Abstract
Mouse Hus1 encodes an evolutionarily conserved DNA damage response protein. In this study we examined how targeted deletion of Hus1 affects cell cycle checkpoint responses to genotoxic stress. Unlike hus1− fission yeast (Schizosaccharomyces pombe) cells, which are defective for the G2/M DNA damage checkpoint, Hus1-null mouse cells did not inappropriately enter mitosis following genotoxin treatment. However, Hus1-deficient cells displayed a striking S-phase DNA damage checkpoint defect. Whereas wild-type cells transiently repressed DNA replication in response to benzo(a)pyrene dihydrodiol epoxide (BPDE), a genotoxin that causes bulky DNA adducts, Hus1-null cells maintained relatively high levels of DNA synthesis following treatment with this agent. However, when treated with DNA strand break-inducing agents such as ionizing radiation (IR), Hus1-deficient cells showed intact S-phase checkpoint responses. Conversely, checkpoint-mediated inhibition of DNA synthesis in response to BPDE did not require NBS1, a component of the IR-responsive S-phase checkpoint pathway. Taken together, these results demonstrate that Hus1 is required specifically for one of two separable mammalian checkpoint pathways that respond to distinct forms of genome damage during S phase.
The presence of a DNA lesion in the genome of a eukaryotic cell triggers activation of complex, highly coordinated DNA damage response pathways. Historically, these mechanisms have been referred to as checkpoints, owing to their important role in causing cell cycle arrest following genotoxic stress. The same pathways also fulfill additional functions, including regulation of apoptosis, telomeres, DNA repair, and transcriptional programs (69). Such regulatory mechanisms ensure the integrity of genetic information and thereby maintain genomic stability. In the absence of these surveillance functions, mutations can accumulate and lead to various cellular defects, including tumor development in mammals (26).
Mammalian cells use two partially separable DNA damage signaling pathways to respond to distinct forms of genome damage. Double-stranded DNA breaks (DSB) induce a pathway that centers on the phosphatidyl 3-kinase-related protein kinase Atm, which phosphorylates and activates downstream checkpoint proteins, such as the transcription factor p53 and the protein kinase Chk2 (30). Atm is dispensable for responses to other forms of genotoxic stress, including the bulky DNA lesions caused by UV light or the inhibition of DNA synthesis caused by hydroxyurea (HU). Instead, UV and HU activate a second mammalian checkpoint pathway headed by Atr (ataxia telangiectasia and Rad3 related) (69). In UV- or HU-treated cells, Atr phosphorylates and activates the downstream kinase Chk1 (37, 67). Similarly, in Xenopus laevis extracts, xAtr phosphorylates xChk1 in response to UV, as well as in response to inhibition of DNA replication (23, 27). It must be noted that there is evidence for significant cross talk between the Atm- and Atr-dependent signaling cascades, as both pathways become activated by ionizing radiation (IR) to some extent, and Atm and Atr share some of the same downstream substrates (69).
Additional key mammalian DNA damage response factors are the evolutionarily conserved checkpoint proteins Hus1, Rad1, Rad9, and Rad17. Hus1, Rad1, and Rad9 physically associate, forming the so-called 9-1-1 complex (53, 57). Each of these proteins bears predicted structural similarity with proliferating cell nuclear antigen (see reference 56 and references cited therein), a homotrimeric sliding clamp that is loaded onto DNA by the five-subunit replication factor C (RFC) complex (43). Interestingly, Rad17 shares homology with RFC proteins and associates with several RFC subunits, forming a putative clamp loader (36). Rad17 also interacts with the 9-1-1 complex (48) and is required for the DNA damage-induced association of Rad9 with chromatin (71), consistent with a model in which DNA damage triggers Rad17-dependent loading of the 9-1-1 complex onto DNA. Reduction of Rad17 expression by RNA interference (71) or targeted deletion of Hus1 (59) causes dramatic defects in genotoxin-induced Chk1 phosphorylation by Atr. However, the precise role of the proposed checkpoint sliding clamp in DNA damage signaling remains unclear.
In response to genome damage, the mammalian DNA damage response signaling cascades prevent entry into S phase (the G1/S checkpoint), slow progression through S phase (the intra-S or S-phase checkpoint), and block entry into mitosis (the G2/M checkpoint). The experiments described in this study deal with the latter two mechanisms. Genome damage during S phase prompts a down-regulation of DNA synthesis. This occurs through a checkpoint-dependent process, as first evidenced by the finding that cells mutated for Atm fail to inhibit DNA synthesis in response to IR, a phenomenon known as radio-resistant DNA synthesis (reviewed in references 34 and 49). Several Atm substrates, including Nbs1 (19, 35, 61, 68), Brca1 (62), Chk2 (15), and Smc1 (32, 65) are also required for repression of DNA synthesis after IR and act cooperatively in parallel signaling pathways downstream of Atm (16). Recent studies suggest that a second, Atm-independent checkpoint pathway also functions in S phase, regulating DNA synthesis when cells suffer other types of genome damage besides DSB (22) or following transient blockage of DNA replication (17). Experiments in both yeast and mammalian systems suggest that S-phase checkpoints function by inhibiting replicon initiation at late-firing replication origins in the presence of DNA damage (34, 49, 50, 52).
The G2/M DNA damage checkpoint blocks mitosis in cells with genome damage, thereby preventing the transmission of DNA lesions to daughter cells. Both Atm (30) and Atr (9, 11) are necessary for this cell cycle checkpoint response to IR. Downstream substrates of these kinases are required for the initiation (37, 62, 64) or maintenance (5, 28) of G2-phase arrest after genome damage. In fission yeast (Schizosaccharomyces pombe), Hus1, Rad1, Rad9, Rad17, and the Atr homolog Rad3, among others, are also required for proper functioning of the G2/M DNA damage checkpoint (8). In both fission yeast and mammals, this checkpoint functions at least in part through control of Cdk1, a key regulator of the G2/M transition (44).
Many of the molecular details regarding the mammalian intra-S and G2/M DNA damage checkpoints remain poorly understood. We previously reported the generation of cells lacking mouse Hus1 by gene targeting, and here we utilize these reagents to gain insights into the molecular mechanisms of mammalian cell cycle checkpoint responses. In short-term viability assays, Hus1-deficient fibroblasts demonstrate heightened sensitivity to UV and HU but show only slight sensitivity to IR (58). In this study, we examined whether Hus1-null cells possess a cell cycle checkpoint defect that might underlie this genotoxin hypersensitivity. Though dispensable for the G2/M DNA damage checkpoint, Hus1 was found to be essential for an S-phase cell cycle checkpoint that inhibits DNA synthesis in response to particular genotoxins. Defects in this S-phase checkpoint correlated with genotoxin sensitivity in Hus1-null cells, suggesting that this Hus1-dependent intra-S checkpoint may be an important determinant of genotoxin sensitivity in mammalian cells and thus also a potential therapeutic target.
MATERIALS AND METHODS
Cell culture, genotoxin treatments, and cell survival assays.
Previously described Hus1+/+ p21−/− and Hus1−/− p21−/− mouse embryonic fibroblasts (MEFs) (58), as well as the corresponding complemented cell pools (59), were cultured in Dulbecco's modified Eagle medium (Dulbecco's MEM) supplemented with 10% fetal bovine serum, 1.0 mM l-glutamine, 0.1 mM MEM nonessential amino acids, streptomycin sulfate (100 μg/ml), and penicillin (100 U/ml). Simian virus 40-transformed control (GM00637) and NBS (GM15989) human fibroblasts were obtained from the Coriell Cell Repositories and cultured in Eagle's MEM supplemented with 15% fetal bovine serum, 1.0 mM l-glutamine, 0.1 mM MEM nonessential amino acids, streptomycin sulfate (100 μg/ml), and penicillin (100 U/ml). For UV irradiation, the culture medium was removed, cells were washed once with phosphate-buffered saline (PBS) and exposed to 254-nm-wavelength UV light in an XL-1500 Spectrolinker (Spectronics Corp), and fresh medium was placed onto the cells. For IR treatment, cells were irradiated with a 137Cs source (Mark 1 Irradiator; J. L. Sheperd and Sons) at a dose rate of approximately 2 Gy/min. For benzo(a)pyrene dihydrodiol epoxide (BPDE) (NCI carcinogen repository) or bleomycin (Sigma) treatment, the genotoxin was added directly to the medium, and cells were then incubated for 1 h, after which time the genotoxin-containing medium was removed, cells were washed once with PBS, and fresh medium was added. For short-term viability assays, 105 cells per six-well dish well were cultured for 3 days following dimethyl sulfoxide (DMSO) or BPDE treatment and then harvested by trypsinization, incubated with trypan blue dye, and counted. For clonogenic survival assays, cells were plated on 10-cm-diameter dishes, treated with DMSO or BPDE, and then cultured in normal medium. After 14 days, the cells were fixed, stained with crystal violet, and photographed.
