cells (Fig. 4, 6, and 7). These results indicate that de novo synthesis of the respective missing factors reconstituted the NMD pathway in each of the strains analyzed, leading to a reduction in t1/2s of preexisting nonsense-containing mRNAs. Several considerations suggest that the observed reductions in mRNA levels reflect a uniform switch in mRNA t1/2s, from >35 min to approximately 7 min (Fig. 7), as opposed to the average of the behaviors of multiple mRNA populations. If, for example, the steady-state populations of ade2-1 and pgk1-UAG-2 mRNAs remained stable and only the newly synthesized mRNA decayed rapidly, then the newly synthesized population would have to comprise at least 85% of the total mRNA of each species to accommodate the observed changes in absolute levels. Such massive accumulation of the newly synthesized mRNA species would be highly unlikely in light of (i) the constraints imposed by their short t1/2 (22) and (ii) the apparent differences in the rates of transcription of the ADE2 and PGK1 genes (31).
Further support for the notion that the ade2-1 and pgk1-UAG-2 mRNA populations are uniformly destabilized after UPF1, NMD2, or UPF3 induction is obtained from the sucrose gradient analysis of Fig. 5. In strains in which NMD was inactive, ade2-1 mRNA was found to cosediment with polyribosomes. Induction of the NMD pathway for 30 min, however, led to a quantitative and selective disappearance of the transcripts harboring early nonsense codons from the polysome fractions. If two separate subpopulations of mRNA existed (i.e., stable preinduction mRNA and unstable postinduction mRNA), we should have detected the residual stable species on Northern blots performed after 30 min of galactose induction.
What comprises a substrate for nonsense-mediated decay of yeast mRNAs?
Yeast mRNAs containing premature translation termination codons are rapidly degraded via the NMD pathway when several criteria are met. The termination codon in question must occur within the first two-thirds to three-quarters of the mRNA coding region and be 5′ proximal to an essential sequence element (the downstream element [64, 66, 89]). Moreover, the nonsense-containing mRNA needs to be translated (90), and several factors essential to the NMD process need to be present and functional (12, 24, 27, 44, 46). The nonsense codon that promotes mRNA destabilization can occur within a conventional coding region (49, 66) or be derived from an upstream open reading frame (12), present within an unprocessed intron (25), recognized only during out-of-frame translation caused by leaky scanning (81), or be the normal termination codon in an mRNA with an extended 3′ untranslated region (UTR) (61). Since NMD has been shown to occur without significant prior shortening of the mRNA poly(A) tail (60), it has been suggested that the decay-initiating event can occur very early in the functional lifetime of the mRNA (35). It has been unclear, however, whether this is obligatory or if an mRNA qualifies as an NMD substrate at any time during its cellular life cycle.
One perspective, originally derived from data in mammalian cells, suggests that spatial relationships reflect temporal relationships, i.e., that the apparent nuclear proximity of NMD (55, 56) and the deposition of factors essential for NMD during pre-mRNA splicing (34, 40, 47, 48, 51) must reflect a decay process that occurs during an early round of translation or not at all (9, 76). Experiments utilizing regulated expression of the Gpx1 mRNA suggest that this rule may only apply to transcripts for which NMD is nucleus associated (78). The notion of a limited opportunity for decay of nonsense-containing transcripts has also been considered in yeast, where it has been shown that the RNA-binding protein Hrp1p associates with Upf1p and with the downstream element of a PGK1 nonsense-containing transcript, and that these associations appear to promote NMD (18). The known roles of Hrp1p in mRNA processing and export (28, 38, 57) led to a model in which Hrp1p association with newly synthesized mRNA would be disrupted by the initial round of translation but maintained if translation were interrupted by premature termination (18). In the latter event, Hrp1p bound to an mRNA was postulated to stimulate rapid mRNA decay as a consequence of its ability to interact with the Upf1p-containing surveillance complex (13, 14, 18).
