Abstract
The Chinese hamster dihydrofolate reductase (DHFR) origin of replication consists of a broad zone of potential initiation sites scattered throughout a 55-kb intergenic spacer, with at least three sites being preferred (ori-β, ori-β′, and ori-γ). We previously showed that deletion of the most active site or region (ori-β) has no demonstrable effect on initiation in the remainder of the intergenic spacer nor on the time of replication of the DHFR locus as a whole. In the present study, we have now deleted ori-β′, both ori-β and ori-β′, an 11-kb region just downstream from the DHFR gene, or the central ∼40-kb core of the spacer. The latter two deletions together encompass >95% of the initiation sites that are normally used in this locus. Two-dimensional gel analysis shows that initiation still occurs in the early S phase in the remainder of the intergenic spacer in each of these deletion variants. Even removal of the 40-kb core fails to elicit a significant effect on the time of replication of the DHFR locus in the S period; indeed, in the truncated spacer that remains, the efficiency of initiation actually appears to increase relative to the corresponding region in the wild-type locus. Thus, if replicators control the positions of nascent strand start sites in this complex origin, either (i) there must be a very large number of redundant elements in the spacer, each of which regulates initiation only in its immediate environment, or (ii) they must lie outside the central core in which the vast majority of nascent strand starts occur.
In the chromosomes of bacteria, bacterial plasmids, viruses, and lower eukaryotes such as Saccharomyces cerevisiae, replication initiates at genetically defined sites known as replicators, which serve as recognition signals for cognate sequence-specific DNA binding protein complexes termed initiators (34, 42). In the chromosomes of metazoans, however, many replication origins consist of broad zones of closely spaced initiation sites, some of which are used with greater efficiency than others (for reviews, see references 14, 19, and 30).
An example of the latter is the early-firing Chinese hamster dihydrofolate reductase (DHFR) origin, which resides in the 55-kb spacer between the convergently transcribed DHFR and 2BE2121 genes (Fig. 1A) (17, 18, 22, 58). Neutral/neutral (7) and neutral/alkaline (49) two-dimensional (2-D) gel studies on several adjacent and overlapping restriction fragments have demonstrated that there are at least 20 nascent strand start sites (and probably many more) scattered throughout the 55-kb spacer (Fig. 1) (23). However, more-quantitative assays showed that the central 40-kb core, and particularly two subregions within it known as ori-β and ori-γ, are preferred (Fig. 1) (23, 43, 44). The results of an in-gel renaturation analysis of early labeled restriction fragments are shown in Fig. 1A (44), as is the frequency of initiation measured in a quantitative early labeled fragment hybridization (ELFH) assay that used very small origin-containing nascent DNA as a probe (23). The results of a PCR-based small nascent strand abundance assay that focused on a 12-kb subregion of the spacer encompassing ori-β, which identified an additional, somewhat less active, site or region immediately downstream (termed ori-β′) (41) are also shown in Fig. 1A. Interestingly, the one silent locus detected by the high-resolution ELFH assay lies in the approximate center of the spacer just upstream from a prominent matrix attachment region (Fig. 1A) (16) and coincides remarkably well with the least-active region as measured in the lower resolution in-gel renaturation study (44) and by neutral/neutral 2-D gel analysis (23).
FIG. 1.
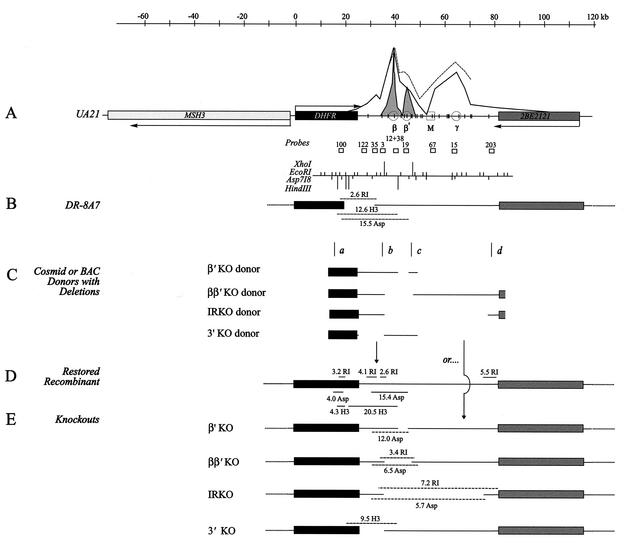
Engineering deletions in the DHFR initiation zone. (A) Map of a 190-kb region encompassing the DHFR locus, showing the positions of the DHFR, 2BE2121 (26), and MSH3 genes and the directions of transcription. The intergenic region constitutes the DHFR origin, which contains a centered matrix attachment region (M), indicated by the box (16). The gray curve is a tracing of the data obtained in a PCR-based small nascent strand abundance analysis of the 12-kb sequence encompassing the ori-β and ori-β′ regions (41), while the open curves traced with solid and dashed lines represent the distribution of initiation sites as measured by high-resolution ELFH (23) and in-gel renaturation experiments (44), respectively. Below are shown EcoRI, Asp718, and (incomplete) HindIII and XhoI maps of the intergenic spacer, along with probes used in the Southern and 2-D gel analyses (see text). The vertical marks on the linear axis correspond to initiation sites identified by 2-D gel analysis (23). (B) The DHFR-deficient variant, DR-8A7, which has only one copy of the DHFR locus and has suffered a radon-induced 14-kb deletion encompassing the 3′ end of the DHFR gene (35). The resulting deletion junction fragments are indicated above and below the map (compare with Fig. 2). (C) Cartoons representing the constructs that were used to introduce deletions into the DHFR locus. Homologous recombination at sites a and b results in restoration of the wild-type locus (as shown in panel D), while recombination at sites a and c or d (depending on the donor) restores the gene and at the same time deletes the indicated sequence from the endogenous locus (indicated in panel E). (D) Map and diagnostic wild-type fragments regenerated by recombination at sites a and b. (E) Maps of KO cell lines generated by recombination between sites a and either c or d (depending on the donor), showing diagnostic fragments detected in the Southern analyses pictured in Fig. 2 (see text).
The seeming complexity of the DHFR origin raises the question of whether its activity is controlled by classic replicator-initiator interactions. If so, the proposed replicators could serve as recognition sites for proteins such as the origin-recognition complex (6), which was first identified in S. cerevisiae and which facilitates loading of other initiation proteins such as CDC6, CDC45, cdt1, and the minichromosome maintenance proteins (reviewed in reference 5). Homologues of these proteins have now been identified in several higher eukaryotes (5, 28), including Chinese hamsters (2; M. Alexandrow and J. L. Hamlin, unpublished data). In the latter system, at least minichromosome maintenance proteins 2 and 5 generally localize to the intergenic region in the early S period and then appear to translocate to flanking regions as cells move through the S period (2), mimicking the behavior of their counterparts in S. cerevisiae (4).
In one extreme model that corresponds most closely to the paradigm established for simpler systems, initiation complexes could load at one or a small number of master replicators residing within the DHFR origin itself. The proposed replicator(s) would presumably correspond to one or more of the most active initiation sites or regions in the spacer (i.e., ori-β, ori-β′, and/or ori-γ). After loading, the initiation complexes could translocate randomly up or down the template via a helicase activity before priming nascent strands, the latter activity displaying some preference for particular sequences in the template. This mechanism could lead to the dispersive mode of nascent strand initiation suggested by the 2-D gel approaches, but it could also explain why the regions around ori-β, ori-β′, and/or ori-γ (the suggested loading sites) are preferred. In an alternative multireplicator model, ori-β, ori-β′, and ori-γ would represent the most active subset of at least 20 redundant replicators distributed throughout the spacer that attract initiation proteins with different efficiencies and which control initiation only in their immediate environments. Clearly, variations on these scenarios can be imagined. With any model, the efficiencies of individual replicators could be modulated by additional factors such as chromatin architecture and disposition relative to genes, matrix attachment regions, and boundary elements, etc.