G2/M DNA damage checkpoint assay.
MEFs at a density of 106 cells per 10-cm-diameter dish were mock or genotoxin treated and, after a 30-min incubation, the cells were then incubated in medium containing nocodazole (0.1 μg/ml; Sigma) for 5 h. Cells were harvested by trypsinization, washed in PBS, and fixed in 70% ethanol at −20°C. Cell staining was performed as described by others (60). Briefly, cells were permeabilized in 0.25% Triton X-100, incubated with antiphosphohistone H3 antibody (Upstate Biotechnology) overnight at 4°C, washed, incubated with fluorescein-conjugated goat-anti-rabbit immunoglobulin G (Jackson Immunoresearch), washed, treated with RNase A, and stained with propidium iodide. Flow cytometry was performed on a FACSCalibur flow cytometer (Becton Dickinson), and data were acquired and analyzed using CellQuest software (Becton Dickinson).
Radioresistant DNA synthesis assay.
Cells were plated at a density of 0.33 × 105 per well of a six-well dish and labeled for 24 h in medium containing [methyl-14C]thymidine (10 nCi/ml; 55.4 mCi/mmol; NEN Life Science Products, Inc.) to provide an internal control for total DNA content. The cells were washed once with PBS and then incubated for 24 h in nonradioactive medium. After genotoxin treatment, cells were incubated for 30 min and then labeled for 1 h in medium containing [methyl-3H]thymidine (2.5 μCi/ml; 2.0 Ci/mmol; NEN Life Science Products, Inc). The radioactive medium was then removed, and unincorporated nucleotides were removed by washing the cells three times in ice-cold 5.0% trichloroacetic acid. The cells were then solubilized in 0.3 N NaOH, followed by neutralization with glacial acetic acid. Radioactivity was quantitated with a liquid scintillation counter, and the 3H/14C ratio was calculated after correction for channel crossover between emissions from the two isotopes.
Antibodies and immunoblotting.
Rabbit anti-Chk1 (FL-476) was purchased from Santa Cruz Biotechnology and rabbit antiphosphoserine 345 mouse Chk1 (catalogue no. 2341) was purchased from Cell Signaling Technology. Total cell lysates were prepared in Triton lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5% Triton X-100, 10 mM NaF, 1 mM Na3VO4, 1× protease inhibitor cocktail [Boehringer Mannheim]). Soluble protein was recovered after centrifugation at 10,000 × g for 10 min, and protein concentrations were determined by Bradford assay (Bio-Rad). Aliquots (50 to 100 μg) of total cell lysate were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (NEN). Immunoblotting was performed by standard methods and signal detection was by chemiluminescence (Pierce).
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay.
Fluorescein-labeling of DNA strand breaks and fluorescence-activated cell sorting (FACS) analysis were performed using a cell death detection kit (Roche) according to the manufacturer's instructions. After drug treatments (performed as described above), the growth medium containing nonadherent cells was removed from the culture dishes and retained. Remaining adherent cells were detached by trypsinization and combined with the nonadherent population. The pooled cells were fixed for 1 h in 4% paraformaldehyde in PBS (pH 7.4) and then permeabilized in ethanol overnight at 4°C. After two washes in PBS, the cells were resuspended in labeling mix containing fluorescein dUTP and terminal deoxynucleotidyltransferase for 1.5 h. The labeled cells were washed once in PBS and then resuspended in PBS prior to FACS analysis. Appropriate negative controls (unlabeled cells) were included to test for autofluorescence on the FL1-H channel.
RESULTS
Mouse Hus1 is dispensable for the G2/M DNA damage checkpoint.
Fission yeast hus1+ is required for cell cycle arrest prior to mitosis in cells treated with DNA replication inhibitors or DNA damaging agents such as UV or radiomimetic drugs (14). To address whether mouse Hus1 is similarly required for proper functioning of the G2/M DNA damage checkpoint, we examined whether Hus1-deficient cells were capable of arresting the cell cycle in G2 phase following genotoxic stress. Although Hus1-deficient MEFs fail to proliferate in culture, likely as a consequence of spontaneous chromosomal abnormalities which occur in the absence of Hus1 (58), deletion of p21 rescues this growth defect, allowing for serial culture of Hus1−/− p21−/− MEFs and examination of the role of Hus1 in the G2/M checkpoint. The integrity of the G2/M checkpoint was assessed by culturing mock- or genotoxin-treated cells in the presence of the microtubule inhibitor nocodazole. Nocodazole disrupts the mitotic spindle and traps cells in mitosis, allowing for identification of cells that have passed through the G2/M checkpoint. The duration of G2 phase is approximately 4 h in the Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs (R. Weiss, unpublished observations). Therefore, in the experiments described below, a 5-h nocodazole treatment was used to trap cells in mitosis, ensuring that we examined all cells that were in G2 phase at the time of genotoxin treatment. Mitotic cells were quantitated by flow cytometry, after staining with antibodies specific for the mitotic marker serine10-phosphorylated histone H3 (60), an approach capable of detecting the defective G2/M checkpoint response to IR in cells lacking Atm (62).
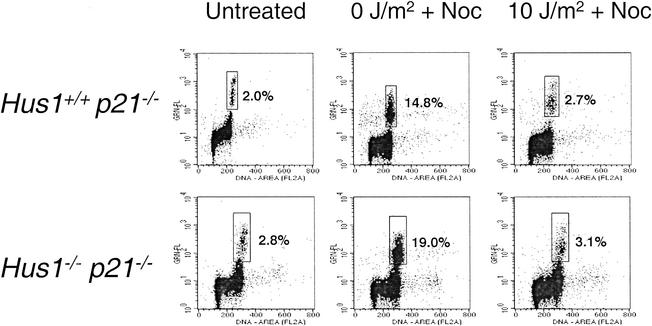
Representative results from an analysis of the G2/M checkpoint response to UV irradiation are shown in Fig. 1. An asynchronous, untreated culture of Hus1+/+ p21−/− MEFs contained a small fraction of serine10-phosphorylated histone H3-positive mitotic cells (2.0% of the total population), and this subpopulation was significantly enriched after treatment with nocodazole, increasing to 14.8% after 5 h. Similar results were obtained with unirradiated Hus1−/− p21−/− MEFs, with the small proportion of mitotic cells in untreated cultures (2.8%) increasing to 19.0% after nocodazole treatment. UV irradiation of Hus1+/+ p21−/− cells triggered cell cycle arrest, as evidenced by the finding that only 2.7% of UV-irradiated Hus1+/+ p21−/− cells were in mitosis after nocodazole treatment, compared to 14.8% for the unirradiated control. Thus, as expected Hus1+/+ p21−/− MEFs possess an intact G2/M DNA damage checkpoint that prevents entry into mitosis after UV irradiation. Importantly, Hus1−/− p21−/− cells also reduced entry into mitosis after UV treatment (3.1% of Hus1−/− p21−/− cells in mitosis after UV irradiation compared to 19.0% for the matched unirradiated control). These results suggest that, unlike fission yeast hus1+, mouse Hus1 is not required for the G2/M checkpoint that responds to UV.
FIG. 1.
