Abstract
ERM (ezrin/radixin/moesin) proteins recognize the cytoplasmic domains of adhesion molecules in the formation of the membrane-associated cytoskeleton. Here we report the crystal structure of the radixin FERM (4.1 and ERM) domain complexed with the ICAM-2 cytoplasmic peptide. The non-polar region of the ICAM-2 peptide contains the RxxTYxVxxA sequence motif to form a β-strand followed by a short 310-helix. It binds the groove of the phosphotyrosine-binding (PTB)-like subdomain C mediated by a β–β association and several side-chain interactions. The binding mode of the ICAM-2 peptide to the FERM domain is distinct from that of the NPxY motif-containing peptide binding to the canonical PTB domain. Mutation analyses based on the crystal structure reveal the determinant elements of recognition and provide the first insights into the physical link between adhesion molecules and ERM proteins.
Keywords: cell adhesion/cytoskeleton/recognition/signal transduction/X-ray
Introduction
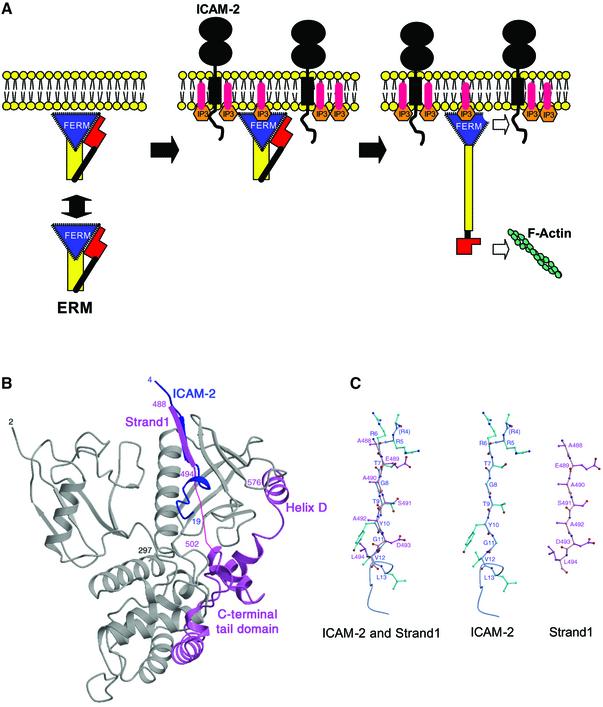
In eukaryotic cells, actin filaments are concentrated in a layer beneath the plasma membrane called the cell cortex. Actin rearrangements within the cortex provide the molecular basis for changes in cell shape and for cell locomotion, cell adhesion and cell–cell communication. Cross-linker proteins that bind both membrane proteins and actin filaments mediate the formation of the membrane-associated cytoskeleton. One such protein is radixin, which is a member of ERM (ezrin/radixin/moesin) proteins localized at cell-surface structures such as microvilli, ruffling membranes and cell-adhesion sites (Sato et al., 1991, 1992; Franck et al., 1993; Amieva and Furthmayr, 1995; Serrador et al., 1997).
ERM proteins are close homologues with ∼75% sequence identity (Arpin et al., 1994; Takeuchi et al., 1994; Bretscher, 1999; Mangeat et al., 1999; Tsukita and Yonemura, 1999) and have three domains: an N-terminal FERM (band 4.1 protein and ERM homology) domain consisting of ∼300 residues (radixin residues 1–297), a central helical domain of ∼200 residues (radixin 310–470) and a C-terminal tail domain of ∼100 residues (radixin 477–583). The FERM domain of ERM proteins is responsible for binding to the cytoplasmic parts of adhesion molecules, while the C-terminal tail domain binds F-actin (Turunen et al., 1994). FERM domains have been found in several other proteins localized at plasma membranes (Chishti et al., 1998) and are currently thought to form a protein module to mediate protein–protein and protein–membrane interactions.
ERM proteins partition between the cell cortex and the cytosol. In the cytosol, ERM proteins exist as a masked form in which the FERM domain binds the C-terminal tail domain to mutually mask the binding sites for their binding partners (Andreoli et al., 1994; Magendantz et al., 1995; Gary and Bretcher, 1995; Hirao et al., 1996). This masked molecule is activated by the binding of phosphatidylinositol 4,5-bisphosphate (PIP2) to the FERM domain (Niggli et al., 1995; Hirao et al., 1996; Nakamura et al., 1999; Yonemura et al., 2002). Recent crystal structures of the radixin FERM domain complexed with the head group of PIP2, inositol-(1,4,5)-trisphosphate (IP3), have revealed that the IP3 binding induces local conformational changes in the FERM domain to release the C-terminal tail domain (Hamada et al., 2000). The unmasked state of ERM proteins is stabilized by phosphorylation of the C-terminal tail domain (reviewed by Tsukita and Yonemura, 1999; Bretscher et al., 2000).