In a previous study (36), a reach-out-and-knockout (ROKO) homologous recombination strategy was utilized to delete ori-β, which was considered at the time to be the predominant initiation site or region in the spacer and therefore might serve as a master replicator (10, 12, 13, 52). However, deletion of ori-β had no detectable effect on initiation in the remainder of the spacer nor on the time of replication of the DHFR locus in the S period (36). Thus, we concluded that, while ori-β is the most efficient initiation site in the spacer, it does not constitute a master, nonredundant replicator for the DHFR locus. This finding raised the possibility that other preferred sites (e.g., ori-β′ or ori-γ) might correspond to a master replicator. Alternatively, ori-β′ and/or ori-γ might represent a redundant replicator that assumes the role of master when ori-β is deleted by relief of origin interference, a phenomenon that occurs between neighboring autonomously replicating sequence (ARS) elements in S. cerevisiae (9).
In the present study, we have used the ROKO approach to delete various parts of the intergenic spacer to determine whether any critical, nonredundant, cis-regulatory elements lie within the spacer itself. We find that even a large deletion encompassing the central 40-kb core fails to inhibit initiation in the remainder of the spacer or dramatically change the time of replication of the locus in the S period. Remarkably, this deletion encompasses >90% of the sites that support initiation in the native locus, including ori-β, ori-β′, and ori-γ. Thus, if classic replicators control the positions of start sites in this complex origin, they either control initiation only in their immediate environments or lie outside the central core in which the great majority of nascent strand starts occur. Our data further suggest that the efficiency of initiation at individual sites may be influenced by the length of the spacer, which, by definition, is controlled by local transcriptional activity.
(This work represents a part of the requirements for the Ph.D. degree for X. Li from the Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, Charlottesville, Va.)
MATERIALS AND METHODS
Preparation of donor constructs for the ROKO strategy.
BAC-KZ381, BAC-KP454, and pWe15-KZ381 were constructed by transferring the NotI-resected insert from pWe16-KZ381 or pWe16-KP454 (46) into the respective vector. BAC-KZ/KP was engineered from the corresponding bacterial artificial chromosomes (BACs), resulting in an insert extending from map position (mp) 10- to 85-kb (Fig. 1A) (L. D. Mesner, unpublished data). The ββ′ knockout (KO) donor is missing the 12-kb sequence extending from mp 36 to 48, and it was derived by recircularizing an XhoI partial digest of BAC-KZ/KP. The intergenic region KO (IRKO) donor was constructed by deleting a 12-kb XhoI fragment (Fig. 1A) from pWe15-KZ381 (mp 36 to 48) and replacing it with a 6-kb KpnI-NotI fragment (mp 78 to 84) from KP454, resulting in a deletion extending from mp 36 to 77. The KZ381 β′ KO donor is missing ∼4 kb extending from mp 43 to 47 and was constructed essentially as described previously (36), except that this region in the subcloned 12-kb XhoI fragment (Fig. 1A) was replaced (via a HindIII complete digestion) with a 4-kb neomycin resistance (Neor) gene flanked by loxP sites. By introducing the cre recombinase in trans, the Neor marker was subsequently removed (36). The DHFR 3′-KO donor was constructed by circularizing a 37-kb BamHI fragment obtained by partial digestion of BAC-KZ381, resulting in an ∼12-kb deletion extending between mp 24 and 36 (Fig. 1A). (All clones and maps are available upon request.)
Cell culture and synchronization protocols.
The DR-8A7 cell line is a DHFR-deficient variant of the hemizygous CHO cell line UA21 (57) and has a deletion of the 14-kb region extending downstream from approximately the center of the last intron of the DHFR gene into the intergenic zone (mp 20 to 34) (Fig. 1A) (35). DR-8A7 cells were propagated on minimal essential medium (MEM) supplemented with nonessential amino acids, 100 μM hypoxanthine, 16 μM thymidine (57), and 10% fetal clone II (HyClone) and were maintained in an atmosphere of 5% CO2. Homologous recombinants that restored the missing part of the gene were propagated on MEM containing nonessential amino acids and 10% fetal clone II. To prepare synchronized populations, cells were arrested in G0 by starving them for isoleucine for 36 h and they were released into complete MEM containing 200 μM mimosine (Sigma) for 12 h to collect them at the G1/S boundary (17, 48). The drug was washed out and replaced with drug-free medium to allow entry into the S phase.
Transfection, selection, and characterization of potential homologous recombinants.
NotI-digested donor constructs were delivered to the DHFR-deficient DR-8A7 cell line by electroporation as previously described (36). After transfection, cells were seeded at a density of ∼2 × 106 per 10-cm-diameter plate and were maintained in F12 medium containing 10% fetal clone I for 36 to 48 h. The medium was then changed to F12 lacking thymidine, hypoxanthine, and glycine, and surviving (DHFR+) colonies were isolated ∼10 days later. Genomic DNA samples were purified by standard procedures (31), digested with the relevant restriction enzymes (see figure legends), and separated on 0.6% agarose gels for Southern blotting onto HybondN+ (Amersham) and hybridization with the appropriate probes (see figure legends) as previously described (21). The probes utilized in this study were as follows: probe 100, a 1.2-kb XbaI/KpnI fragment; probe 122, a 0.4-kb BamHI/EcoRI fragment; probe 35, a 1.0-kb KpnI/EcoRI fragment; probe 3, a 2.0-kb EcoRI/BamHI fragment; probe 12, a 0.3-kb BamHI/PvuII fragment; probe 38, a 0.4-kb PvuII/XmnI fragment; probe 19, a 1.1-kb HindIII/KpnI fragment; probe 67, a 0.7-kb EcoRI/XbaI fragment; probe 15, a 0.8-kb HindIII/BamHI fragment; and probe 203, a 0.25-kb XbaI/BstEII fragment. Based on the patterns of diagnostic fragments, colonies were selected that had undergone clean double homologous recombination events at a site in the DHFR gene and at the correct general location in the intergenic region downstream (Fig. 1 and text). The restored cell line represents a control that underwent recombination exchanges near sites a and b, thus restoring the wild-type arrangement.
2-D gel analysis (7).
After 12 h in mimosine, cell lines were released into the S period by washing them with and returning them to prewarmed MEM containing 10% fetal clone II (supplemented with thymidine and hypoxanthine for DR-8A7 cells). At selected intervals thereafter (see figure legends), cells were harvested and replication intermediates were prepared by digesting matrix-attached DNA with EcoRI as described previously (21). Replication intermediates were analyzed by neutral/neutral 2-D gel electrophoresis exactly as described previously (21).
FISH-based replication timing analysis (36, 40).
The determination of replication timing by fluorescence in situ hybridization (FISH) was performed as described previously (36, 56), except that prior to fixation, cells were swollen in 40 mM KCl and 5 mM glycine (pH 9.5) instead of the typical neutral swelling regimen (61; L. D. Mesner, P. A. Dijkwel, and J. L. Hamlin, unpublished data). Only those alkaline FISH samples that showed no appreciable loss in the number of cells compared to neutral FISH samples were analyzed.