A second, related model for yeast NMD postulates that mRNA decay is triggered by a ribosome's failure to terminate adjacent to a properly configured 3′ UTR (30, 36). This “faux-UTR” model suggests that proper termination of translation and normal rates of mRNA decay only occur in the context of interactions between a terminating ribosome and a specific RNP domain or set of factors localized 3′ to the stop codon (6, 30, 36). The mammalian and surveillance complex models both imply that at least some nonsense-containing mRNAs are only capable of being degraded during their initial rounds of translation, after which they acquire immunity to NMD. However, the data presented here demonstrate that, at least in yeast, NMD is not limited to an early round of mRNA translation and can occur at any time during an mRNA's life cycle. This suggests that (i) there are no NMD-essential factors that are shed during readthrough translation in NMD-deficient strains (45, 52, 82, 83), (ii) factors that are shed can reassociate with an mRNA while it remains in the cytoplasm, or (iii) marking is not critical for NMD of ade2 and pgk1 mRNAs and that their mode of decay more closely approximates the tenets of the faux-UTR model (36).
The continual availability of substrates for the yeast NMD apparatus implies that their decay occurs in the cytoplasm.
The mere presence of a premature nonsense codon within a transcript is not sufficient to promote its degradation. Destabilization of yeast nonsense-containing mRNAs requires their translation, a conclusion that follows from observations that NMD, its principal factors, and decay intermediates are all localized to polysomes (1, 2, 53, 64, 73, 81, 90; see also Fig. 5) and that decay can be antagonized by drugs or mutations that interfere with protein synthesis (70, 90) or by tRNAs that suppress termination (20, 49). While a role for translation has generally been regarded to imply a cytoplasmic event, recent results with mammalian cells (33, 63) demand firmer evidence for such a spatial assignment. Experiments providing additional substantiation include those showing that (i) mutations of the UPF/NMD genes not only lead to the stabilization of nonsense-containing mRNAs but also promote nonsense suppression (45, 52, 79, 82, 83), (ii) dominant-negative Nmd2p is only active when localized to the cytoplasm (24), and (iii) Upf1p interacts with the polypeptide release factors Sup35p (eRF3) and Sup45p (eRF1) to modulate termination (13, 14, 79). The data of this paper provide yet another indication of the cytoplasmic nature of yeast NMD: nonsense-containing transcripts are capable of being degraded long after they have been synthesized and exported to the cytoplasm. In the absence of evidence for reassociation of these transcripts with nuclei, their continual availability to the NMD apparatus is not consistent with a nuclear, or nucleus-associated, decay pathway.
Acknowledgments
This work was supported by a grant (GM27757) to A.J. from the National Institutes of Health and a predoctoral NRSA fellowship (GM18043) to A.M. from the National Institutes of Health.
We thank the members of the Jacobson laboratory for their helpful advice and editorial comments and are especially indebted to David Mangus for stimulating insights.
Alan B. Maderazo and Jonathan P. Belk contributed equally to this work.
REFERENCES
- 1.Atkin, A., L. Schenkman, M. Eastham, J. Dahlseid, M. Lelivelt, and M. Culbertson. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272:22163-22172. [DOI] [PubMed] [Google Scholar]
- 2.Atkin, A., N. Altamura, P. Leeds, and M. Culbertson. 1995. The majority of yeast UPF1 colocalizes with polyribosomes in the cytoplasm. Mol. Cell. Biol. 6:611-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beelman, C., A. Stevens, G. Caponigro, T. LaGrandeur, L. Hatfield, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 4.Belk, J., F. He, and A. Jacobson. 1999. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA 5:1055-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benard, L., K. Carroll, R. Valle, and R. Wickner. 1998. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 18:2688-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonetti, B., L. Fu, J. Moon, and D. M. Bedwell. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251:334-345. [DOI] [PubMed] [Google Scholar]
- 7.Carter, M., S. Li, and M. Wilkinson. 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15:5965-5975. [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, J., M. Fogel-Petrovic, and L. Maquat. 1990. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol. 10:5215-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, J., and L. Maquat. 1993. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol. 13:1892-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, K., and A. M. Pyle. 1995. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA 1:391-406. [PMC free article] [PubMed] [Google Scholar]
- 11.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 12.Cui, Y., K. Hagan, S. Zhang, and S. Peltz. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9:423-436. [DOI] [PubMed] [Google Scholar]
- 13.Czaplinski, K., M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng, H. A. Perlick, H. C. Dietz, M. D. Ter-Avanesyan, and S. W. Peltz. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czaplinski, K., M. Ruiz-Echevarria, C. Gonzalez, and S. Peltz. 1999. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays 21:685-696. [DOI] [PubMed] [Google Scholar]
