Abstract
Protein kinase CK2 is a ubiquitous protein kinase implicated in proliferation and cell survival. Its regulatory β subunit, CK2β, which is encoded by a single gene in mammals, has been suspected of regulating other protein kinases. In this work, we show that knockout of the CK2β gene in mice leads to postimplantation lethality. Mutant embryos were reduced in size at embryonic day 6.5 (E6.5). They did not exhibit signs of apoptosis but did show reduced cell proliferation. Mutant embryos were resorbed at E7.5. In vitro, CK2β−/− morula development stopped after the blastocyst stage. Attempts to generate homozygous embryonic stem (ES) cells failed. By using a conditional knockout approach, we show that lack of CK2β is deleterious for mouse ES cells and primary embryonic fibroblasts. This is in contrast to what occurs with yeast cells, which can survive without functional CK2β. Thus, our study demonstrates that in mammals, CK2β is essential for viability at the cellular level, possibly because it acquired new functions during evolution.
Protein kinase CK2 is a pleiotropic and highly conserved protein kinase with more than 300 substrates described to date. It seems to be involved in controlling a large panel of normal cellular functions such as gene expression, protein synthesis, cell cycle, and proliferation, as well as pathological processes such as carcinogenesis and viral tumorigenesis (12, 33). Recently, its function in protecting cells against apoptosis has been reported (1).
CK2 is a tetrameric holoenzyme generally composed of two catalytic subunits, α and α′, and two regulatory β subunits which combine to form an αα′β2, α2β2, or α′2β2 heterotetramer. The catalytic CK2 subunits α and α′ belong to the eukaryotic protein kinase superfamily. In contrast, the regulatory β subunit is a unique protein encoded by a single gene in mammals (3) and does not belong to a known protein family.
CK2β has several functions in the holoenzyme complex. Reconstitution experiments with recombinant purified subunits have demonstrated that CK2β modulates the activity of CK2. Depending on the substrate, CK2β activates or down-regulates the activity of the catalytic subunit (24). CK2β also confers stability to the holoenzyme complex (18) and seems to mediate interaction with a number of substrates (19).
The crystal structure elucidations of the isolated CK2β subunit (5) and of the holoenzyme complex (28) indicate that the β subunit exists as a dimer and is the building block of the CK2 holoenzyme bridging the two catalytic subunits. The crystal structure is also consistent with the suggested flexible role of the β subunit as a docking partner for other protein kinases and other interacting partners in the cell (28).
Functional and biochemical studies have indicated that fractions of both the catalytic and regulatory subunits may exist separately. A population of CK2α that binds to protein phosphatase 2A is free of CK2β (16). Moreover, CK2β fractions devoid of the catalytic subunit, but probably involved in complexes with other proteins, have been described in extracts of mouse brain and testis (11). Isolated CK2β has been shown to interact with and modulate the activities of other serine/threonine kinases such as A-Raf and c-Mos (4, 6). CK2β is a positive regulator of A-Raf in vitro (14), whereas CK2β-c-Mos interaction negatively regulates the catalytic activity of c-Mos (6). Taken together, these observations suggest a regulatory function for CK2β in signaling networks involving several protein kinases.
The in vivo role of CK2 in yeast has been studied by using genetic approaches. Knockout of the gene encoding one of the two catalytic CK2 α subunits in Saccharomyces cerevisiae revealed a functional redundancy of the two subunits. Knocking out both catalytic subunits is lethal (29). This was not the case for the regulatory CK2β subunit. In S. cerevisiae, which possesses two different CK2β subunits (CKB1 and CKB2), the deletion of either one or both regulatory subunits has no consequence on viability and growth but results in salt sensitivity to Na+ and Li+ (2). Moreover, deletion of CKB2 causes a partial block in the adaptation to the G2/M checkpoint arrest induced by DNA damage, suggesting a role for CK2β in the maintenance of cell viability during arrest (36). In Schizosaccharomyces pombe, the deletion of the CK2β gene results in a more severe phenotype, i.e., slow growth, sensitivity to cold, and abnormalities in cell shape (34).
In mice, only CK2α′ has been knocked out thus far. This has been shown to result in globozoospermia, due to a specific role for CK2α′ during late spermatogenesis. In other tissues, CK2α could obviously compensate for the loss of α′, supporting the functional redundancy of CK2α and α′ already observed in yeast (38).
To gain insight into the functional and developmental roles of the regulatory β subunit of CK2 in mammals, we generated embryonic stem (ES) cells and mice with either a conditional CK2β allele or a CK2β null allele by applying gene targeting and the Cre/loxP system. Here we report that in contrast to what occurs in yeast, the protein kinase CK2β protein is essential for cell viability in mice and is therefore imperatively required during early embryonic development.
MATERIALS AND METHODS
Gene targeting, ES cell manipulation, and mouse breeding.