RESULTS
Construction of the intergenic KO cell lines by the ROKO approach (36).
To engineer deletions into the intergenic region in loco, the 3′ end of the DHFR gene in the DR-8A7 variant is restored to the wild type by homologous recombination with a donor construct that provides (i) the missing part of the gene, (ii) a small region of homologous overlap, and (iii) part of the spacer from which the relevant downstream target has been removed (Fig. 1B and C). Since DR-8A7 has no functional copy of the DHFR gene, it cannot survive on minimal medium lacking thymidine, hypoxanthine, and glycine (57) unless the missing 3′ sequences are restored. Homologous exchanges near sites a and b yield a wild-type restored derivative (Fig. 1B and D) (∼1/106 transfected cells) while exchanges near a and a position downstream from the desired deletion (e.g., near site c or d) restore the gene and also effect the deletion (Fig. 1E) (1 to 3% of DHFR+ cells).
In the present study, donor constructs were either BACs or cosmids from which sequences encompassing the desired region had been removed (see Materials and Methods). Donors with the following deletions were constructed (Fig. 1C): (i) a 4.4-kb sequence encompassing the ori-β′ locus; (ii) a 12-kb sequence encompassing both ori-β and ori-β′; (iii) the 40-kb central core of the intergenic spacer, in which >90% of initiations occur in the native locus (compare the IRKO deletion in Fig. 1C to the ELFH data in Fig. 1A); and (iv) a 12-kb sequence containing the segment of the intergenic region deleted in the DR-8A7 cell line. After transfecting each donor BAC or cosmid into DR-8A7 by electroporation, potential intergenic deletion variants were selected on MEM and characterized by Southern blotting and hybridization with the relevant probes. Control cells were UA21, which contains a single wild-type locus, and the starting dhfr-deficient DR-8A7 recipient. Usually, two different digests were run on the same gel and hybridized with a mixture of two probes, at least one of which illuminates a diagnostic fragment in each digest of the individual cell lines. Examples of Southern analyses are shown in Fig. 2.
FIG. 2.
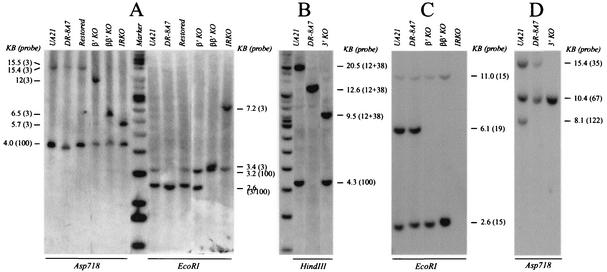
Southern analysis confirms successful restoration of the DHFR gene and the relevant deletions in DHFR-proficient survivors. Relevant restriction digests of DNA from the indicated cell lines were separated on a 0.6% agarose gel along with a 1-kb ladder and high-molecular-weight standard (Invitrogen), transferred to a HybondN+ membrane (Amersham), and hybridized with the indicated probes to detect fragments expected for the desired deletions. Diagnostic fragments for each successful recombinant are indicated in Fig. 1B, D, and E and are discussed in the text. The probes utilized are pictured in Fig. 1A and specified in Materials and Methods.
In an Asp718 digest (Fig. 2A), a mixture of probes 3 and 100 detects the following fragments (Fig. 1A and E): (i) 15.4- and 4.0-kb wild-type fragments in UA21 and the wild-type restored recombinant; (ii) 15.5-kb junction and 4.0-kb wild-type fragments in DR-8A7; (iii) 12.0-kb junction and 4.0-kb wild-type fragments in the ori-β′ KO (β′ KO); (iv) 6.5-kb junction and 4.0-kb wild-type fragments in the β and β′ KO; and (v) 5.7-kb junction and 4.0-kb wild-type fragments in the IRKO. In an EcoRI digest (Fig. 2A), a mixture of probes 3 and 100 detects the following: (i) wild-type 3.2- and 2.6-kb fragments in UA21 and the restored cell line; (ii) 2.6-kb wild-type and 2.6-kb junction fragments in DR-8A7; (iii) 3.2- and 2.6-kb wild-type fragments in the β′ KO; (iv) 3.4-kb junction and 3.2-kb wild-type fragments in the β and β′ KO; and (v) 7.2-kb junction and 3.2-kb wild-type fragments in the IRKO cell line. For the 3′-end deletion (Fig. 2B), a mixture of probes 100 and 12 and 38 on a HindIII digest detects 20.5- and 4.3-kb wild-type fragments in UA21, a 12.6-kb HindIII junction fragment in DR-8A7, and 9.5-kb junction and 4.3-kb wild-type fragments in the 3′-end KO.
To demonstrate that the desired regions were actually deleted by successful double homologous recombination events, genomic DNAs were digested with either EcoRI or Asp718 and were hybridized with small probes complementary to the deleted region in each case (maps and probe positions are given in Fig. 1A). For example, in the results shown in Fig. 2C, a mixture of probes 19 and 15 recognizes 6.1-kb (probe 19) and 11.0- and 2.6-kb (probe 15) EcoRI fragments in wild-type UA21 and DR-8A7 cells. As predicted, probe 19 fails to illuminate the 6.1-kb fragment in the β′ and β/β′ KOs, and probes 19 and 15 do not detect either the 11.0-, 6.1-, or 2.6-kb fragments in the IRKO deletion variant. For analysis of the 3′ KO, an Asp718 digest was hybridized with a mixture of probes 35, 67, and 122. As seen in the results shown in Fig. 2D, UA21 DNA displays the predicted 15.4-, 10.4-, and 8.1-kb Asp718 fragments, DR-8A7 lacks the 8.1-kb fragment, and the 3′ KO lacks both the 8.1- and 15.4-kb Asp718 fragments. Southern analyses with several other restriction enzymes and probes confirmed that clean restorations of the gene and deletion of the relevant sequences had occurred, resulting in the KO cell lines diagramed in Fig. 1E (Mesner, unpublished).
Deletion of ori-β′ alone or in combination with ori-β has no demonstrable effect on initiation in the remainder of the intergenic zone.
We first asked whether ori-β′ might correspond to the master replicator in the region or to a redundant master replicator that takes over in the absence of ori-β by relief of origin interference (9). To test these possibilities, the first recombinant cell lines constructed were the ori-β′ and ori-β and ori-β′ KO cell lines (β′ KO and ββ′ KO) (Fig. 1E). Any effects on initiation in the remainder of the spacer resulting from these deletions were assessed by neutral/neutral 2-D gel analysis of replication intermediates isolated from synchronized cells 90, 180, and 360 min after entry into the S phase (the S phase lasts ∼8 h in these cell lines). EcoRI digests of replication intermediates were separated on a 2-D gel, transferred to a membrane, and analyzed with the relevant hybridization probes. As shown in Fig. 3A and B, different arcs are traced in these gels depending upon whether a fragment is replicated passively by single forks entering from either side or if it contains an initiation site (7).
FIG. 3.
