Abstract
The biological actions of estrogens are mediated via two distinct intranuclear estrogen receptor (ER) proteins, ERα and ERβ. We have used an in vitro chromatin assembly and transcription system to compare the transcriptional activities of the two ERs in the context of chromatin, the physiological template for transcription by RNA polymerase II. We find that under conditions where many biochemical activities of the receptors are similar (e.g. ligand binding, chromatin binding, chromatin remodeling and co-activator recruitment), liganded ERα is a much more potent transcriptional activator than ERβ with chromatin templates, but not with naked DNA. This difference is attributable to the N-terminal A/B region of ERα, which contains a transferable activation function that facilitates transcription specifically with chromatin templates. Interestingly, chromatin selectively restricts ligand-dependent transcriptional activation by ERβ under some conditions (e.g. with a closed chromatin architecture), while allowing it under other conditions (e.g. with an open chromatin architecture). Collectively, our results define an important role for chromatin in determining signaling outcomes mediated by distinct subtypes of signal-transducing transcriptional activator proteins.
Keywords: activation domain/chromatin/estrogen/estrogen receptor/transcription
Introduction
Estrogens, such as the predominant naturally occurring estrogen 17β-estradiol (E2), play critical roles in many physiological processes in both females and males, including normal growth, development and cell type-specific gene regulation in tissues of the reproductive tract, central nervous system and skeleton (Couse and Korach, 1999; Nilsson et al., 2001; Pettersson and Gustafsson, 2001). In addition, estrogens play integral roles in hormone-dependent diseases, such as breast cancer and osteoporosis (Couse and Korach, 1999; Sommer and Fuqua, 2001). The biological actions of estrogens are mediated via two distinct intranuclear estrogen receptor (ER) proteins, ERα and ERβ, which belong to a large conserved superfamily of nuclear receptor proteins (Couse and Korach, 1999; Nilsson et al., 2001; Pettersson and Gustafsson, 2001). The ERs are widely distributed throughout the body, displaying distinct but overlapping expression patterns in a variety of tissues (Couse and Korach, 1999; Nilsson et al., 2001; Pettersson and Gustafsson, 2001). ERα is expressed primarily in the uterus, liver, kidney and heart, whereas ERβ is expressed primarily in the ovary, prostate, lung, gastrointestinal tract and bladder. Co-expression of both receptors occurs in the mammary glands, epididymis, thyroid, adrenals, bone and certain regions of the brain. Pharmacologically, the ERs are targets for estrogen antagonists which are used therapeutically to treat breast cancers and other endocrine-related diseases (Sommer and Fuqua, 2001).
ERα and ERβ are distinct proteins encoded by separate genes located on different chromosomes (Couse and Korach, 1999; Nilsson et al., 2001). The length of human ERα has been established definitively as 595 amino acids. In contrast, the length of human ERβ has been revised several times based on the discovery of additional upstream translation start codons and reports of new sequence information that alter the predicted length of the N-terminal A/B region (Pettersson and Gustafsson, 2001). A survey of DNA sequence database information, representing distinct genomic and cDNA sequences reported by at least nine independent research groups, suggests that the 530 amino acid form of human ERβ represents the most common form (see Supplementary table I and references therein, available at The EMBO Journal Online) and, hence, is the form of ERβ used in the studies described herein. Possible distinct roles for the other N-terminal variants of ERβ have yet to be fully explored.
Despite the differences in their lengths, ERα and ERβ share a conserved structural and functional organization with other members of the nuclear receptor superfamily, including domains responsible for ligand binding, dimerization, DNA binding and transcriptional activation (Nilsson et al., 2001) (see Figure 1A). The DNA-binding domains (DBDs) of ERα and ERβ are highly homologous (96%), allowing both receptors to bind to the same estrogen response elements (EREs) and regulate similar sets of genes (Klinge, 2001). The ligand-binding domains (LBDs) are also conserved (58% homology), as suggested by the similar affinities of the two ERs for E2 (Kuiper et al., 1998). In spite of these similarities, ERα and ERβ exhibit different affinities and responses with subsets of natural EREs and pharmacological ligands (Kuiper et al., 1998; Klinge, 2001; Meyers et al., 2001). In addition to their DBDs and LBDs, both ERs contain transcription activation functions (AFs), which allow the receptors to stimulate the transcription of estrogen-regulated genes. ERα contains two potent AFs, an N-terminal, ligand-independent activation function (AF-1) and a C-terminal, ligand-dependent activation function (AF-2) (Nilsson et al., 2001). Both AFs in ERα are required for synergistic transcriptional activation, but can also function independently with certain cell type and promoter specificities (Tzukerman et al., 1994). Like ERα, ERβ also contains an AF-2, but appears to have a weaker AF-1 which may possess repressive activity (McInerney et al., 1998; Cowley and Parker, 1999; Hall and McDonnell, 1999; Delaunay et al., 2000).
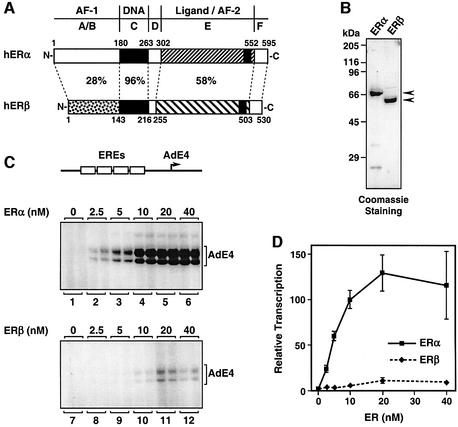
Fig. 1. ERβ is a weak transcriptional activator with chromatin templates. (A) Schematic diagrams of human ERα (1–595) and human ERβ (1–530) showing percentage homology between the different receptor functional domains. The domains include the DNA-binding domain (‘DNA’), ligand-binding domain (‘Ligand’) and two transcriptional activation functions (‘AF-1’ and ‘AF-2’). (B) SDS–PAGE analysis of purified, recombinant ERα and ERβ expressed in insect cells. FLAG-tagged ERs were expressed by using recombinant baculovirus vectors and purified by anti-FLAG M2 affinity chromatography. Equal amounts of the receptor proteins were run on 10% acrylamide–SDS gels with subsequent staining using Coomassie Brilliant Blue R-250. The sizes of molecular mass markers are shown. (C) Assessment of ERα and ERβ transcriptional activities in receptor dose–response experiments using an in vitro chromatin assembly and transcription system. A plasmid template containing four EREs upstream of the adenovirus E4 promoter (pERE; top) was assembled into chromatin using the S190 extract in the presence of increasing amounts of purified ERα or ERβ, as indicated (in this experiment, all reactions that contained ER also contained E2). The chromatin samples were subjected to in vitro transcription analysis in duplicate using a HeLa cell nuclear extract, and the resulting RNA products were analyzed by primer extension (bottom). (D) Quantification by PhosphorImager analysis of multiple experiments like those shown in (C). Each point represents the mean ± SEM for three or more separate determinations.
