Abstract
Nuclear receptors are ligand-modulated transcription factors. On the basis of the completed human genome sequence, this family was thought to contain 48 functional members. However, by mining human and mouse genomic sequences, we identified FXRβ as a novel family member. It is a functional receptor in mice, rats, rabbits, and dogs but constitutes a pseudogene in humans and primates. Murine FXRβ is widely coexpressed with FXR in embryonic and adult tissues. It heterodimerizes with RXRα and stimulates transcription through specific DNA response elements upon addition of 9-cis-retinoic acid. Finally, we identified lanosterol as a candidate endogenous ligand that induces coactivator recruitment and transcriptional activation by mFXRβ. Lanosterol is an intermediate of cholesterol biosynthesis, which suggests a direct role in the control of cholesterol biosynthesis in nonprimates. The identification of FXRβ as a novel functional receptor in nonprimate animals sheds new light on the species differences in cholesterol metabolism and has strong implications for the interpretation of genetic and pharmacological studies of FXR-directed physiologies and drug discovery programs.
Cholesterol metabolism is a tightly regulated enzymatic pathway. Deregulation of this pathway leads to accumulation of excess cholesterol and can result in diseases such as atherosclerosis and gallstone formation (10). The homeostatic balance between uptake and elimination of cholesterol is accomplished by regulation of three pathways: de novo cholesterol synthesis from acetate, uptake of cholesterol from the intestine, and elimination of cholesterol through the synthesis of bile acids.
Cholesterol catabolism into bile acids is controlled by transcriptional feedback and feedforward mechanisms that are mediated by members of the nuclear receptor family. Activation of cholesterol breakdown into bile acids is mediated by liver X receptor alpha (LXRα; NR1H3), a nuclear receptor that binds oxysterols formed during the synthesis and metabolism of cholesterol (14, 18, 28). Together with another nuclear receptor, liver receptor homologue 1 (NR5A2), LXRα stimulates transcription of cholesterol 7α-hydroxylase (CYP7A1), an enzyme catalyzing the rate-limiting step of this pathway. Suppression of cholesterol degradation into bile acids is triggered by the farnesoid X receptor (FXR; NR1H4), which binds to and is activated by bile acids (21, 27, 33). Ligand-bound FXR activates transcription of the short heterodimer partner (NR0B2) and subsequently downregulates transcription of CYP7A1 (11, 19). However, there are species differences in the regulation of cholesterol metabolism and sensitivity to dietary cholesterol. While rodents respond to cholesterol feeding with induction of CYP7A1, humans and rabbits appear to lack this response and are left more sensitive to the cholesterolemic effects of dietary cholesterol (13, 35, 36). This difference was directly attributed to the ability of LXR to regulate CYP7A in different species (4, 24).
Nuclear hormone receptors form a family of evolutionarily conserved sequences, and a standardized nomenclature has been proposed (26). Six subfamilies are currently recognized; the nuclear receptors FXR and LXRα and -β form the subgroup NR1H together with the Drosophila ecdysone receptor (EcR), which recognizes the metamorphosis steroid ecdysone as its natural ligand (16). A set of 48 human nuclear hormone receptors was known well before the human genome was sequenced. In the Drosophila genome, only 21 nuclear hormone receptors were predicted (1), whereas for Caenorhabditis elegans, more than 270 sequences homologous to the nuclear hormone receptor family were identified (32). Publication of the sequence of the human genome allowed the identification of all genes homologous to the nuclear receptor family of proteins. We and others identified three novel nuclear receptor sequences apart from the 48 known nuclear receptors, all potentially representing pseudogenes in humans (6, 20, 29).
Given the known species-specific difference in cholesterol metabolism, our attention was particularly drawn to a pseudogene showing strong homology to FXR (NR1H4) that we and others designated FXRβ (NR1H5). Here we demonstrate that FXRβ is, in contrast to previous perception, a novel functional member of the NR1H subgroup of nuclear receptors. It constitutes a functional nuclear hormone receptor in all of the mammalian species investigated except primates and humans, where it encodes a nonfunctional protein. In mice, FXRβ is able to activate transcription and its endogenous ligand was identified as the cholesterol precursor lanosterol. Since lanosterol is an intermediate in the cholesterol biosynthetic pathway, this nuclear receptor may represent an additional level at which cholesterol levels are controlled in nonprimate mammals.
MATERIALS AND METHODS
Bioinformatic analysis.
A database of all nuclear receptor protein sequences was constructed and used to search the daily human genome updates (GenBank) by employing the BLASTX tool with default parameters (2). Two FXRβ-containing bacterial artificial chromosomes (BACs) were identified: GenBanknew accession no. AC026039 and EMBLnew accession no. AL390235.
Molecular cloning of FXRβ.