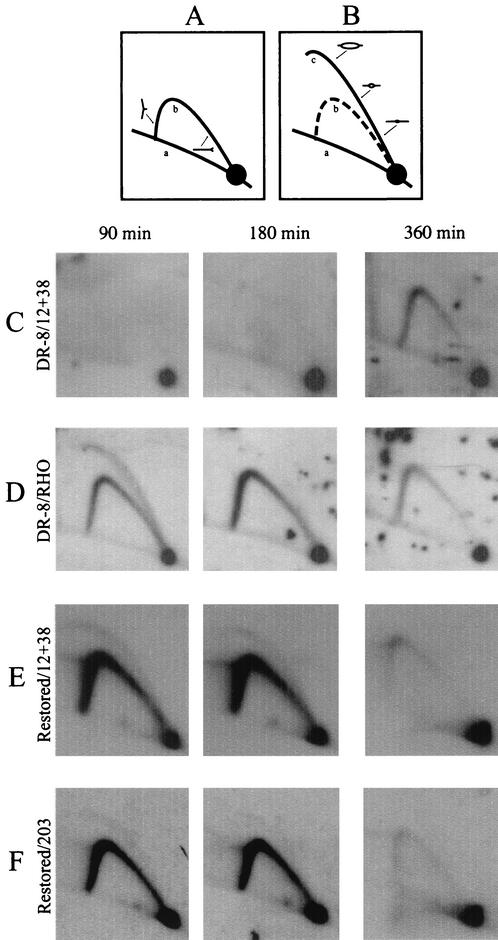
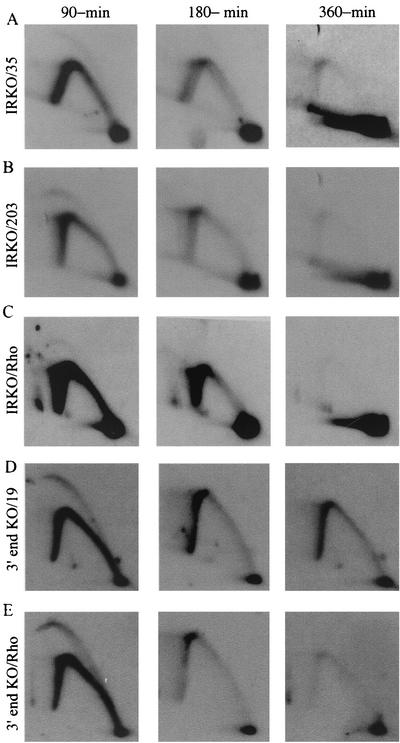
Neutral/neutral 2-D gel patterns of origin-positive and -negative cell lines. (A and B) Principle of the neutral/neutral 2-D gel method. A restriction digest containing replication intermediates is separated in the first dimension gel (left to right) largely according to molecular mass and in the second dimension according to both mass and shape (top to bottom). After transfer to a membrane, the digest is hybridized with a probe specific for the fragment of interest. The patterns traced by a fragment replicated either passively by a single fork or actively from an internal, centered origin are shown in panels A and B, respectively. (C to E) DR-8A7 and the restored derivative were synchronized at the G1/S boundary with mimosine, and samples were collected 90, 180, and 360 min after drug removal. After purification of EcoRI-digested replication intermediates, separation on a 2-D gel, and transfer to a membrane, the digests were hybridized with probes 12 and 38 (rows C and E) or a probe specific for a 6.5-kb fragment in the rhodopsin origin (row D) (20) (see Fig. 1A and Materials and Methods for probe positions). The average film exposure time with an intensifying screen for all of the hemizygous cell lines studied here was 10 to 21 days at −70°C.
Figure 3C to F shows replication intermediates detected in the intergenic region of the starting DR-8A7 cell line and a restored wild-type derivative of DR-8A7 (Fig. 1B and D). In DR-8A7, virtually no replication intermediates can be detected in a 6.2-kb fragment encompassing the ori-β region at 90 and 180 min into the S phase when hybridized with probes 12 and 38 (Fig. 3C and 1A). These data are in agreement with earlier studies showing that early-firing origin activity is inhibited in this locus as a consequence of the deletion encompassing the 3′ end of the gene and part of the intergenic region (36). By 360 min, a relatively intense single fork arc and the absence of a detectable bubble arc indicate that this region is replicated late in the S phase, probably by forks from distant active origins (Fig. 3C, compare to Fig. 3A) (see Discussion).
In contrast, when the transfer shown in Fig. 3C was stripped and rehybridized with a probe for the control, early-replicating rhodopsin origin (27), a composite pattern is detected at 90 min consisting of a complete bubble arc and a single fork arc (Fig. 3D) (20). This pattern is characteristic of fragments residing in broad initiation zones that fire in the early S phase (including the wild-type DHFR locus), since such fragments sometimes sustain internal initiation events but more frequently are replicated passively by forks from neighboring initiation sites in the same zone (17, 18, 20, 22, 58). Very few bubbles can be detected in the rhodopsin origin at 180 min, indicating that initiation is largely complete by this time, but a single fork arc persists in the region until ∼360 min. This is because the rhodopsin origin, like the wild-type DHFR origin, is inefficient overall, firing only 30 to 40% of the time in any given cell cycle (17, 20, 22); thus, in many cells, the locus is replicated passively well into the S period by forks from active origins lying upstream or downstream from the locus.
When DR-8A7 is converted to the wild-type arrangement in the restored derivative (Fig. 1B and D), the DHFR origin reverts to the early-firing phenotype characteristic of the wild-type locus and the rhodopsin early-firing control, as indicated by the patterns obtained with probes 12 and 38 for the ori-β locus and probe 203 for a region lying downstream from the 2BE2121 gene (Fig. 3E and F) (20). (Note that the transfers shown in Fig. 3E and F are overexposed relative to that shown in Fig. 3D, but the overall patterns are similar.) Thus, in studies discussed below on the effects of intergenic deletions, the rhodopsin locus will be utilized as the early firing internal control. Importantly, faithful restoration of the truncated gene in DR-8A7 with a concomitant deletion of a downstream target (as in Fig. 1E) should restore the origin to the early-firing phenotype only if deletion of the target has no effect on initiation. If a deletion renders the DHFR origin either inactive or late firing, the cell line should display the late-replicating phenotype that characterizes DR-8A7.
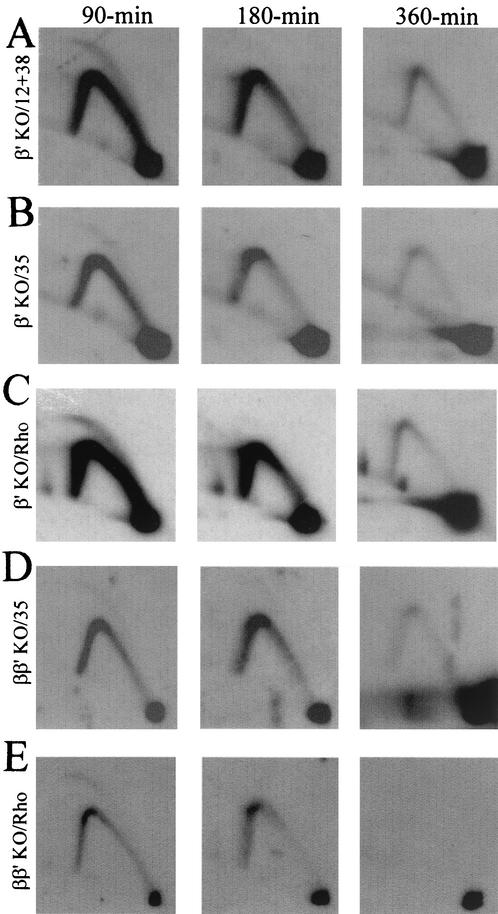
We next examined replication intermediates from the β′ KO cell line by the 2-D gel method (Fig. 4). The pattern detected with a mixture of probes 12 and 38, which illuminates the ori-β-containing fragment, is remarkably similar to that detected when the same transfer was stripped and rehybridized with a probe specific for the rhodopsin locus (compare Fig. 4A and C; also compare to the ori-β locus in the wild-type restored control in Fig. 3E): initiation is seen at 90 min and is largely complete by 180 min while single forks indicative of passive replication persist until about 360 min into the S phase. Probe 35, which is specific for a fragment in the 5′ end of the initiation zone (Fig. 1A), also detects an early-firing pattern in this region of the β′ KO cell line, although the intensity of the bubble arc relative to the single fork arc is lower than at ori-β (compare Fig. 4A and B), just as it is in the wild-type locus (23). Even more interesting is the observation that when ori-β and ori-β′ are coordinately deleted in the ββ′ KO cell line (Fig. 4D), probe 35 again displays an early replicating pattern similar to the rhodopsin origin in the same cells (Fig. 4E) and to that of the β′ KO cell line (Fig. 4B).