Hus1-deficient cells are capable of G2-phase arrest following UV irradiation. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were left completely untreated or were mock (0 J/m2) or UV (10 J/m2) irradiated and then cultured in nocodazole (Noc)-containing medium for 5 h. Cells were harvested, stained with propidium iodide and with an antibody specific for the mitotic marker phosphohistone H3, and analyzed by flow cytometry. Plots show staining intensity for propidium iodide (x axis) versus antiphosphohistone H3 (y axis). The percentage of cells in the boxed region corresponding to the mitotic fraction is indicated.
G2/M cell cycle checkpoint function in Hus1-null and control cells was then tested in response to other genotoxins. These experiments utilized pools of Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs stably transduced with retroviruses encoding either Hus1 or, as a control, GFP (59). Similar to the results described above for parental Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs, all three stable cell pools (Hus1+/+ p21−/− +GFP), Hus1−/− p21−/−+GFP, and Hus1−/− p21−/−+Hus1) demonstrated reduced transit into mitosis following UV irradiation (Table 1). In similar experiments using IR as the DNA damaging agent, all three cell pools again exhibited normal G2/M cell cycle checkpoint responses. Finally, activation of the G2/M DNA damage checkpoint was assessed in cells treated with BPDE, which forms bulky guanine adducts (29). Surprisingly, BPDE was a relatively poor inducer of the G2/M DNA damage checkpoint, and many cells proceeded into mitosis after BPDE exposure. Nevertheless, the lack of G2-phase arrest following BPDE treatment was independent of Hus1 status.
TABLE 1.
G2/M DNA damage checkpoint function in Hus1-deficient MEFsa
| Treatment | % Mitotic cells (% of Noc control)
|
||
|---|---|---|---|
| Hus1+/+ p21−/− + GFP | Hus1−/− p21−/− + GFP | Hus1−/− p21−/− + Hus1 | |
| Untreated | 2.55 | 1.90 | 2.13 |
| Noc | 17.77 (100) | 13.70 (100) | 15.81 (100) |
| UV (10 J/m2) + Noc | 2.57 (14.5) | 1.95 (14.2) | 2.01 (12.7) |
| 10 Gy of IR + Noc | 2.36 (13.3) | 1.64 (12.0) | 0.90 (6.7) |
| 0.75 μM BPDE + Noc | 5.77 (32.5) | 5.34 (39.0) | 8.13 (51.4) |
The indicated MEF lines were left untreated or treated with genotoxins and, after 30 min, placed into medium containing 0.1-μg/ml nocodazole (Noc). After 5 h in nocodazole, cells were harvested, stained for DNA and phosphohistone H3, and analyzed by FACS as described in Materials and Methods. The percentage of mitotic cells is indicated. The values in parentheses are the percentage of mitotic cells for that sample relative to results for the corresponding mock-treated control (Noc).
In the experiments described in Fig. 1 and Table 1, it was possible that a small subpopulation of the cells examined for G2/M checkpoint function was in late S phase at the time of genotoxin treatment. To eliminate this possibility we repeated the G2/M checkpoint analyses using a protocol in which cells were harvested 1 h after genotoxin treatment and without nocodazole. The results of these experiments, which detected only cells that were in G2 phase at the time of genotoxin treatment, are shown in Table 2. Consistent with the previous analyses (Fig. 1 and Table 1), the results in Table 2 indicate that there is no G2/M checkpoint defect in response to UV, IR, or BPDE in Hus1−/− p21−/− cells. Overall, our results demonstrate that mouse Hus1 is not essential for the G2/M DNA damage checkpoint.
TABLE 2.
G2/M DNA damage checkpoint function in Hus1-deficient MEFsa
| Treatment | % Mitotic cells (% of untreated control)
|
||
|---|---|---|---|
| Hus1+/+ p21−/− + GFP | Hus1−/− p21−/− + GFP | Hus1−/− p21−/− + Hus1 | |
| Untreated | 2.77 (100) | 2.63 (100) | 2.45 (100) |
| UV (5 J/m2) | 0.65 (23.5) | 0.40 (15.2) | 0.46 (18.8) |
| 5 Gy of IR | 0.35 (12.6) | 0.30 (11.4) | 0.12 (4.9) |
| 10 Gy of IR | 0.30 (10.8) | 0.16 (6.1) | 0.02 (1.0) |
| 0.25 μM BPDE | 1.89 (68.2) | 1.37 (52.1) | 1.54 (62.9) |
The indicated MEF lines were left untreated or treated with genotoxins. One hour later, cells were harvested, stained for DNA and phosphohistone H3, and analyzed by FACS as described in Materials and Methods. The percentage of mitotic cells is indicated. The values in parentheses are the percentage of mitotic cells for that sample relative to results for the corresponding untreated control.
Hus1 functions in an intra-S DNA damage cell cycle checkpoint.
An intra-S cell cycle checkpoint oversees DNA replication and slows DNA synthesis in cells with genome damage. Hus1 homologs from S. pombe and Saccharomyces cerevisiae participate in S-phase responses to genome damage (14, 38, 41). Therefore, we examined whether mouse Hus1 might regulate DNA synthesis in response to DNA damage in mammalian cells. DNA synthesis was quantitated 1 h following mock- or genotoxin-treatment by measurement of 3H-thymidine incorporation. The DNA damaging agent used in initial experiments was BPDE, which is known to activate a caffeine-sensitive, ATM-independent S-phase DNA damage checkpoint in mammalian cells (22). As expected, Hus1+/+ p21−/− MEFs showed a dose-dependent decrease in [3H]thymidine incorporation after BPDE treatment (Fig. 2A). Importantly, Hus1−/− p21−/− MEFs were defective for this checkpoint response. For example, treatment of Hus1+/+ p21−/− MEFs with 70 nM BPDE caused DNA synthesis to be reduced to 68.0% of that for untreated control cells, whereas Hus1−/− p21−/− MEFs treated with the same dose showed DNA synthesis at 97.0% of untreated control levels. At higher BPDE doses, DNA synthesis was significantly reduced in cells of both genotypes, possibly due to Hus1-independent checkpoint responses or physical stalling of replicative polymerases due to blocking lesions.
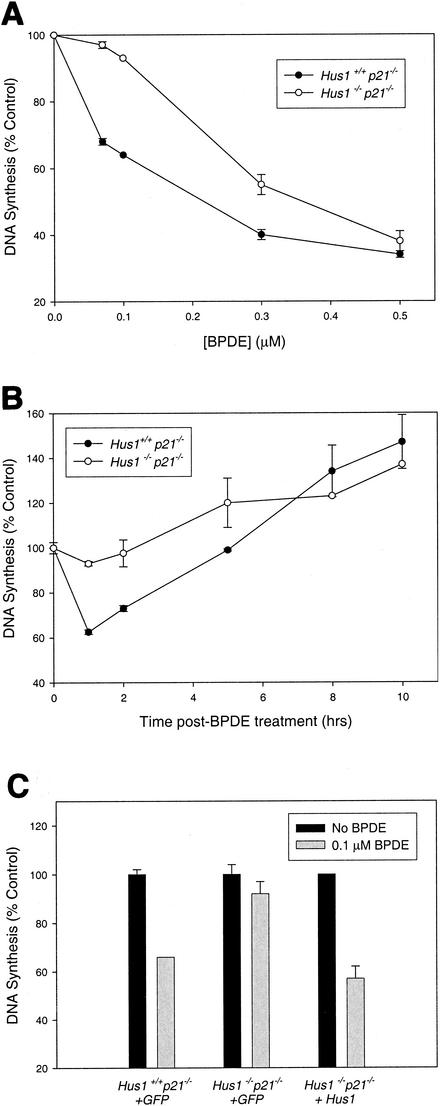
FIG. 2.