The murine FXRβ (mFXRβ) genome locus was isolated by screening the BAC library Easy-to-Screen DNA Pools BAC Mouse ES (release II) from Incyte Genomics by using primers 5′ TGGGGTCCTTTGTTTTCCA3′ and 5′GTGAAATGGACATGTACATGCG. FXRβ-containing BACs were purchased, and the locus was sequenced by using PCR products generated with the above-described primers. Flanking regions were sequenced by primer walking. FXRβ cDNAs from mice and other mammalian species except humans were isolated by using adult liver and testis RNAs from the respective species and the above-described primers. For amplification of primate FXRβ sequences, genomic DNAs and RNAs from the respective species were derived from the collections at the S. Pääbo laboratory. Full-length cDNAs were isolated by rapid amplification of cDNA ends with a GeneRacer kit (Invitrogen). Human FXRβ (hFXRβ) cDNAs were isolated by reverse transcription (RT)-PCR of adult human testis RNA (Clontech) with primers 5′ CCAGACCAACCTATTCTTCCTCGAGAAATAAGGGAC 3′ and 5′ TGGGGTCCTTTGTTTTCCAAGTGCTAAGTATTTCTG 3′ in 35 PCR cycles followed by 30 cycles of nested PCR.
Expression analysis.
RT-PCR expression analysis of mFXRβ was performed with primers 5′ TCATCCAGCACCAGATCTGGGAAAG 3′ and 5′ GTCCTTTGTTTTCCACATGCGAAGG 3′ and that of mFXR was performed with primers 5′ GGGATGTTGGCTGAATGTTTGTTAACTG3′ and 5′ TCACTGCACATCCCAGATCTCACAG3′ in 35 and 28 cycles PCR cycles, respectively, with a panel of adult (Clontech) and embryonic (Quantum Appligene) RNAs.
Plasmids.
The gateway cloning system (Life Technologies) was used to insert the ligand binding domains (LBDs) of hFXR (amino acids 187 to 472) and mFXRβ (amino acids 186 to 505) into pCMV-BD (Stratagene) and that of human retinoic X receptor alpha (hRXRα; amino acids 197 to 461) into pCMV-AD (Stratagene) to generate mammalian two-hybrid vectors. pCMV-BD-hFXR and pCMV-BD-mFXRβ express their respective FXR LBDs as fusion proteins with the Gal4 DNA binding domain (DBD). The hRXRα LBD is expressed fused with the activation domain (AD) of NF-κB. As a Gal4-responsive luciferase reporter, pFR-LUC (Stratagene) was used. Full-length mFXRβ (GenBank accession no. AY094586) was inserted into expression vector pCMV-AD containing an NF-κB AD (Stratagene) and expression vector pcDNA4/TO (Invitrogen). Luciferase reporter plasmids were constructed by inserting synthetic response elements (REs) into the XhoI site of vector pGL2-Promoter (Promega). Oligonucleotides were designed containing two REs separated by 10 nucleotides. The REs consist of two consensus half sites with the sequence AGGTCA as inverted repeats (IRs), everted repeats (ERs), or direct repeats (DRs) with zero- to eight-nucleotide spacers between the half sites and flanking XhoI sites.
The I-BABP RE reporter construct contains two tandem copies of the oligonucleotide CCCCAGGGTGAATAACCTCGGGGCTCTGTCCCTCCAATCCCAGGGTGAATAACCTCGGGA from the human I-BABP promoter (12) ligated between the KpnI and BglII sites of pGL2-Promoter (Promega). For the homogeneous time-resolved fluorescence (HTRF) assay, the mFXRβ LBD (amino acids 186 to 505) and the hLXRα LBD (amino acids 155 to 447) were fused to glutathione S-transferase (GST), His tagged, and recombined into the pACGHLT polylinker (Pharmingen) with a gateway cassette (Life Technologies). A GST fusion of the hFXR LBD (amino acids 187 to 476) and a His-tagged NR interaction domain of human Tif2 (hTif2; amino acids 548 to 878) were recombined into pDest15 and pDest17 (Life Technologies).
Transient transfections.
Human embryonic kidney (HEK) 293 cells were maintained at 37°C in 5% CO2 in Dulbecco modified Eagle medium (DMEM; Invitrogen) containing 10% fetal calf serum. Cotransfection was performed in Optimem (Invitrogen) with 50 ng of the appropriate expression vector, 50 ng of the reporter construct, and 10 ng of a thymidine kinase-Renilla luciferase control vector with Lipofectamine (Invitrogen) for 4 h. Cells were then treated with ligands and grown in DMEM for 18 h. Ligands and ketoconazole (Sigma) were dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C. Lanosterol was freshly dissolved in ethanol at 35°C. When treated with lanosterol, cells were grown in the presence of 1% DMSO for 2.5 h to permeabilize cell membranes and facilitate uptake (38) and then kept in DMEM for 16 h. After ligand treatment, cells were harvested and luciferase assays were performed with the dual-luciferase reporter assay as described by the manufacturer's (Promega) manual. Transfection data were normalized to the Renilla control and expressed as mean numbers of relative light units from triplicate assays ± the standard deviations (SDs).
Coactivator recruitment assay.
The mFXRβ and hLXRα LBDs were used in the Pharmingenes Baculovirus Expression vector system. The hFXR LBD and hTif2 were expressed in Escherichia coli. The resulting proteins were GST or His affinity purified as recommended by Pharmacia and QIAGEN. His-hTif2 was biotinylated (Roche). A biotinylated mSCR1 peptide (amino acids 680 through 704) was purchased (SynPep Corp.). Proteins and compounds were mixed with 1 nM europium cryptate-anti-GST antibody and 100 ng of streptavidin-allophycocyanin in black 384-well microtiter plates (Nunc Fluotrac200). The reaction mixture volume was 25 μl. Plates were shaken for 2 min at 800 rpm and incubated for 60 min at room temperature. Measurement of fluorescence from both fluorophores at 665 nm (acceptor signal) and 615 nm (donor signal) occurred in a time window of 50 to 400 μs after excitation on a Victor V HTRF reader. Signals were plotted as ratios of light intensity at 665 nm versus light intensity at 615 nm. For analysis of dose-response curves, ratios were plotted against log concentrations and 50% effective concentrations (EC50s) were calculated by Prism (GraphPad Software Inc.)