A CK2β genomic clone was isolated from a λ phage 129/Sv mouse genomic library (Stratagene). The overall structure of the mouse CK2β gene locus was confirmed as described previously (3). For the gene-targeting vector, 9.3 kb of the CK2β gene locus were used. The vector contained a 1.95-kb 5′ homologous region, followed by a loxP site, a new HindIII recognition sequence, 2.35-kb genomic sequences containing the first two exons, a phosphoglycerate kinase-neo cassette flanked by two loxP sites within the second intron, and a 5-kb 3′ homologous region. The targeting vector was linearized by NotI digestion and capped with a hairpin oligonucleotide (37). Newly established ES cells, AT1, derived from 129/SvPas@Ico mice (Charles River Laboratories), were transfected by electroporation with the CK2β targeting vector. Clones were selected with G418 (250 μg/ml) and ganciclovir (2 μM). Clones with a correct recombination event were used to obtain germ line-transmitting chimeras after aggregation of ES cells with OF1 (Ico:OF1/Caw) morula stage embryos. Chimeras were crossed with C57Bl/6J@Ico wild-type mice. Heterozygous offsprings were crossed with EIIa-Cre transgenic mice (22) on a C57Bl/B6 background and further backcrossed to C57Bl/B6 mice. In parallel, selected ES cell clones were transfected with the Cre recombinase expression vector pIC-Cre (10) and tested for sensitivity to G418 (310 μg/ml). Chimera were generated independently from G418-sensitive ES cells.
Southern blot and PCR analysis.
Genomic DNA was extracted from gene-targeted ES cells, mouse tails, or embryonic tissue. Southern blot hybridization was done with HindIII-, EcoRI-, or XhoI-digested genomic DNA by using a 5′ external, 3′ external, or internal probe, respectively (Fig. 1B and C). The Gene Images random prime labeling and CDP-Star detection system (Amersham Pharmacia) was used. For PCR analysis, the following primers were used: primer 1, 5′-GAGGGCATAGTAGATATGAATCTG-3′; primer 2, 5′-ATTTCTGAGATCGAGGCCAGTCTG-3′; primer 3, 5′-ATGAGTAGCTCTGAGGAGGTG-3′; primer 4, 5′-GGATAGCAAACTCTCTGAG-3′; Cre forward, 5′-ATGTCCAATTTACTGACCGTACAC-3′; and Cre reverse, 5′-CGCATAACCAGTGAAACAGCATTG-3′. The absence of the CK2β wild-type alleles in early embryonic stages was demonstrated by a PCR using primers 4, 5 (5′-TGGCCTTGAACTCCTGGCAG-3′), and 6 (5′-TACCTCTGGGTGACCACTAGG-3′). The binding sites for primers 5 and 6 are located 246 and 162 bp, respectively, upstream of primer 4 in the CK2β wild-type allele sequence. The binding site for primer 5 was deleted in the CK2β− allele.
FIG. 1.
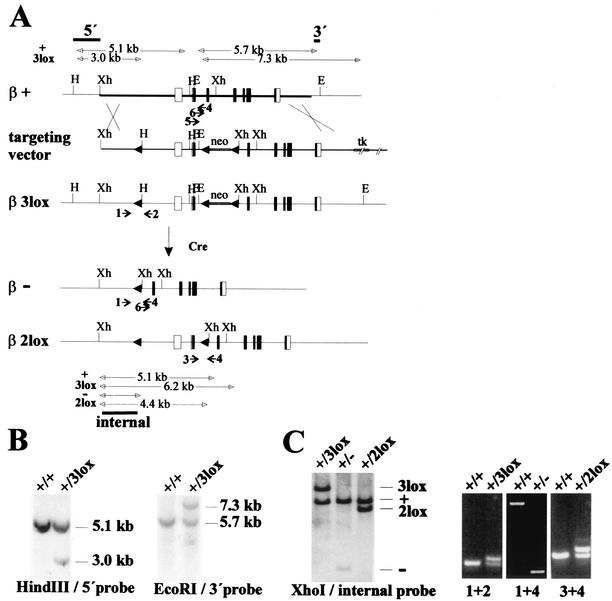
Classical and conditional knockout of CK2β. (A) Gene-targeting strategy and deletion events after Cre expression. Maps of the CK2β wild-type allele, CK2β+, the targeting vector, the CK2β3lox allele after gene targeting, and the resulting CK2β− and CK2β2lox alleles after Cre expression. Rectangles represent the seven exons of the CK2β gene. The area of the CK2β+ genomic region used for gene targeting is indicated by the thick horizontal line. loxP sites are indicated by black arrowheads. The positions of the 5′ and 3′ external probes and the internal probe used for Southern blot hybridization and the fragments detected for the different alleles are indicated above the CK2β+ allele and below the CKβ2lox allele by black bars. The positions of primers (numbered 1 to 6) for PCR analysis are indicated by arrows below the CK2β+, CK2β3lox, CK2β−, and CK2β2lox alleles. Restriction enzyme cutting sites relevant for the detection of the different alleles are marked as follows: H, HindIII; E, EcoRI; and Xh, XhoI. (B) Identification of CK2β3lox/+ ES cell clones. The correct targeting event was confirmed by the appearances of a 3.0-kb HindIII fragment after hybridization with the 5′ probe and of a 7.3-kb EcoRI fragment after hybridization with the 3′ probe, whereas the wild-type allele gave rise to 5.1- and 5.7-kb bands, respectively. The genotypes are indicated above the lanes. (C) Strategy to discriminate CK2β alleles by Southern blot hybridization (left) and PCR analysis (right). For Southern blot hybridization, genomic DNA was digested with XhoI and hybridized with the internal probe, which revealed fragments of specific sizes for the four different alleles. For PCR analysis, two of the four primers (designated 1 to 4) were used in three different combinations, as indicated below the lanes. Fragments of different sizes, depending on the alleles present, were obtained. The genotypes are indicated above the lanes. In the case of the PCR with primers 1 and 4 and the CK2β+/− genotype, competition between the 2.6- and 0.3-kb products from the CK2β+ and CK2β− alleles, respectively, led to amplification of only the small fragment specific for the CK2β− allele. Figure 4 shows the results of PCR with primers 5 and 6.
Western blot analysis.