Hus1 is required for inhibition of DNA synthesis following BPDE treatment. (A) DNA synthesis in control and Hus1-null cells following BPDE treatment. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were treated with the indicated dose of BPDE and then assayed for DNA synthesis 1 h later by measurement of radiolabeled thymidine incorporation. The percentage of DNA synthesis for treated samplesrelative to that for the corresponding untreated control cells is shown. Data points indicate the mean for two samples, with error bars representing the range. (B) Kinetics of the BPDE-induced S-phase checkpoint. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were treated with 100 nM BPDE for 1 h and then cultured in normal medium. DNA synthesis was measured at the indicated times. (C) Inhibition of DNA synthesis following BPDE treatment of complemented Hus1-null cells. DNA synthesis was measured in the indicated cell pools 1 h following treatment with 100 nM BPDE.
The kinetics of the BPDE-induced S-phase checkpoint were then assessed. When treated with a low dose of BPDE, Hus1+/+ p21−/− MEFs showed a striking, yet transient, inhibition of DNA synthesis (Fig. 2B). At 1 h after treatment with 100 nM BPDE, the level of DNA synthesis was 62.6% of that for untreated control cultures. At subsequent time points, the level of DNA synthesis gradually increased and reached the 100% level at approximately 5 h posttreatment. Hus1−/− p21−/− MEFs failed to execute the initial inhibition of DNA synthesis immediately after BPDE treatment and then showed gradually increasing levels of DNA synthesis at subsequent time points. Thus, Hus1-null cells are defective specifically for the rapid and transient repression of DNA synthesis that occurs after BPDE treatment.
To verify that the S-phase checkpoint defect in Hus1−/− p21−/− MEFs was due to the absence of Hus1, we tested whether reintroduction of Hus1 into Hus1−/− p21−/− MEFs would complement this phenotype (Fig. 2C). As expected, cell pools stably transduced with the GFP-expressing retrovirus behaved similarly to the parental cells with respect to BPDE-induced inhibition of DNA synthesis. By 1 h after treatment with 100 nM BPDE, Hus1+/+ p21−/−+GFP cells had reduced DNA synthesis to 66.0% of that for untreated control cells, while Hus1−/− p21−/−+GFP MEFs maintained high rates of DNA replication (92.0% of untreated control levels). Importantly, restoration of Hus1 expression in Hus1−/− p21−/− MEFs, via a Hus1-expressing retroviral vector, resulted in a level of DNA synthesis after BPDE treatment that was 57.0% of that for untreated control cells. At present we cannot rule out an effect of p21 deficiency on these findings, as p21 has been implicated in the control of S-phase progression (45). Nevertheless, the finding that reconstitution of Hus1 expression in Hus1-null MEFs fully complemented the S-phase checkpoint defect verifies that Hus1 is necessary for a BPDE-induced S-phase checkpoint.
Hus1 is required for BPDE-induced Chk1 phosphorylation.
Previously, we showed that the BPDE-induced S-phase checkpoint can be eliminated by overexpression of dominant-negative Chk1 or by preincubation of cells with a chemical inhibitor of Chk1 (22). Because our results demonstrated a role for Hus1 in BPDE-induced S-phase arrest and since Hus1 was previously implicated in genotoxin-induced Chk1 phosphorylation (59), we suspected that Hus1 might be an upstream regulator of Chk1 in a BPDE-responsive checkpoint signaling cascade. This hypothesis was tested by comparing BPDE-induced Chk1 phosphorylation in Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs. Total cell extracts were immunoblotted with antibodies specific for either activated, serine 345-phosphorylated Chk1 (phospho-Chk1) or total Chk1. Hus1+/+ p21−/− cells treated with 100 nM BPDE showed transient Chk1 phosphorylation which peaked at 1 h after treatment (Fig. 3A), coincident with maximal inhibition of DNA synthesis (see Fig. 2B). In marked contrast, treatment of Hus1−/− p21−/− MEFs with 100 nM BPDE did not elicit Chk1 phosphorylation at 1 h after treatment. Increased levels of phospho-Chk1 did become apparent in Hus1−/− p21−/− MEFs at later time points following 100 nM BPDE treatment but never reached the peak levels observed in Hus1+/+ p21−/− cells. Importantly, BPDE-induced Chk1 phosphorylation was restored to near-normal levels in Hus1−/− p21−/− MEFs that were stably transduced with a Hus1-expressing retrovirus (Fig. 3B). These results indicate that Hus1 is required for optimal BPDE-induced Chk1 phosphorylation.
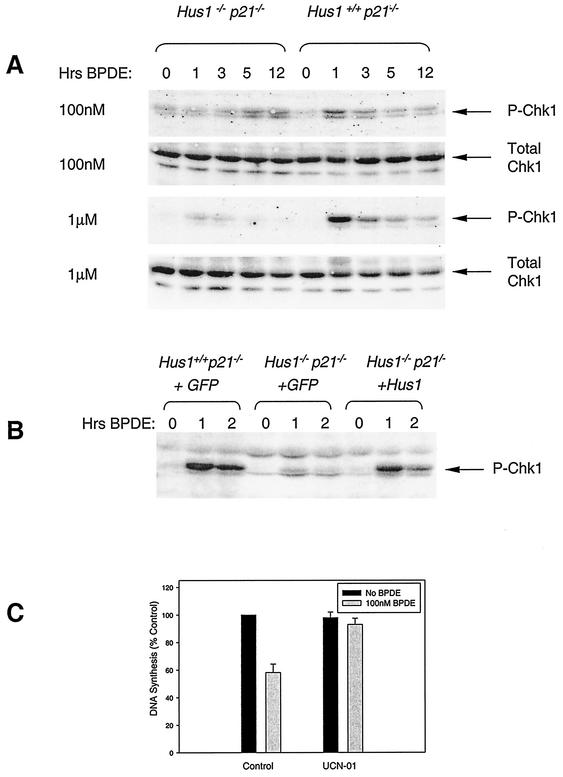
FIG. 3.
BPDE-induced Chk1 phosphorylation is impaired in Hus1-null cells. (A) Evaluation of Chk1 phosphorylation following BPDE treatment of cells. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were treated with 0.1 or 1.0 μM BPDE, or with DMSO as a control, and then cultured in normal medium. At the indicated times, total cell lysates were prepared and immunoblotted with antibodies specific for phosphoserine345-Chk1 or total Chk1. (B) BPDE-induced Chk1 phosphorylation in complemented Hus1-null cells. Hus1+/+ p21−/−+GFP, Hus1−/− p21−/−+GFP, and Hus1−/− p21−/−+Hus1 MEFs were treated with 0.1 μM BPDE or DMSO as a control and then assayed for Chk1 phosphorylation as described above. (C) Blockage of the S-phase checkpoint response to BPDE by the Chk1 inhibitor UCN-01. Hus1+/+ p21−/− MEFs were treated for 1 h with 100 nM UCN-01 or DMSO as a control. UCN-01- or DMSO-treated cells were then given 100 nM BPDE (or no genotoxin) for 1 h prior to measurement of DNA synthesis. Data points indicate the mean for two samples, with error bars representing the range.
The defect in BPDE-induced Chk1 activation in Hus1-null cells was also readily apparent following treatment of cells with a higher BPDE dose (Fig. 3A). Exposure of Hus1+/+ p21−/− MEFs to 1 μM BPDE induced a rapid and transient appearance of phospho-Chk1, and this occurred to a greater extent than in response to 100 nM BPDE, demonstrating that Chk1 phosphorylation in Hus1-expressing cells was dose dependent. Hus1−/− p21−/− MEFs showed only a very slight increase in phospho-Chk1 levels after treatment with 1 μM BPDE. The delayed accumulation of phospho-Chk1 observed in Hus1-null cells treated with a lower BPDE dose was not observed after high-dose BPDE treatment, possibly because of cell death associated with high-dose BPDE treatment of Hus1−/− p21−/− cells at late time points (see below). Although Chk1 phosphorylation in response to high-dose BPDE treatment was impaired in Hus1-deficient cells, both Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs showed repression of DNA synthesis under these conditions (Fig. 2A). These findings are consistent with our previous results, indicating that high doses of BPDE cause S-phase arrest independently of Chk1 (22), possibly due to stalling of replicative polymerases at sites of DNA damage (31). Thus, the Hus1- and Chk1-dependent S-phase checkpoint pathway mediates transient S-phase arrest elicited specifically by low doses of BPDE.