As suggested by their domain structures, ERα and ERβ function as ligand-regulated, DNA-binding transcription factors (Couse and Korach, 1999; Nilsson et al., 2001). Their transcriptional activities are dependent on a variety of co-regulatory proteins (i.e. co-activators and co-repressors) that are recruited by the receptors to estrogen-regulated promoters embedded in chromatin through direct or indirect interactions (Nilsson et al., 2001). To date, a wide array of factors have been shown to interact with and enhance the transcriptional activities of ERα and ERβ (Klinge, 2000; Nilsson et al., 2001). A large subset of these factors interacts directly with the LBD in a ligand- and AF-2 dependent manner, including the steroid receptor co-activator (SRC) family of proteins and the Mediator-like complexes (e.g. TRAP, DRIP and ARC) (Klinge, 2000; Nilsson et al., 2001). Other factors, such as the histone acetyltransferase (HAT) p300/CBP and the histone methyltransferase (HMT) CARM-1, are recruited to the ERs primarily via interactions with the SRC proteins (Klinge, 2000; Nilsson et al., 2001). Several studies have shown that ERα and ERβ can bind to the SRCs with similar affinities (Tremblay et al., 1997; Cowley and Parker, 1999; Kraichely et al., 2000). Moreover, in transient transfection studies, SRC and p300/CBP were found to enhance the ligand-dependent transcriptional activity of both receptors (Smith et al., 1996; Tremblay et al., 1997; Klinge, 2000). SRCs have also been shown to interact with the N-terminal regions of ERα and ERβ, an interaction that may mediate synergy between AF-1 and AF-2 (Webb et al., 1998, 1999; Tremblay et al., 1999; Benecke et al., 2000; Metivier et al., 2001). A smaller subset of the ER-interacting factors have been shown to bind primarily to the N-terminal A/B region of the receptors (Klinge, 2000). These include the RNA-binding protein p68/72, which is found in a complex containing the AF-1-specific RNA co-activator SRA, as well as SRC proteins, and may be specific for ERα (Endoh et al., 1999; Lanz et al., 1999; Watanabe et al., 2001).
The fact that many nuclear receptor co-activators possess intrinsic histone-modifying activities suggests that chromatin is a major factor in determining transcriptional outcomes for hormone-regulated genes (Kraus and Wong, 2002). The packaging of promoters into chromatin results in a general repression in transcription (Kadonaga, 1998). Cofactors with histone-modifying and chromatin-remodeling activities function with nuclear receptors to overcome chromatin-mediated repression and activate transcription by RNA polymerase II (RNA pol II) (Kraus and Wong, 2002). In previous biochemical studies, we demonstrated the importance of chromatin in determining estrogen-regulated transcriptional outcomes mediated by ERα (Kraus and Kadonaga, 1998). Specifically, we were only able to recapitulate accurately ligand- and co-activator-dependent transcription by ERα with chromatin templates, but not naked DNA (Kraus and Kadonaga, 1998). Previous cell-based assays have shown that ERα and ERβ have different transcriptional activities in certain ligand, cell type and promoter contexts (Paech et al., 1997; Barkhem et al., 1998; Kuiper et al., 1998; McInerney et al., 1998; Cowley and Parker, 1999; Jones et al., 1999; Delaunay et al., 2000; Saville et al., 2000; Meyers et al., 2001). To explore the molecular mechanisms for these differences in further detail, including a possible role for chromatin, we have used a biochemical approach, including an in vitro chromatin assembly and transcription system. We find that ERα is a more potent transcriptional activator than ERβ with chromatin templates, but not with naked DNA. This difference is attributable to the N-terminal A/B region of ERα, which contains an AF that facilitates transcription specifically with chromatin templates. Collectively, our results define an important role for chromatin in determining signaling outcomes mediated by distinct subtypes of signal-transducing transcriptional activator proteins.
Results
ERα and ERβ are not equally potent transcriptional activators with chromatin templates
To compare the transcriptional activities of human ERα and ERβ, we used a biochemical approach, including a previously described in vitro chromatin assembly and transcription system that accurately recapitulates the known ligand-dependent transcriptional activities of nuclear receptors (Kraus and Kadonaga, 1998). FLAG epitope-tagged versions of human ERα and ERβ were expressed in Sf9 insect cells using recombinant baculoviruses and subsequently were purified using anti-FLAG M2 affinity chromatography (Figure 1B). The purified receptors exhibited similar levels of E2 binding at the saturating hormone concentrations used in our in vitro assays (i.e. >10 nM) (see Supplementary figure 1). In addition, the purified receptors showed similar apparent binding affinities for the Xenopus vitellogenin A2 ERE, as assessed by gel mobility shift assays (see Supplementary figure 2). Thus, the purified ERα and ERβ proteins exhibited similar ligand binding and DNA binding activities under the conditions used in our assays, allowing us to compare directly the transcriptional activities of the two receptors in a carefully controlled manner.
We compared the transcriptional activities of ERα and ERβ in a chromatin environment using an in vitro chromatin assembly and transcription system. The plasmid template pERE, which contains four copies of the Xenopus vitellogenin A2 ERE upstream of the adenovirus E4 promoter (Figure 1C, top), was assembled into chromatin using a Drosophila chromatin assembly extract (the S190) in the presence of E2 and increasing amounts of the receptor proteins. The templates were then transcribed using a HeLa cell nuclear extract as a source of the RNA pol II transcriptional machinery. As shown previously, ERα was a potent stimulator of transcription with chromatin templates, typically producing a 25- to 50-fold activation over basal transcription that was saturable at higher receptor concentrations (Figure 1C, lanes 1–6, and D). In contrast, ERβ was a weak activator with chromatin templates, typically producing a 3- to 7-fold activation over basal transcription that was also saturable at higher receptor concentrations (Figure 1C, lanes 7–12, and D). Thus, under assay conditions where ERα and ERβ exhibit similar binding to ligand and DNA, there is a large difference in their transcriptional activities.
Chromatin mediates the different transcriptional activities of ERα and ERβ
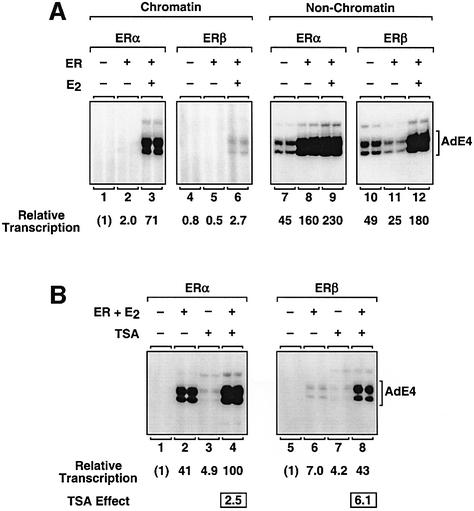
To explore the role of chromatin as a possible mediator of the different transcriptional activities of ERα and ERβ, we performed experiments comparing the activities of the two receptors with chromatin and non-chromatin (i.e. mock-assembled or naked DNA) templates (Figure 2A). As expected, the basal levels of transcription (i.e. without ERα or ERβ) with the non-chromatin templates were ∼40- to 50-fold higher than with the chromatin templates (compare lanes 1 and 4 with lanes 7 and 10). As shown above, liganded ERα was a much stronger activator than liganded ERβ with chromatin templates (Figure 2A, lanes 3 and 6). Surprisingly, when examined using non-chromatin templates, liganded ERα and ERβ showed similar levels of transcriptional activation (lanes 9 and 12). Thus, liganded ERα and ERβ exhibit different transcriptional responses that are regulated selectively by chromatin. Unliganded ERα and ERβ also showed different effects on transcription with the non-chromatin templates; unliganded ERα stimulated a 3- to 4-fold activation of transcription (lanes 7 and 8), whereas unliganded ERβ caused a 2-fold repression of transcription (lanes 10 and 11). These differences with non-chromatin templates are being pursued in more detail in other studies.