RESULTS
Identification of primate FXRβ as a pseudogene.
In an effort to identify novel nuclear hormone receptor proteins after the publication of the human genome sequence, we performed exhaustive database searches with the BLAST algorithm (2). As one result, we and others (6, 20, 29) detected a novel member of the NR1H subgroup, termed FXRβ (NR1H5), on the basis of its homology to FXR. The novel gene has conserved intron positions compared to FXR, which, together with the sequence conservation, argues in favor for a functional nuclear receptor. In addition to previously published results (20), we identified several splice variants of FXRβ in RNA from human testis (data not shown). However, by sequencing those cDNAs and three independent genome loci, we found that hFXRβ contains two stop codons in exon 11 and three frame shifts at exon-intron junctions (Fig. 1A, arrows). The extremely low transcript levels determined by nested RT-PCR and the presence of two stop codons and three frame shifts in the cDNA strongly argue that FXRβ is a pseudogene in the human lineage.
FIG. 1.
FXRβ genomic structures and molecular cloning. (A) Schematic representation of FXRβ genomic structure. mFXRβ and hFXRβ loci show similar exon-intron structures. Here the murine locus is displayed, consisting of at least 11 exons spanning 26 kb of genomic DNA. Striped boxes represent the DBD, gray boxes represent the LBD, the asterisk represents the stop codon, and the flag represents the start codon. The 5′ untranslated region is encoded by exon 1, while exon 11 encodes the 3′ untranslated region (black boxes). Human stop codons and frame shifts are indicated by vertical arrows. Untranslated regions are not completely sequenced for the human locus. (B) Sequence of mFXRβ. Shown are cDNA and protein sequences of mFXRβ (GenBank accession no. AY094586). Start and stop codons are bold, and the poly(A) signal is underlined.
To assess whether FXRβ constitutes a functional protein in other primate species, we performed PCR amplification of genome loci and analysis of the chimpanzee, gorilla, orangutan, and rhesus monkey FXRβ sequences. In all cases, stop codons and frame shifts were identified at positions similar to those in the human mRNA and no transcripts could be detected by RT-PCR from liver and testis RNA sources (data not shown). Thus, FXRβ seems to be a nonfunctional gene in these primate species as well.
FXRβ codes for a functional receptor in other mammals.
RT-PCR amplification of liver RNA and subsequent sequence analysis of nonprimate mammals, including mice, rats, rabbits, and dogs, revealed FXRβ mRNAs showing continuous open reading frames (ORFs). These mRNAs therefore potentially encode functional nuclear receptor proteins. RT-PCR and rapid amplification of cDNA ends of mFXRβ verified 10 coding exons translating into a protein of 505 amino acids (Fig. 1A and B). The 5′ untranslated region is encoded by exon 1, while exon 11 encodes the 3′ untranslated region and is flanked by a poly(A) signal.
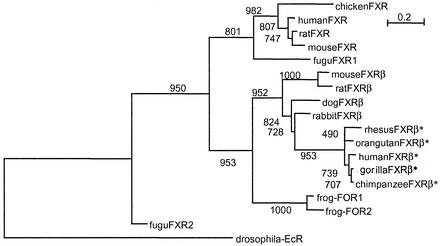
All isolated FXRβ sequences form a robust phylogenetic group well separated from the FXR cluster (Fig. 2). The FXR orthologs recently isolated from Xenopus laevis, FOR1 and -2 (31), are included with high confidence in the FXRβ branch, indicating that these two genes are, in fact, FXRβ orthologs. We identified in silico two different ORFs from the draft sequence of the pufferfish (Fugu rubripes) genome that show similarity to the FXR genes. Fugu rupies FXR1 is included with good confidence in the FXR lineage. FXR2 is building a separate deep branch at the root of the tree and appears to be ancestral to the duplication that led to the clusters of FXR and FXRβ. Mutation rates are higher for FXRβ than for FXR, as represented by respective branch lengths on the phylogenetic tree (Fig. 2).
FIG. 2.
Phylogenetic tree containing FXR and FXRβ genes from different species. The phylogenetic tree was calculated on the basis of protein alignments by using PHYLIP (phylogeny interference package, 1993; distributed by J. Felsenstein, University of Washington, Seattle, Wash.) on the basis of the following sequences (accession numbers): chicken FXR (AF492497), hFXR (U68233), rat FXR (U18374), mFXR (NM009108), Fugu FXR1 (S001361), mFXRβ (AY094586), frog FOR2 (AF456452), frog FOR1 (AF4564519), Fugu FXR2 (S000248), and drosophila EcR (M74078). Numbers indicate the bootstrap support (out of 1,000) for the respective nodes. Asterisks indicate putatively nonfunctional proteins in primates.