These data suggested a role for Hus1 upstream of Chk1 in an S-phase checkpoint pathway. To test whether direct inhibition of Chk1 would perturb the S-phase checkpoint in mouse fibroblasts, we examined the effects of the Chk1 inhibitor UCN-01 (7, 20) on BPDE-induced inhibition of DNA synthesis (Fig. 3C). Hus1+/+ p21−/− cells were pretreated for 1 h with either 100 nM UCN-01 or DMSO as a control. Control and UCN-01-treated cultures then received 100 nM BPDE for 1 h, after which time relative rates of DNA synthesis were assessed by measurement of [3H]thymidine incorporation. In Hus1+/+ p21−/− cultures that did not receive UCN-01, BPDE elicited an inhibition of DNA synthesis to 58.2% of that for untreated control cells. While UCN-01 had no effect on basal rates of DNA synthesis (data not shown), cells that were pretreated with UCN-01 maintained high levels of DNA synthesis (93.0% of control levels) following BPDE exposure. Results from experiments with UCN-01 must be interpreted with caution, because this compound may affect other kinases besides Chk1. Nevertheless, these findings are consistent with the notion that Chk1 activation is necessary for BPDE-induced down-regulation of DNA synthesis. Overall, these results suggest that Hus1-dependent Chk1 phosphorylation and activation is required for a BPDE-responsive S-phase checkpoint pathway.
Hus1-null cells are hypersensitive to BPDE.
During the course of these experiments, we noticed dramatic effects of high (1 μM) doses of BPDE on the morphology of Hus1-deficient cells. Figure 4A shows photographs of Hus1+/+ p21−/− and Hus1−/− p21−/− cells 24 h after treatment with 1 μM BPDE. BPDE-treated Hus1+/+ p21−/− cells appeared enlarged and flattened, a morphology commonly observed in growth-arrested fibroblasts. Strikingly, Hus1−/− p21−/− MEFs treated with the same BPDE dose were rounded and detached from the culture surface. Because the appearance of BPDE-treated Hus1−/− p21−/− MEFs was characteristic of apoptotic cells, we performed TUNEL staining of Hus1+/+ p21−/− and Hus1−/− p21−/− cells after treatment with 1 μM BPDE or DMSO as a control. As shown in the FACS profiles in Fig. 4B, only 2.2% of Hus1+/+ p21−/− cells were TUNEL positive 24 h after BPDE treatment. In contrast, 33% of the Hus1−/− p21−/− cells were TUNEL positive after the same treatment, indicating that the bulky DNA adducts generated by BPDE induce apoptosis in Hus1-deficient cells.
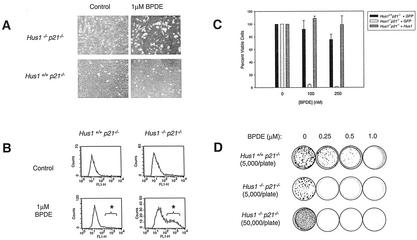
FIG. 4.
Hus1-deficient cells are hypersensitive to BPDE. (A) Photographs of mock- and BPDE-treated MEF cultures. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were mock treated or incubated in 1.0 μM BPDE for 1 h and then photographed after 24 h. (B) Measurement of apoptosis in BPDE-treated cultures. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were treated with 1.0 μM BPDE and, after 24 h, harvested, subjected to TUNEL staining as described in Materials and Methods, and analyzed by FACS. Asterisks indicate the TUNEL-positive cell populations. (C) Short-term viability of BPDE-treated cells. Hus1+/+ p21−/−+GFP, Hus1−/− p21−/−+GFP, and Hus1−/− p21−/−+Hus1 MEFs were treated with the indicated dose of BPDE. Cell viability was measured after 72 h and is plotted as the percent viable cells relative to results for mock-treated control cultures. Data points indicate the mean for three samples, with error bars representing the standard deviation. (D) Clonogenic survival of BPDE-treated cells. Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs were plated on 10-cm-diameter dishes at the indicated density, treated with BPDE, and then cultured in normal medium. After 14 days, the cells were fixed, stained with crystal violet, and photographed.
Consistent with increased programmed cell death in BPDE-treated Hus1−/− p21−/− MEFs, cell viability assays demonstrated dramatic hypersensitivity to BPDE in Hus1-deficient cells. In short-term viability assays (Fig. 4C), 92.0% of Hus1+/+ p21−/−+GFP MEFs were viable 72 h after treatment with 100 nM BPDE, whereas the same treatment reduced viability to 4.4% in Hus1−/− p21−/−+GFP cultures (t test, P = 0.0004). Importantly, complementation of the Hus1−/− p21−/− MEFs with a Hus1-expressing retrovirus restored normal BPDE sensitivity. Hus1-null MEFs also showed impaired clonogenic survival following BPDE exposure (Fig. 4D). Hus1+/+ p21−/− MEFs demonstrated a dose-dependent decrease in survival after BPDE treatment, but numerous colonies persisted after exposure to 250 nM BPDE. In contrast, Hus1−/− p21−/− MEFs displayed dramatically heightened sensitivity to BPDE, with survival completely eliminated by the 250 nM BPDE dose. Although untreated Hus1−/− p21−/− cells showed reduced clonogenic survival relative to Hus1+/+ p21−/− control cells, plating 10-fold more Hus1−/− p21−/− MEFs did not result in the appearance of surviving colonies after BPDE treatment. Collectively, these data show that Hus1-null cells are hypersensitive to BPDE.
The Hus1-dependent S-phase checkpoint responds primarily to BPDE and UV, but not IR.
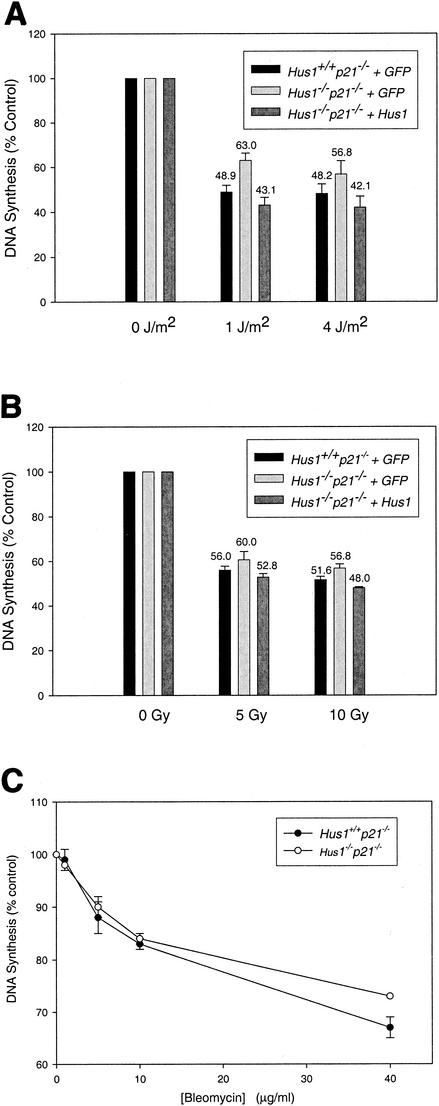
To determine if the Hus1-dependent S-phase checkpoint activated by BPDE also responds to other genotoxins, we tested whether Hus1 was required for inhibition of DNA synthesis following treatment of cells with UV or IR. When treated with UV, Hus1+/+ p21−/−+GFP MEFs reduced DNA synthesis to 48.9% of unirradiated control levels (Fig. 5A). Similar to results with BPDE-treated cells, Hus1−/− p21−/−+GFP MEFs were not fully functional for this checkpoint response and maintained a relatively higher level of DNA synthesis after UV irradiation (63.0% of that for unirradiated controls; t test, P = 0.0052). This checkpoint defect was fully corrected in complemented Hus1−/− p21−/−+Hus1 MEFs, in which UV elicited a reduction in DNA synthesis to 43.1% of that for unirradiated control cells. It is unclear why the S-phase checkpoint defect in UV-irradiated Hus1-null cells is not as dramatic as that observed following BPDE treatment. However, UV is known to exert a broad range of effects on cells, including activation of stress signaling pathways for instance, that could contribute to S-phase arrest independently of Hus1. IR also elicited a significant inhibition of DNA synthesis in Hus1+/+ p21−/−+GFP MEFs, with 5 Gy of IR inducing reduction of DNA synthesis to 56.0% of that for untreated control cells (Fig. 5B). Interestingly, Hus1−/− p21−/−+GFP cells showed only a very slight defect in this S-phase checkpoint response, with DNA synthesis reduced after 5 Gy of IR to 60.0% of that for unirradiated control cells (t test, P = 0.1259). This mild defect, though not statistically significant, was corrected in Hus1−/− p21−/− +Hus1 MEFs, which responded to IR by reducing DNA synthesis to 52.8% of unirradiated control levels. Consistent with these observations, Hus1−/− p21−/−+GFP MEFs also did not display significant defects in the capacity to repress DNA synthesis in response to the radiomimetic drug bleomycin (Fig. 5C). Thus, mouse Hus1 is required primarily for S-phase cell cycle checkpoint responses to DNA lesions caused by genotoxins like BPDE and UV, and to a much lesser extent to DSB. Notably, this parallels the genotoxin sensitivity profile of Hus1-deficient cells, which display hypersensitivity to UV (58) and BPDE (this study) but show only slight sensitivity to IR (58).
