Abstract
The mouse mammary tumor virus (MMTV) promoter has been used as a model to study how the glucocorticoid receptor (GR) remodels chromatin to allow other transcription factors to bind and activate transcription. To dissect the precise role of nuclear factor 1 (NF1) in chromatin remodeling and transcriptional activation, we used linker-scanning mutants of transcription factor binding sites on the MMTV promoter. We compared the NF1 mutant MMTV promoter in the context of transiently transfected templates (transient transfection) and templates organized as chromatin (stable transfection) to understand the effect of chromatin on factor binding and transcription. We show that on a transiently transfected template, mutation in the NF1 binding site reduces both basal and hormone-dependent transcription. This suggests that NF1 is required for transcription in the absence of organized chromatin. We also found that binding of NF1 on a transiently transfected template is independent of mutation in hormone response elements or the octamer transcription factor (OTF) binding site. In contrast, the binding of OTF proteins to a transiently transfected template was found to be dependent on the binding of NF1, which may imply that NF1 has a stabilizing effect on OTF binding. On a chromatin template, mutation in the NF1 binding site does not affect the positioning of nucleosomes on the promoter. We also show that in the absence of NF1 binding, GR-mediated chromatin remodeling of nucleosome B is reduced and hormone-dependent activation of transcription is abolished. Further, we demonstrate that NF1 is required for both the association of BRG1 chromatin remodeling complex and the GR on the promoter in vivo. These results suggest the novel possibility that NF1 may participate in chromatin remodeling activities in addition to directly enhancing transcription and that in the absence of its binding site the GR is unable to effectively bind the promoter and recruit the remodeling complex.
The eukaryotic genome is structurally organized into nucleosomes to form chromatin (55). During gene expression, there is a need for distinct multiprotein complexes to modulate higher-order chromatin structure (20, 30), modify nucleosomal structures (45), and bind to regulatory sequences to initiate transcription. Sequence-specific DNA binding factors represent a large group of regulatory proteins that bind to the upstream regulatory regions of promoters and enhancer regions to mediate transcription (26). The organization of DNA in chromatin influences the ability of these factors to interact with their cognate recognition sites (51, 54). Steroid receptors are a class of transcription factors that can interact with the repressive chromatin structure and remodel the chromatin to allow other transcription factors to bind (29). The potential role of steroid receptors in chromatin remodeling and transcription is exemplified by the glucocorticoid receptor (GR)-mediated transactivation of the mouse mammary tumor virus (MMTV) promoter (17).
The MMTV promoter is organized into a phased array of six nucleosomes (A to F) when stably integrated into mammalian cells (22, 49). The second nucleosome (nucB) is positioned over the binding sites for the GR and nuclear factor 1 (NF1). Adjacent to nucB are the binding sites for octamer transcription factor (OTFs) and the TFIID complex (36). Binding of NF1 has been shown to be essential for GR-mediated transactivation of the MMTV promoter (7, 10, 13). In the absence of glucocorticoid, the chromatin structure of the promoter excludes the binding of NF1 (2). Upon hormone administration, GR recruits an ATP-dependent remodeling complex (BRG1-BAF or SWI-SNF) to remodel the chromatin (25, 41). The remodeling process converts the closed conformation of the MMTV promoter to an open one without altering the nucleosomal positioning. The remodeling of the promoter permits NF1 binding and the assembly of a transcription initiation complex (4, 17).
NF1 is a multifunctional transcription factor which displays the unusual property of regulating not only the initiation of transcription of viral and endogenous inducible promoters (12, 27, 37, 56) but also the regulation of DNA replication of several viral genomes (14, 15, 44). With respect to transcription, NF1 represents the archetypical transcription factor in GR-mediated transactivation of the MMTV promoter (24, 28). However, recent studies suggest that NF1 may contribute under specific circumstances to the MMTV promoter being maintained in an open conformation prior to hormonal stimulation (35, 43). This suggests that the function of NF1 may not be exclusively restricted to formation of the transcriptional initiation complex (7).
In this study, we analyzed the function of NF1 binding in GR-mediated activation of the MMTV promoter by comparing the contribution of NF1 binding to transiently transfected and stable (chromatin) templates. Previously, it has been shown that transiently transfected MMTV promoter does not assume the highly organized chromatin structure of stable replicating MMTV promoter (4). We used linker scanner (LS) mutants of the MMTV promoter with mutations in NF1, OTF, and GR binding sites. Our results show that binding of NF1 to the MMTV promoter is required for both transcription and chromatin remodeling processes. Furthermore, we demonstrate that NF1 participates in the recruitment of the BRG1 complex to the promoter and stabilization of the GR's association with the promoter in vivo. Thus, NF1 has a dual function in GR-mediated transactivation of the MMTV promoter: one as a classical transcription factor and another as a factor involved in the alteration of the chromatin.
MATERIALS AND METHODS
Plasmids.
Construction of LS mutants has been described previously (9, 10) (Fig. 1). For a wild-type plasmid, pLS-Wt or pMMTV-CAT (11) was used. The wild-type construct pLS-Wt was created by ligating a 4.5-kb SstI fragment of pmNF1 with a 0.75-kb SstI fragment of pmPhre. The pMMTV-CAT construct contains the MMTV promoter linked to the chloramphenicol acetyltransferase (CAT) gene, and the pLS-Wt and LS mutant constructs have the promoter fused to the thymidine kinase (TK) gene as reporters.
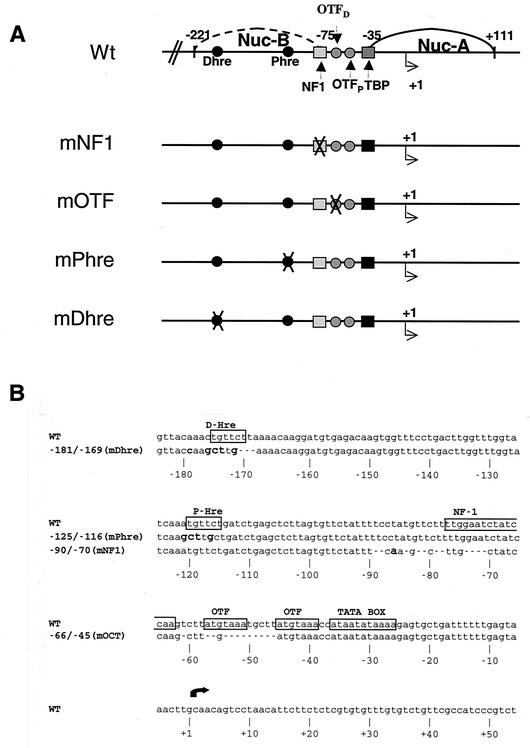
FIG. 1.