Ubiquitous tissue expression of mFXRβ.
To investigate the potential function of the novel receptor, we analyzed mFXRβ transcripts in embryonic and adult tissues. Five splice variants of mFXRβ were isolated by RT-PCR from adult liver and testis RNA. Isoforms differentially splice out parts of exon 3, 8, or 10 and thereby differ in the regions encoding the DBD and the LBD (Fig. 3A). mFXRβ is ubiquitously and strongly expressed during embryonic development, and its expression pattern largely overlaps that of FXR (Fig. 3B). This mRNA distribution suggests a function of mFXRβ during embryonic development. In the adult, however, mFXRβ expression is more tissue specific and restricted mainly to the liver, reproductive tissues, and heart. Coexpression with FXR is observed in the liver and intestine, main tissues of FXR expression and function, but not in the kidney (8).
FIG. 3.
Transcription of mFXRβ. (A) Splice variants of mFXRβ. Five splice variants were isolated and sequenced (GenBank accession no. AY094586 through AY094590). Striped boxes indicate exons encoding the DBD, and gray boxes indicate exons encoding the LBD. Numbers indicate exons. Shown are only coding regions. (B) Tissue distribution of mFXRs. mFXR and mFXRβ transcripts were determined by RT-PCR amplification of nonhomologous sequences derived from the LBDs of the respective receptors. β-Actin served as an internal control. RNAs from indicated embryonic and adult tissues were used as templates. Lane M, DNA size marker. d, day.
The IR1 and ER2 REs are recognized by mFXRβ.
Nuclear receptors bind specifically to repeats of single DNA hexamers called REs (23). They bind either as monomers, homodimers, or heterodimers with RXR. As heterodimers, they bind to REs consisting of a repeated core motif that can be an IR, a DR, or an ER. To determine the DNA binding properties of mFXRβ, we fused mFXRβ to a constitutively active NF-κB AD and screened against a panel of luciferase reporter constructs containing synthetic consensus IR, DR, and ER elements in transient cotransfection assays. Transactivation of the luciferase reporter gene was observed with the synthetic IR1 and ER2 elements, as well as the FXR RE from the I-BABP promoter, which is itself an IR1 element (12) (Fig. 4A). The specific interaction with typical FXR REs suggests that at least part of the FXR target genes may also be regulated by FXRβ.
FIG. 4.
mFXRβ binds selectively to nuclear REs and forms heterodimers with RXRα. (A) DNA REs recognized by mFXRβ. HEK 293 cells were cotransfected with expression plasmids containing mFXRβ fused to an NF-κB AD (black bars) or an empty vector (white bars) and the indicated plasmid from a panel of luciferase reporter constructs containing synthetic IR, ER, or DR REs with different spacers between the half sites (zero to eight nucleotides). Luciferase activities are means of triplicate experiments, and the SDs are indicated. (B) Interaction of mFXRβ with RXRα is enhanced by 9-cis-RA. HEK 293 cells were cotransfected with the reporter vector pFR-Luc and RXRα fused to an NF-κB AD (pCMV-AD-hRXRα) in combination with either pCMV-BD (white bars), FXR fused to the Gal4 DBD (pCMV-BD-hFXR; gray bars) or an mFXRβ fusion with the Gal4 DBD (pCMV-BD-mFXRβ; black bars). Cells were treated with 1 μM 9-cis-RA (+) or an equivalent volume of DMSO (−). Cells from triplicate experiments were assayed for luciferase activity, and means ± SDs are shown. RLU, relative light units.
mFXRβ heterodimerizes with RXRα.
Since nuclear receptors are known to interact with their REs as either homodimers or RXR heterodimers (23), we examined the ability of mFXRβ to interact with RXR in response to its ligand, 9-cis-retinoic acid (9-cis-RA). For a mammalian two-hybrid assay, the LBDs of mFXRβ and hFXR were fused to the Gal4 DBD while hRXRα was fused to an NF-κB AD and cotransfected in HEK 293 cells with a luciferase reporter construct. Compared to the control, we observed the expected constitutive interaction between the hFXR and hRXRα LBDs (Fig. 4B). Addition of 9-cis-RA further enhanced this interaction. Compared to hFXR, mFXRβ exhibited a significantly stronger constitutive interaction with RXRα that was also potentiated by 9-cis-RA. These data indicate a constitutive mFXRβ-RXRα interaction and a responsiveness of the complex formed to the RXR ligand 9-cis-RA.
Lanosterol promotes coactivator recruitment by mFXRβ.