FIG. 5.
The Hus1-dependent S-phase checkpoint also responds to UV but not to IR. DNA synthesis was measured in the indicated cell pools after mock treatment or treatment with UV (A), IR (B), or bleomycin (C). (A and B) Data points indicate the mean for three samples, with error bars representing the standard deviation. (C) Data points indicate the mean for two samples, with error bars representing the range.
NBS1 is dispensable for the S-phase checkpoint that responds to BPDE.
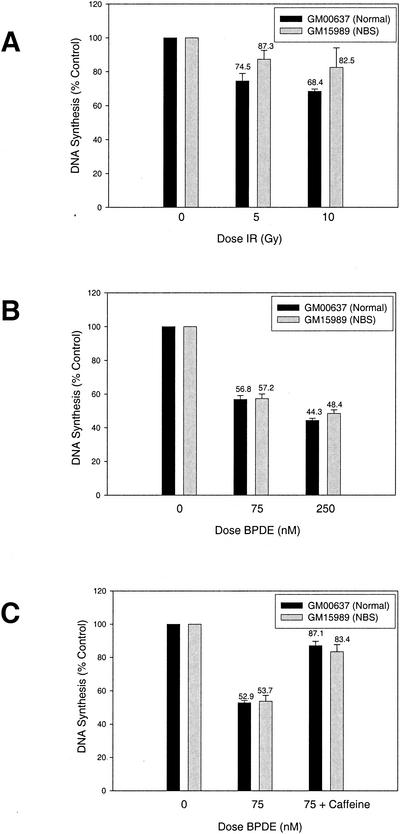
NBS1, the gene mutated in the human cancer predisposition syndrome Nijmegen breakage syndrome, is required for the S-phase checkpoint response to IR (47). NBS1 becomes phosphorylated by the upstream kinase ATM in cells treated with IR but also becomes phosphorylated in UV- or HU-treated cells in an ATM-independent manner (19, 35, 61, 68). Thus, NBS1 could be a common target for multiple S-phase checkpoint pathways that initially act in parallel to respond to distinct genotoxins. Alternatively, NBS1 might be required specifically for the S-phase checkpoint that responds to DSB-inducing agents like IR and radiomimetics, a notion that would be consistent with reports that the budding yeast homolog of NBS1 functions in a complex that is required to slow S phase in response to IR but not UV (12, 21). To address these possibilities, we compared S-phase checkpoint responses in NBS and normal control human fibroblasts after treatment with IR or BPDE. Normal human cells showed a dose-dependent inhibition of DNA synthesis after IR treatment, with 5 Gy IR causing a reduction in DNA synthesis to 74.5% of that for unirradiated control cells (Fig. 6A). Consistent with published results, NBS cells were partially defective for this response and maintained DNA synthesis after irradiation at 87.3% of control levels (t test, P = 0.0312). BPDE treatment also potently elicited inhibition of DNA synthesis in the normal control fibroblasts, with 75 nM BPDE causing a reduction in DNA synthesis to 56.8% of that for untreated control cells (Fig. 6B). Notably, DNA synthesis was inhibited to a nearly identical extent in BPDE-treated NBS cells, to 57.2% of control levels (t test, P = 0.8568). To verify that this repression of DNA synthesis represented an active, checkpoint-dependent process, we tested whether this response could be abolished by pretreatment of the human fibroblast lines with caffeine, an inhibitor of Atm and Atr that inactivates S-phase checkpoint responses (2, 24, 51). Pretreatment of control and NBS cells with caffeine blocked the S-phase checkpoint response to BPDE and in both cultures resulted in high levels of DNA synthesis after BPDE exposure (Fig. 6C). These results demonstrate that the caffeine-sensitive, Hus1-dependent pathway that mediates the intra-S DNA damage checkpoint response to BPDE does not require NBS1 and further establish that two separable pathways mediate checkpoint responses to distinct types of genome damage during S phase.
FIG. 6.
A caffeine-sensitive, NBS1-independent S-phase checkpoint pathway inhibits DNA synthesis in response to BPDE. (A and B) S-phase checkpoint responses to IR and BPDE in NBS cells. DNA synthesis was measured in GM00637 (control) or GM15989 (NBS) simian virus 40-transformed human fibroblasts after IR (A) or BPDE (B) treatment. (C) Effects of caffeine on the S-phase checkpoint response to BPDE in control and NBS cells. DNA synthesis was measured in BPDE-treated GM15989 and GM00637 fibroblasts that had been preincubated in 5 mM caffeine. Data points indicate the mean for three samples, with error bars representing the standard deviation.
DISCUSSION
In response to genotoxic stress, eukaryotic cells arrest cell cycle progression and induce DNA repair. During S phase, a DNA damage checkpoint responds to genome damage by actively repressing DNA replication. In this study, we demonstrate that inhibition of DNA synthesis in response to bulky DNA lesions caused by BPDE requires the murine DNA damage response gene Hus1. S-phase stress response mechanisms act in part to prevent mutagenic misreplication of damaged template DNA (18). In addition, the same pathways function to prevent spontaneous and genotoxin-induced gross chromosomal rearrangements (33). Thus, S-phase DNA damage responses are critical for the maintenance of genomic integrity. Consistent with this important role, mutations in genes required for S-phase checkpoint function, including ATM, NBS1, and BRCA1, result in genetic instability and cancer predisposition in humans.
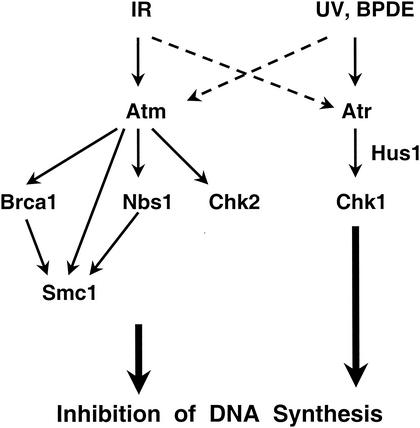
While Hus1 was found to be essential for the inhibition of DNA synthesis in response to BPDE-adducted DNA and also to contribute to the repression of DNA synthesis after UV, Hus1-null cells showed largely normal S-phase checkpoint responses to DSB-inducing agents such as IR and bleomycin. These findings show that two partially separable mammalian checkpoint pathways respond to distinct forms of DNA damage during S phase. Based on our results, we propose an updated scheme to describe the organization of mammalian intra-S checkpoint pathways (Fig. 7). An Atm-dependent pathway mediates S-phase checkpoint responses to DSB, while a Hus1-dependent and Atm-independent mechanism acts in parallel to respond primarily to other forms of genome damage. Multiple Atm substrates, including Brca1, Nbs1, Chk2, and Smc1, are believed to cooperate in mediating the S-phase checkpoint response to IR (16, 32, 65). Nbs1, like other Atm substrates, not only becomes phosphorylated in response to IR but also in UV- and HU-treated cells (19, 35, 61, 68). Although this raises the possibility that Nbs1 is a common target for several S-phase checkpoint pathways, NBS1 was found to be dispensable for the inhibition of DNA synthesis in BPDE-treated cells. These data further establish the fact that mammalian cells utilize at least two genetically distinct checkpoint pathways during S phase, possibly as a means to effectively sense different DNA lesions and subsequently induce appropriate repair mechanisms.