Cells were lysed in 25 mM Tris-HCl (pH 8.5), 1 mM dithiothreitol, and 100 mM NaCl by sonication, and lysates were cleared by centrifugation. Forty micrograms of lysates was separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. CK2 subunits were detected by using monoclonal anti-CK2α and anti-CK2β antibodies (Calbiochem) and a polyclonal antibody against the C terminus of CK2β. Protein-antibody complexes were visualized by a chemiluminescence Western blotting detection system (Applied Biosystems). As the control, an anti-actin antibody (monoclonal anti-actin β antibody; clone AC-15; Sigma) was used.
Manipulation of embryos.
Embryos were generated by natural matings. Eight-cell stage embryos were cultured in gelatinized dishes in ES cell medium. Primary mouse embryonic fibroblasts were isolated from embryonic day 14.5 (E14.5) embryos as described by Hogan et al. (17).
Histological analysis.
Bromodeoxyuridine (BrdU) labeling of cells from embryos in the S phase of the cell cycle was performed according to the protocol described by Hakem et al. (15). One hour after intraperitoneal injection of BrdU (100 μg/gram of body weight), pregnant females at E6.5 and E7.5 were sacrificed. Deciduae were embedded in paraffin, and sections were processed for immunohistochemistry. Endogenous peroxidases were quenched by incubating the sections for 20 min in methanol-1% H2O2. The sections were further incubated with rat monoclonal anti-BrdU (ImmunologicalsDirect.com) at a 1:75 dilution. A biotinylated secondary rabbit antibody against rat immunoglobulins (1:300) and ABC peroxidase complex (Dako) were used for immunostaining. The signal was visualized in 5 to 10 min by using a horseradish peroxidase reaction with diaminobenzidine (Dako). Sections were stained (10 s) with hematoxylin. For detection of apoptotic cells, dewaxed and rehydrated sections of deciduae at E6.5 and E7.5 were incubated for 5 min with a Hoechst dye solution (2 μg/ml), washed twice in phosphate-buffered saline, and viewed on a Zeiss fluorescence microscope.
Retroviral infections.
The cre coding region was subcloned into retroviral vectors pMSCVpuro (Clontech) and pBabepuro (26). Cre retroviral supernatants were generated by transfection of BOSC23 cells (30). A total of 5 × 104 ES cells and 105 primary embryonic fibroblasts/well were plated in 24-well and 6-well dishes, respectively, and infected with retroviral Cre-pMSCV-puro and Cre-pBabe-puro supernatants, respectively. Puromycin selection (1.5 μg/ml) was started 2 days postinfection. At 4, 6, and 9 days postinfection, cells were fixed and stained with 0.3% methylene blue-0.1% basic fuchsin in methanol and harvested for DNA isolation.
RESULTS
Generation of CK2β knockout ES cells and mice.
To generate classical and conditional CK2β knockout ES cells and mice, we used a gene-targeting vector that contained a loxP site upstream of the promoter region, a neo cassette for positive selection flanked by two other loxP sites in the second intron of the gene, and a thymidine kinase cassette for negative selection (Fig. 1A). In this way, loxP sites flanked the first two exons, including the translation start site in exon 2. Sixteen of the 376 ES cell clones screened had the correct homologous recombination event (Fig. 1B). The resulting CK2β allele was designated CK2β3lox. Germ line-transmitting chimeras and heterozygous CK2β3lox/+ offsprings were obtained. To generate mice with classical CK2β− and conditional CK2β2lox knockout alleles, the CK2β3lox/+ mice were bred to heterozygous EIIa-Cre transgenic mice, which express Cre in zygotes (22) (Fig. 1A and C). Among the 74 offsprings, two had an allele where the region between the two loxP sites, comprising the CK2β promoter region, the first two exons, and the neo cassette, was deleted, leaving behind the CK2β null allele, CK2β−. In 1 of the 74 offsprings, a mixture of the CK2β− and the conditional CK2β2lox alleles, where only the neo cassette was deleted, was present (Fig. 1A and C). Both the CK2β− and the CK2β2lox alleles were transmitted through the germ line. Subsequently, heterozygous CK2β+/− and CK2β2lox/+ mouse lines without the Cre transgene were established.
In parallel, Cre was transiently expressed in the CK2β3lox/+ ES cells. Of the 288 colonies screened, 17 neomycin-sensitive colonies were obtained. Thirteen colonies had the CK2β− allele, and four had the CK2β2lox allele, resulting in CK2β+/− and CK2β2lox/+ ES cells. A second, independent, heterozygous mouse line with the CK2β− allele was established by using these CK2β+/− ES cells. The CK2β− allele generated by expressing Cre in either mice or ES cells no longer contained the neo cassette, thereby preventing any possible side effect of the neo gene.
To target the wild-type allele in CK2β+/− ES cells, these cells were subjected to a second round of gene targeting and transient expression of Cre. By this method, CK2β3lox/− and CK2β2lox/− ES cells were successively obtained.