To date, three adhesion molecules have been characterized as possible membrane partners of ERM proteins. These are a cell-surface hyaluronate receptor protein, CD44 (Tsukita et al., 1994; Serrador et al., 1997), a cell-surface glycoprotein of the sialomutin family, CD43 (Yonemura et al., 1993; Serrador et al., 1998) and the immunoglobulin-superfamily membrane proteins, ICAM-1 (Heiska et al., 1998), -2 (Helander et al., 1996; Heiska et al., 1998) and -3 (Serrador et al., 1997). These ICAMs mediate leukocyte and endothelial binding to β2-integrins. ICAM-1, -2 and -3 bind lymphocyte function-associated antigen-1 (LFA-1) via the N-terminal one or two immunoglobulin-fold domain(s), and ICAM-1 binds complement receptor 3/Mac-1 via the third immuno globulin-fold domain. Thus, ICAMs do play an important role in cell–cell communication in the immune system.
How cross-linker proteins recognize adhesion molecules continues to be an area of intensive study. CD44 and CD43 have cytoplasmic domains consisting of 70 and 124 residues, respectively. In contrast, ICAMs have short cytoplasmic tail peptides as their cytoplasmic portions. For example, the mouse ICAM-2 tail consists of 28 residues. Recently, possible ERM-binding regions of CD43 and CD44 have been identified in their juxtamembrane regions consisting of 31 and 19 residues, respectively (Yonemura et al., 1998). All these regions contain repeated basic residues, although no significant sequence homology has been found. Interestingly, these positively charged regions are in sharp contrast with the highly negatively charged regions of cadherins for β-catenin binding (Huber and Weis, 2001). Because such a cluster of basic residues is a common feature of juxtamembrane regions of many adhesion molecules and receptors, it remains unclear how ERM proteins discriminate between them. Here, we report the first crystal structure of the FERM domain bound to the ICAM-2 cytoplasmic tail. The structure reveals how the FERM domain recognizes the cytoplasmic tail through interactions with subdomain C, which is folded into a phosphotyrosine-binding (PTB)-like domain, but exhibits a binding specificity distinct from canonical PTB domains.
Results and discussion
Structure determination
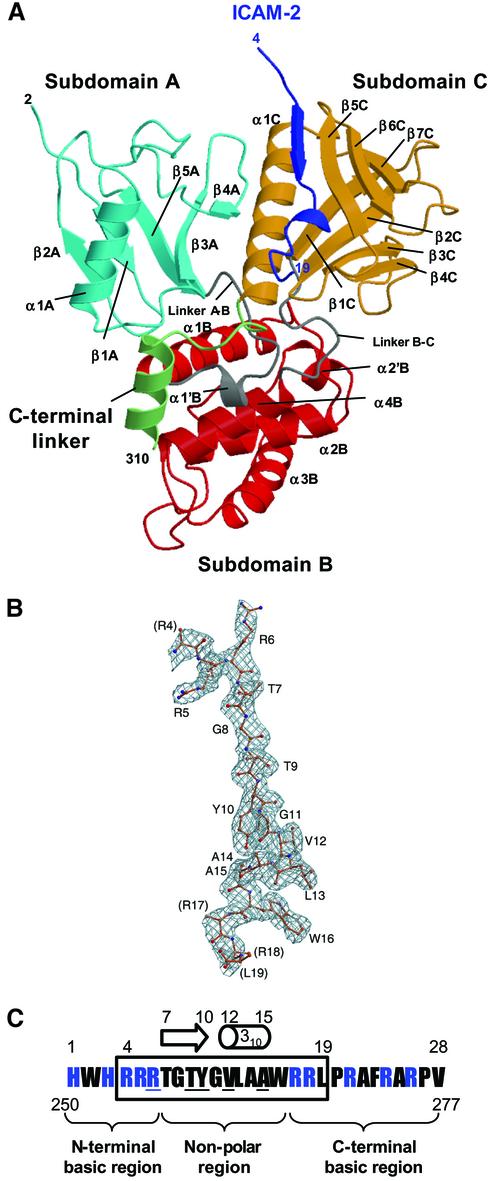
The mouse radixin FERM domain containing residues 1–310 was purified as described (Hamada et al., 2001). The synthesized mouse ICAM-2 peptide is composed of 28 residues of the whole cytoplasmic tail (residues 250–277). Crystals of the radixin FERM domain bound to the ICAM-2 peptide containing one molecular complex per asymmetric unit were obtained. The structure was determined by molecular replacement using the free radixin FERM domain (Hamada et al., 2000) and refined to 2.4 Å resolution to an R value of 22.9% (a free R value of 24.0%). The current model includes 2–310 residues of the FERM domain and 16 residues (residues 4–19) of the ICAM-2 peptide, hereafter referred to as the FERM–ICAM-2 complex (Figure 1A). Most of the 16 residues are well defined on the electron density maps (Figure 1B). The numbering scheme of the ICAM-2 peptide is shown in Figure 1C. No model of the peptide was built for the three N-terminal residues (residues 1–3) and the nine C-terminal residues (residues 20–28), which were poorly defined in the current map, probably because of disordering.