Fig. 2. ERα and ERβ exhibit different transcriptional responses with chromatin and non-chromatin templates. In vitro transcription reactions were performed as described for Figure 1C. ERα, ERβ, E2 and TSA were added as indicated. The relative transcription values listed for these and all other transcription experiments shown herein represent the mean from three or more separate determinations. All SEMs are <15%, but, more typically, <10%, of the mean value shown. (A) In vitro transcription experiments comparing the activities of ERα and ERβ with chromatin and non-chromatin (i.e. mock-assembled) templates. (B) Effect of TSA on the transcriptional activities of ERα and ERβ with chromatin templates.
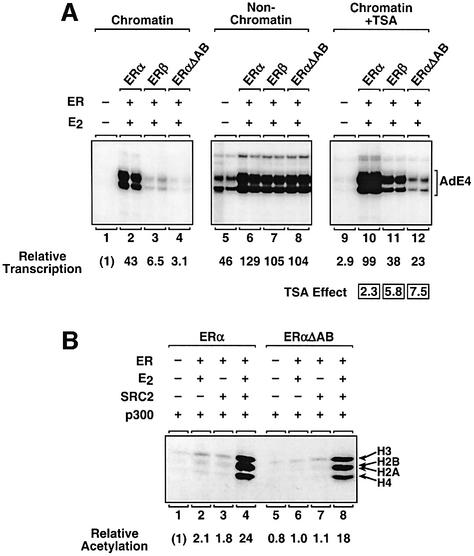
To examine further the role of chromatin in determining the different transcriptional responses of ERα and ERβ, we used the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) (Figure 2B). In this system, TSA blocks deacetylation by the endogenous HDACs in the S190 and HeLa extracts. The end result is a bulk increase in histone acetylation and a chromatin template that is less restrictive to transcription, as illustrated by a 3- to 5-fold increase in basal transcription (e.g. compare lanes 1 and 3, and lanes 5 and 7). With liganded ERα, TSA stimulated a modest (2.5-fold) increase in receptor-dependent transcription (lanes 2 and 4). With liganded ERβ, however, TSA stimulated a more robust (6-fold) increase in receptor-dependent transcription (lanes 6 and 8). The modest response of liganded ERα to TSA is not due simply to a saturation of RNA pol II transcription in these assays, as higher levels of transcription can be observed under other conditions (i.e. with liganded ERα using non-chromatin templates). These experiments with TSA confirmed our initial observations that ERα and ERβ exhibit different transcriptional responses that are regulated selectively by chromatin. In the experiments described below, we investigated the underlying biochemical and molecular basis for this effect.
ERα and ERβ bind to and remodel chromatin templates equally well
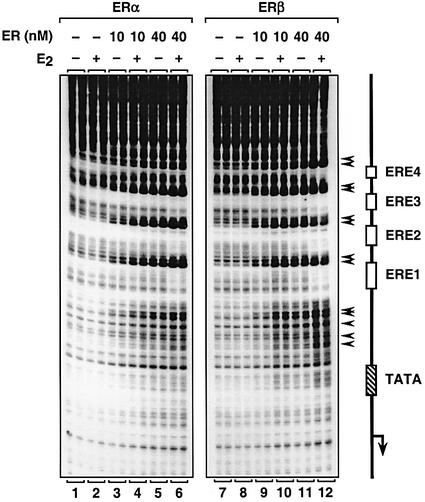
One explanation that might account for the observed differences in the transcriptional activities of liganded ERα and ERβ with chromatin templates is impaired binding of ERβ to chromatin. Although we found that both ERα and ERβ bound equally well to an ERE in gel mobility shift assays (Supplementary figure 2), we directly compared the ability of the two receptors to bind to a chromatin template containing EREs (pERE) by DNase I primer extension footprinting assays. As shown in Figure 3, the addition of ERα resulted in a reproducible pattern of DNase I hypersensitivity (black arrows) and protection in the area around the EREs (compare lanes 1 and 2 with lanes 3–6), as shown previously (Kraus and Kadonaga, 1998). The extent of DNase I hypersensitivity and protection was dependent on the receptor concentration and was approximately the same with or without E2, indicating that the binding of purified ERα to chromatin templates was largely ligand independent. The results obtained with ERβ were similar to those obtained with ERα in both the pattern and extent of digestion (compare lanes 7–12 with lanes 1–6), indicating that both ERα and ERβ bind to the Xenopus vitellogenin A2 ERE in templates assembled into chromatin with similar apparent binding affinities. Thus, it is unlikely that differences in the binding of ERα and ERβ to the chromatin templates account for the differences in the transcriptional activities of the receptors in our assays since we used the same chromatin templates for the transcription and footprinting assays.
Fig. 3. ERα and ERβ bind with similar apparent binding affinities to EREs in chromatin. In vitro DNase I footprinting experiments with chromatin templates. The plasmid template pERE was assembled into chromatin using the S190 extract in the presence or absence of ERα, ERβ and E2, as indicated. The chromatin samples were then subjected to DNase I primer extension footprinting analysis in duplicate. A schematic representation of the pERE template is shown to the right, including the location of the EREs, TATA box and transcription initiation site. The major DNase I-hypersensitive sites are indicated by arrows.
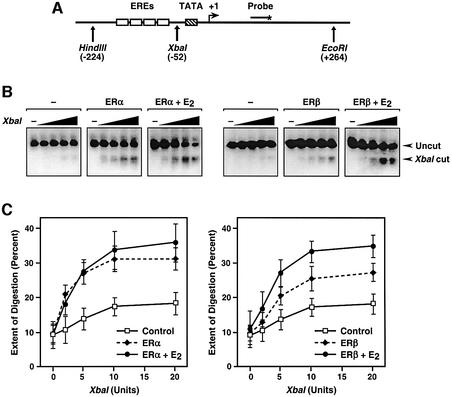
The binding of nuclear receptors to chromatin induces localized alterations in chromatin structure (i.e. chromatin remodeling) required for subsequent transcription by RNA pol II (Kraus and Wong, 2002). To examine the possibility that the observed differences in the transcriptional activities of liganded ERα and ERβ with chromatin templates were due to impaired receptor-dependent chromatin remodeling by ERβ, we performed restriction endonuclease accessibility assays (Figure 4). Briefly, S190-assembled chromatin, with or without ER and E2, was digested with increasing amounts of the restriction endonuclease XbaI, which cuts at –52 in pERE (relative to the 3′ most transcription initiation site), between the EREs and the TATA box (see template schematic in Figure 4A). The samples were deproteinized, digested with the restriction endonucleases HindIII and EcoRI to set 5′ and 3′ boundaries surrounding the promoter, and analyzed by Southern blotting with a probe that hybridizes between the XbaI and EcoRI sites (Figure 4A and B). The extent of XbaI digestion under each condition was quantified and plotted graphically (Figure 4C). Both liganded ERα and ERβ stimulated similar increases in XbaI accessibility. Although the effects with ERα were largely ligand independent, the effects with ERβ were enhanced by ∼2-fold with E2 (Figure 4C, compare the left and right panels). Collectively, the results of the footprinting assays and the restriction enzyme accessibility assays indicate that liganded ERα and ERβ exhibit similar chromatin binding and remodeling activities in spite of their dramatically different transcriptional activities with chromatin templates.