Schematic and sequence of the MMTV Wt and mutant promoter constructs. (A) Schematic of Wt and LS mutant constructs. In the wild-type pMMTV-CAT construct the MMTV promoter is fused to the coding region of a CAT gene, and the wild-type construct pLS-Wt and the LS mutant promoters are fused to the coding region of the herpesvirus TK gene. (B) Sequence of Wt and the LS mutant constructs. The LS mutant constructs used in this work include promoter constructs with mutations in the NF1 binding site (half site; mNF1), octamer binding site (mOTF), and proximal (mPhre) and distal (mDhre) HREs. The top line of the sequence represents wild-type sequences and the bottom line shows mutant sequences. Gaps in the mutant sequences denote deletions, and bold characters denote mismatches.
Cell culture and transfections.
C127 mouse mammary carcinoma cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 mg of penicillin-streptomycin/ml, and 10 mM HEPES. Cells were treated with a synthetic glucocorticoid, dexamethasone (DEX) (10−7 M). Transfections were performed by the calcium phosphate method or the Lipofectamine PLUS method (Invitrogen, Carlsbad, Calif.) (5, 36) as described by the manufacturer. For stable transfections, cells were cotransfected with promoter constructs (pLS-Wt or pmNF1 LS mutant constructs), and pRSV-neo and transformants were selected with 1,200 μg of G418 (Invitrogen)/ml. The positive clones were screened by PCR and assessed for their copy number.
RNA isolation, primer extension, and reverse transcriptase-PCR.
Cells were treated or untreated for 4 h with DEX (10−7 M), and total RNA was isolated using Trizol reagent (Invitrogen) as described by the manufacturer. Primer extension analysis of total RNA was performed with 32P-end-labeled specific oligonucleotide primers for MMTV mRNA (oligo-22, 5′TCT GGA AAG TGA AGG ATA AGT GAC GA 3′; bp +60 to +84) or 18S rRNA (oligo, 5′ TTA GCT TCC TTA GCT CCT GAA AAT 3′) as described previously (36). To assay transcription from a transient template, primer extension was performed on 100 μg of RNA treated with DNase I (Ambion, Austin, Tex.) for 30 min at 37°C. As a transfection control, a pGL3-control plasmid (Promega, Madison, Wis.) was cotransfected in these transfections, and the transcripts were assayed by reverse transcriptase reaction followed by PCR with luciferase-specific oligonucleotides as previously described (35) by using a 32P-end-labeled GL2 oligo (5′CTT TAT GTT TTT GGC GTC TTC CA 3′) and GLR oligo (5′ CTA GGC TTT TGC AAA AAG CTT GG 3′). Primer extension products and PCR products were analyzed on 8% polyacrylamide denaturing gels and exposed to PhosphorImager screens for analysis.
In vivo analysis of transcription factor loading.
For transcription factor loading on a transient template, nuclei were isolated as previously described (3) and digested with DraI (1,000 U/ml) and exonuclease III (1,000 U/ml) to detect 5′ boundaries of the transcription factors on the MMTV promoter. DNA was purified, and single-stranded overhangs were removed with mung bean nuclease. All samples were digested to completion with AlwNI prior to reiterative primer extension with Taq DNA polymerase and a 32P-labeled oligo-22 oligonucleotide. For detecting the 3′ boundaries of the transcription factors on the MMTV promoter, the nuclei were digested with HpaII (2,000 U/ml) and exonuclease III (1,000 U/ml). After the removal of single-strand overhangs with mung bean nuclease, the samples were digested to completion with HaeIII and the samples were amplified by reiterative primer extension with 32P-labeled oligo-38 oligonucleotide (5′ GGT TTA AAT AAG TTT ATG GTT ACA AAC TG 3′). The purified extension products were analyzed on an 8% polyacrylamide denaturing gel.
For transcription factor loading on a stable template, isolated nuclei were digested with SstI (3,000 U/ml) and lambda exonuclease (300 U/ml) as previously described (42). The samples were purified and digested to completion with AlwNI prior to reiterative primer extension with a 32P-labeled oligo TK-1 (5′AAG CCA TAC GCG CTT CTA CAA G 3′; bp +170 to +195). Extension products were purified and analyzed on an 8% polyacrylamide denaturing gel.
In vivo restriction enzyme hypersensitivity analysis.
Isolated nuclei were digested with DraI (1,000 U/ml), SstI (100 U/ml), or FokI (400 U/ml). DNA was purified and then digested to completion with AlwNI (for DraI- and SstI-treated DNA) and SstI (for FokI-treated DNA). The samples were analyzed by reiterative primer extension with a 32P-labeled oligo TK-1, and the products were separated on 8% polyacrylamide denaturing gels.
MNase analysis.
Isolated nuclei were resuspended in 200 μl of wash buffer containing 1 mM CaCl2 and digested with 0 to 75 U of micrococcal nuclease (MNase; Worthington Biochemicals, Lakewood, N.J.)/ml for 5 min at 30°C (16). The reaction was stopped by adding 40 μl of 100 mM EDTA (pH 8.0), 10 mM EGTA (pH 7.5). The samples were purified and digested to completion with BamHI prior to Southern blotting or reiterative primer extension. Control genomic DNA was prepared by digesting purified genomic DNA with 1 U of MNase/ml for 5 min at 30°C and redigested with BamHI. Twenty micrograms of MNase-digested DNA was separated on a 1.5% agarose gel and transferred to Hybond N+ membrane (Amersham-Pharmacia, Piscataway, N.J.). The blot was probed with a 120-bp BamHI-FspI fragment, which was radiolabeled with 32P by random priming (Ready-to-go beads; Amersham-Pharmacia). To resolve the micrococcal products with high resolution, reiterative PCR was performed on these samples with Taq DNA polymerase and 32P-labeled oligo-22. The products were separated on an 8% polyacrylamide denaturing gel.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis was carried out using the ChIP assay kit from Upstate Biotechnologies with minor modifications of the protocol. Two million cells were plated on 100-mm plates and treated the next day with DEX for 1 h. The cells were lysed, the DNA was fragmented by sonication, and 10 μl of the chromatin solution was saved as input. Ten micrograms of antibody (anti-BRG1 [Santa Cruz Biotech], anti-GR [made from myeloma cells; FIGR; American Type Culture Collection]), mouse immunoglobulin G (IgG; Santa Cruz Biotech), or rabbit IgG (Santa Cruz Biotech) was added to tubes containing 900 μl of chromatin solution. Following incubation with antibody, the antibody complexes were captured using protein A-agarose beads. The beads were pelleted and washed. The chromatin was extracted and reverse cross-linked, and the DNA was purified. PCR was performed on these DNA samples using oligo-38 and 32P-end-labeled oligo-22 and analyzed on 6% nondenaturing polyacrylamide gels. For Wt and mNF1 input samples, 1/100 and 1/800, respectively, of the total sample was used to reflect the fact that the Wt cell line contained five copies and the mNF1 cell line contained 80 copies of the MMTV promoter per genome. For Wt and mNF1 immunoprecipitated samples, 1/10 and 1/80, respectively, of the total sample were used. Following electrophoresis the gels were exposed to PhosphorImager screens and analyzed with ImageQuant software.