Fig. 1. Overall structure of the radixin FERM domain bound to the ICAM-2 tail peptide. (A) Views of the radixin FERM domain bound to the ICAM-2 peptide by ribbon representations. The ICAM-2 peptide is shown in blue. The radixin FERM domain consists of subdomains A (light blue), B (red) and C (brown). The linkers A–B (residues 83–95) and B–C (residues 196–203) are colored in gray and the C-terminal linker in green. (B) The ICAM-2 peptide model in a 2Fo–Fc electron density map countered at the 1σ level. The amino acid residues are indicated with labels of one-letter codes. Labels in parentheses indicate the terminal residues whose side chains were not defined in the map. (C) The 28-residue peptide synthesized based on the sequence of the mouse ICAM-2 cytoplasmic tail was used for the structural work. Basic residues are in blue. This peptide has two basic regions and a non-polar region between them. The 16 residues of the peptide defined on the current map are boxed. Key residues in binding to the radixin FERM domain are underlined (see text). The short β-strand (residues 7–10) and one 310 helix (resides 12–15) are indicated with an arrow and a cylinder.
Overall structure of the FERM domain
The radixin FERM domain contains three subdomains, A (the N-terminal 82 residues, light blue in Figure 1A), B (residues 96–195, red) and C (residues 204–297, brown). Subdomain A has a typical ubiquitin fold, and subdomain B has an α-helix bundle structure classified as an acyl-coenzyme A binding protein-like fold. Subdomain C folds into a standard seven-stranded β-sandwich with one long α-helix (α1C), classified as a PTB or pleckstrin-homology (PH) domain. These subdomain structures are essentially the same as those of the free and IP3-bound forms of the radixin FERM domain (Hamada et al., 2000). The average root mean square (r.m.s.) deviations in Cα atoms obtained from the pair-wise superposition of each subdomain in the ICAM-2- and the IP3-bound forms were found to be 0.58 Å, 0.86 Å and 0.89 Å for subdomains A, B and C, respectively. Subdomain B has a short flexible loop (residues 160–165) between helices α2′B and α3B displaying an exceptionally local deviation (∼5 Å). In subdomain C, the C-terminal helix α1C moves toward the bound ICAM-2 peptide by 1 Å, which would be an induced fit by peptide binding. The C-terminal residues 298–310 (green in Figure 1A) of the current structure form a stable conformation, including an additional helix (residues 300–310). A proline residue Pro297 followed by helix α1C directs this segment to run along the interface between subdomain A and linker A–B.
The overall r.m.s. deviations of the ICAM-2 bound FERM domain from the free form are relatively large (1.35 Å), compared with the small deviations in each subdomain. In these global structural comparisons, the major deviation has been found to be due to subdomain A, which shifts its mutual position relative to subdomains B and C with a rotation of 3.5° around the A–B linker. The movement of helix α1C may trigger the movement of sub domain A, although the structure of the FERM–ICAM-2 complex does not preclude crystal-packing effects on subdomain A movement. The principal structural changes in the FERM domains described above are also seen in comparison with the moesin FERM–C-tail complex (Pearson et al., 2000), giving an average r.m.s. deviation of 1.55 Å in Cα atom positions. One major difference producing this large deviation is the conformational differences in subdomains B and C, which contain the helix α2B and loop α2B–α2′B and β-sheet β5C–β6C–β7C. These differences are caused by induced fits upon binding to the C-terminal tail domain in the moesin complex, as discussed previously (Hamada et al., 2000).