- 15.Daar, I., and L. Maquat. 1988. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol. Cell. Biol. 8:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, A., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 17.Freist, W., H. Sternbach, and F. Cramer. 1996. Phenylalanyl-tRNA synthetase from yeast and its discrimination of 19 amino acids in aminoacylation of tRNA(Phe)-C-C-A and tRNA(Phe)-C-C-A(3′NH2). Eur. J. Biochem. 240:526-531. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, C., M. Ruiz-Echevarria, S. Vasudevan, M. Henry, and S. Peltz. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 3:489-499. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman, S., R. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 20.Gozalbo, D., and S. Hohmann. 1990. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr. Genet. 17:77-79. [DOI] [PubMed] [Google Scholar]
- 21.Hagan, K., M. Ruiz-Echevarria, Y. Quan, and S. Peltz. 1995. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 15:809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hargrove, J. L., and F. H. Schmidt. 1989. The role of mRNA and protein stability in gene expression. FASEB J. 3:2360-2370. [DOI] [PubMed] [Google Scholar]
- 23.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.He, F., and A. Jacobson. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437-454. [DOI] [PubMed] [Google Scholar]
- 25.He, F., S. Peltz, J. Donahue, M. Rosbash, and A. Jacobson. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc. Natl. Acad. Sci. USA 90:7034-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, F., A. Brown, and A. Jacobson. 1996. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2:153-170. [PMC free article] [PubMed] [Google Scholar]
- 27.He, F., A. Brown, and A. Jacobson. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry, M., C. Z. Borland, M. Bossie, and P. A. Silver. 1996. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 142:103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrick, D., R. Parker, and A. Jacobson. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilleren, P., and R. Parker. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33:229-260. [DOI] [PubMed] [Google Scholar]
- 31.Holstege, F., E. Jennings, J. Wyrick, T. Lee, C. Hengartner, M. Green, T. Golub, E. Lander, and R. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, C., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 34.Ishigaki, Y., X. Li, G. Serin, and L. E. Maquat. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607-617. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson, A., and S. W. Peltz. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65:693-739. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson, A., and S. W. Peltz. 2000. Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae, p. 827-847. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control, 2nd ed. Cold Spring Harbor Laboratory Press, N.Y.
- 37.Jeon, C., and K. Agarwal. 1996. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl. Acad. Sci. USA 93:13677-13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, and P. A. Silver. 1997. Hrp1p, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler, O., and L. A. Chasin. 1996. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol. Cell. Biol. 16:4426-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, V. N., N. Kataoka, and G. Dreyfuss. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293:1832-1836. [DOI] [PubMed] [Google Scholar]
- 41.Kugler, W., J. Enssle, M. W. Hentze, and A. E. Kulozik. 1995. Nuclear degradation of nonsense mutated beta-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia? Nucleic Acids Res. 23:413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 43.LaGrandeur, T., and R. Parker. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, B., and M. Culbertson. 1995. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl. Acad. Sci. USA 92:10354-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leeds, P., J. Wood, B. Lee, and M. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leeds, P., S. Peltz, A. Jacobson, and M. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2203-2314. [DOI] [PubMed] [Google Scholar]
- 47.Le Hir, H., M. J. Moore, and L. E. Maquat. 2000. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 14:1098-1108. [PMC free article] [PubMed] [Google Scholar]
- 48.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Losson, R., and F. Lacroute. 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA 76:5134-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lozano, F., B. Maertzdorf, R. Pannell, and C. Milstein. 1994. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 13:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836-1839. [DOI] [PubMed] [Google Scholar]
- 52.Maderazo, A., F. He, D. Mangus, and A. Jacobson. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mangus, D., and A. Jacobson. 1999. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods 17:28-37. [DOI] [PubMed] [Google Scholar]
- 54.Maquat, L., A. Kinniburgh, E. Rachmilewitz, and J. Ross. 1981. Unstable beta-globin mRNA in mRNA-deficient beta0 thalassemia. Cell 27:543-553. [DOI] [PubMed] [Google Scholar]
- 55.Maquat, L. 1995. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453-465. [PMC free article] [PubMed] [Google Scholar]
- 56.Maquat, L. E. 2000. Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation, p. 849-868. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control, 2nd ed. Cold Spring Harbor Laboratory Press, N.Y.