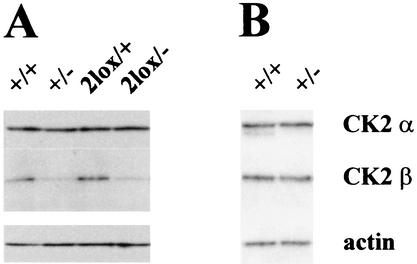
All ES cell lines obtained were checked for CK2β expression by Western blot analysis using two different antibodies (see Materials and Methods) to assess whether manipulations in the CK2β locus interfered with CK2β expression (Fig. 2A). ES cells with either the CK2β3lox (not shown) or CK2β2lox allele expressed levels of CK2β that were identical to those expressed by wild-type ES cells. In contrast, the level of CK2β expression in CK2β+/− and in CK2β2lox/− ES cells was reduced by approximately half while the level of CK2α expression was unchanged. Moreover, no truncated form of the protein could be detected with the two antibodies used in this study. These results show that the introduction of loxP sites into the CK2β gene does not interfere with normal CK2β expression and demonstrate the loss of function by the CK2β− allele. Interestingly, when tissue extracts with the highest level of CK2β expression from adult mice, namely, brain (Fig. 2B) and testis (not shown) tissues, were analyzed, the CK2β expression levels of heterozygous CK2β+/− mice and wild-type mice were the same.
FIG. 2.
Western blot analysis of CK2α and CK2β. ES cell extracts (A) and brain tissue extracts from adult mice (B) with different genotypes as indicated above the lanes were analyzed for CK2α and CK2β expression. For CK2β detection, the same result was obtained with two different antibodies. To ensure equal amounts of loading, an anti-actin antibody was used.
CK2β−/− mice die shortly after implantation.
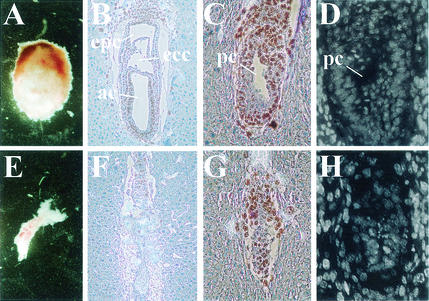
Heterozygous CK2β+/− mice were phenotypically normal and fertile. However, when CK2β+/− mice were intercrossed, no homozygous CK2β−/− mice were detected at birth among the 167 offsprings (Table 1). In addition, a smaller-than-expected number of heterozygous offsprings were obtained. From heterozygous intercrossings with lethality of homozygous knockout mice, a 1:2 ratio of wild-type to heterozygous mice is normally expected. We obtained a 1:1.4 ratio (71 wild-type versus 96 heterozygous mice [Table 1]), which is a significantly lower ratio according to a chi-square test (P < 0.01). When embryos from intercrossed CK2β+/− mice from E8.5 to E11.5 were analyzed, no CK2β−/− embryos were detected (Table 1), suggesting that loss of CK2β function leads to early embryonic lethality. Interestingly, 32% of the deciduae, an abnormally high percentage, were found to be empty, indicating the eventual resorption of CK2β−/− embryos. Consequently, earlier embryonic stages were investigated (Fig. 3). When whole deciduae were dissected, small, obviously degenerated embryos were observed at E7.5 (Fig. 3E). Since their genotypes could not be determined due to contamination with maternal material, they were histologically analyzed and scored as being either of wild-type or abnormal histology. Twelve of 17 embryos had the normal primitive streak stage morphology (Fig. 3B). Abnormal E7.5 embryos accounted for 29% of the analyzed embryos (5 of 17 [Fig. 3F]). Proliferating cells were visualized by BrdU incorporation. In the embryos with normal morphology, BrdU-positive cells were seen throughout the embryonic and extraembryonic tissues, whereas in the abnormal embryos, only a few BrdU-positive cells were seen. These abnormal embryos were presumably homozygous CK2β−/− embryos, and examination of E6.5 embryos confirmed this hypothesis. At E6.5, 31% of the embryos (5 of 16) were much smaller than the rest and had obviously stopped development at an earlier stage (Fig. 3G). BrdU incorporation in sections of these embryos was decreased compared with that in the embryos with normal morphology. BrdU-positive cells were mostly seen inside the extraplacental cone, which contains a mixture of maternal and embryonic cells. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays showed minimal apoptosis in these retarded embryos at E6.5, comparable to that in E6.5 wild-type embryos (data not shown). By using Hoechst staining, we confirmed that most cells of either normal or retarded E6.5 embryos displayed uniform nuclear staining and did not show the condensed and fragmented nuclei typical of apoptotic cells (Fig. 3D and H). Also, at E7.5, no considerable apoptosis could be detected (data not shown). These results indicate that homozygosity for the CK2β null allele results in embryonic lethality and resorption around E7.5 preceded by growth retardation, suggesting that embryos lacking CK2β cease to develop shortly after implantation.
TABLE 1.
Genotypes of live-born mice and embryos from CK2β+/− intercrossingsa
| Offspring or embryo | No. of offspring or embryo with genotype:
|
||
|---|---|---|---|
| CK2β+/+ | CK2β+/− | CK2β−/− | |
| Live-born mice | 71 | 96 | 0 |
| E11.5 | 4 | 8 | 0 |
| E10.5 | 17 | 41 | 0 |
| E9.5 | 3 | 13 | 0 |
| E8.5 | 6 | 10 | 0 |
| E3.5 (blastocysts) | 24 | 62 | 17 |
| E.2.5 (morula) | 10 | 14 | 12 |
| No hatching | 0 | 1 | 2 |
| ICM present at day 7 | 9 | 13 | 1 |
| ICM absent at day 7 | 1 | 0 | 9 |
Live-born mice and E11.5 and E10.5 embryos were genotyped by Southern blot hybridization, and other embryos were genotyped by PCR. Morula were genotyped by PCR after 7 days of culture. The absence of the CK2β wild-type allele was demonstrated by PCR using three primers (see Materials and Methods and Fig. 4I). Progeny and embryos were obtained from two independent founder lines. The genetic background of the mice is 129/SvPas-C57BL/6J (first to fifth backcross generation). The presented data were consistent among the two independently generated mouse lines and all backcross generations.