FXR, LXRα, and LXRβ are activated by bile acids (21, 27, 33) and oxysterols (14, 18), respectively, all of which are intermediates of the cholesterol metabolic pathway. Since these receptors are evolutionarily related to FXRβ, we tested compounds that are related to the respective ligands of these homologs for binding to mFXRβ. This set of around 100 compounds includes cholesterol metabolites, steroid hormones, and other bile acid metabolites. We employed an HTRF ligand-sensing assay to measure the interaction between the mFXRβ LBD and a short peptide derived from the coactivator mSRC-1 induced by a ligand. Interestingly, lanosterol, an intermediate of the de novo cholesterol biosynthetic pathway, showed the greatest affinity in the ligand-sensing assay. Lanosterol induces interaction between mSRC-1 and the mFXRβ LBD with an EC50 of ∼1 μM (Fig. 5A). Similar affinities are reported for the physiological ligands of other lipid receptors (14, 18, 34). Thus, lanosterol is a likely candidate for an endogenous agonist ligand that induces coactivator recruitment by mFXRβ. The FXR-specific compound GW4064 (22) has very poor affinity for mFXRβ (Fig. 5B), suggesting different ligand specificities for both receptors. Furthermore, lanosterol displays a remarkable specificity for mFXRβ as it binds neither to the closest relative of mFXRβ, FXR (Fig. 5B), nor to the more distant relative LXRα (data not shown). In addition, mFXRβ shows somewhat promiscuous ligand binding as it binds a quite diverse spectrum of chemical structures with lower affinity (Fig. 5C). Again, these compounds do not bind to FXR (data not shown). Finally it is important to note that various bile acids that, in part, can serve as endogenous ligands for FXR have no significant affinity for mFXRβ (Fig. 5D). This again indicates that there is no overlap between FXR and FXRβ in terms of specificity for natural ligands
FIG. 5.
Identification of ligands for mFXRβ. (A) Lanosterol selectively induces SRC1 binding to mFXRβ at physiological concentrations. GST-mFXRβ LBD and biotinylated mSRC1 peptide were mixed, and the effect of lanosterol and the FXR agonist GW4064 on the interaction was determined by HTRF assay. Cofactor interaction is induced by lanosterol with an EC50 of 1 μM, whereas GW4064 only acts as a weak agonist. Signals are plotted as means of triplicates, and error bars indicate SDs. (B) GST-hFXR LBD was mixed with biotinylated, His-tagged hTif2, and the effects of lanosterol and GW4064 on the respective interaction were assayed. The FXR agonist GW4064 induces cofactor interaction with an EC50 of 70 nM, whereas lanosterol does not show any effect. (C) A diverse set of compounds induces cofactor interaction with mFXRβ but not FXR. Shown are EC50s and the relative efficacy of SRC1 recruitment (the efficacy of lanosterol was set to 100%) calculated from dose-response experiments for a selected group of compounds found to be active for FXRβ. (D) Cholates and cholic acids do not induce significant SRC1 binding to mFXRβ, whereas lanosterol does induce SRC1 recruitment. GST-mFXRβ LBD and biotinylated mSRC1 peptide were mixed. The effects of various compounds at a concentration of 40 μM on the interaction was determined by HTRF assay. Ratios of signals versus DMSO controls are plotted as means of triplicates, and SDs are indicated. CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; UDCA, ursodeoxycholic acid; CA, cholic acid.
Lanosterol activates mFXRβ as an agonist ligand.
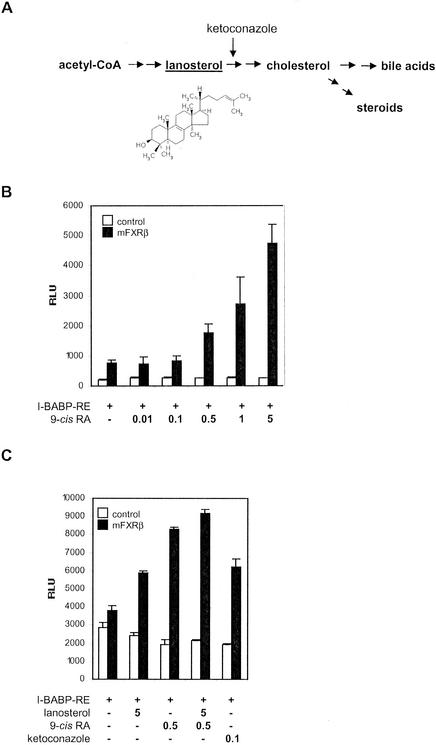
Finally, we sought to investigate the transcriptional activities of mFXRβ on the I-BABP RE-containing reporter gene construct in a cellular context and the modulation of these activities by 9-cis-RA and lanosterol. Lanosterol is an intermediate of the cholesterol biosynthetic pathway (Fig. 6A). It is a precursor of the meiosis-activating sterols FF-MAS and T-MAS, which are synthesized from lanosterol by the cytochrome P-450 enzyme CYP51 (30). In transient transfection assays, we cotransfected a full-length mFXRβ expression plasmid with the I-BABP reporter construct. mFXRβ expression results in constitutive activation of reporter gene activity compared to the vector control (Fig. 6B). Addition of 9-cis-RA results in a dose-dependent increase in luciferase activity only in the presence of mFXRβ. This supports the notion that mFXRβ can exert its transcriptional activity as a heterodimer with RXRα. Furthermore, treatment of cells with 5 μM lanosterol significantly stimulated reporter gene expression (Fig. 6C). This activation was potentiated by the simultaneous addition of 9-cis-RA. These data strongly suggest that mFXRβ is, indeed, a functional hormone receptor, with lanosterol as its physiological agonist ligand.
FIG. 6.