FIG. 4.
Deletion of ori-β′ alone or ori-β and ori-β′ has no apparent effect on initiation in the remainder of the spacer region. The ori-β′ and ori-β/ori-β′ KO cell lines were synchronized and processed as described in the legend to Fig. 3 and Materials and Methods. The images shown in panels A to C were derived from successive hybridizations and stripping of the same blot, as were the images shown in panels D and E (22).
Critical, nonredundant, genetic elements required for early-firing origin activity do not reside in the 50-kb region extending between the 3′ ends of the DHFR and 2BE2121 genes.
Thus, neither ori-β nor ori-β′ corresponds to an essential replicator responsible for controlling initiation in the DHFR origin as a whole. Furthermore, these two sites or regions do not appear to correspond to redundant master replicators that interfere with each other. However, the possibility remained that these two regions are redundant with yet another master element elsewhere in the spacer (possibly ori-γ). We addressed this issue directly by constructing the IRKO cell line, which has a deletion of the central 40-kb core of the intergenic spacer, including ori-β, ori-β′, and ori-γ, and in which >90% of initiations normally occur as assessed from the quantitative ELFH data (Fig. 1A) (23). The positions of this deletion and the diagnostic fragments are shown in Fig. 1C and E.
The IRKO cell line was synchronized and sampled as described previously, and EcoRI digests were separated on a 2-D gel. The digests were blotted and hybridized successively with probes 35 and 203, which are specific for 4.1- and 5.5-kb fragments near the ends of the intergenic spacer in the wild-type configuration but recognize 4.1-kb wild-type and 7.2-kb junction fragments in the truncated spacer in the IRKO variant. In the IRKO variant, probes 35 and 203 each detect bubble arcs in their cognate fragments in the 90-min sample (compare to the bubble arc in the rhodopsin control on the same blot) (Fig. 5C). Furthermore, replication of the locus is largely completed by 360 min into the S phase, which would not be predicted if early-firing origin activity had been severely compromised by the IRKO deletion (i.e., as in DR-8A7 cells) (Fig. 3C). Thus, we can conclude that critical, nonredundant sequences required to fire this origin in the early S phase do not reside within the 40-kb central core.
FIG. 5.
Deletion of the central 40-kb core of the DHFR origin or sequences lying immediately downstream from the DHFR gene does not noticeably change the pattern of replication intermediates in the remainder of the intergenic spacer. The IRKO (panels A to C) and 3′ end (panels D and E) deletion variant cell lines were synchronized and sampled as described in the legend to Fig. 3. Replication intermediates were prepared by using EcoRI to digest the DNA, and the resulting digests were hybridized with the indicated probes. In the IRKO variant, probe 35 detects a wild-type 4.1-kb fragment, but probe 203 illuminates a 7.2-kb deletion junction fragment. In the 3′ end deletion variant, probe 19 recognizes a 6.1-kb wild-type EcoRI fragment encompassing ori-β′.
We then asked whether any critical genetic element(s) required to activate this origin resides in the 12-kb region of the spacer lying just downstream from the DHFR gene. This region corresponds to the part of the spacer that is deleted in DR-8A7, whose origin is inactive, but is retained in the IRKO deletion variant, whose origin is active (Fig. 1A, B, and E). However, as shown in Fig. 5D and E, removing the region between the poly(A) sites in the DHFR gene and the 5′ boundary of the IRKO deletion does not evoke a demonstrable change in the early-firing origin activity: probe 19 detects the classic early replicating pattern in an EcoRI fragment containing ori-β′, which displays the same high level of replication bubbles as does ori-β in the wild-type locus (17, 18, 22, 58).
The efficiency of initiation in the spacer appears to increase when the 40-kb core is deleted.
Thus, deletion of a region containing the majority of start sites from the intergenic region in the IRKO cell line did not noticeably reduce origin activity in the remainder of the spacer according to the criterion of 2-D gel analysis. Additional probings did not detect initiation within the body of the DHFR gene itself, suggesting that the initiation zone in the IRKO variant did not spread outward (data not shown). This raised the question whether the efficiency of initiation in the truncated spacer was affected by the loss of the 40-kb core.
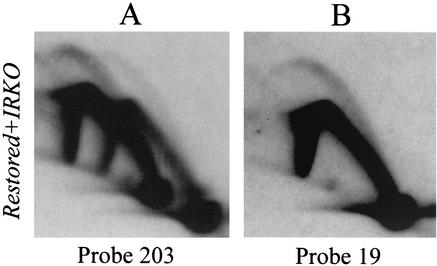
This was first assessed by directly comparing the intensities of the bubble arcs in the spacers of the IRKO and wild-type cell lines in a 2-D gel-mixing experiment. IRKO cells and the restored derivative were synchronized with mimosine by the regimen described above and were sampled 90 min after release into the S period. Fluorescence-activated cell sorter analysis indicated that the two cell lines were synchronized almost identically by this regimen (data not shown). Equal numbers of each cell type were combined, and replication intermediates were prepared from the mixture by using EcoRI to digest the DNA. After separation on a 2-D gel and transfer to a membrane, the digest was hybridized with probe 203, which recognizes a 5.5-kb fragment in the wild-type locus and a 7.2-kb deletion junction fragment in the IRKO cell line formed by the fusion of the 5.5- and 2.6-kb wild-type versions (Fig. 1A). Because of the difference in size, the two fragments could be separated enough to distinguish the intermediates arising from each. Importantly, the two wild-type fragments that were fused in the IRKO deletion reside near the ends of the spacer in the wild-type locus, which normally support fewer initiation events than the 40-kb core (Fig. 1A).
The results of this analysis with probe 203 are shown in Fig. 6A. The intensities of both the bubble and fork arcs in the 7.2-kb deletion junction fragment in the IRKO cell line are clearly higher than they are in the 5.5-kb wild-type fragment from the restored wild-type derivative, even though the nonreplicated 1n spots are comparable. To estimate the level of replication intermediates in the IRKO junction fragment compared to a fragment in the active central core in the restored wild-type locus, the same blot was stripped and rehybridized with probe 19; this probe recognizes a 6.1-kb EcoRI fragment encompassing ori-β′ in the wild-type locus of the restored cell line. This same fragment is deleted in the IRKO cell line (Fig. 1C). As shown in Fig. 6B, the intensities of the bubble and fork arcs detected with probe 19 in the ori-β′ locus in the restored derivative are approximately equal to those detected with probe 203 in the deletion junction fragment in the IRKO variant, even though the intensities of the 1n spots are approximately equal. In other words, the junction fragment in the truncated spacer now appears as active as does a fragment in the most-active region of the wild-type locus. Although some of the differences in the intensities of the bubble arcs in the 7.2- and 5.5-kb fragments shown in Fig. 6A can be attributed to a size difference (45), the magnitude of this difference suggests that the frequency of initiation in the truncated spacer may have actually increased over that of the corresponding sequences in the wild-type locus.