Peptide structure and recognition
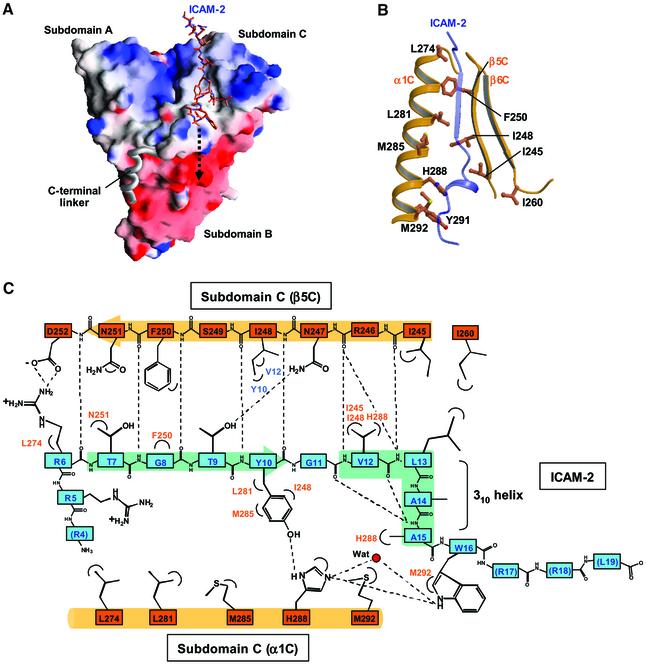
As shown in Figure 1C, the ICAM-2 peptide consists of three regions: the positively charged N-terminal and C-terminal regions separated by the non-polar region. Both the N-terminal and C-terminal regions are flexible, while the non-polar region forms a short β-strand (residues 7–10) followed by a short 310 helix (residues 12–15). The structured peptide region fills a long shallow groove on the molecular surface of subdomain C (Figure 2A). This peptide-binding groove is formed by hydrophobic side chains from helix α1C and strand β5C (Figure 2B). Trp242 from loop β4C–β5C also participates in formation of the groove. Eleven residues (residues 6–16) of the ICAM-2 peptide are in contact with subdomain C and bury 1761 Å2 of the total accessible surface area. The β-strand of the bound peptide forms an anti-parallel β–β association with strand β5C from subdomain C (Figure 2C). The binding mode of ICAM-2 to the FERM domain is comparable to those of phosphotyrosine-containing peptides bound to PTB domains, which will be discussed later. In our complex structure, eight main-chain–main-chain hydrogen bonds stabilize the ICAM-2 binding; five at the β-strand and three at the 310 helix of the ICAM-2 peptide. These three hydrogen bonds contribute to stabilizing the short 310 helix structure, along with two intra-molecular main-chain–main-chain hydrogen bonds (Figure 2C).
Fig. 2. The ICAM-2 tail peptide recognition by the radixin FERM domain. (A) Surface electrostatic potentials of the radixin FERM domain viewed from the same direction as in Figure 11A. Positive (blue, +14 kT/e) and negative (red, –14 kT/e) potentials are mapped on the van der Waals surfaces. The ICAM-2 peptide found in the complex crystal is shown in a stick model. The disordered C-terminal basic region is indicated by an arrow of dotted lines. (B) The ICAM-2 binding groove on subdomain C is formed primarily by hydrophobic residues from helix α1C and strand β5C. The bound ICAM-2 peptide is shown in a transparency ribbon model. (C) Schematic representation of the interactions between the ICAM-2 peptide (blue) and subdomain C (brown). Hydrogen bonds are shown by broken lines. (D) The ICAM-2 peptide found in the FERM–ICAM-2 complex is shown in a stick model (light blue) with their interacting residues from subdomain C (brown). Hydrogen bonds are shown by dotted lines. (E) A close-up view of the hydrophobic and hydrogen bonding interactions between the ICAM-2 peptide and the FERM domain mediated by His288 from subdomain C.