RESULTS
In vivo binding of NF1 to transiently transfected LS mutant and wild-type MMTV promoter plasmids.
Transcriptional activation of the MMTV promoter is accomplished by the action of a variety of transcription factors that are thought to act cooperatively. To begin to understand the interplay between these various factors, we have examined the binding of NF1 to a transiently transfected MMTV promoter that is devoid of binding sites for a subset of transcription factors. In the first set of experiments we examined NF1 binding in vivo, on a transient template, in the absence of several cis elements on the promoter by using LS mutant constructs (9) (Fig. 1A). These mutants were directed against the NF1 binding site (mNF1), one OTF site (mOTF), the proximal hormone response element (HRE) (mPhre), and the distal HRE (mDhre). The mPhre has a point mutation, while the mDhre, mNF1, and mOTF constructs have 3-, 11-, and 12-bp deletions, respectively (Fig. 1B).
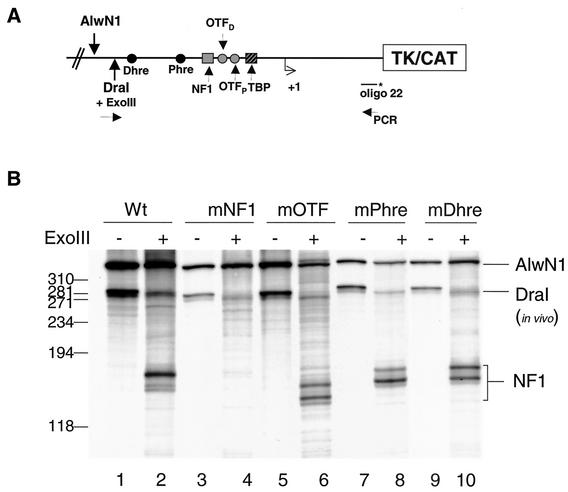
As anticipated, the exonuclease III-in vivo footprinting assays detected the binding of NF1 on the wild-type promoter (Fig. 2B, lane 2) but not the NF1 mutant (Fig. 2B, lane 4). In contrast, NF1 binding appeared independent of binding of the OTF proteins and the mutation in either of the HREs, as indicated by binding of NF1 at the mOTF, mPhre, and mDhre templates (Fig. 2B, compare lanes 2, 6, 8, and 10). In addition, and consistent with previous reports, the NF1 binding was independent of hormone treatment when binding on transient templates was examined (data not shown) (4). These assays established that NF1 binding was not influenced by the loss of binding sites to other transcription factors within the MMTV promoter.
FIG. 2.
Binding of NF1 on a transient template does not require the binding of OTFs or the HREs. MMTV constructs were transfected into C127 cells for 24 h and nuclei were isolated. The nuclei were digested with DraI and ExoIII, and the purified DNA was then digested to completion by AlwNI. Ten micrograms of DNA was amplified by reiterative PCR using Taq polymerase and 32P-end-labeled oligo-22. The PCR products were separated on an 8% denaturing polyacrylamide gel. (A) Schematic of the MMTV promoter as a transient template showing the restriction enzyme and PCR primer sites. (B) Samples from Wt (pMMTV-CAT), mNF1, mOTF, mPHre, and mDHre LS mutant construct transfections. The 5′ boundary of NF1 is indicated.
Mutation of the NF1 binding sites results in the loss of OTF binding to the MMTV promoter.
Having established that NF1 binding was not dependent on the binding of OTF proteins, we next asked if OTFs could bind in the absence of a functional NF1 binding site. The organization of the MMTV promoter is such that the binding sites for the TATA binding protein (TBP) and OTFs are 3′ to the binding site of NF1 (Fig. 2A). When in vivo footprinting by ExoIII is performed in a direction so that NF1 is the first factor in a series of factors, a single predominant boundary corresponding to NF1 is observed (3). The NF1 protein has a high affinity for its binding site, which creates a barrier for the ExoIII digestion. This is evident in the Wt samples where we cannot footprint any other factors (OTFs and TBP) from this direction of ExoIII digestion (Fig. 2B, lane 2). Conversely, one would expect to detect the presence of OTF boundaries in the absence of NF1 binding, i.e., in the mNF1 template (Fig. 2B, lane 4). We observed no boundaries for OTFs in the mNF1 template. This suggests that NF1 stabilizes the interaction of OTFs to their binding sites on the promoter.
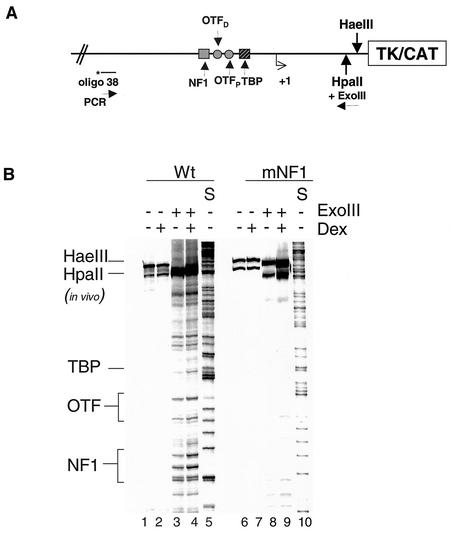
To support this hypothesis, we performed a second ExoIII-in vivo footprinting experiment. It was observed previously that, in contrast to NF1, OTF and TBP are weak binders and hence do not impede the digestion of template by ExoIII (36). Since TBP and OTFs bind downstream of the NF1 site, ExoIII digestion from an entry site downstream of the OTF and TBP binding site enables us to detect all three factors binding to the promoter (Fig. 3A). In the case of the Wt transient template, the 3′ boundaries of NF1 and OTFs are evident in both the presence and absence of hormone treatment (Fig. 3B, lanes 3 and 4). Binding of TBP shows the expected hormone-dependent loading on the Wt template. However, on an mNF1 transient template, there is no detectable binding of the OTFs or TBP proteins (Fig. 3B, lanes 8 and 9). Again, this suggests that binding of the OTF proteins may require an intact NF1 binding site within the promoter.
FIG. 3.
Binding of NF1 to the MMTV promoter is required for the binding of OTF proteins. MMTV constructs (pMMTV-CAT and mNF1 mutant) were transfected into mouse C127 cells for 24 h, and the cells were treated with DEX for 1 h. Isolated nuclei were subjected to limited digestion with HpaII and ExoIII, and the purified DNA was digested to completion with HaeIII. Reiterative PCR was performed with 20 μg of these samples by using 32P-labeled oligo-38. The PCR products were separated on an 8% denaturing polyacrylamide gel. (A) Schematic of the MMTV promoter as a transient template showing the restriction enzyme and PCR primer sites. (B) Samples from Wt (pMMTV-CAT) and mNF1 construct transfections. Lanes 5 and 10 are G sequencing tracks. The 3′ boundaries of NF1, OTF, and TBP are indicated.