FIG. 3.
Knockout of CK2β results in postimplantation lethality. (A to D) Embryos with normal morphology. (E to H) Embryos with abnormal morphology. Shown are dissected E7.5 embryos (A and E), BrdU labeling and hematoxylin staining of saggital sections of E7.5 (B and F) and E6.5 (C and G) embryos, and Hoechst staining of E6.5 embryos (D and H). In normal embryos at E7.5 (B), the amniotic (ac), exocoelomic (ecc), and extraplacental (epc) cavities can be seen; at E6.5 (C), the preamniotic canal (pc) is visible (20). In abnormal embryos at E6.5 (G), development is stopped: the embryos are smaller, and no preamniotic canal, only an ICM with a small, probably blastocoelic, cavity, can be seen.
CK2β−/− blastocysts do not develop an inner cell mass (ICM) in vitro.
When blastocysts (E3.5) from intercrossed heterozygous CK2β+/− mice were analyzed, we found at this early embryonic stage that 16.5% of them were nullizygous embryos (Table 1). These were not morphologically different from the embryos with a CK2β+/− or CK2β+/+ genotype. Again, an apoptosis test (TUNEL staining) did not reveal any significant difference between CK2β−/− and CK2β+/+ blastocysts (data not shown).
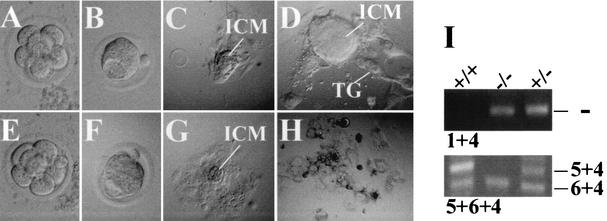
To further define the time point when CK2β−/− embryos stop developing, we collected eight-cell stage embryos (morulae) from heterozygous intercrossings at E2.5 and cultured them for 7 days (Table 1 and Fig. 4). All collected embryos developed normally without morphological differences until the blastocyst stage (Fig. 4B and F). This is in agreement with the in vivo data. The majority of the blastocysts hatched from the zona pellucida on the fourth day in culture (Fig. 4C), attached to the plastic surface of the culture dish, and continued to proliferate. At day 7 in culture, a large ICM which lay above the trophectoderm-derived giant trophoblast cells had formed (Fig. 4D). The majority of these proliferating embryos were of the wild-type CK2β+/+ or heterozygous CK2β+/− genotype as determined by PCR (Fig. 4I). In contrast, another group of embryos stopped proliferating, with no ICM present and trophoblast cells having degenerated by day 7 (Fig. 4H). Eight of the 9 nonproliferative embryos were found to be homozygous CK2β−/−. These results demonstrate that CK2β−/− blastocyst development is impaired in vitro and support the observation that in vivo development of putative CK2β−/− embryos stops after blastocyst implantation.
FIG. 4.
Failure of CK2β−/− morula outgrowth in vitro. (A to D) Typical outgrowth of CK2β+/+ and CK2β+/− morulae. (E to H) Typical outgrowth of CK2β−/− morulae. Panels A and E show cells at the morula stage, and panels B and F show cells at the blastocyst stage. No difference between individual embryos was seen at the morula and blastocyst stages. Outgrowth after 4 (C and G) and 7 (D and H) days in culture are shown. In contrast to CK2β+/+ and CK2β+/− embryos (D), CK2β−/− embryos (H) did not develop an ICM, and they display vacuolated trophoblastic cells (TG). (I) Genotype determination of cells at the end of the blastocyst outgrowth experiment by PCR. Primers 1 and 4 detected the CK2β− allele. Primers 5 and 4 amplified a 246-bp fragment from the CK2β wild-type allele only, not from the CK2β knockout allele, since the primer 5 binding site was lost after Cre deletion. Primers 6 and 4 amplified a 162-bp fragment from both the CK2β wild-type and knockout alleles (see Fig. 1A for locations of the primers), i.e., the absence of the 246-kb fragment indicates the absence of the CK2β wild-type allele.
Knockout of CK2β results in a cell-autonomous defect.
Furthermore, we obtained evidence that loss of CK2β is deleterious at the cellular level. Two attempts to generate ES cells homozygous for a CK2β null allele failed. In the first attempt, we tried to obtain CK2β−neoR/−neoR ES cell lines by culturing CK2β+/−neoR ES cells in a high concentration of G418 (1.4 mg/ml) (27). The necessary CK2β+/−neoR ES cells were generated in parallel to CK2β+/− and CK2β2lox/+ ES cells after transient Cre expression (Fig. 1A) from the third expected Cre recombination event (data not shown). However, only heterozygous CK2β+/−neoR clones resistant to the high G418 concentration were obtained.
The second attempt to generate homozygous CK2β knockout ES cells, i.e., by gene targeting of the wild-type allele of CK2β+/− ES cells using the same targeting vector that was before followed by transient expression of Cre, also failed. This second targeting of the wild-type gene led to the obtention of CK2β3lox/− ES cells at a frequency similar to that in the first round of gene targeting. Subsequent transient Cre expression gave rise only to neomycin-sensitive ES cell clones with a CK2β2lox/− genotype. The other two deletion events, which should result in ES cells nullizygous for a functional CK2β allele either with or without the neo cassette, were not detected. The frequency of detection for CK2β2lox/− ES cell clones (2 of 288) was similar to that for CK2β2lox/+ ES cell clones obtained in the first round of gene targeting and transient Cre expression (4 of 288). This indicates that it might not be possible to obtain homozygous CK2β knockout ES cells, because they do not survive.