FIG. 7.
Two parallel signaling pathways mediate S-phase checkpoint responses to distinct forms of genome damage. See text for details.
The BPDE-induced S-phase checkpoint can be eliminated by pretreating cells with the Chk1 inhibitor UCN-01 or by overexpression of a dominant-negative form of Chk1 (22). Consistent with the notion that Hus1 and Chk1 function in the same S-phase checkpoint pathway, Hus1-null MEFs showed a dramatic defect in BPDE-induced Chk1 phosphorylation on serine 345, an activating modification mediated by Atr (37, 67). The downstream effectors of this Hus1- and Chk1-dependent pathway remain largely unknown. In response to UV, activated Chk1 phosphorylates and promotes the degradation of Cdc25A, an important positive regulator of Cdk2, and this likely plays a key role in controlling the G1/S transition as well as S-phase progression (15, 40, 42). By analogy to the Atm-dependent S-phase checkpoint pathway, in which multiple downstream factors act in parallel, there likely are additional Atr and/or Chk1 substrates that contribute to the regulation of DNA replication in cells with genome damage.
The molecular mechanism of the Hus1-mediated intra-S checkpoint remains undefined. Potentially, the checkpoint could operate by inhibiting origin firing or by reducing the rate of chain elongation. In both yeasts and higher eukaryotes, checkpoint-mediated regulation of DNA synthesis following DNA damage is achieved primarily through control of DNA replication initiation, usually by inhibition of late firing origins of replication in damaged cells (34, 49, 50, 52, 55). Accordingly, inhibition of DNA replication in cells treated with low doses of BPDE is associated with reduced initiation of DNA synthesis (3, 6, 10, 31). Whether failure of this specific regulatory mechanism is the basis for the S-phase checkpoint defect in Hus1-null cells will require further investigation. However, Chk1 has been implicated in the regulation of late replication origins after transient blockage of DNA synthesis (17) and in a delayed response to IR (70). Therefore, it appears most likely that the Hus1- and Chk1-mediated intra-S DNA damage checkpoint responds to BPDE and UV by inhibiting initiation of DNA synthesis. Following DNA damage, S. cerevisiae mutants with S-phase checkpoint defects additionally demonstrate irreversible replication fork collapse at high frequency (39, 55). An analogous role for mouse Hus1 in the stabilization of stalled replication forks is also possible.
Despite the fact that fission yeast hus1+ plays a prominent role in preventing cells with genome damage from entering mitosis (14), we failed to identify a requirement for mouse Hus1 in a G2/M DNA damage checkpoint. There are several possible explanations for this finding. First, it could be that Hus1 acts specifically during S phase in mammalian cells. Consistent with this possibility, the bulky DNA adducts produced by BPDE triggered a strong Hus1-dependent S-phase checkpoint response but were relatively poor at eliciting G2-phase arrest. Thus, DNA replication may be required for BPDE-induced lesions to be sensed and to trigger checkpoint signaling. A similar mechanism has been suggested for the cellular response to DNA interstrand cross-links, which also fail to elicit G2-phase arrest in normal mammalian cells and induce a checkpoint response only after passage of cells into S phase (1). Conversely, IR induces a strong G2/M checkpoint response. Because DSB would be particularly problematic for a dividing cell, the mammalian G2/M checkpoint machinery might respond primarily to these lesions while allowing other forms of genome damage to be passed on to daughter cells for repair during the next cell cycle. Accordingly, Hus1, which appears to be dispensable for many responses to DSB, might not have a prominent role in the G2/M DNA damage checkpoint. Although UV irradiation generates bulky DNA lesions such as thymidine dimers, it also potently activated a G2/M checkpoint in our experiments. However, UV is known to have multiple biological consequences, including production of strand breaks and induction of membrane signaling. These additional UV-induced events might elicit G2-phase arrest through distinct mechanisms (4, 46).
A role for mouse Hus1 in the G2/M checkpoint might be expected given that Atr and Chk1, which act in the same pathway as Hus1 (59), are required for G2-phase arrest in response to diverse genotoxins (9, 11, 37, 54). Indeed, it is somewhat surprising that Hus1-null cells show defects in DNA damage-induced Chk1 phosphorylation, which is believed to be required for the initiation of the G2/M DNA damage checkpoint (37, 54, 67), yet are capable of G2-phase arrest in response to genotoxic stress. However, Chk1 serine 345 phosphorylation still occurs in Hus1-null cells, albeit to a greatly reduced extent. This limited Chk1 phosphorylation after DNA damage might be sufficient to activate the G2/M checkpoint but not the S-phase checkpoint. Alternatively, redundant mechanisms might mask a role for mouse Hus1 in the G2/M checkpoint. Potentially, the recently identified Hus1 paralog, Hus1B (25), could mediate the G2/M DNA damage checkpoint in the absence of Hus1. However, unlike Hus1, Hus1B does not directly interact with Rad9, suggesting that it is not functionally equivalent to Hus1 (25). Moreover, Hus1B expression is undetectable by Northern blotting in Hus1+/+ p21−/− and Hus1−/− p21−/− MEFs (R. S. Weiss and P. Leder, unpublished data). Alternatively, functional redundancy could be provided by Atm, which is known to mediate a G2/M DNA damage checkpoint. While Atm ordinarily responds specifically to DSB, in the absence of Hus1 other types of genotoxic stress besides IR might lead to DSB as a form of secondary damage, perhaps from inappropriate processing of primary genotoxin-induced lesions. This secondary damage might activate Atm and induce the G2/M checkpoint. Whether such a mechanism accounts for the apparently normal G2/M checkpoint response to UV in Hus1-null cells could be addressed through the generation and analysis of MEFs nullizygous for both Hus1 and Atm. Why this redundancy would be manifested at the G2/M checkpoint but not during the S phase, when Hus1-null cells have an explicit checkpoint defect, is not clear. One possibility is that the pathways involving Atm and Hus1 feed into the same effectors at the G2/M checkpoint but have distinct targets during S phase.
We observed a strong correlation between functioning of the Hus1-dependent S-phase checkpoint and genotoxin sensitivity. Hus1-null cells are hypersensitive to agents that normally elicit a Hus1-dependent S-phase checkpoint response, such as BPDE and UV, but show only slight sensitivity to IR, which triggers a predominantly Hus1-independent S-phase checkpoint (58; also this report). Therefore, the Hus1-mediated S-phase checkpoint mechanism could be an important determinant of genotoxin sensitivity in mammalian cells. Notably, the sensitivity of yeast checkpoint mutants to genotoxic stress has been attributed specifically to defects in S-phase functions (13, 14, 55). In higher eukaryotes, however, the relationship between checkpoint responses and genotoxin sensitivity is poorly understood. With regard to the S-phase checkpoint that responds to IR, a correlation with radiosensitivity has not been observed (66). However, recent work indicates that a G2/M checkpoint defect alone likewise does not confer genotoxin sensitivity, leading to the suggestion that multiple checkpoint defects within a cell may act synergistically to cause sensitivity (63). If this is generally true, it may be that Hus1-null cells harbor another, as-yet-unidentified, cell cycle regulatory defect. Nonetheless, the Hus1-dependent checkpoint pathway described here represents another therapeutic target that potentially could be exploited to selectively sensitize tumor cells to anticancer treatments.