NF1, OTF, and Dhre mutants show a similar decline in hormone-inducible expression.
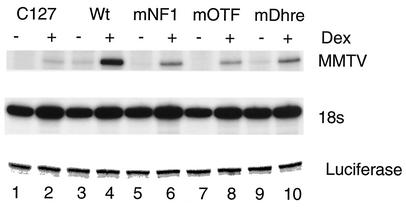
In the next series of experiments, we examined the role of NF1 binding in MMTV transcription by using the promoters at which NF1 binding had been studied. We evaluated the requirement for NF1 in the context of the full promoter (Wt), in the absence of OTF, or the GR at the distal HRE, and also in the absence of NF1's cognate binding site. In the C127 cells transfected with pLS-Wt, there is a low level of MMTV transcript expressed in the absence of the hormone and there is a marked increase in expression upon hormone treatment, as assayed by primer extension (Fig. 4, lanes 3 and 4). In the mNF1 mutant-transfected cells, there is a lower level of both basal and induced MMTV RNA (Fig. 4, compare lanes 4 and 6). Although the mNF1 mutant promoter is still hormone inducible, the induced level of expression is 30% of that of the Wt promoter (Fig. 4, lanes 5 and 6). There are similar reductions in both the basal and hormone-induced levels of MMTV transcripts in mOTF and mDhre mutant template-transfected cells (Fig. 4, lanes 8 to 10), suggesting that they are required for transcription on the MMTV promoter. We also observed the presence of the MMTV transcript in C127 cells (Fig. 4, lanes 1 and 2) since they have endogenous copies of MMTV in their genome.
FIG. 4.
Expression from a transient MMTV template in the absence of NF1 binding is reduced. MMTV constructs (pLS-Wt and LS mutant constructs) along with a pGL3-control plasmid were transfected into C127 cells for 24 h. The cells were treated with DEX for 4 h and RNA was isolated. (A) Primer extension was performed with 100 μg of total RNA with oligo-22 and with 2 μg of RNA with an 18S-specific primer. For transfection efficiency, message from pGL3-control (luciferase) was analyzed by PCR with primer sets (GLR and GL2) using first-strand cDNA. The products were run on 8% denaturing polyacrylamide gels.
Chromatin remodeling of MMTV promoter requires the binding of NF1.
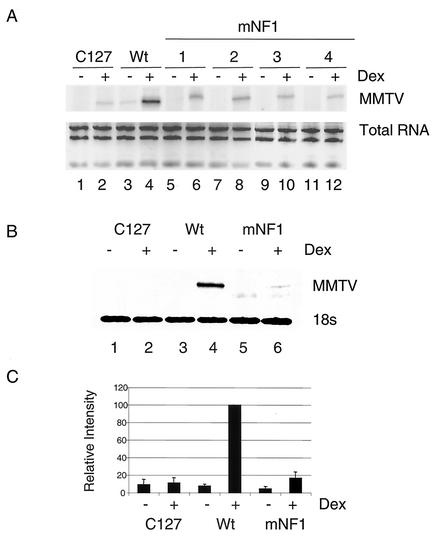
Previous analyses have demonstrated that transiently transfected templates do not fully replicate the GR transactivation process, as it occurs on promoters within chromosomes (17). Specifically, these templates do not require chromatin remodeling to allow binding of transcription factors such as NF1 and OTF (36). To investigate the function of the NF1 mutant within the context of chromatin, we established stable lines containing pLS-Wt and mNF1 LS mutant constructs in a C127 cell line. Figure 5 shows the results of an assay with Wt- and mNF1-containing clones for MMTV expression after DEX treatment by using primer extension. As expected, there was a marked reduction in MMTV message after DEX treatment in all four clones (number 1 to 4) containing the mNF1 construct (Fig. 5A). In Fig. 5B and C, we show MMTV expression as assayed in cells containing Wt and mNF1 (clone 1) constructs, which were used for further studies (see Fig. 6, 7, and 8). The mNF1 construct-containing cell line (clone 1) has 15% of the induced level of MMTV mRNA of the Wt construct-containing cell line (Fig. 5B and C). We also determined the number of copies of the MMTV promoter per genome in Wt and mNF1 to be 5 and 80, respectively (data not shown).
FIG. 5.
Expression from a stable MMTV promoter requires the binding of NF1. RNA was isolated from C127, stable clones carrying Wt (pLS-Wt), and NF1 LS mutant promoters, which were treated with DEX for 4 h. (A) Primer extension was performed with 20 μg of total RNA from cell lines containing Wt and mNF1 clones (no. 1 to 4) with oligo-22. The products were run on 8% denaturing polyacrylamide gel. Total RNA samples were run on 1% agarose-Tris-borate-EDTA gel and stained with ethidium bromide. (B) Primer extension was performed with total RNA from Wt and mNF1(no. 1) cell lines with oligo-22 (20 μg of RNA) and 18S-specific primer (2 μg of RNA). (C) Values represent the average of three independent experiments (shown in panel B) after normalization with 18S product. The induced level of expression from the Wt promoter was set to 100.
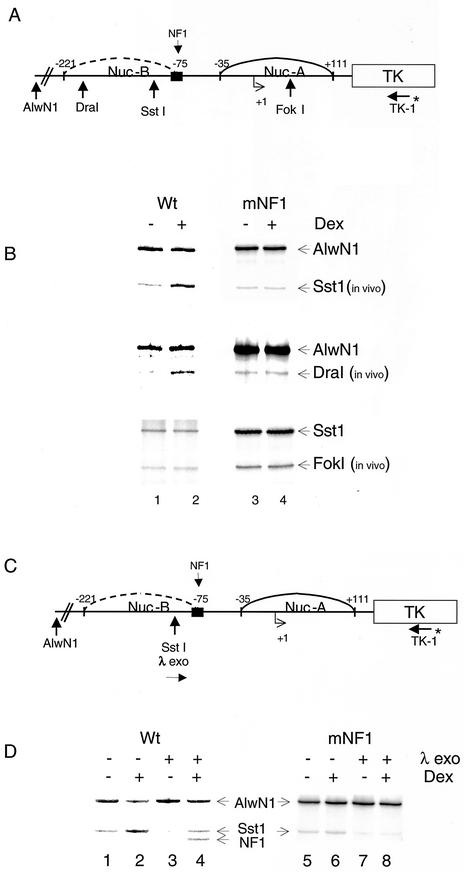
FIG.6.