Fig. 4. Liganded ERα and ERβ stimulate similar levels of chromatin remodeling upon binding to chromatin templates. (A) Schematic diagram of the pERE template, showing the location of the EREs, TATA box, transcription initiation site, oligonucleotide probe and restriction endonuclease cleavage sites. (B) In vitro restriction endonuclease accessibility experiments. The plasmid template pERE was assembled into chromatin using the S190 extract in the presence or absence of ERα, ERβ and E2, as indicated. The chromatin samples were then subjected to digestion using increasing concentrations of the restriction endonuclease XbaI. After deproteinization, the templates were digested with HindIII and EcoRI to give common ends to the DNA fragments. The samples were analyzed by agarose gel electrophoresis with subsequent Southern blotting. The signals were quantified by PhosphorImager analysis and expressed as percentage digestion by XbaI. (C) Quantification of multiple experiments like those shown in (B). Each point represents the mean ± SEM for three or more separate determinations.
ERα and ERβ recruit SRC–p300 complexes to chromatin templates with similar efficiencies, but exhibit moderately different sensitivities to SRC and p300 co-activator activities
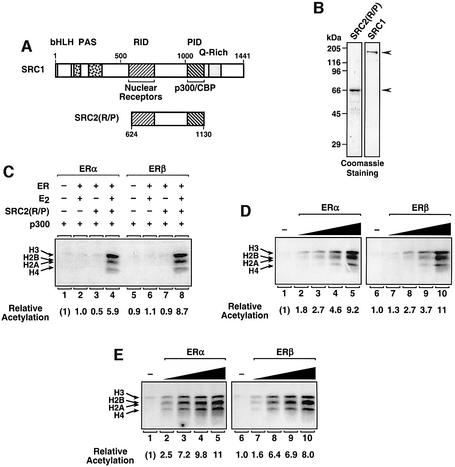
Since the differences we observed between the transcriptional activities of ERα and ERβ were not clearly explained by differences in binding to chromatin or receptor-mediated chromatin remodeling, we explored other possible mechanisms. The recruitment of bridging and histone-modifying co-activators (e.g. SRC proteins and p300/CBP, respectively) has been shown to play a critical role in estrogen-dependent transcription with chromatin (Kim et al., 2001); therefore, we examined whether ERα and ERβ might differ in their abilities to recruit these factors to free DNA or chromatin templates. In gel mobility shift assays, increasing amounts of a fragment of SRC2 containing the nuclear receptor and p300/CBP interaction domains (RID and PID, respectively), referred to as SRC2(R/P) (see Figure 5A and B), supershifted an ER–ERE complex in a ligand-dependent manner with similar efficiencies for ERα and ERβ, indicating similar affinities of the two receptors for the co-activator fragment (Supplementary figure 3).
Fig. 5. Liganded ERα and ERβ recruit p300 HAT activity to chromatin templates via SRC with similar efficiencies. (A) Schematic diagram of SRC1 and SRC2(R/P). Specific regions of the SRC proteins are indicated: basic helix–loop–helix region (bHLH), Per-Arnt-Sim domain (PAS), nuclear receptor interaction domain (RID), p300/CBP interaction domain (PID), and glutamine-rich region (Q-rich). The residues included in the SRC2(R/P) polypeptide are indicated. (B) SDS–PAGE analysis of purified, recombinant SRC1 and SRC2(R/P). The purified proteins were run on 12% acrylamide–SDS gels with subsequent staining using Coomassie Brilliant Blue R-250. The sizes of molecular mass markers are shown. (C) Targeted histone acetylation assays using SRC2(R/P). The plasmid template pERE was assembled into chromatin by salt gradient dialysis. Unincorporated (i.e. free) histones were removed by sucrose gradient centrifugation. The salt-dialyzed chromatin was used in acetylation reactions containing [3H]acetyl-CoA, as well as the following factors, as indicated: p300, SRC2(R/P), ERα, ERβ and E2. After incubation, the reactions were subjected to electrophoresis on 15% polyacrylamide–SDS gels with subsequent fluorography. The 3H-labeled core histone bands were excised from the gel and quantified by liquid scintillation counting. The core histones (H2A, H2B, H3 and H4) and relative acetylation levels are indicated. (D and E) Targeted histone acetyl ation assays performed in the presence of increasing amounts of ERα or ERβ (a range of 12.5–100 nM), as indicated. All samples that contained ER also contained p300, E2 and SRC2(R/P) (D) or SRC1 (E).
To examine co-activator recruitment in the context of chromatin, we used a previously described assay in which the ability of ER to recruit p300 HAT activity via SRC2 (R/P) was assessed by the receptor-dependent acetylation of nucleosomal histones in the presence of [3H]acetyl-CoA. As shown in Figure 5C, ERα-dependent acetylation of nucleosomal histones by p300 required both ligand (lanes 3 and 4) and the SRC2(R/P) fragment (lanes 2 and 4). Similar results were observed with ERβ (lanes 5–8). For a better comparison of the efficiencies with which ERα and ERβ recruit p300 to chromatin templates, we performed receptor dose–response experiments using the HAT recruitment assay (Figure 5D). ERα and ERβ recruited p300 via SRC2(R/P) with similar efficiencies (compare lanes 1–5 with lanes 6–10). Since SRCs have been shown to bind to the N-terminal region of ERα, but not ERβ, through a C-terminal domain not present in SRC2(R/P) (Webb et al., 1998, 1999), we repeated the assay shown in Figure 5D using full-length SRC1. As we observed with SRC2(R/P), ERα and ERβ recruited p300 via SRC1 with similar efficiencies (Figure 5E; compare lanes 1–5 with lanes 6–10). Thus, by three different assays (Figure 5D and E; Supplementary figure 3), liganded ERα and ERβ did not differ substantially in the ability to recruit p300 and/or SRCs.
Next, we examined the transcriptional activities of ERα and ERβ in response to exogenously added p300, SRC2 (R/P) and SRC1 in dose–response transcription studies with chromatin templates (Supplementary figure 4). In all cases, with the exception of ERα with SRC1, the exogenously added factors enhanced receptor-dependent transcription. Compared with ERβ, ERα showed a moderately increased sensitivity (i.e. half-maximal effective concentration or EC50) to both p300 and SRC2(R/P), exhibiting an ∼3-fold lower EC50 in both cases (i.e. a dose shift to the left). However, compared with ERα, ERβ showed a greater responsiveness (i.e. fold activation in the presence of saturating concentrations of the co-activators) to p300 (2- versus 7-fold), SRC2(R/P) (1.5- versus 4-fold, respectively) and SRC1 (no enhancement versus 3-fold) in spite of the fact that ERα gave a greater maximal level of transcription with or without the exogenously added co-activators in all cases. Thus, although ERα was slightly more sensitive to lower concentrations of the co-activators, ERβ gave a greater response to the co-activators, possibly due to its intrinsically weak transcriptional activity in these assays. Nevertheless, the small differences that we found with these particular co-activators are probably insufficient to account for the large differences in the maximal transcriptional activities observed with the two different ERs.