FIG. 6.
The efficiency of initiation in the truncated spacer in the IRKO cell line appears to have increased relative to the same sequences in the wild-type locus. The IRKO and restored wild-type control cell lines were synchronized as described previously, and samples were taken in the early S phase (90 min). Equal numbers of cells from each cell line were mixed, and replication intermediates were prepared by using EcoRI to digest the DNA. The transfer of the resulting 2-D gel was first hybridized with probe 203, which detects a wild-type 5.5-kb EcoRI fragment lying downstream from the 2BE2121 gene in the restored cell line (Fig. 1D) and a 7.2-kb deletion junction fragment in the IRKO variant (Fig. 1). The blot was then stripped and rehybridized with probe 19, which detects a 6.1-kb EcoRI fragment containing ori-β′ in the wild-type restored control that is deleted in the IRKO variant.
A quantitative replication timing assay suggests that deletion of the central 40-kb core of the DHFR origin has little effect on the time of replication of the DHFR locus.
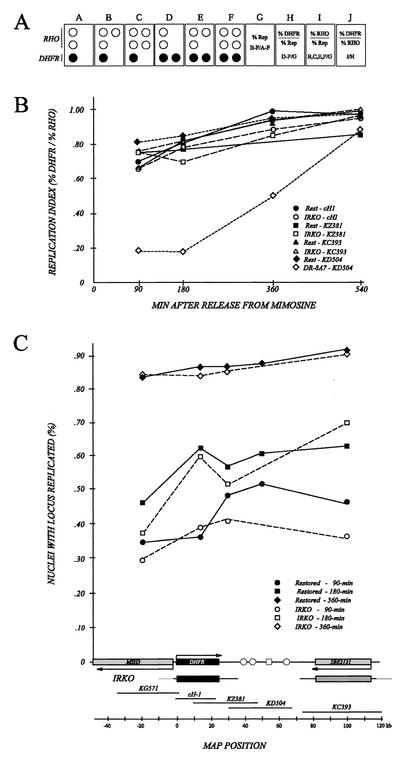
To address the question of efficiency of initiation more quantitatively, the IRKO variant was examined by using a replication timing assay in which the number of copies of a locus of interest in a given cell (and thus its replication status) is compared to the copy number of an internal, early replicating standard (Fig. 7A) (36). In this assay, cells were immobilized on microscope slides and hybridized with a mixture of probes specific for the relevant loci. In the present case, a cosmid from the early-firing rhodopsin control locus was labeled with biotinylated dATP and was mixed with digoxigenin-labeled cosmids spanning various sections of the DHFR locus (Fig. 7C). The probe for rhodopsin was detected with streptavidin-Texas Red, while the DHFR-specific probes were detected with sheep anti-digoxigenin and fluorescein isothiocyanate-conjugated donkey anti-sheep immunoglobulin G (36, 56). By counting the numbers of each of the fluorescent patterns shown in Fig. 7A (panels A to F), one arrives at a replication index, which is the ratio of cells that have replicated the DHFR locus versus the early-firing rhodopsin control (Fig. 7A, panel J). If both markers replicate at the same time in a given cell line, a replication index near unity will result, while late-replicating loci will display lower replication indices (36). Although this method does not measure effects on initiation per se, if a region normally replicates early because it contains an early-firing origin, the locus as a whole will remain early replicating only if its origin still fires in the early S phase. Thus, the assay will be sensitive to changes that cause an early-firing origin to fire later or not at all, and the magnitude of the delay can be quantified.
FIG. 7.
The time of replication of the DHFR locus in the IRKO deletion variant does not differ significantly from that observed in wild-type cells. (A) Principle of the FISH-based replication timing assay (see text). (B) Synchronized cells were sampled at 90, 180, 360, and 540 min after release from mimosine, fixed, and spread on microscope slides. The spreads were hybridized with a mixture of a rhodopsin (RHO) probe and a second probe from the DHFR locus (see figure and text), and the numbers of each of the patterns shown in panel A were determined by fluorescence microscopy for each cell line. A replication index for each sample was calculated as shown in panel A (J) and discussed in the text. Data were expressed either as a replication index as a function of time in the S period (B) or as percent nuclei with the locus replicated (C). The cosmids used to detect various parts of the DHFR domain are indicated below panel C.
As an example, the DR-8A7 and restored cell lines were subjected to this analysis in synchronized cell populations with cosmid KD504 (which is centered in the intergenic region) as a hybridization probe. The late-replicating phenotype of DR-8A7 detected by 2-D gel analysis is clearly recapitulated here, with the replication index of DR-8A7 approaching that of rhodopsin (i.e., unity) only after ∼540 min in the S period (Fig. 7B, compare to the restored cell line). When the cosmids cH1, KZ381, and KC393 were used to probe the IRKO cell line, which lacks >90% of the start sites utilized in wild-type cells, each of the regions represented by these cosmids can be seen to replicate at approximately the same time as the early replicating rhodopsin control. (Note that the DHFR origin appears to fire somewhat later than rhodopsin, even in the wild-type restored control, because we have scored rhodopsin as having replicated even if only one of the two loci has doubled [as in Fig. 7A, panels B and E]. If only those cells that had doubled both rhodopsin loci had been scored as having replicated, the single DHFR locus would appear to replicate earlier than rhodopsin.)
In Fig. 7C, the data for the IRKO and restored cell lines are expressed as percentages of cells that have duplicated a given marker as a function of time. This plot shows that all regions of the DHFR locus examined here replicated at approximately the same time in the IRKO cell line as in the wild-type restored control. (Note that there is no data point for the cosmid KD504 in the IRKO cell line, since the cognate sequences have been deleted.) This plot also shows that, although the DHFR origin is early firing in wild-type cells, it takes somewhere between 180 and 360 min to replicate all copies of the locus (as shown in Fig. 3E and F). The FISH data are consistent with the observation that the DHFR locus is often replicated passively by forks from distant origins at later times in the S phase. We have also examined each of the deletion variants in unsynchronized exponentially growing populations by the FISH-based timing assay. With the exception of DR-8A7, every deletion variant examined by this criterion replicates the DHFR locus at approximately the same time as in the wild-type restored control (Mesner, unpublished).
DISCUSSION
In most bacteria, plasmids, viruses, and yeast, well-defined genetic elements (replicators) serve as loading sites for the replication initiation complex, thereby directing nascent strand priming to positions very near the replicator itself. In cases where there is a single replicator for the entire genome (as in bacteria, plasmids, and most viruses), the genome is equivalent to a replicon, and with very rare exceptions, deletion of the replicator results in a failure to initiate DNA synthesis (reviewed in reference 42). In S. cerevisiae and Schizosaccharomyces pombe, the situation is somewhat more complex, because the genomes contain multiple linear chromosomes, each consisting of many replicons and each of which contains its own genetic replicator (defined originally as ARS elements) (11, 53). Replicators in S. cerevisiae are ∼100 bp in length and consist of a very highly conserved AT-rich ARS consensus sequence flanked by necessary but ill-defined auxiliary sequences (37, 51). S. cerevisiae origins are distributed at 30- to 40-kb intervals in the genome (50) but rarely in the bodies of genes (51, 60). In S. pombe, replicators are somewhat larger (∼500 bp long) and multipartite, with no obvious common sequence elements other than adenine tracts (38, 39).