Chromatin remodeling of the MMTV promoter requires the binding of NF1. (A) Schematic of the MMTV promoter as a stable template, indicating the sites for restriction enzyme and PCR primer. (B) Stable cell lines containing Wt (pLS-Wt) or mNF1 constructs were treated with DEX for 1 h. Isolated nuclei were subjected to limited digestion with restriction endonucleases (DraI, SstI, and FokI). Purified DNA was digested to completion with AlwNI (for DraI- and SstI-digested samples) and with SstI (for FokI samples). Twenty micrograms of DNA was amplified by reiterative primer extension using Taq DNA polymerase and 32P-labeled oligo TK-1. The PCR products were then separated on 6% denaturing polyacrylamide gels. (C) Schematic of MMTV promoter as a stable template showing the entry site for λ exonuclease, sites for restriction enzymes, and PCR primer. (D) As described in the legend for panel B, isolated nuclei were subjected to limited digestion with SstI with or without λ exonuclease, and purified DNA was digested to completion with AlwNI. Twenty micrograms of DNA was amplified by reiterative primer extension by using Taq DNA polymerase and 32P-labeled oligo TK-1, and the products were analyzed on a 6% polyacrylamide gel. The 5′ boundary of NF1 is indicated.
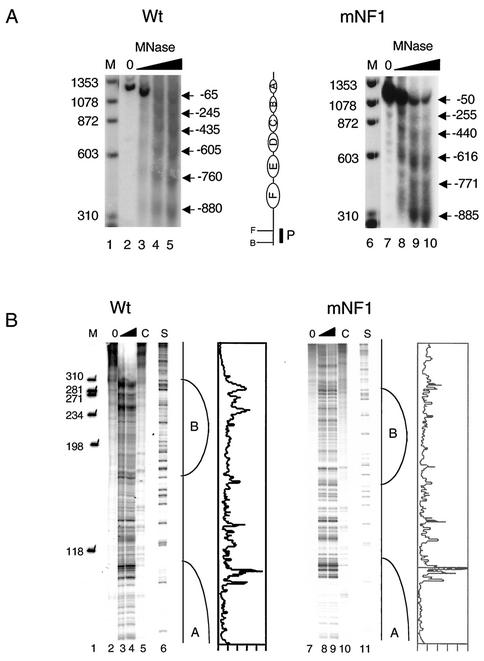
FIG. 7.
Organization of the nucleosome on the MMTV promoter is not altered in the absence of NF1 binding. Nuclei were isolated from stable cell lines containing Wt (pLS-Wt) or mNF1 constructs and digested with MNase (0 to 75 U/ml), and the purified DNA was digested to completion with BamHI. (A) Southern blot hybridization of DNA treated with increasing amounts of MNase and probed with a 32P-radiolabeled 120-bp BamHI-FspI fragment. Sizes of the bands are represented with reference to +1 of the MMTV promoter. The schematic represents the position of six nucleosomes (denoted A to F), restriction sites, and position of the probe used for Southern blot hybridization (B and F represent BamHI [−1208] and FspI [−1096], respectively). (B) Reiterative PCR with MNase-treated DNA of Wt- and mNF1-containing cells with 32P-labeled oligo-22. Lanes 5 and 10 (denoted with C) indicate genomic DNA digested with MNase. Lanes 6 and 11 are G sequencing tracks. The nucleosome boundaries (A and B) are indicated. Line graphs, representing Wt (lane 4) and mNF1 (lane 9) band intensity, were created using the Molecular Dynamics PhosphorImager software.
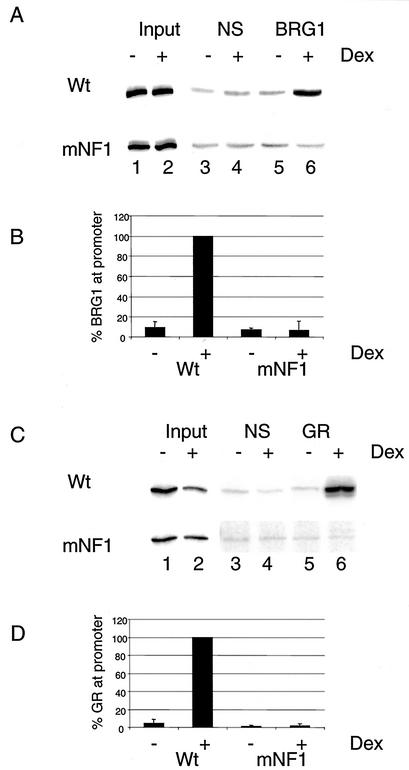
FIG. 8.
BRG1 and GR do not associate with the promoter in the absence of NF1 binding. Cells from stable cell lines containing Wt (pLS-Wt) or mNF1 constructs were treated without or with DEX for 1 h and analyzed by ChIP assay with BRG1 and GR antibodies. The cells were sonicated and immunoprecipitated with nonspecific (rabbit IgG) or BRG1 antibodies (A) or nonspecific (mouse IgG) or GR antibodies (C). The MMTV promoter sequence was detected by PCR using oligo-38 and 32P-end-labeled oligo-22. The PCR products were run on nondenaturing polyacrylamide gels and exposed to PhosphorImager screens for further analysis. The association of BRG1 (B) or GR (D) with Wt and NF1 templates was quantified from two independent experiments and by setting the association with the wild-type promoter in the presence of DEX to 100.
We wanted to examine the effect of mutation in the NF1 binding site on chromatin remodeling, since in the absence of NF1 binding transcription is eliminated from a stable template. Within the MMTV promoter, the second nucleosome (nucB) encompasses the binding site of NF1, as well as the binding sites for GR. The glucocorticoid-mediated disruption of nucB results in access of the transcription factors to the promoter and renders the promoter hypersensitive to digestion by restriction endonucleases in this region (Fig. 6A). Exposure of the pLS-Wt cells to DEX for 1 h resulted in chromatin remodeling of the MMTV promoter, as assayed by in vivo restriction endonuclease hypersensitivity (Fig. 6B). For this assay, we used DraI and SstI enzymes that cleave in the nucB region of the MMTV promoter (Fig. 6B, lanes 1 and 2). In contrast to the Wt promoter and congruent with the expression profile, on the mNF1 template there is no increase in hypersensitivity upon DEX treatment (Fig. 6B, lanes 3 and 4). This suggests that binding of NF1 is required for GR-mediated chromatin remodeling of the MMTV promoter of the nucB region. To investigate the effect of NF1 mutation on other parts of the MMTV promoter, we used FokI enzyme, which cleaves in the nucA region (+50) (23). There was no difference in cleavage upon hormone treatment in both the Wt and the mNF1 promoter-containing cells (Fig. 6B, compare lanes 1 and 2 with lanes 3 and 4 for FokI digestion). We also confirmed these results using three additional clones containing the mNF1 construct (data not shown).