Synergism between AF-1 and AF-2 of ERα is required for transcriptional activation with chromatin templates, but not naked DNA
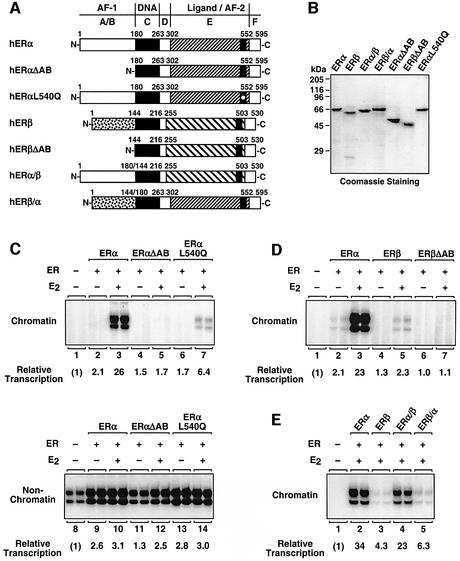
Since our initial comparative functional assays did not provide an obvious explanation for the large transcriptional differences between ERα and ERβ, we considered intrinsic differences in the transcriptional activation functions of the two receptors (i.e. AF-1 and AF-2). For these studies, we expressed and purified the panel of mutant ERs shown in Figure 6A and B. First, we examined the contribution of the two AFs in ERα since ERα is the more transcriptionally potent of the two ERs with chromatin templates. Deletion of the N-terminal A/B region of ERα, which contains AF-1, completely abrogated ligand-dependent transcription by the receptor with chromatin templates (Figure 6C, compare lanes 3 and 5). Likewise, a single point mutation in ERα (Leu540Gln) that greatly reduces AF-2 activity (Wrenn and Katzenellenbogen, 1993) also caused a dramatic reduction in ligand-dependent transcription by the receptor with chromatin templates (Figure 6C, compare lanes 3 and 7). Thus, both AF-1 and AF-2 are required for efficient ERα-dependent transcription with chromatin templates in these particular promoter and cell (i.e. HeLa cell extract) contexts. In addition, our results indicate that AF-1 and AF-2 function synergis tically in ERα-dependent transcription with chromatin templates (Figure 6C, lanes 3, 5 and 7), but not naked DNA (Figure 6C, lanes 10, 12 and 14). Thus, synergism between AF-1 and AF-2 is an important mechanism for transcriptional activation by ERα in a transcriptionally repressive chromatin environment.
Fig. 6. The A/B region of ERα, but not ERβ, contains a transferable activation function required for transcriptional activation with chromatin templates. (A) Schematic diagrams of wild-type and variant ERα and ERβ proteins. The specific residues present in each ER variant are indicated. (B) SDS–PAGE analysis of purified, recombinant wild-type and variant ERα and ERβ proteins expressed in insect cells. The proteins were expressed, purified and analyzed as described in Figure 1B. The sizes of molecular mass markers are shown. (C–E) Transcriptional activities of wild-type and variant ERs. In vitro transcription reactions were performed as described for Figure 1C. Wild-type and variant ERs and E2 were added as indicated.
Previous studies have suggested that the ERβ N-terminal A/B region contains a repression function that might attenuate the transcriptional activity of the receptor in certain promoter and cell contexts (Hall and McDonnell, 1999). To determine whether such a repression function might be contributing to the weak activity of ERβ in our assays, we generated a version of ERβ lacking the A/B region (ERβΔAB) and tested its transcriptional activity with chromatin templates. We reasoned that if the ERβ A/B region contained a repression function, deletion of the A/B region would lead to increased activity. As shown in Figure 6D (lanes 5 and 7), this was not the case; in fact, ERβΔAB had reduced activity. Thus, the A/B region of ERβ does not contain a repression function that would account for the weak transcriptional activity of the receptor in our assays.
The A/B region of ERα, but not ERβ, contains a transferable activation function required for transcriptional activation with chromatin templates
We hypothesized that the A/B region of ERα might contain a determinant that distinguishes between the strong transcriptional activity of ERα and the weak transcriptional activity of ERβ with chromatin based on the following: (i) the low level of amino acid sequence homology between the A/B regions of ERα and ERβ (see Figure 1A); (ii) the functional equivalence of ERα and ERβ AF-2 activities in AF-2-dependent functional assays (e.g. Figure 5 and Supplementary figure 3); (iii) the dependence of ERα on AF-1 for efficient transcription with chromatin templates (Figure 6C); and (iv) previous studies suggesting a role for the A/B region in determining ERα and ERβ activities in cell-based assays (McInerney et al., 1998; Cowley and Parker, 1999; Hall and McDonnell, 1999; Jones et al., 1999; Delaunay et al., 2000). To test this hypothesis directly, we performed domain swap experiments with the A/B regions of ERα and ERβ. Replacement of the A/B region of ERβ with the same region from ERα produced an ERβ variant with greatly increased transcriptional activity in the context of chromatin (Figure 6E, lanes 3 and 4). In contrast, the reciprocal domain swap produced an ERα variant with weak transcriptional activity in the context of chromatin (lanes 2 and 5). Thus, the A/B region of ERα, but not ERβ, contains a strong transferable activation function that supports transcription with chromatin templates.
To explore further the role of the A/B region in ERα-dependent transcription, we compared the activity of ERαΔAB with the activities of ERα and ERβ in additional assays. The results from the domain swap experiments suggested that deletion of the ERα A/B region might produce a receptor with weak transcriptional responses more similar in magnitude to the responses of ERβ than ERα. This was indeed the case in the transcription assays shown in Figure 7A, comparing receptor activity with chromatin templates (lanes 1–4), non-chromatin templates (lanes 5–8) and chromatin templates in the presence of TSA (lanes 9–12). Furthermore, in HAT recruitment assays, ERαΔAB, like ERβ, was able to recruit p300 HAT activity with an efficiency similar to ERα in spite of its weak overall transcriptional activity (Figures 5A and 7B). Thus, as predicted, ERαΔAB showed responses in the transcription and HAT assays that were similar in magnitude to the responses of ERβ, suggesting that the ERα A/B region contains a determinant that distinguishes between the strong transcriptional activity of ERα and the weak transcriptional activity of ERβ.
Fig. 7. Deletion of the N-terminal A/B region of ERα generates a receptor whose transcriptional responses are similar in magnitude to the responses obtained with ERβ. (A) In vitro chromatin assembly and transcription assays comparing the transcriptional activities of ERα, ERβ and ERαΔAB. The assays were performed as described for Figure 1C. The ER proteins, E2 and TSA were added as indicated. Note that the exposure time for the ‘non-chromatin’ gel was reduced relative to the other two gels to show better the effects on basal transcription. (B) Targeted histone acetylation assays comparing the activities of ERα and ERαΔAB. The assays were performed as described for Figure 5C. Similar assays comparing the activities of ERα and ERβ can be found in Figure 5C.