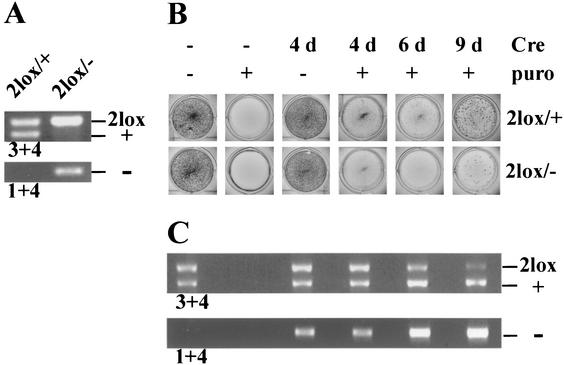
To support this hypothesis, we infected CK2β2lox/− ES cells and, as the control, CK2β2lox/+ ES cells with a Cre-expressing retrovirus (Fig. 5). The retroviral vector pMSCVpuro that we used allowed for selection of infected cells with puromycin. In the infected CK2β2lox/+ cells, puromycin-resistant colonies formed and grew and the Cre recombinase was active and converted the 2lox allele into the null allele (Fig. 5C). In contrast, infected CK2β2lox/− cells continuously disappeared during the selection process (Fig. 5B). The few puromycin-resistant cells available for analysis still had the CK2β2lox/− genotype, i.e., in a low percentage of cells, Cre was obviously not expressed or not active. Similar results were obtained when primary embryonic fibroblasts derived from CK2β2lox/+ and β2lox/− mice were infected with the retroviral vector pBabepuro (data not shown). These results strongly suggest that in CK2β2lox/− cells, Cre converts the 2lox allele into the null allele, leading to a CK2β−/− genotype and a strong cellular defect that results in immediate cell death.
FIG. 5.
Cre expression in CK2β2lox/+ and CK2β2lox/− ES cells. (A) Genotype determination of CK2β2lox/+ and CK2β2lox/− ES cells by PCR with primers 3 and 4 and 1 and 4 (Fig. 1A). (B) CK2β2lox/+ and CK2β2lox/− ES cells were infected with Cre-expressing retrovirus cultured for 4, 6, or 9 days as indicated and stained. Puromycin selection (puro) was applied as indicated. (C) Genotype determination, by PCR, of the CK2β2lox/+ ES cells shown in panel B. In the infected CK2β2lox/+ ES cells, the CK2β− allele (−) could be detected 4 days after infection. Nine days after infection, the CK2β2lox allele (2lox) had almost disappeared. Data shown are representative of four independent experiments.
DISCUSSION
To study the in vivo function of protein kinase CK2, we generated ES cells and, subsequently, mice with a knockout allele of the regulatory CK2β subunit. We chose an approach which allowed the generation of a classical (CK2β−) and a conditional (CK2β2lox) knockout allele in parallel by use of the Cre/loxP system.
Live mice hemizygous for the CK2β− allele developed normally and were fertile. However, it should be noted that the number of heterozygous offsprings obtained was lower than expected, meaning that some heterozygous offspring embryos eventually do not survive. Since CK2β+/− ES cells express a considerably lower level of the protein compared with that expressed by their wild-type counterparts, the low number of heterozygous offsprings could indicate that an appropriate amount of CK2β is required for normal embryonic development. Indeed, live mice hemizygous for the null allele had levels of CK2β expression that were not significantly different from those of wild-type mice, suggesting a compensatory mechanism that adjusts the CK2β protein level during development in the majority of, but not all, cases. In contrast, no homozygous mutant offsprings were obtained from heterozygote intercrosses nor were CK2β−/− embryos detected at E8.5 to E11.5 of development. However, CK2β−/− morula and blastocyst stages were found which were morphologically identical to wild-type stages. Morphological and histological analysis of embryos from heterozygote intercrosses at E6.5 and E7.5 revealed that approximately 30% of these embryos were much smaller than the rest and were abnormal, indicating that the CK2β−/− embryos develop normally until the blastocyst stage and are able to implant but die shortly afterwards.
On the one hand, the observed embryonically lethal phenotype was expected, since ES cells (Fig. 2A) and all of the embryonic stages of different species investigated thus far (reviewed in reference 12) express CK2 ubiquitously. In addition, CK2 is a pleiotropic protein kinase whose activity is probably required to phosphorylate many substrates whose functions are critical in cell proliferation. On the other hand, knocking out CK2β in yeast did not lead to a lethal phenotype. However, the situation in yeast is obviously different from that in higher organisms. In mice, as shown here, and in Caenorhabditis elegans, as recently shown by RNA interference experiments (8), functional loss of CK2β is deleterious to embryonic development. This implies that when CK2β is lost any residual CK2α activity is not enough to support normal development. In addition, the substrate specificities of CK2α and the CK2 holoenzyme complex are different (25). This might lead to an unbalanced phosphorylation of CK2α- and holoenzyme-specific substrates in CK2β−/− cells and to changes in the phosphorylation pattern of substrates relevant for cell survival. Furthermore, CK2β might have acquired other tasks in higher organisms that it does not perform in yeast, which might be reflected by the fact that CK2β can interact with many cellular proteins (9, 21). It is worth noting that in the two yeast species that have been analyzed, different phenotypes were observed when CK2β was knocked out. S. pombe, which is more closely related to higher eukaryotic cells than is S. cerevisiae, displayed a more severe phenotype (2, 34).