Coincident with the GR-induced remodeling of the nucB region of the MMTV promoter is the hormone-dependent formation of a preinitiation complex (PIC), which can be seen by examining loading of NF1 using in vivo foot printing analysis. For this assay we used λ exonuclease and SstI as the entry site enzyme (Fig. 6C). NF1 binding was observed on Wt templates in the presence of hormone (Fig. 6D, lane 4). As predicted, there was no binding of NF1 on the mNF1 promoter (Fig. 6D, lane 8).
Mutation in the NF1 binding site does not alter nucleosomal organization of the promoter.
When introduced as a stable template, the MMTV promoter assumes a phased array of six nucleosomes (Fig. 7A) (22, 49). We wanted to determine if the mutation in the NF1 binding site, specifically the 11-bp deletion in creating the mNF1 template, had any effect on the positioning of these nucleosomes. To determine the nucleosomal organization within these cells, we used MNase digestion in combination with Southern blotting and reiterative primer extension. In this way, we were able to obtain both a low- and a high-resolution mapping of the promoters. Regularly positioned nucleosomes occupied both the Wt and the mNF1 stable promoters, as seen with Southern blotting (Fig. 7A). To confirm the nucleosome positions identified by Southern blotting, finer PCR-based mapping of the nucA-B junction was carried out (Fig. 7B). We did not find any difference in the digestion pattern between the Wt and mNF1 templates. This suggests that the binding of NF1 is not required for the nucleosomal organization of the promoter.
The NF1 binding site is required for GR-mediated BRG1 recruitment to the MMTV promoter.
Previously, our laboratory have established that GR-mediated chromatin remodeling of the MMTV promoter requires the participation of the BRG1 complex in vivo (25). The preceding data establish that the binding of NF1 to the promoter is also required for chromatin remodeling. In the next set of experiments, we sought to examine potential connections between NF1 binding and the recruitment of the BRG1 complex. We used a ChIP assay with an antibody specific to BRG1 to investigate the association of the BRG1 complex with the mutant promoter. In the cells containing the Wt stable promoter, BRG1 associated with the promoter in a hormone-dependent manner (Fig. 8A, lanes 5 and 6), leading to an approximately 10-fold increase in the association of BRG1 with the Wt promoter upon hormone treatment (Fig. 8B). Interestingly, we did not see any specific association of the BRG1 protein on the NF1 mutant promoter (Fig. 8A, lanes 5 and 6, and B). These results demonstrate that the lack of chromatin remodeling and transcriptional activation seen when the NF1 site is mutated may result, at least in part, from the failure to effectively recruit the BRG1 complex to the promoter.
NF1 is required for the association of GR on the MMTV promoter.
Since BRG1 could not associate with the MMTV promoter in the absence of NF1 binding, we next wanted to examine the association of GR with this template. To investigate the association of GR with the Wt and mNF1 stable promoters, we used a ChIP assay with an antibody specific to GR. As expected, in cells containing Wt stable promoter a hormone-dependent association of GR can be observed (Fig. 8C, lanes 5 and 6, and D). Similar to what we observed with the recruitment of BRG1, we failed to see any specific association of GR on the mNF1 stable promoter (Fig. 8C, lanes 5 and 6, and D). These results suggest that an intact NF1 binding site is also required for the stable interaction of GR on a chromatin MMTV promoter and are thus consistent with the failure to recruit the chromatin remodeling complex and efficiently initiate transcription.
DISCUSSION
The MMTV promoter has been extensively studied as a model for GR-mediated chromatin remodeling and transactivation. Upon ligand binding, the GR facilitates binding of transcription factors, such as NF1, OTFs, and TBP, to the promoter by remodeling the chromatin. The binding site of NF1 has been shown to have a dual function in the transcriptional stimulation by GR: one as an upstream element for basal, hormone-independent promoter activity and another as an element which interacts synergistically with the HREs in a hormone-dependent promoter activity. Although NF1 has been shown to be important for GR-mediated transactivation of the MMTV promoter, it remains unclear whether the transactivation function of NF1 is a result of its contribution to the chromatin remodeling process that precedes transcription. To dissect the role of NF1 in the two processes, chromatin remodeling and transcription (recruitment of PIC), we analyzed the contribution of NF1 towards transcription and DNA binding in the absence of an organized chromatin structure, i.e., a transiently transfected template, and in the presence of chromatin, i.e., a stable template. Transiently transfected DNA does not assume the highly organized chromatin structure characteristic of stable, replicating templates.
On a transient template, we demonstrated that in vivo binding of NF1 was not dependent on the binding of GR to the distal or proximal HREs. We also showed that a mutation in the NF1 binding site destabilized the interaction of OTFs to its sites. This is consistent with the results from an in vitro DNase I footprinting experiment with these LS mutants, which showed that NF1 stabilized the interaction of OTFs to its sites (8). To correlate factor binding with transcription, we also showed that a mutation in the NF1 binding site resulted in decreased transcription from a transient template. Similarly, a mutation in one of the OTF binding sites resulted in reduced expression (both basal as well as induced) from the promoter. This is consistent with previous work showing that OTFs play a role in the basal transcription machinery (33, 48). Because our experiments indicate that NF1 might stabilize the binding of the OTFs on the promoter, it is not clear if the mutation in the NF1 site has a direct effect on transcription or if the reduction in transcription is due to the absence of OTFs on the promoter. Nevertheless NF1 seems to play a role in transcription on a transient MMTV template, which suggests that NF1 contributes to transcription in the absence of chromatin remodeling.
When the MMTV template was organized within chromatin, we showed that NF1 binding is required for GR-dependent transcription. Consistent with the requirement for transcription, we also demonstrated that binding of NF1 is required for GR-mediated chromatin remodeling in vivo. As a control, we determined the nucleosomal positioning on the mutant promoter to confirm that the 11-bp deletion in the NF1 mutant promoter is not the cause of altered chromatin remodeling in our experiments. Indeed, deletion of 11 bp of DNA would not be expected to change nucleosomal organization, since this would not disturb the helical periodicity of the DNA. Our experiments demonstrate no alteration in nucleosomal positioning on the mutant promoter. Taken together, our results suggest that the binding of NF1 to the promoter is required for GR-mediated chromatin remodeling but is not needed for the spatial organization of nucleosomes at the promoter. This is consistent with the recent description in Saccharomyces cerevisiae of synergism between the DNA binding domain of NF1 and the GR, which is necessary for chromatin remodeling of the MMTV promoter (47).