Lanosterol activates mFXRβ. (A) Lanosterol is an intermediate of the cholesterol metabolism pathway. Cholesterol is synthesized from acetyl coenzyme A (acetyl-CoA) via lanosterol and catabolized into bile acids and steroids. Ketoconazole inhibits CYP51, the enzyme catalyzing the demethylation of lanosterol. (B) mFXRβ is activated by 9-cis-RA. HEK 293 cells were cotransfected with an mFXRβ full-length expression vector (black bars) or an empty vector (white bars) and the I-BABP RE luc reporter. Cells were treated with the indicated micromolar concentrations of 9-cis-RA. Mean luciferase activities and SDs determined from triplicate experiments are shown. (C) mFXRβ is activated by lanosterol. HEK 293 cells were cotransfected with an mFXRβ full-length expression vector (black bars) or an empty vector (white bars) and the I-BABP RE luc reporter and treated with the indicated micromolar concentrations of lanosterol, 9-cis-RA, and ketoconazole. Mean luciferase activities and SDs determined from triplicate experiments are shown. RLU, relative light units.
However, as lanosterol is a highly water-insoluble compound and may only be able to enter cells to a small extent during the transfection assay, we sought to independently confirm the observed lanosterol effect. Inhibition of CYP51, the enzyme that demethylates lanosterol as the first step in the conversion to cholesterol, by ketoconazole (25, 37; Fig. 6A) should lead to accumulation of lanosterol within the cell. We therefore cotransfected HEK 293 cells with the full-length mFXRβ expression vector and the I-BABP reporter construct and treated the cells with ketoconazole. We indeed observed transcriptional activation by mFXRβ above the basal level (Fig. 6C). This effect could be potentiated by the addition of lanosterol and 9-cis-RA to the medium. Together, these results provide convincing evidence that lanosterol acts as an agonist ligand for mFXRβ both in vitro and in vivo.
DISCUSSION
Nuclear hormone receptors are ligand-modulated transcription factors that mediate the transcriptional effects of endogenous ligands such as steroid, thyroid, and retinoid hormones (9). The importance of nuclear receptors as targets for the development of medicines raised strong interest in the identification of novel functional nuclear receptors in the human genome. Bioinformatic approaches did not identify novel functional receptors in the human genome in addition to the 48 previously known nuclear receptors. Only three additional pseudogenes were identified, including an FXR homologue designated FXRβ (6, 20, 29).
We demonstrate in our report that in other mammalian species like mice, rats, rabbits, and dogs, the gene for FXRβ has the capacity to code for a functional nuclear receptor protein. In a phylogenetic analysis, all isolated FXRβ sequences form a phylogenetic group well separated from the FXR cluster. Since the FXR homologs from X. laevis, FOR1 and -2 (31), are, in fact, FXRβ orthologs, our data indicate the existence of an FXRβ-type gene already in the common ancestors of amphibians and mammals. We identified in silico two different ORFs from the pufferfish (F. rubripes) genome, one of which is included in the FXR lineage while the other is building a separate deep branch at the root of the tree. Since no sequence with similarity to FXRβ could be detected in the pufferfish genome, the phylogenetic data indicate an origin of the FXRβ gene as part of the postulated second wave of gene duplications during tetrapod (land-living vertebrate) evolution that probably led to the diversification of receptors (7). FXRβ later lost its function as a secondary event. However, it is unknown when during the evolution of primates the gene lost its function, since our analysis focused on great apes (gorillas, chimpanzees, and orangutans) and just one Old World monkey (rhesus monkey) was added. Further analyses of other branches of the primate lineage are necessary to solve this issue. To our knowledge, FXRβ represents the first member of the nuclear hormone receptor family constituting a functional receptor protein in several mammalian species but a pseudogene in the primate lineage. It is therefore possible that other identified human pseudogenes in this family have functional counterparts in primate or nonprimate species as well.
FXRβ was identified through its sequence similarity to FXR and was shown in our phylogenetic analysis to code for an independent nuclear receptor protein. However, when examining transcript levels in mouse tissues, we observed significant coexpression of FXR and FXRβ. Both receptors are ubiquitously expressed in embryos, which argues for a role of both receptors during embryonic development. In addition, they are coexpressed in a subset of tissues in adult organisms. FXR has been shown to be involved in the regulation of cholesterol-bile acid metabolism and in the regulation of bile acid transporters in the intestine, which is accompanied by high expression levels in tissues such as those of the liver, kidneys, and intestine (8). Coexpression with mFXRβ in the liver and intestine but not in the kidneys may indicate at least partly overlapping functions. The expression of mFXRβ in tissues such as the testis and ovary suggests additional roles in reproductive functions.
Like most nonsteroidal receptors, such as LXR and FXR, mFXRβ binds specific DNA elements as a heterodimer with RXRα, the 9-cis-RA receptor. Furthermore, we show that mFXRβ can function through consensus IR1 and ER2 REs and is active as well on an FXR RE originating from the I-BABP promoter (12). Most published FXR target genes contain one or more IR1-type elements in the proximal promoter or distal enhancers that function as an FXR RE and are required for transcriptional activation (3, 11, 12, 17, 19, 21). Our finding that mFXRβ recognizes consensus, as well as natural, IR1 elements strongly suggests the existence of common target genes for both FXR and FXRβ. Moreover, FXRβ activates transcription through ER2 but not ER8 elements, derivatives of which were recently described as targets for FXR (15). This finding, on the other hand, indicates possible differences in target genes between the two receptors.