Discussion
ERα and ERβ are not functionally equivalent as transcriptional activators with chromatin templates
Nuclear signaling by estrogens is mediated by two distinct receptor proteins, ERα and ERβ (Nilsson et al., 2001). Previous studies have indicated that although ERα and ERβ share significant sequence, structural and functional homologies, they may exhibit different activities with regard to ligand binding, DNA binding, co-regulator interactions and transcriptional activation in certain ligand-, cell- and gene-specific contexts (see Introduction). With regard to transcriptional activity, ERα generally, but not exclusively, has been found to be a more potent transcriptional activator than ERβ in cell-based assays (McInerney et al., 1998; Cowley and Parker, 1999; Hall and McDonnell, 1999), although the molecular mechanisms underlying this difference are unclear. This is due, in part, to the fact that parameters such as receptor concentration, DNA or chromatin binding, receptor–co-regulator interactions and chromatin remodeling are difficult to control or assess quantitatively in intact cells. To address the mechanistic basis for the different transcriptional activities of ERα and ERβ, we have used a biochemical approach to examine and compare the transcriptional activities of ERα and ERβ under experimental conditions where differences in these various parameters are minimized. Furthermore, we have been able to assay the same preparations of receptor in different types of assays (e.g. ligand binding, chromatin footprinting, chromatin remodeling, HAT recruitment and transcription), allowing for greater internal consistency in our experiments than has been achieved previously.
Our results indicate that biochemically pure, ligand-activated human ERα and ERβ do not have equivalent transcriptional activities in the context of chromatin (Figures 1C and 2A). Yet, the same preparations of the receptors have similar E2-binding capacities at saturating hormone concentrations (Supplementary figure 1) and similar apparent binding affinities for a ‘perfect’ ERE, namely the ERE from the Xenopus vitellogenin A2 gene (Supplementary figure 2). Likewise, the purified E2-bound receptors (i.e. the transcriptionally active forms) are similar with respect to their apparent binding affinities for EREs in chromatin (Figure 3), abilities to stimulate chromatin remodeling (Figure 4), efficiencies in recruiting p300 HAT activity to a chromatin template via SRCs (Figure 5; see also Supplementary figure 3) and transcriptional activities with naked DNA (Figure 2A). Thus, under conditions where many biochemical activities of ERα and ERβ are similar, the two receptors still exhibit large differences in their transcriptional activities with chromatin templates (Figures 1C and 2A). Although it is likely that differences in ligand binding, DNA binding and cell context are important in determining outcomes in the estrogen signaling pathway with synthetic ER ligands, natural EREs and physiological target tissues (Klinge, 2001; Nilsson et al., 2001), it is only with the use of a biochemical system where these and other parameters are controlled that the central role for chromatin in exposing the intrinsic differences in the transcriptional activities of ERα and ERβ can be observed.
Chromatin exposes the different transcriptional activities of ERα and ERβ
Various mechanisms may account for the different gene regulatory activities of ERα and ERβ noted by us herein and by others. For example, in some gene contexts, the sequence of an ERE might lead to higher affinity binding of one ER subtype and, hence, a greater role for that ER subtype in the regulation of a particular gene (Klinge, 2001). Likewise, in certain pharmacological contexts, the structure of the ligand could lead to greater activation or inhibition of one ER subtype, selectively enhancing or inhibiting the activity of that ER subtype (Kuiper et al., 1998; Meyers et al., 2001). Herein, we identify a previously uncharacterized regulatory mechanism for distinguishing between the intrinsic transcriptional activities of ERα and ERβ, namely chromatin structure at estrogen-regulated promoters. The in vitro chromatin assembly system that we used in our experiments generates a chromatin template that is very restrictive to transcription (note the very low levels of basal transcription). When this restrictive structure was ‘loosened’ (e.g. by the addition of TSA), the transcriptional activity of ERβ increased dramatically (Figure 2B). TSA in this system does not disassemble the chromatin template, as nucleosomal arrays are still observed upon micrococcal nuclease digestion (data not shown), and basal transcription, although elevated, is considerably less than that observed with naked DNA (Figure 2). Our results suggest that chromatin has the potential to act as a ‘molecular rheostat’, allowing ligand-dependent transcriptional activation by ERβ under some conditions (e.g. open chromatin architecture), while restricting it under other conditions (e.g. closed chromatin architecture). Thus, two different ER subtypes within the same cell could differentially control the expression of the wide array of estrogen-regulated genes in the chromatin environment of the nucleus. It will be interesting to explore this and other possible mechanisms further in future studies using cell-based assays.
Chromatin is a dynamic polymer that exhibits structural alterations both locally (e.g. by the addition of linker histones and through changes in the association of specific chromatin domains with the nuclear matrix) and globally (e.g. during the cell cycle and DNA replication) (Kadonaga, 1998). As such, transcriptional activation by ERβ could be restricted by chromatin to particular regions of the genome or to certain times during the cell cycle. Previous studies with glucocorticoid receptor and progesterone receptor support the idea that the nature of different chromatin environments can influence outcomes in steroid hormone-mediated signaling within the same cell (Archer et al., 1994; Lambert and Nordeen, 1998). Interestingly, the same general principles regarding the regulatory effects of chromatin also apply to ERα, as the assembly of higher order chromatin structures through the addition of the linker histone H1 can act to repress ERα transcriptional activity (Cheung et al., 2002). Note that the role of chromatin in distinguishing between the intrinsic transcriptional activities of the two ERs may be limited to ligand and promoter contexts where both AF-1 and AF-2 are required for full activation (i.e. with a classical ERE, as we have used herein; see Figure 6C), since ERβ has been shown to be a more potent activator than ERα with some non-classical EREs and synthetic ligands (see for example Paech et al., 1997; Barkhem et al., 1998; Kuiper et al., 1998; Jones et al., 1999; Meyers et al., 2001).
The ERα A/B region contains a transferable activation function that facilitates transcription with chromatin templates
Previous studies have shown that the amino acid sequence differences between the A/B regions of ERα and ERβ can contribute to the different transcriptional activities of the two receptors in various cell type and promoter contexts (McInerney et al., 1998; Cowley and Parker, 1999; Hall and McDonnell, 1999; Jones et al., 1999; Delaunay et al., 2000). Our results suggest that the A/B regions are also important in distinguishing between the different transcriptional activities of the two ERs in certain chromatin contexts. Specifically, we found that the A/B region of ERα, but not ERβ, contains a transferable activation function (AF-1) that facilitates transcription with chromatin templates (Figure 6E), but not naked DNA (Figure 7A). This finding raises several important questions. For example, what makes the ERα AF-1 a more potent activation domain than the ERβ AF-1? What are the underlying molecular determinants of a ‘chromatin-dependent’ AF? The answers to these questions are probably found in the protein–protein interactions that are specific to the A/B region of one ER subtype or the other.
A small number of ER-interacting factors that bind primarily to the N-terminal A/B regions of the receptors and function as transcriptional co-activators have been identified (Klinge, 2000). These include the RNA-binding protein p68/72, which is found in a complex containing the AF-1-specific RNA co-activator SRA, as well as SRC proteins (Endoh et al., 1999; Lanz et al., 1999; Watanabe et al., 2001). Interestingly, p68/p72 shows selectivity for ERα with regard to both receptor binding and transcriptional enhancement (Watanabe et al., 2001), as would be expected of an ERα- and AF-1-selective chromatin-dependent activation domain. Similar selectivity for the binding of TBP to the ERα A/B region has also been reported (Warnmark et al., 2001b). Whether these or other as yet unidentified factors contribute to the distinct transcriptional activities of ERα and ERβ with chromatin templates has not yet been determined.