To gain mechanistic insight into the inability of the GR to remodel the mNF1 promoter as chromatin, we examined the hormone-dependent recruitment of GR and BRG1 to the promoter. Unexpectedly, these studies demonstrated that the association of BRG1 and the GR itself with the promoter required the NF1 binding site. We hypothesize that this requirement for NF1 binding may be to facilitate or stabilize GR interactions with the promoter. Although it is well known that on a stable template NF1 requires GR to remodel the chromatin to bind to its site, our experiments suggest that simultaneous participation of NF1 is required for GR to associate with the promoter, to recruit BRG1 remodeling complex, and to initiate transcription. We propose that either (i) there is cooperativity between NF1 and the GR such that NF1 is required for the GR to functionally interact with the chromatin or (ii) that NF1 is required to stabilize an open conformation of nucB required for efficient binding of the BRG1 complex. These possibilities are compatible with a prior proposal that NF1 may stabilize progesterone receptor binding to the MMTV promoter assembled into minichromosomes in vitro and promote the efficient binding of further receptor molecules (19).
Previous studies support a direct role for NF1 in the chromatin remodeling process that is initiated by the GR (31). The fact that NF1 binds to a transient template and open chromatin template constitutively suggests that binding of NF1 to the promoter is tightly linked to the promoter being in a remodeled or open configuration (4, 35). NF1 is also known to bind other inducible cellular promoters that employ chromatin remodeling for their activation, such as the dioxin-responsive CYP1A1 promoter (56). The most direct support of this concept comes from the recent observations linking NF1 in the recruitment of the BRG1 complex to the CSF-1 promoter (37). Interestingly, while the case for NF1 participating in the remodeling process is strong, in vitro studies suggest that the GR can remodel chromatin with the SWI/SNF complex in the absence of NF1 (21, 46). This apparent difference between the in vitro and in vivo assays may reflect, in part, distinct experimental conditions employed with respect to DNA and protein concentrations and the participation of other molecules. Alternatively, it may indicate support for the hypothesis that NF1 functions as an architectural protein by stabilizing the binding of GR and BRG1 complex on the promoter or by anchoring the promoter in a remodeled state in vivo. Indeed, while in vivo the GR is known to bind the promoter transiently and a cycling mechanism on the promoter has been proposed, GR dynamics in vitro have not been as well described (38).
Several possibilities can be advanced to explain the stabilizing and cooperative effect of NF1 on the GR-mediated chromatin remodeling process. (i) Binding of NF1 to the promoter may anchor the complex to the nuclear matrix. NF1 and OTF have been shown to have affinity to the nuclear matrix (34, 50). It is very well characterized that adenoviral DNA replication occurs at distinct subnuclear foci, suggesting a potential link to nuclear matrix (14). The nuclear matrix is considered to be a scaffold to which the replication proteins are recruited. The nuclear matrix affinities of NF1 and OTF may also contribute to the increase in the stability of the PIC (39). The GR has also been shown to interact with the nuclear matrix (18, 32). (ii) NF1 may stabilize the remodeling complex on the promoter by interacting with core histones. NF1 was shown to interact with histone H3 and was suggested to restructure chromatin of target promoters and replication origins (1, 40). (iii) Binding of NF1 to the promoter may stabilize the chromatin remodeling process by removal of the repressive linker H1 histone (6). The position of NF1 binding site on the MMTV promoter is near the 3′ boundary of nucB, which is in close proximity to the histone H1 binding site. Therefore, there may be a competition between H1 and NF1 binding to the promoter.
In vivo analyses of MMTV promoters with and without an intact NF1 binding site suggest that NF1 acts as a dual-function transcriptional activator. It possesses a significant transcriptional activation potential and works in synergy with the GR and other transcription factors. In addition, it may play an architectural role in the chromatin remodeling of the MMTV promoter by stabilizing or facilitating the association of the BRG1 complex and GR with the promoter. This is particularly exciting as it defines a new role for NF1 in the GR-mediated transcriptional activation of the promoter. Previously, NF1 was envisioned to act downstream of the remodeling process such that the GR would recruit the remodeling complex to the promoter and remodel chromatin, which would then allow NF1 and the PIC to assemble and initiate transcription. These new data demonstrate that NF1 is required for the initiating step of chromatin remodeling, and in the absence of its binding site the GR is unable to effectively bind and recruit the remodeling complex to the promoter. In this way, these data are consistent with the growing body of evidence that intimately links the remodeling and transcriptional activation properties of nuclear receptors (52, 53). The capacity to assess the contributions of an organized chromatin structure to hormone-mediated transcriptional activation of the MMTV promoter should facilitate an evaluation of the precise mechanism(s) by which NF1 contributes to the GR-mediated recruitment of the BRG1 complex and chromatin remodeling process.
Acknowledgments
We thank E. Buetti for providing the LS mutant constructs. We are also grateful to B. Deroo and J. Mason for critical review of the manuscript and members of the Chromatin and Gene Expression Section, Laboratory of Reproductive and Developmental Toxicology, for helpful suggestions.
REFERENCES
- 1.Alevizopoulos, A., Y. Dusserre, M. Tsai-Pflugfelder, T. von der Weid, W. Wahli, and N. Mermod. 1995. A proline-rich TGF-beta-responsive transcriptional activator interacts with histone H3. Genes Dev. 9:3051-3066. [DOI] [PubMed] [Google Scholar]
- 2.Archer, T. K., M. G. Cordingley, R. G. Wolford, and G. L. Hager. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol. 11:688-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, T. K., and H. L. Lee. 1997. Visualization of multicomponent transcription factor complexes on chromatin and nonnucleosomal templates in vivo. Methods 11:235-245. [DOI] [PubMed] [Google Scholar]
- 4.Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee, R. N., G. C. Banks, K. W. Trotter, H. L. Lee, and T. K. Archer. 2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21:5417-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnick, E. H., M. Bustin, V. Marsaud, H. Richard-Foy, and G. L. Hager. 1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 20:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruggemeier, U., L. Rogge, E. L. Winnacker, and M. Beato. 1990. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 9:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buetti, E. 1994. Stably integrated mouse mammary tumor virus long terminal repeat DNA requires the octamer motifs for basal promoter activity. Mol. Cell. Biol. 14:1191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buetti, E., and B. Kuhnel. 1986. Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J. Mol. Biol. 190:379-389. [DOI] [PubMed] [Google Scholar]