The defect of CK2β−/− embryos which we observed during mouse development was obviously a general defect of both embryonic and extraembryonic cells. In vitro, the embryonic ICM was absent, and in vivo, development of the embryo was arrested before E6.5, a time point where the highest level of mitotic activity during mouse embryonic development is found (35). Although extraembryonic trophoblast cells formed a layer in vitro, they displayed large vacuoles. This could indicate that implantation in vivo might be impaired, leading to degeneration and resorption of the embryo, which was also observed. This general growth defect of CK2β−/− cells is corroborated by the conditional knockout of CK2β in ES cells and embryonic fibroblasts upon infection with a Cre-expressing retrovirus. Cells with a CK2β−/− genotype obviously did not survive.
The fact that CK2β−/− embryos exist in vivo and develop normally until the blastocyst stage might be explained by a maternal effect owing to the presence of CK2β mRNA and protein in the zygote. CK2β protein is not readily degraded in the holoenzyme complex (23) and is rather stable; in addition, CK2-mediated phosphorylation might be sustained from the egg stage until the blastocyst stage. However, shortly after implantation, when the embryo undergoes its first burst of proliferation, the deleterious defect becomes obvious.
When we analyzed blastocysts, E6.5 embryos, and E7.5 embryos for apoptotic cells, we could not detect any apoptosis. CK2 has recently been shown to be able to prevent apoptosis (7, 13), but this effect was mainly correlated with CK2α overexpression. In contrast, CK2β has been shown to be essential for cell proliferation. By using microinjection of antibodies or antisense treatment directed against the CK2β subunit in fibroblasts, Pepperkok et al. found a proliferation defect with arrest at G0/G1 and G1/S phases (31, 32). Our data imply that during embryogenesis CK2β is mainly involved in proliferation rather than in the prevention of apoptosis.
In summary, we showed that knocking out CK2β results in a cell-autonomous defect in two types of proliferating cells; in vitro-cultured ES cells and primary embryonic fibroblasts which were depleted of CK2β do not survive. In vivo, CK2β−/− embryos can develop until the blastocyst stage but die shortly after implantation. In vitro-cultured embryos with a CK2β−/− genotype degenerate shortly after hatching. Together, these results reveal the importance of CK2β in the maintenance of cell proliferation and/or cell viability.
Because of the cell-autonomous defect in CK2β−/− cells, mice with the conditional CK2β allele might serve as a useful tool, upon breeding with suitable Cre transgenic mice, for cell lineage ablation and for the study of the origin, function, and fate of particular cell lineages. Moreover, the conditional cell death phenotype may be used to study the structure-function relationship of CK2β with exogenous expression of wild-type or mutant proteins in cell rescue experiments.
Acknowledgments
We thank R. Oschwald for the isolation of the CK2β genomic clone from a 129/Sv mouse genomic λ library and its characterization, H. Westphal for providing the EIIa-Cre transgenic mice, I. Marechal, S. Bama, H. Jørgensen, and M. Ritskes-Hoitinga for their efficient help with mouse breeding, and M. Thomas for critically reading the manuscript.
The project was funded by grants from the Danish Research Council (no. 21-01-0098 and 51-00-0239 to B.B.), the Ministère des Affaires Etrangères (to T.B. and B.B.), the Association pour la Recherche sur le Cancer (to M.V. and C.C.), and the INSERM, the CEA, and the Fondation pour la Recherche Médicale (to C.C.).
REFERENCES
- 1.Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. [DOI] [PubMed] [Google Scholar]
- 2.Bidwai, A. P., J. C. Reed, and C. V. C. Glover. 1995. Cloning and disruption of CKB1, the gene encoding the 38-kDa β subunit of Saccharomyces cerevisiae casein kinase II (CKII). J. Biol. Chem. 270:10395-10404. [DOI] [PubMed] [Google Scholar]
- 3.Boldyreff, B., and O. G. Issinger. 1995. Structure of the gene encoding the murine protein kinase CK2 β subunit. Genomics 29:253-256. [DOI] [PubMed] [Google Scholar]
- 4.Boldyreff, B., and O. G. Issinger. 1997. A-Raf kinase is a new interacting partner of protein kinase CK2 β subunit. FEBS Lett. 403:197-199. [DOI] [PubMed] [Google Scholar]
- 5.Chantalat, L., D. Leroy, O. Filhol, A. Nueda, M. J. Benitez, E. M. Chambaz, C. Cochet, and O. Dideberg. 1999. Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J. 18:2930-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, M., D. Li, E. G. Krebs, and J. A. Cooper. 1997. The casein kinase II β subunit binds to Mos and inhibits Mos activity. Mol. Cell. Biol. 17:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desagher, S., A. Osen-Sand, S. Montessuit, E. Magnenat, F. Vilbois, A. Hochmann, L. Journot, B. Antonsson, and J. C. Martinou. 2001. Phosphorylation of Bid by casein kinase I and II regulates cleavage by caspase 8. Mol. Cell 8:601-611. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325-330. [DOI] [PubMed] [Google Scholar]
- 9.Grein, S., K. Raymond, C. Cochet, W. Pyerin, E. M. Chambaz, and O. Filhol. 1999. Searching interaction partners of protein kinase CK2β subunit by two-hybrid screening. Mol. Cell. Biochem. 191:105-109. [PubMed] [Google Scholar]