An alternative possibility to explain the different transcriptional activities of ERα and ERβ with chromatin templates would be a repressive function in the ERβ A/B region, as has been suggested previously (Hall and McDonnell, 1999). Again, such an activity would probably require the selective binding of a factor to the ERβ A/B region that could repress transcription in the context of chromatin. However, in our studies, we did not observe a repressive activity for the ERβ A/B region (Figure 6D). It is possible that such an activity might only be observed in the context of an ERα–ERβ heterodimer. Indeed, we have observed repression of ERα-mediated transcription with chromatin templates upon the addition of ERβ (E.Cheung and W.L.Kraus, unpublished data). Yet, this seems unlikely to account for the different transcriptional activities that we see with ER homodimers.
In contrast to AF-1, cofactor interactions involving the AF-2s of ERα and ERβ are unlikely to account for the different transcriptional activities observed for the two receptors with chromatin templates. AF-2-dependent co-activators, such as the SRC family of proteins and the TRAP complex (especially the receptor-binding TRAP220 subunit), have been shown to bind equally well or more strongly to ERβ than ERα (Cowley and Parker, 1999; Burakov et al., 2000; Warnmark et al., 2001a; Kang et al., 2002; see also Figure 5 and Supplementary figure 3). Likewise, the enhancing effect of the TRAP complex on ER-mediated transcription was shown to be greater for ERβ than ERα, although these studies were done with naked DNA (Kang et al., 2002). Furthermore, we observed only modest differences in the transcriptional responses of ERα and ERβ to p300, SRC2(R/P) and SRC1 (Supplementary figure 4), which are unlikely to be of sufficient magnitude to account for the large differences in the transcriptional activities of the two receptors with chromatin templates. Finally, we have shown that the AF-1 of ERα can synergize with the AF-2 of either ERα or ERβ with chromatin templates (Figure 6C and E), suggesting that under our assays conditions both AF-2s are functionally similar. Collectively, our studies suggest that differences between the AF-1s, not the AF-2s, of ERα and ERβ underlie the different transcriptional activities of the two receptors in the context of chromatin.
Materials and methods
Synthesis and purification of recombinant proteins
FLAG-tagged hERα(1–595), hERβ(1–530), hER derivatives and full-length SRC1 were expressed in Sf9 cells and purified as described previously (Kraus and Kadonaga, 1999; Thackray and Nordeen, 2002). The chimeric hER cDNAs have been described elsewhere (McInerney et al., 1998). The cDNAs for the other hER derivatives were constructed by PCR. His6-tagged human p300 was expressed in Sf9 cells and purified as described previously (Kraus and Kadonaga, 1999). His6-tagged mouse SRC2(R/P), which contains the receptor and p300/CBP interaction domains of the protein (amino acids 624–1130), was expressed in Escherichia coli and purified as described previously (Kim et al., 2001).
Chromatin assembly and analysis
The plasmid template pERE contains four copies of the Xenopus vitellogenin A2 gene ERE located upstream of the adenovirus E4 core promoter (Kraus and Kadonaga, 1998). Chromatin assembly reactions were carried out as described previously using an extract derived from Drosophila embryos (the S190) (Kraus and Kadonaga, 1999). Purified ER proteins, E2 and TSA were added during the chromatin assembly reactions at concentrations of 10 nM, 100 nM and 10 µM, respectively, unless indicated otherwise. Purified p300, SRC2(R/P) and SRC1 proteins were added after the chromatin assembly reactions were complete, followed by a 15 min incubation at 27°C to allow interaction of the factors with ER and the chromatin templates. DNase I primer extension footprinting assays and restriction endonuclease accessibility assays were performed as described previously (Pazin and Kadonaga, 1998; Cheung et al., 2002). For the restriction endonuclease accessibility assays, the data were quantified by PhosphorImager analysis (Molecular Dynamics). Both assays were run a minimum of three separate times to ensure reproducibility.
In vitro transcription assays
In vitro transcription reactions with chromatin templates were performed as described previously using a HeLa cell nuclear extract (Kraus and Kadonaga, 1998, 1999). The RNA products were analyzed by primer extension (Kraus and Kadonaga, 1999). Transcription with mock-assembled (i.e. non-chromatin) templates was performed as described previously (Kraus and Kadonaga, 1999). The data from the transcription experiments were quantified by PhosphorImager analysis (Molecular Dynamics). All transcription reactions were carried out in duplicate, and each experiment was performed three or more times to ensure reproducibility. Note that the final concentrations of factors, E2 and TSA in the transcription reactions were approximately one-third of the concentrations listed for the chromatin assembly reactions.
Targeted histone acetylation assays
Targeted histone acetylation assays with chromatin templates were performed as described previously (Kim et al., 2001). Briefly, aliquots of salt-dialyzed chromatin assembled using pERE were incubated with [3H]acetyl-CoA (5 µM) and various combinations of ER proteins (100 nM), p300 (40 nM), SRC2(R/P) (40 nM), SRC1 (10 nM) and E2 (1 µM) for 30 min at 27°C in a final volume of 40 µl under reaction conditions described previously (Kim et al., 2001). The reactions were subjected to SDS–PAGE and fluorography, and the 3H-labeled histone bands were excised from the gel and quantified by liquid scintillation counting. The assays were run at least three separate times to ensure reproducibility.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Steve Nordeen, John Lis, Mi Young Kim, Mari Acevedo and Kathy Lee for critical reading of this manuscript, Benita Katzenellenbogen for the chimeric ER constructs, Steve Nordeen for the recombinant SRC1 baculovirus, and Mi Young Kim for mutant ERα proteins and help with the HAT recruitment assays. This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (DK58110) to W.L.K., and a postdoctoral fellowship from the Susan G.Komen Breast Cancer Foundation to E.C.