- 10.Buetti, E., B. Kuhnel, and H. Diggelmann. 1989. Dual function of a nuclear factor I binding site in MMTV transcription regulation. Nucleic Acids Res. 17:3065-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cato, A. C., R. Miksicek, G. Schutz, J. Arnemann, and M. Beato. 1986. The hormone regulatory element of mouse mammary tumour virus mediates progesterone induction. EMBO J. 5:2237-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, H. M., W. H. Fischer, T. F. Osborne, and M. J. Comb. 1991. NF-I proteins from brain interact with the proenkephalin cAMP inducible enhancer. Nucleic Acids Res. 19:2721-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordingley, M. G., A. T. Riegel, and G. L. Hager. 1987. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell 48:261-270. [DOI] [PubMed] [Google Scholar]
- 14.de Jong, R. N., and P. C. van der Vliet. 1999. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236:1-12. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis, M. L. 1988. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell 52:635-638. [DOI] [PubMed] [Google Scholar]
- 16.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor activation of the I kappa B alpha promoter within chromatin. Mol. Biol. Cell 12:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene 20:3039-3046. [DOI] [PubMed] [Google Scholar]
- 18.Deroo, B. J., C. Rentsch, S. Sampath, J. Young, D. B. DeFranco, and T. K. Archer. 2002. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 22:4113-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Croce, L., R. Koop, P. Venditti, H. M. Westphal, K. P. Nightingale, D. F. Corona, P. B. Becker, and M. Beato. 1999. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell 4:45-54. [DOI] [PubMed] [Google Scholar]
- 20.Eissenberg, J. C. 2001. Decisive factors: a transcription activator can overcome heterochromatin silencing. Bioessays 23:767-771. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher, T. M., N. Xiao, G. Mautino, C. T. Baumann, R. Wolford, B. S. Warren, and G. L. Hager. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fragoso, G., S. John, M. S. Roberts, and G. L. Hager. 1995. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 9:1933-1947. [DOI] [PubMed] [Google Scholar]
- 23.Fragoso, G., W. D. Pennie, S. John, and G. L. Hager. 1998. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol. Cell. Biol. 18:3633-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fryer, C. J., and T. K. Archer. 2001. Analyzing the contributions of chromatin structure in nuclear hormone receptor activated transcription in vivo. Methods Mol. Biol. 176:283-296. [DOI] [PubMed] [Google Scholar]
- 25.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 26.Garvie, C. W., and C. Wolberger. 2001. Recognition of specific DNA sequences. Mol. Cell 8:937-946. [DOI] [PubMed] [Google Scholar]
- 27.Graves, R. A., P. Tontonoz, S. R. Ross, and B. M. Spiegelman. 1991. Identification of a potent adipocyte-specific enhancer: involvement of an NF-1-like factor. Genes Dev. 5:428-437. [DOI] [PubMed] [Google Scholar]
- 28.Hager, G. L. 2001. Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus. Prog. Nucleic Acid Res. Mol. Biol. 66:279-305. [DOI] [PubMed] [Google Scholar]
- 29.Hager, G. L., T. M. Fletcher, N. Xiao, C. T. Baumann, W. G. Muller, and J. G. McNally. 2000. Dynamics of gene targeting and chromatin remodelling by nuclear receptors. Biochem. Soc. Trans. 28:405-410. [PubMed] [Google Scholar]
- 30.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 31.Hsiao, P. W., B. J. Deroo, and T. K. Archer. 2002. Chromatin remodeling and tissue-selective responses of nuclear hormone receptors. Biochem. Cell. Biol. 80:343-351. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann, S. H., S. Okret, A. C. Wikstrom, J. A. Gustafsson, and J. H. Shaper. 1986. Binding of the glucocorticoid receptor to the rat liver nuclear matrix. The role of disulfide bond formation. J. Biol. Chem. 261:11962-11967. [PubMed] [Google Scholar]
- 33.Kim, M. H., and D. O. Peterson. 1995. Oct-1 protein promotes functional transcription complex assembly on the mouse mammary tumor virus promoter. J. Biol. Chem. 270:27823-27828. [PubMed] [Google Scholar]
- 34.Kim, M. K., L. A. Lesoon-Wood, B. D. Weintraub, and J. H. Chung. 1996. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Mol. Cell. Biol. 16:4366-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinyamu, H. K., C. J. Fryer, K. B. Horwitz, and T. K. Archer. 2000. The mouse mammary tumor virus promoter adopts distinct chromatin structures in human breast cancer cells with and without glucocorticoid receptor. J. Biol. Chem. 275:20061-20068. [DOI] [PubMed] [Google Scholar]
- 36.Lee, H. L., and T. K. Archer. 1994. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol. Cell. Biol. 14:32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 38.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 39.Mul, Y. M., and P. C. Van der Vliet. 1992. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 11:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, K., and N. Mermod. 2000. The histone-interacting domain of nuclear factor I activates simian virus 40 DNA replication in vivo. J. Biol. Chem. 275:1645-1650. [DOI] [PubMed] [Google Scholar]
- 41.Muller, W. G., D. Walker, G. L. Hager, and J. G. McNally. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mymryk, J. S., and T. K. Archer. 1994. Detection of transcription factor binding in vivo using lambda exonuclease. Nucleic Acids Res. 22:4344-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mymryk, J. S., and T. K. Archer. 1995. Dissection of progesterone receptor-mediated chromatin remodeling and transcriptional activation in vivo. Genes Dev. 9:1366-1376. [DOI] [PubMed] [Google Scholar]
- 44.Nagata, K., R. A. Guggenheimer, and J. Hurwitz. 1983. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl. Acad. Sci. USA 80:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 46.Ostlund Farrants, A. K., P. Blomquist, H. Kwon, and O. Wrange. 1997. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol. 17:895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prado, F., R. Koop, and M. Beato. 2002. Accurate chromatin organization of the mouse mammary tumor virus promoter determines the nature of the synergism between transcription factors. J. Biol. Chem. 277:4911-4917. [DOI] [PubMed] [Google Scholar]
- 48.Prefontaine, G. G., M. E. Lemieux, W. Giffin, C. Schild-Poulter, L. Pope, E. LaCasse, P. Walker, and R. J. Hache. 1998. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol. Cell. Biol. 18:3416-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richard-Foy, H., and G. L. Hager. 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 6:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, J. M., H. Y. Chen, and J. R. Davie. 1994. Nuclear factor 1 is a component of the nuclear matrix. J. Cell. Biochem. 55:252-263. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, I. C., J. L. Workman, T. J. Schuetz, and R. E. Kingston. 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 5:1285-1298. [DOI] [PubMed] [Google Scholar]
- 52.Wade, P. A., and A. P. Wolffe. 1999. Transcriptional regulation: SWItching circuitry. Curr. Biol. 9:R221-R224. [DOI] [PubMed] [Google Scholar]
- 53.Wolffe, A. P. 1997. Chromatin remodeling regulated by steroid and nuclear receptors. Cell Res. 7:127-142. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe, A. P., G. Almouzni, K. Ura, D. Pruss, and J. J. Hayes. 1993. Transcription factor access to DNA in the nucleosome. Cold Spring Harb. Symp. Quant. Biol. 58:225-235. [DOI] [PubMed] [Google Scholar]
- 55.Wolffe, A. P., and D. Guschin. 2000. Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129:102-122. [DOI] [PubMed] [Google Scholar]
- 56.Wu, L., and J. P. Whitlock, Jr. 1992. Mechanism of dioxin action: Ah receptor-mediated increase in promoter accessibility in vivo. Proc. Natl. Acad. Sci. USA 89:4811-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]