Our results show that mFXRβ is a nuclear hormone receptor that is activated by lanosterol, an intermediate in the cholesterol biosynthetic pathway. Importantly, mFXRβ is not activated by bile acids, the endogenous ligands for its closest homolog, FXR. The ligand-bound nuclear receptors FXR and LXR are known to modulate cholesterol breakdown through regulation of CYP7A1, the rate-limiting enzyme of this pathway. However, there are observed differences between species in the regulation of cholesterol homeostasis. While rodents seem to be able to adapt to large fluctuations in sterol input with little change in plasma cholesterol concentrations, humans do experience alterations in plasma cholesterol levels when sterol balances change (5). These observed differences have so far been explained by a rapid catabolism in rodents caused by high CYP7A1 activity, which is balanced by a high rate of endogenous cholesterol synthesis. In humans, lower enzyme levels, together with a lack of inducibility of CYP7A1 by dietary input of cholesterol, seem to lead to a greater susceptibility to high levels of cholesterol intake. As FXRβ is absent in humans but serves as a nuclear receptor in rodents, it may add an additional layer of regulation to cholesterol biosynthesis in rodents that is absent in primates and in this way contribute to the observed species-specific differences.
Finally, our understanding of cholesterol metabolic pathways in humans, which in part is deduced from pharmacological studies and compound testing in nonprimate mammals, may have to be revisited because of our finding of a novel and additional nuclear receptor, FXRβ, that is a pseudogene in primates.
Acknowledgments
We thank R. Bürgermeister, M. Post, K. Fedel, K. Matena, I. Balogh, V. Fichter, and S. Ellwanger for excellent technical assistance and S. Heck and R. Löbbert for critical suggestions.
REFERENCES
- 1.Adams, M. D., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 2:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ananthanarayanan, M., N. Balasubramanian, M. Makishima, D. J. Mangelsdorf, and F. J. Suchy. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276:28857-28865. [DOI] [PubMed] [Google Scholar]
- 4.Chiang, J. Y., R. Kimmel, and D. Stroup. 2001. Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα). Gene 262:257-265. [DOI] [PubMed] [Google Scholar]
- 5.Dietschy, J. M., S. D. Turley, and D. K. Spady. 1993. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J. Lipid Res. 34:637-659. Review. [PubMed] [Google Scholar]
- 6.Enmark, E., and J. A. Gustafsson. 2001. Comparing nuclear receptors in worms, flies and humans. Trends Pharmacol. Sci 22:611-615. [DOI] [PubMed] [Google Scholar]
- 7.Escriva, H., R. Safi, C. Hanni, M. C. Langlois, P. Saumitou-Laprade, D. Stehelin, A. Capron, R. Pierce, and V. Laudet. 1997. Ligand binding was acquired during evolution of nuclear receptors. Proc. Natl. Acad. Sci. USA 94:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman, B. M., E. Goode, J. Chen, A. E. Oro, D. J. Bradley, T. Perlmann, D. J. Noonan, L. T. Burka, T. McMorris, W. W. Lamph, et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687-693. [DOI] [PubMed] [Google Scholar]
- 9.Giguere, V. 1999. Orphan nuclear receptors: from gene to function. Endocr. Rev. 20:689-725. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, J. L., and M. S. Brown. 1990. Regulation of the mevalonate pathway. Nature 343:425-430. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 12.Grober, J., I. Zaghini, H. Fujii, S. A. Jones, S. A. Kliewer, T. M. Willson, T. Ono, and P. Besnard. 1999. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274:29749-29754. [DOI] [PubMed] [Google Scholar]
- 13.Horton, J. D., J. A. Cuthbert, and D. K. Spady. 1995. Regulation of hepatic 7α-hydroxylase expression and response to dietary cholesterol in the rat and hamster. J. Biol. Chem. 270:5381-5387. [DOI] [PubMed] [Google Scholar]
- 14.Janowski, B. A., P. J. Willy, T. R. Devi, J. R. Falck, and D. J. Mangelsdorf. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature 383:728-731. [DOI] [PubMed] [Google Scholar]
- 15.Kast, H. R., B. Goodwin, P. T. Tarr, S. A. Jones, A. M. Anisfeld, C. M. Stoltz, P. Tontonoz, S. Kliewer, T. M. Willson, and P. A. Edwards. 2002. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J. Biol. Chem. 277:2908-2915. [DOI] [PubMed] [Google Scholar]
- 16.Koelle, M. R., W. S. Talbot, W. A. Segraves, M. T. Bender, P. Cherbas, and D. S. Hogness. 1991. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67:59-77. [DOI] [PubMed] [Google Scholar]