Unlike bacteria, yeast can usually tolerate the deletion of one or more of these replicators because the region will eventually be synthesized passively by forks from active origins lying upstream or downstream (15). A similar situation characterizes ARS elements that are less than 100% active (which appears to be most of them) (51) and which must also be replicated passively in some cell cycles. One is led to the conclusion that there are usually no fixed termini between origins so that the boundaries of replicons are remarkably fluid (a notable exception is a replication fork barrier at the 3′ end of ribosomal DNA [rDNA] genes) (8).
The multiple origins in mammalian genomes are distributed at ∼15- to 300-kb intervals (32). With a few exceptions (for examples, see references 1, 29, and 55), most mammalian origins consist of broad zones of closely spaced, potential initiation sites rather than narrow zones closely flanking a replicator (see reference 19 for a review). Interestingly, homologues of most of the proteins that facilitate initiation at yeast ARS elements have been identified in mammalian cells (5, 28), but there is very little data on the nature of the sequences with which they interact. Within the DHFR initiation zone, ori-β, ori-β′, and ori-γ are clearly preferred initiation sites (Fig. 1A) and thus would be likely candidates to serve as replicators. However, they share no easily recognizable sequence motifs that could correspond to initiator recognition elements (P. J. Mosca, L. D. Mesner, and H.-B. Lin, unpublished data). Furthermore, the data presented here and in a previous study (36) show clearly that none of these sites is uniquely required for controlling initiation in this locus. Even when all three are deleted in the IRKO variant, the locus still replicates at approximately the same time in the S period. The latter observation rules out the possibility that ori-β, ori-β′, and ori-γ correspond to redundant master replicators that can take over in each other's absence. The lack of a significant effect of the 3′ deletion (Fig. 5D and E) on initiation also ruled out the possibility that a uniquely required element resides in the part of the spacer deleted in the origin-negative DR-8A7 cell line. The IRKO deletion also retains ∼5 kb downstream from the 2BE2121 gene that are normally used infrequently for initiation (Fig. 1A and E) (23) but which could theoretically contain a critical genetic element. Although we have not yet deleted this region, any critical element within it cannot be sufficient for initiation. This follows because this region is retained in the DR-8A7 and DG22 deletion variants, which lack the 3′ and 5′ ends of the DHFR gene, respectively, and whose origins are inactive (36; S. Saha, Y. Shan, L. D. Mesner, and J. L. Hamlin, unpublished data). Indeed, a distal ancillary element such as the locus control region identified in the β-globin domain (1a) could also reside either in the 5′ or 3′ regions deleted in DG22 and DR-8A7, respectively.
Because we have not removed the entire spacer in a single deletion variant, we have not formally ruled out the possibility that a small number of critical but redundant genetic elements reside within the origin itself. The minimum number would be three (i.e., one residing in the 40-kb core, one in the 11 kb immediately downstream from the DHFR gene, and a third in the 5-kb region downstream from 2BE2121). However, we believe that most of the observations made about this locus so far conform to a somewhat different model that relies in part on the yeast paradigm. In these simple eukaryotes, deletion of an active replicator from a chromosome can have two consequences: (i) the activity of a nearby less-active or silent replicator can increase because of relief of origin interference (9, 59), resulting in an insignificant change in replication timing of the locus, or (ii) the region has to wait to be replicated passively by forks from a distant active origin, which would result in a measurable change in replication timing (the magnitude depending upon the distance between the deletion and the nearest origin) (15, 51). Since we have not detected a significant change in replication timing as a consequence of any of the deletions in the DHFR origin, it appears that the region is being replicated from start sites in the immediate vicinity of each intergenic deletion. Coupled with the observation that initiation occurs at virtually every position tested in the spacer (whole or truncated), it is likely that potential replicators are distributed at very frequent intervals in the spacer, some of which are more active than others (at least 20 sites, but probably many more) (23).
It follows from our data that each of these proposed replicators must control initiation only in its immediate environment, since deletion of the prominent start sites has no apparent negative effect on initiation at other sites in the spacer. This again raises the question of whether there are bona fide replicators in mammalian genomes or whether initiation proteins can bind to large numbers of degenerate sequences almost at random, with epigenetic factors largely determining the efficiency of utilization. In this regard, mutagenesis studies on ectopically integrated copies of ori-β have shown that its activity as an origin can be partially suppressed by certain deletions near the peak of initiation shown in Fig. 1A (3). In this study, initiation activity was assessed by the PCR-based nascent strand abundance assay, which allowed relatively high resolution analysis of the region surrounding ori-β. In this experimental situation, there appear to be at least some sequence requirements for binding initiation complexes, in agreement with studies on the β-globin locus in human cells (1). However, sequence analysis of ∼30 kb of the intergenic region has uncovered no obvious simple repeated elements analogous to the ARS consensus sequence that could correspond to the 20 or more other potential replicators in the DHFR intergenic spacer (L. D. Mesner, A. Pemov, and P. J. Mosca, unpublished data) (also see reference 24). A fascinating question for the future is whether (and if so, how) the mammalian counterparts of replication initiation proteins that were originally identified in yeast recognize the large spectrum of initiation sites that characterize this and many other mammalian origins. It is conceivable that some or all of these proteins have lost much of their specificity for defined sequence elements during evolution.
Another surprising finding is that deletion of the 40-kb core of the spacer, in which the majority of initiation normally occurs, does not dramatically affect the time of replication of the remainder of the spacer nor the flanking genes (Fig. 7). The usual time of replication appears to be approximately maintained by an increase in the efficiency of initiation in the truncated spacer. Thus, regions that were not very active in the native locus (the outer edges of the spacer) assume a more-prominent role as substrates for initiation in the absence of the core. Somehow, loss of the central core is transmitted to the remaining sequences, probably by epigenetic factors. One obvious possibility is the closer juxtaposition of the two genes in the IRKO variant (i.e., reduction in the size of the intergenic spacer). In this regard, it is interesting that origins whose start sites or regions are thought to be very circumscribed (e.g., lamin B2 [29] and globin [1]) are situated in very narrow intergenic regions. Perhaps transcription itself or chromatin remodeling specific to active transcription units affects the activity of neighboring origins in the intergenic spacers. In fact, there is a precedent for the effects of transcription on replication initiation in the rDNA loci in developing Xenopus laevis (33). Prior to the mid-blastula transition when transcription commences, initiation sites are distributed throughout the rDNA repeats (including the inchoate genes). Once transcription begins, initiation is confined to the intergenic spacers. It is not clear whether transcription actually increases the efficiency of initiation in the spacer or merely prevents it in the body of active genes.
Another explanation for the apparent increase in the efficiency of initiation in the truncated IRKO spacer might be that there is a defined number of initiators for each permissive initiation zone, whether large or small, with the consequence that the concentration in a small zone would be higher. A third possibility is that initiation complexes take advantage of the open configurations of promoters to load onto the template, and are then delivered to intergenic origins either by looping, as has been suggested to explain the activities of enhancers (25, 47, 54), or by delivery to the spacer via the transcription machinery itself.
Acknowledgments
We thank Lawrence Chasin and Adelaide Carothers for providing us with the UA21 and DR-8A7 cell lines, respectively. We are also appreciative of the cheerful and expert technical assistance provided by Carlton White and Kevin Cox. We thank the other members of the Hamlin laboratory for excellent input during the course of this project.