Acknowledgments
We thank Juanita Campos-Torres for extensive assistance with the FACS analysis and Anindya Dutta, Sylvia Lee, Boris Reizis, and Tom Wolkow for helpful discussions and comments on the manuscript.
This work was supported by American Cancer Society postdoctoral fellowship PF-98-131-01 and NCI training grant CA 09361-20 (R.S.W.), the Howard Hughes Medical Institute (P.L.), and NIH grant RO1 ES009558 (C.V.).
REFERENCES
- 1.Akkari, Y. M., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasina, A., B. D. Price, G. A. Turenne, and C. H. McGowan. 1999. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 9:1135-1138. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, G. T., V. McGovern, N. Ossanna, and H. Rosenthal. 1982. Concentration dependent alterations of DNA replication initiation and elongation by benzo[a]pyrene diol epoxide. Carcinogenesis 3:473-480. [DOI] [PubMed] [Google Scholar]
- 4.Bulavin, D. V., Y. Higashimoto, I. J. Popoff, W. A. Gaarde, V. Basrur, O. Potapova, E. Appella, and A. J. Fornace, Jr. 2001. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411:102-107. [DOI] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Busbee, D. L., C. O. Joe, J. O. Norman, and P. W. Rankin. 1984. Inhibition of DNA synthesis by an electrophilic metabolite of benzo[a]pyrene. Proc. Natl. Acad. Sci. USA 81:5300-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby, E. C., D. F. Leistritz, R. T. Abraham, L. M. Karnitz, and J. N. Sarkaria. 2000. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60:2108-2112. [PubMed] [Google Scholar]
- 8.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 9.Cliby, W. A., C. J. Roberts, K. A. Cimprich, C. M. Stringer, J. R. Lamb, S. L. Schreiber, and S. H. Friend. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordeiro-Stone, M., J. C. Boyer, B. A. Smith, and W. K. Kaufmann. 1986. Effect of benzo[a]pyrene-diol-epoxide-I on growth of nascent DNA in synchronized human fibroblasts. Carcinogenesis 7:1775-1781. [DOI] [PubMed] [Google Scholar]
- 11.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 12.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enoch, T., A. M. Carr, and P. Nurse. 1992. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6:2035-2046. [DOI] [PubMed] [Google Scholar]
- 15.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 16.Falck, J., J. H. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290-294. [DOI] [PubMed] [Google Scholar]
- 17.Feijoo, C., C. Hall-Jackson, R. Wu, D. Jenkins, J. Leitch, D. M. Gilbert, and C. Smythe. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D. C.
- 19.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 20.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275:5600-5605. [DOI] [PubMed] [Google Scholar]
- 21.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 22.Guo, N., D. V. Faller, and C. Vaziri. 2002. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 13:77-86. [PubMed] [Google Scholar]
- 23.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall-Jackson, C. A., D. A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18:6707-6713. [DOI] [PubMed] [Google Scholar]
- 25.Hang, H., Y. Zhang, R. L. Dunbrack, Jr., C. Wang, and H. B. Lieberman. 2002. Identification and characterization of a paralog of human cell cycle checkpoint gene HUS1. Genomics 79:487-492. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell, L. H., and M. B. Kastan. 1994. Cell cycle control and cancer. Science 266:1821-1828. [DOI] [PubMed] [Google Scholar]
- 27.Hekmat-Nejad, M., Z. You, M. C. Yee, J. W. Newport, and K. A. Cimprich. 2000. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10:1565-1573. [DOI] [PubMed] [Google Scholar]
- 28.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 29.Jernstrom, B., and A. Graslund. 1994. Covalent binding of benzo[a]pyrene 7,8-dihydrodiol 9,10-epoxides to DNA: molecular structures, induced mutations and biological consequences. Biophys. Chem. 49:185-199. [DOI] [PubMed] [Google Scholar]
- 30.Kastan, M. B., and D. S. Lim. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell. Biol. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 31.Kaufmann, W. K., J. C. Boyer, B. A. Smith, and M. Cordeiro-Stone. 1985. DNA repair and replication in human fibroblasts treated with (+/−)-r-7,t-8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Biochim. Biophys. Acta 824:146-151. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. T., B. Xu, and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 34.Larner, J. M., H. Lee, and J. L. Hamlin. 1997. S phase damage sensing checkpoints in mammalian cells. Cancer Surv. 29:25-45. [PubMed] [Google Scholar]
- 35.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 36.Lindsey-Boltz, L. A., V. P. Bermudez, J. Hurwitz, and A. Sancar. 2001. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc. Natl. Acad. Sci. USA 98:11236-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 38.Longhese, M. P., R. Fraschini, P. Plevani, and G. Lucchini. 1996. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol. 16:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 40.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 41.Marchetti, M. A., S. Kumar, E. Hartsuiker, M. Maftahi, A. M. Carr, G. A. Freyer, W. C. Burhans, and J. A. Huberman. 2002. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. USA 99:7472-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molinari, M., C. Mercurio, J. Dominguez, F. Goubin, and G. F. Draetta. 2000. Human Cdc25A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mossi, R., and U. Hubscher. 1998. Clamping down on clamps and clamp loaders: the eukaryotic replication factor C. Eur. J. Biochem. 254:209-216. [PubMed] [Google Scholar]
- 44.O'Connell, M. J., N. C. Walworth, and A. M. Carr. 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10:296-303. [DOI] [PubMed] [Google Scholar]
- 45.Ogryzko, V. V., P. Wong, and B. H. Howard. 1997. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol. Cell. Biol. 17:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orren, D. K., L. N. Petersen, and V. A. Bohr. 1995. A UV-responsive G2 checkpoint in rodent cells. Mol. Cell. Biol. 15:3722-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 48.Rauen, M., M. A. Burtelow, V. M. Dufault, and L. M. Karnitz. 2000. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J. Biol. Chem. 275:29767-29771. [DOI] [PubMed] [Google Scholar]
- 49.Rowley, R., E. N. Phillips, and A. L. Schroeder. 1999. The effects of ionizing radiation on DNA synthesis in eukaryotic cells. Int. J. Radiat. Biol. 75:267-283. [DOI] [PubMed] [Google Scholar]
- 50.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 51.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375-4382. [PubMed] [Google Scholar]
- 52.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 53.St. Onge, R. P., C. M. Udell, R. Casselman, and S. Davey. 1999. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell 10:1985-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takai, H., K. Tominaga, N. Motoyama, Y. A. Minamishima, H. Nagahama, T. Tsukiyama, K. Ikeda, K. Nakayama, M. Nakanishi, and K. Nakayama. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes Dev. 14:1439-1447. [PMC free article] [PubMed] [Google Scholar]
- 55.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 56.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volkmer, E., and L. M. Karnitz. 1999. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 274:567-570. [DOI] [PubMed] [Google Scholar]
- 58.Weiss, R. S., T. Enoch, and P. Leder. 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 14:1886-1898. [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, R. S., S. Matsuoka, S. J. Elledge, and P. Leder. 2002. Hus1 acts upstream of Chk1 in a mammalian DNA damage response pathway. Curr. Biol. 12:73-77. [DOI] [PubMed] [Google Scholar]
- 60.Widrow, R. J., and C. D. Laird. 2000. Enrichment for submitotic cell populations using flow cytometry. Cytometry 39:126-130. [DOI] [PubMed] [Google Scholar]
- 61.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 62.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, B., S. T. Kim, D. S. Lim, and M. B. Kastan. 2002. Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 65.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zdzienicka, M. Z. 1996. Mammalian X ray sensitive mutants: a tool for the elucidation of the cellular response to ionizing radiation. Cancer Surv. 28:281-293. [PubMed] [Google Scholar]
- 67.Zhao, H., and H. Piwnica-Worms. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 70.Zhou, X. Y., X. Wang, B. Hu, J. Guan, G. Iliakis, and Y. Wang. 2002. An ATM-independent S-phase checkpoint response involves CHK1 pathway. Cancer Res. 62:1598-1603. [PubMed] [Google Scholar]
- 71.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]