- 17.Laffitte, B. A., H. R. Kast, C. M. Nguyen, A. M. Zavacki, D. D. Moore, and P. A. Edwards. 2000. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem. 275:10638-10647. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann, J. M., S. A. Kliewer, L. B. Moore, T. A. Smith-Oliver, B. B. Oliver, J. L. Su, S. S. Sundseth, D. A. Winegar, D. E. Blanchard, T. A. Spencer, and T. M. Willson. 1997. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 272:3137-3140. [DOI] [PubMed] [Google Scholar]
- 19.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 20.Maglich, J. M., A. Sluder, X. Guan, Y. Shi, D. D. McKee, K. Carrick, K. Kamdar, T. M. Willson, and J. T. Moore. 2001. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2:RESEARCH0029. [Online.] http://www.genomebiology.com/ [DOI] [PMC free article] [PubMed]
- 21.Makishima, M., A. Y. Okamoto, J. J. Repa, H. Tu, R. M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf, and B. Shan. 1999. Identification of a nuclear receptor for bile acids. Science 284:1362-1365. [DOI] [PubMed] [Google Scholar]
- 22.Maloney, P. R., D. J. Parks, C. D. Haffner, A. M. Fivush, G. Chandra, K. D. Plunket, K. L. Creech, L. B. Moore, J. G. Wilson, M. C. Lewis, S. A. Jones, and T. M. Willson. 2000. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43:2971-2974. [DOI] [PubMed] [Google Scholar]
- 23.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menke, J. G., K. L. Macnaul, N. S. Hayes, J. Baffic, Y. S. Chao, A. Elbrecht, L. J. Kelly, M. H. Lam, A. Schmidt, S. Sahoo, J. Wang, S. D. Wright, P. Xin, G. Zhou, D. E. Moller, and C. P. Sparrow. 2002. A novel liver X receptor agonist establishes species differences in the regulation of cholesterol 7α-hydroxylase (CYP7a). Endocrinology 143:2548-2558. [DOI] [PubMed] [Google Scholar]
- 25.Nitahara, Y., K. Kishimoto, Y. Yabusaki, O. Gotoh, Y. Yoshida, T. Horiuchi, and Y. Aoyama. 2001. The amino acid residues affecting the activity and azole susceptibility of rat CYP51 (sterol 14-demethylase P450). J. Biochem. 129:761-768. [DOI] [PubMed] [Google Scholar]
- 26.Nuclear Receptors Nomenclature Committee. 1999. A unified nomenclature system for the nuclear receptor superfamily. Cell 97:161-163. [DOI] [PubMed] [Google Scholar]
- 27.Parks, D. J., S. G. Blanchard, R. K. Bledsoe, G. Chandra, T. G. Consler, S. A. Kliewer, J. B. Stimmel, T. M. Willson, A. M. Zavacki, D. D. Moore, and J. M. Lehmann. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365-1368. [DOI] [PubMed] [Google Scholar]
- 28.Peet, D. J., S. D. Turley, W. Ma, B. A. Janowski, J. M. Lobaccaro, R. E. Hammer, and D. J. Mangelsdorf. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 93:693-704. [DOI] [PubMed] [Google Scholar]
- 29.Robinson-Rechavi, M., A. S. Carpentier, M. Duffraisse, and V. Laudet. 2001. How many nuclear hormone receptors are there in the human genome? Trends Genet. 17:554-556. [DOI] [PubMed] [Google Scholar]
- 30.Rozman, D., M. Cotman, and R. Frangez. 2002. Lanosterol 14α-demethylase and MAS sterols in mammalian gametogenesis. Mol. Cell. Endocrinol. 187:179-187. [DOI] [PubMed] [Google Scholar]
- 31.Seo, Y. W., S. Sanyal, H. J. Kim, D. H. Won, J. Y. An, T. Amano, A. M. Zavacki, H. B. Kwon, Y. B. Shi, W. S. Kim, H. Kang, D. D. Moore, and H. S. Choi. 2002. FOR, a novel orphan nuclear receptor related to FXR. J. Biol. Chem. 277:17836-17844. [DOI] [PubMed] [Google Scholar]
- 32.Sluder, A. E., and C. V. Maina. 2001. Nuclear receptors in nematodes: themes and variations. Trends Genet. 17:206-213. [DOI] [PubMed] [Google Scholar]
- 33.Wang, H., J. Chen, K. Hollister, L. C. Sowers, and B. M. Forman. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3:543-553. [DOI] [PubMed] [Google Scholar]
- 34.Willson, T. M., P. J. Brown, D. D. Sternbach, and B. R. Henke. 2000. The PPARs: from orphan receptors to drug discovery. J. Med. Chem. 43:527-550. [DOI] [PubMed] [Google Scholar]
- 35.Quintao, E., S. M. Grundy, and E. H. Ahrens, Jr. 1971. Effects of dietary cholesterol on the regulation of total body cholesterol in man. J. Lipid Res. 12:233-247. [PubMed] [Google Scholar]
- 36.Xu, G., G. Salen, S. Shefer, G. S. Tint, L. B. Nguyen, T. T. Parker, T. S. Chen, J. Roberts, X. Kong, and D. Greenblatt. 1998. Regulation of classic and alternative bile acid synthesis in hypercholesterolemic rabbits: effects of cholesterol feeding and bile acid depletion. J. Lipid Res. 39:1608-1615. [PubMed] [Google Scholar]
- 37.Yoshida, Y., and Y. Aoyama. 1987. Interaction of azole antifungal agents with cytochrome P-450 14DM purified from Saccharomyces cerevisiae microsomes. Biochem. Pharmacol. 36:229-235. [DOI] [PubMed] [Google Scholar]
- 38.Yu, Z. W., and P. J. Quinn. 1998. The modulation of membrane structure and stability by dimethyl sulphoxide. Mol. Membr. Biol. 15:159-168. [DOI] [PubMed] [Google Scholar]