- 57.Minvielle-Sebastia, L., K. Beyer, A. M. Krecic, R. E. Hector, M. S. Swanson, and W. Keller. 1998. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 17:7454-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriarty, P. M., C. C. Reddy, and L. E. Maquat. 1997. The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA 3:1369-1373. [PMC free article] [PubMed] [Google Scholar]
- 59.Moriarty, P. M., C. C. Reddy, and L. E. Maquat. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muhlrad, D., and R. Parker. 1994. Premature translation termination triggers mRNA decapping. Nature 370:578-581. [DOI] [PubMed] [Google Scholar]
- 61.Muhlrad, D., and R. Parker. 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muhlrad, D., C. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 63.Pederson, T. 2001. Is the nucleus in need of translation? Trends Cell Biol. 11:395-397. [DOI] [PubMed] [Google Scholar]
- 64.Peltz, S., C. Trotta, F. He, A. Brown, J. Donahue, E. Welch, and A. Jacobson. 1993. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay, p. 1-10. In A. Brown, M. Tuite, and J. McCarthy (ed.), Protein synthesis and targeting in yeast. Springer-Verlag, Berlin, German.
- 65.Peltz, S., J. L. Donahue, and A. Jacobson. 1992. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5778-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peltz, S., A. Brown, and A. Jacobson. 1993. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 7:1737-1754. [DOI] [PubMed] [Google Scholar]
- 67.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885-1897. [DOI] [PubMed] [Google Scholar]
- 68.Rajavel, K. S., and E. F. Neufeld. 2001. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 21:5512-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rose, M., F. Winston, and P. Heiter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 70.Ruiz-Echevarria, M. J., C. I. Gonzalez, and S. W. Peltz. 1998. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 17:575-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Sanger, F., S. Nicklen, and A. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shirley, R., M. Lelivelt, L. Schenkman, J. Dahlseid, and M. Culbertson. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111:3129-3143. [DOI] [PubMed] [Google Scholar]
- 74.Sikorski, R., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soni, R., J. Carmichael, and J. Murray. 1993. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet. 24:455-459. [DOI] [PubMed] [Google Scholar]
- 76.Stephenson, L., and L. Maquat. 1996. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie 78:1043-1047. [DOI] [PubMed] [Google Scholar]
- 77.Stotz, A., and P. Linder. 1990. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene 95:91-98. [DOI] [PubMed] [Google Scholar]
- 78.Sun, X., P. M. Moriarty, and L. E. Maquat. 2000. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J. 19:4734-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, W., K. Czaplinski, Y. Rao, and S. W. Peltz. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20:880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Welch, E., and A. Jacobson. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18:6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weng, Y., K. Czaplinski, and S. W. Peltz. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16:5491-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weng, Y., K. Czaplinski, and S. W. Peltz. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 16:5477-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.White, T., N. Arnheim, and H. Erlich. 1989. The polymerase chain reaction. Trends Genet. 5:185-189. [DOI] [PubMed] [Google Scholar]
- 85.Yarus, M. 1992. Proofreading, NTPases and translation: successful increase in specificity. Trends Biochem. Sci. 17:171-174. [DOI] [PubMed] [Google Scholar]
- 86.Zecherle, G., S. Whelen, and B. Hall. 1996. Purines are required at the 5′ ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Mol. Cell. Biol. 16:5801-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang, J., X. Sun, Y. Qian, J. P. LaDuca, and L. E. Maquat. 1998. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 18:5272-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, J., X. Sun, Y. Qian, and L. E. Maquat. 1998. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4:801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, S., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15:2231-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang, S., E. Welch, K. Hogan, A. Brown, S. Peltz, and A. Jacobson. 1997. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA 3:234-244. [PMC free article] [PubMed] [Google Scholar]
- 91.Zieler, H. A., M. Walberg, and P. Berg. 1995. Suppression of mutations in two Saccharomyces cerevisiae genes by the adenovirus E1A protein. Mol. Cell. Biol. 15:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]