- 10.Gu, H., J. D. Marth, P. C. Orban, H. Mosmann, and K. Rajewski. 1994. Deletion of a DNA polymerase α gene segment in T cells using cell type-specific gene targeting. Science 265:103-106. [DOI] [PubMed] [Google Scholar]
- 11.Guerra, B., S. Siemer, B. Boldyreff, and O. G. Issinger. 1999. Protein kinase CK2: evidence for a protein kinase CK2β subunit fraction, devoid of the catalytic CK2α subunit, in mouse brain and testicles. FEBS Lett. 462:353-357. [DOI] [PubMed] [Google Scholar]
- 12.Guerra, B., and O. G. Issinger. 1999. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 20:391-408. [DOI] [PubMed] [Google Scholar]
- 13.Guo, C., S. Yu, A. T. Davis, H. Wang, J. E. Green, and K. Ahmed. 2001. A potential role of nuclear matrix-associated protein kinase CK2 in protection against drug-induced apoptosis in cancer cells. J. Biol. Chem. 276:5992-5999. [DOI] [PubMed] [Google Scholar]
- 14.Hagemann, C., A. Kalmes, V. Wixler, L. Wixler, T. Schuster, and U. R. Rapp. 1997. The regulatory subunit of protein kinase CK2 is a specific A-Raf activator. FEBS Lett. 403:200-202. [DOI] [PubMed] [Google Scholar]
- 15.Hakem, R., J. L. De la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmar, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 16.Hériché, J. K., F. Lebrin, T. Rabilloud, D. Leroy, E. M. Chambaz, and Y. Goldberg. 1997. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science 276:952-955. [DOI] [PubMed] [Google Scholar]
- 17.Hogan, B., R. Beddington, F. Costatini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Issinger, O. G., C. Brockel, B. Boldyreff, and J. T. Pelton. 1992. Characterization of the α and β subunits of casein kinase 2 by far-UV CD spectroscopy. Biochemistry 31:6098-6103. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, H. H., M. Hjerrild, B. Guerra, M. Larsen, P. Højrup, and B. Boldyreff. 2001. Phosphorylation of the Fas associated factor FAF1 by protein kinase CK2 and identification of serines 289 and 291 as the in vitro phosphorylation sites. Int. J. Biochem. Cell Biol. 33:577-589. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman, M. H. 1994. The atlas of mouse development. Academic Press, London, England.
- 21.Kusk, M., R. Ahmed, B. Thomsen, C. Bendixen, O. G. Issinger, and B. Boldyreff. 1999. Interactions of protein kinase CK2β subunit within the holoenzyme and with other proteins. Mol. Cell. Biochem. 191:51-58. [PubMed] [Google Scholar]
- 22.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luscher, B., and D. W. Lichfield. 1994. Biosynthesis of casein kinase II in lymphoid cell lines. Eur. J. Biochem. 220:521-526. [DOI] [PubMed] [Google Scholar]
- 24.Meggio, F., B. Boldyreff, O. Marin, F. Marchiori, J. W. Perich, O. G. Issinger, and L. A. Pinna. 1992. The effect of polylysine on casein-kinase-2 activity is influenced by both the structure of the protein/peptide substrates and the subunit composition of the enzyme. Eur. J. Biochem. 205:939-945. [DOI] [PubMed] [Google Scholar]
- 25.Meggio, F., B. Boldyreff, O. G. Issinger, and L. A. Pinna. 1994. Casein kinase 2 down-regulation and activation by polybasic peptides are mediated by acidic residues in the 55-64 region of the β-subunit. A study with calmodulin as phosphorylatable substrate. Biochemistry 33:4336-4342. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen, R. M., D. A. Conner, S. Chao, A. A. T. Geisterfer-Lowrance, and J. G. Seidman. 1992. Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niefind., K., B. Guerra, I. Ermakowa, and O. G. Issinger. 2001. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 20:5320-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmanabha, R., J. L. Chen-Wu, D. E. Hanna, and C. V. Glover. 1990. Isolation, sequencing and disruption of the yeast CKA2 gene, casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:4089-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pepperkok, R., P. Lorenz, R. Jakobi, W. Ansorge, and W. Pyerin. 1991. Cell growth stimulation by EGF: inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase II. Exp. Cell Res. 197:245-253. [DOI] [PubMed] [Google Scholar]
- 32.Pepperkok, R., P. Lorenz, W. Ansorge, and W. Pyerin. 1994. Casein kinase II is required for transition of G0/G1, early G1, and G1/S phases of the cell cycle. J. Biol. Chem. 269:6986-6991. [PubMed] [Google Scholar]
- 33.Pinna, L. A., and F. Meggio. 1997. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog. Cell Cycle Res. 3:77-97. [DOI] [PubMed] [Google Scholar]
- 34.Roussou, I., and G. Draetta. 1994. The Schizosaccharomyces pombe casein kinase II α and β subunits: evolutionary conservation and positive role of the β subunit. Mol. Cell. Biol. 14:576-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snow, M. H. L. 1977. Gastrulation in the mouse: growth and regionalization of the epiblast. J. Embryol. Exp. Morphol. 42:293-303. [Google Scholar]
- 36.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 37.Vittet, D., T. Buchou, A. Schweitzer, E. Dejana, and P. Huber. 1997. Targeted null-mutation in the vascular endothelial-cadherin gene impairs the organization of vascular-like structures in embryonic bodies. Proc. Natl. Acad. Sci. USA 94:6273-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, X., A. P. Toselli, L. D. Russell, and D. C. Seldin. 1999. Globozoospermia in mice lacking the casein kinase II α′ catalytic subunit. Nat. Genet. 23:118-121. [DOI] [PubMed] [Google Scholar]