This work was supported by NIH grant RO1 GM26108.
REFERENCES
- 1.Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrow, M. G., M. Ritzi, A. Pemov, and J. L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702-2708. [DOI] [PubMed] [Google Scholar]
- 3.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 8.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, B. J., and W. L. Fangman. 1993. Initiation at closely spaced replication origins in a yeast chromosome. Science 262:1728-1731. [DOI] [PubMed] [Google Scholar]
- 10.Burhans, W. C., L. T. Vassilev, M. S. Caddle, N. H. Heintz, and M. L. DePamphilis. 1990. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell 62:955-965. [DOI] [PubMed] [Google Scholar]
- 11.Chan, C. S., and B. K. Tye. 1980. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:6329-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 62:29-63. [DOI] [PubMed] [Google Scholar]
- 13.DePamphilis, M. L. 1993. Origins of DNA replication in metazoan chromosomes. J. Biol. Chem. 268:1-4. [PubMed] [Google Scholar]
- 14.DePamphilis, M. L. 1997. The search for origins of DNA replication. Methods 13:211-219. [DOI] [PubMed] [Google Scholar]
- 15.Dershowitz, A., and C. S. Newlon. 1993. The effect on chromosome stability of deleting replication origins. Mol. Cell. Biol. 13:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkwel, P. A., and J. L. Hamlin. 1988. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol. Cell. Biol. 8:5398-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkwel, P. A., and J. L. Hamlin. 1992. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol. Cell. Biol. 12:3715-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkwel, P. A., and J. L. Hamlin. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 15:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dijkwel, P. A., and J. L. Hamlin. 1996. Sequence and context effects on origin function in mammalian cells. J. Cell. Biochem. 62:210-222. [DOI] [PubMed] [Google Scholar]
- 20.Dijkwel, P. A., L. D. Mesner, V. V. Levenson, J. d'Anna, and J. L. Hamlin. 2000. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 256:150-157. [DOI] [PubMed] [Google Scholar]
- 21.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1991. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol. Cell. Biol. 11:3850-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1994. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 22:4989-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dijkwel, P. A., S. Wang, and J. L. Hamlin. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22:3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobbs, D. L., W. L. Shaiu, and R. M. Benbow. 1994. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 22:2479-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dworkin, J., A. J. Ninfa, and P. Model. 1998. A protein-induced DNA bend increases the specificity of a prokaryotic enhancer-binding protein. Genes Dev. 12:894-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foreman, P. K., and J. L. Hamlin. 1989. Identification and characterization of a gene that is coamplified with dihydrofolate reductase in a methotrexate-resistant CHO cell line. Mol. Cell. Biol. 9:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gale, J. M., R. A. Tobey, and J. A. D'Anna. 1992. Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J. Mol. Biol. 224:343-358. [DOI] [PubMed] [Google Scholar]
- 28.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 270:1667-1671. [DOI] [PubMed] [Google Scholar]
- 29.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross-Bellard, M., P. Oudet, and P. Chambon. 1973. Isolation of high-molecular-weight DNA from mammalian cells. Eur. J. Biochem. 36:32-38. [DOI] [PubMed] [Google Scholar]
- 32.Huberman, J. A., and A. D. Riggs. 1968. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 32:327-341. [DOI] [PubMed] [Google Scholar]
- 33.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 34.Jacob, F., and S. Brenner. 1963. Sur la regulation de la synthese du DNA chez les bacteriers: l'hypothese du replicon. C. R. Acad. Sci. 246:298-300. [Google Scholar]
- 35.Jin, Y., T. A. Yie, and A. M. Carothers. 1995. Non-random deletions at the dihydrofolate reductase locus of Chinese hamster ovary cells induced by alpha-particles simulating radon. Carcinogen 16:1981-1991. [DOI] [PubMed] [Google Scholar]
- 36.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 37.Kearsey, S. 1984. Structural requirements for the function of a yeast chromosomal replicator. Cell 37:299-307. [DOI] [PubMed] [Google Scholar]
- 38.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 39.Kim, S. M., and J. A. Huberman. 2001. Regulation of replication timing in fission yeast. EMBO J. 20:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588-590. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornberg, A., and T. A. Baker. 1992. DNA Replication. W. H. Freeman, New York, N.Y.
- 43.Leu, T. H., B. Anachkova, and J. L. Hamlin. 1990. Repetitive sequence elements in an initiation locus of the amplified dihydrofolate reductase domain in CHO cells. Genomics 7:428-433. [DOI] [PubMed] [Google Scholar]
- 44.Leu, T. H., and J. L. Hamlin. 1989. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol. Cell. Biol. 9:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linskens, M. H., and J. A. Huberman. 1990. Ambiguities in results obtained with 2D gel replicon mapping techniques. Nucleic Acids Res. 18:647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma, C., T. H. Leu, and J. L. Hamlin. 1990. Multiple origins of replication in the dihydrofolate reductase amplicons of a methotrexate-resistant Chinese hamster cell line. Mol. Cell. Biol. 10:1338-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meacock, S., R. Pescini-Gobert, J. F. DeLamarter, and R. Hooft van Huijsduijnen. 1994. Transcription factor-induced, phased bending of the E-selectin promoter. J. Biol. Chem. 269:31756-31762. [PubMed] [Google Scholar]
- 48.Mosca, P. J., P. A. Dijkwel, and J. L. Hamlin. 1992. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells. Mol. Cell. Biol. 12:4375-4383. (Erratum, 13:1981, 1993.) [DOI] [PMC free article] [PubMed]
- 49.Nawotka, K. A., and J. A. Huberman. 1988. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol. Cell. Biol. 8:1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newlon, C. S., T. D. Petes, L. M. Hereford, and W. L. Fangman. 1974. Replication of yeast chromosomal DNA. Nature 247:32-35. [DOI] [PubMed] [Google Scholar]
- 51.Newlon, C. S., and J. F. Theis. 1993. The structure and function of yeast autonomously replicating sequences. Curr. Opin. Genet. Dev. 3:752-758. [DOI] [PubMed] [Google Scholar]
- 52.Pelizon, C., S. Diviacco, A. Falaschi, and M. Giacca. 1996. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16:5358-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinchcomb, D. T., K. Struhl, and R. W. Davis. 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282:39-43. [DOI] [PubMed] [Google Scholar]
- 54.Talbot, D., and F. Grosveld. 1991. The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 10:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toledo, F., B. Baron, M. A. Fernandez, A. M. Lachages, V. Mayau, G. Buttin, and M. Debatisse. 1998. oriGNAI3: a narrow zone of preferential replication initiation in mammalian cells identified by 2D gel and competitive PCR replicon mapping techniques. Nucleic Acids Res. 26:2313-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trask, B. J. 1997. Fluorescence in situ hybridization. In B. Birren, E. Green, P. Hiater, and R. Myers (ed.), Genome analysis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Urlaub, G., and L. A. Chasin. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. USA 77:4216-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaughn, J. P., P. A. Dijkwel, and J. L. Hamlin. 1990. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell 61:1075-1087. [DOI] [PubMed]
- 59.Vujcic, M., C. A. Miller, and D. Kowalski. 1999. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol. Cell. Biol. 19:6098-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 61.Yokota, H., G. van den Engh, M. Mostert, and B. J. Trask. 1995. Treatment of cells with alkaline borate buffer extends the capability of interphase FISH mapping. Genomics 25:485-491. [DOI] [PubMed] [Google Scholar]