References
- Archer T.K., Zaniewski,E., Moyer,M.L. and Nordeen,S.K. (1994) The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol. Endocrinol., 8, 1154–1162. [DOI] [PubMed] [Google Scholar]
- Barkhem T., Carlsson,B., Nilsson,Y., Enmark,E., Gustafsson,J. and Nilsson,S. (1998) Differential response of estrogen receptor α and estrogen receptor β to partial estrogen agonists/antagonists. Mol. Pharmacol., 54, 105–112. [DOI] [PubMed] [Google Scholar]
- Benecke A., Chambon,P. and Gronemeyer,H. (2000) Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep., 1, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov D., Wong,C.W., Rachez,C., Cheskis,B.J. and Freedman,L.P. (2000) Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J. Biol. Chem., 275, 20928–20934. [DOI] [PubMed] [Google Scholar]
- Cheung E., Zarifyan,A.S. and Kraus,W.L. (2002) Histone H1 represses estrogen receptor α transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol., 22, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse J.F. and Korach,K.S. (1999) Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev., 20, 358–417. [DOI] [PubMed] [Google Scholar]
- Cowley S.M. and Parker,M.G. (1999) A comparison of transcriptional activation by ERα and ERβ. J. Steroid Biochem. Mol. Biol., 69, 165–175. [DOI] [PubMed] [Google Scholar]
- Delaunay F., Pettersson,K., Tujague,M. and Gustafsson,J.A. (2000) Functional differences between the amino-terminal domains of estrogen receptors α and β. Mol. Pharmacol., 58, 584–590. [DOI] [PubMed] [Google Scholar]
- Endoh H. et al. (1999) Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol., 19, 5363–5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hall J.M. and McDonnell,D.P. (1999) The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology, 140, 5566–5578. [DOI] [PubMed] [Google Scholar]
- Jones P.S., Parrott,E. and White,I.N. (1999) Activation of transcription by estrogen receptor α and β is cell type- and promoter-dependent. J. Biol. Chem., 274, 32008–32014. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. (1998) Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell, 92, 307–313. [DOI] [PubMed] [Google Scholar]
- Kang Y.K., Guermah,M., Yuan,C.X. and Roeder,R.G. (2002) The TRAP/Mediator coactivator complex interacts directly with estrogen receptors α and β through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl Acad. Sci. USA, 99, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Hsiao,S.J. and Kraus,W.L. (2001) A role for coactivators and histone acetylation in estrogen receptor α-mediated transcription initiation. EMBO J., 20, 6084–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge C.M. (2000) Estrogen receptor interaction with co-activators and co-repressors. Steroids, 65, 227–251. [DOI] [PubMed] [Google Scholar]
- Klinge C.M. (2001) Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res., 29, 2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraichely D.M., Sun,J., Katzenellenbogen,J.A. and Katzenellenbogen, B.S. (2000) Conformational changes and coactivator recruitment by novel ligands for estrogen receptor-α and estrogen receptor-β: correlations with biological character and distinct differences among SRC coactivator family members. Endocrinology, 141, 3534–3545. [DOI] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev., 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1999) Ligand- and cofactor-regulated transcription with chromatin templates. In Picard,D. (ed.), Steroid/Nuclear Receptor Superfamily: A Practical Approach. Oxford University Press, Oxford, UK, pp. 167–189.
- Kraus W.L. and Wong,J. (2002) Nuclear receptor-dependent transcription with chromatin—is it all about enzymes? Eur. J. Biochem., 269, 2275–2283. [DOI] [PubMed] [Google Scholar]
- Kuiper G.G., Lemmen,J.G., Carlsson,B., Corton,J.C., Safe,S.H., van der Saag,P.T., van der Burg,B. and Gustafsson,J.A. (1998) Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology, 139, 4252–4263. [DOI] [PubMed] [Google Scholar]
- Lambert J.R. and Nordeen,S.K. (1998) Steroid-selective initiation of chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter is controlled by the site of promoter integration. J. Biol. Chem., 273, 32708–32714. [DOI] [PubMed] [Google Scholar]
- Lanz R.B., McKenna,N.J., Onate,S.A., Albrecht,U., Wong,J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1999) A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell, 97, 17–27. [DOI] [PubMed] [Google Scholar]
- McInerney E.M., Weis,K.E., Sun,J., Mosselman,S. and Katzenellenbogen, B.S. (1998) Transcription activation by the human estrogen receptor subtype β (ERβ) studied with ERβ and ERα receptor chimeras. Endocrinology, 139, 4513–4522. [DOI] [PubMed] [Google Scholar]
- Metivier R., Penot,G., Flouriot,G. and Pakdel,F. (2001) Synergism between ERα transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol. Endocrinol., 15, 1953–1970. [DOI] [PubMed] [Google Scholar]
- Meyers M.J., Sun,J., Carlson,K.E., Marriner,G.A., Katzenellenbogen, B.S. and Katzenellenbogen,J.A. (2001) Estrogen receptor-β potency-selective ligands: structure–activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J. Med. Chem., 44, 4230–4251. [DOI] [PubMed] [Google Scholar]
- Nilsson S. et al. (2001) Mechanisms of estrogen action. Physiol. Rev., 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- Paech K., Webb,P., Kuiper,G.G., Nilsson,S., Gustafsson,J., Kushner,P.J. and Scanlan,T.S. (1997) Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science, 277, 1508–1510. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1998) Transcriptional and structural analysis of chromatin assembled in vitro. In Gould,H. (ed.), Chromatin: A Practical Approach. Oxford University Press, Oxford, UK, pp. 173–194.
- Pettersson K. and Gustafsson,J.A. (2001) Role of estrogen receptor β in estrogen action. Annu. Rev. Physiol., 63, 165–192. [DOI] [PubMed] [Google Scholar]
- Saville B., Wormke,M., Wang,F., Nguyen,T., Enmark,E., Kuiper,G., Gustafsson,J.A. and Safe,S. (2000) Ligand-, cell- and estrogen receptor subtype (α/β)-dependent activation at GC-rich (Sp1) promoter elements. J. Biol. Chem., 275, 5379–5387. [DOI] [PubMed] [Google Scholar]
- Smith C.L., Onate,S.A., Tsai,M.J. and O’Malley,B.W. (1996) CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc. Natl Acad. Sci. USA, 93, 8884–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. and Fuqua,S.A. (2001) Estrogen receptor and breast cancer. Semin. Cancer Biol., 11, 339–352. [DOI] [PubMed] [Google Scholar]
- Thackray V.G. and Nordeen,S.K. (2002) High-yield purification of functional, full-length steroid receptor coactivator 1 expressed in insect cells. Biotechniques, 32, 260–263. [DOI] [PubMed] [Google Scholar]
- Tremblay G.B., Tremblay,A., Copeland,N.G., Gilbert,D.J., Jenkins,N.A., Labrie,F. and Giguere,V. (1997) Cloning, chromosomal localization and functional analysis of the murine estrogen receptor β. Mol. Endocrinol., 11, 353–365. [DOI] [PubMed] [Google Scholar]
- Tremblay A., Tremblay,G.B., Labrie,F. and Giguere,V. (1999) Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activation function AF-1. Mol. Cell, 3, 513–519. [DOI] [PubMed] [Google Scholar]
- Tzukerman M.T., Esty,A., Santiso-Mere,D., Danielian,P., Parker,M.G., Stein,R.B., Pike,J.W. and McDonnell,D.P. (1994) Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol. Endocrinol., 8, 21–30. [DOI] [PubMed] [Google Scholar]
- Warnmark A., Almlof,T., Leers,J., Gustafsson,J.A. and Treuter,E. (2001a) Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERα and ERβ. J. Biol. Chem., 276, 23397–23404. [DOI] [PubMed] [Google Scholar]
- Warnmark A., Wikstrom,A., Wright,A.P., Gustafsson,J.A. and Hard,T. (2001b) The N-terminal regions of estrogen receptor α and β are unstructured in vitro and show different TBP binding properties. J. Biol. Chem., 276, 45939–45944. [DOI] [PubMed] [Google Scholar]
- Watanabe M. et al. (2001) A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J., 20, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Webb P. et al. (1998) Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol., 12, 1605–1618. [DOI] [PubMed] [Google Scholar]
- Webb P. et al. (1999) The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol. Endocrinol., 13, 1672–1685. [DOI] [PubMed] [Google Scholar]
- Wrenn C.K. and Katzenellenbogen,B.S. (1993) Structure–function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J. Biol. Chem., 268, 24089–24098. [PubMed] [Google Scholar]