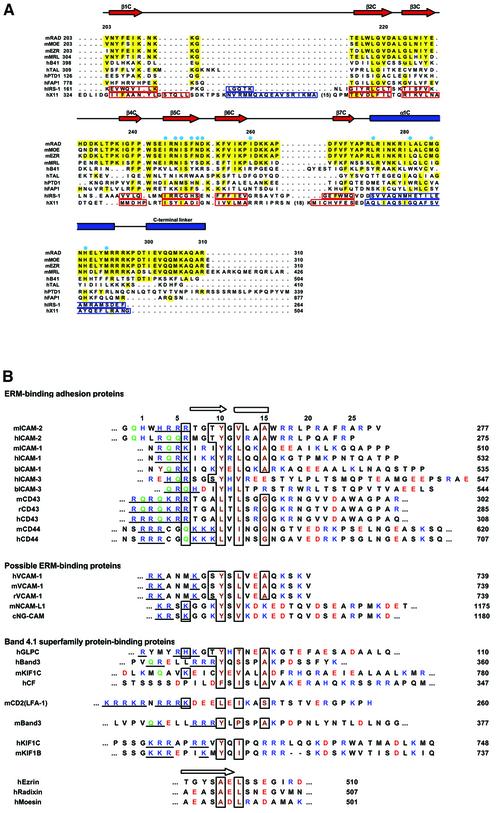
The ICAM-2 peptide contacts both polar and non-polar residues from strand β5C and helix α1C. The contacts contain hydrogen bonding, salt-bridging and hydrophobic interactions involving side chains, which together produce the specificity of the peptide binding (Figure 2C and D). Notably, a stretch of the non-polar residues (residues 10–16) of ICAM-2 peptide forms a compact fold with the short 310-helix and produces several contacts with the groove. Of these residues, Tyr10, Val12 and Ala15 insert their side chains deeply into the groove, allowing for intimate interactions with several non-polar residues from helix α1C and strand β5C. The well-conserved His288 residue (Figure 3A) from helix α1C is located at the center of the multiple contacts with the ICAM-2 residues and thereby seems to play a key role in stabilizing the peptide binding (Figure 2E).
Fig. 3. Sequence alignments of subdomain C and the FERM-binding peptides. (A) Secondary structure elements and sequence alignments of subdomains C of FERM domains with the related PTB domains. The FERM subdomain C of mouse radixin (mRAD) and the related proteins are aligned with the secondary structure elements of the radixin FERM subdomain C at the top: α-helix (blue rectangles) and β-strands (red arrows). Identical residues are highlighted in yellow. The aligned FERM subdomains C are mouse radixin (mRAD), moesin (mMOE), ezrin (mEZR), merlin (mMRL), human band 4.1 (hB41), talin (hTAL), protein tyrosine phosphatase D1 (hPTPD1) and Fas-associated protein tyrosine phosphatase 1 (hFAP1). The aligned PTB domains are the IRS-1 PTB domain (hIRS1), the X11 PTB domain (hX11). The secondary structure elements of these PTB domains are boxed with blue lines (α-helices) and with red lines (β-strands). The mouse FERM domains exhibit 100% identities with those of human and >99.7% identities with other mammalian homologues. Blue circles mark the radixin subdomain C residues interacting with the ICAM-2 peptide. (B) Sequence alignment of the juxtamembrane cytoplasmic regions of adhesion molecules that bind ERM proteins and of the related adhesion molecules. Basic and acidic residues are shown in blue and red, respectively. The basic clusters located at the N-terminal region are underlined. Glutamines frequently appearing in this N-terminal basic region are shown in green. Key residues of the ICAM-2 peptide for binding to the radixin FERM domain are boxed with hydrophobic residues in brown. The sources are mouse (m), rat (r), human (h), bovine (b) and nematode (c). The numbers on the right-hand side of the column indicate the C-terminal residue numbers in the sequences. The N-terminal regions of the C-terminal tail domains of ERM proteins are also aligned at the bottom of the figure with an arrow indicating strand 1.
Using surface plasmon resonance (SPR) measurements, we found that the mouse radixin FERM domain binds the ICAM-2 peptide with high affinity (16.4 nM), as shown in Table I. Based on our complex crystal structure, the binding affinity for several mutated or truncated ICAM-2 peptides was determined by the measurements. These measurements resulted in a weakening of the binding affinity to the FERM domain by 2- to 50-fold and allowed us to identify key residues of the peptide as discussed below.
Table I. Binding affinities of the ICAM-2 peptides for the radixin FERM domain.
One determinant residue for the ICAM-2 binding specificity is Tyr10, which forms a hydrogen bond (2.7 Å) with His288 and is in contact with Ile248, Leu281 and Met285 from subdomain C. This Tyr residue is conserved in all ICAMs from different sources (Figure 3B), and the replacement with an Ala residue indeed significantly causes 16-fold reduction in the binding affinity (ICAM-2/Y10 in Table I). Val12 of the ICAM-2 peptide is conservative and is replaced with a Leu or Ile residue in some ICAMs, indicating the importance of its hydrophobic properties in binding to subdomain C. Leu13 is variant, contacting Ile260 from subdomain C at the molecular surface. A double mutation of Val12 and Leu13 by Ala reduces the binding affinity by 11-fold (ICAM-2/VL12 in Table I).
Trp16 of the ICAM-2 peptide also formed a pos sibly direct hydrogen bond (3.5 Å) as well as water-mediated hydrogen bonds with His288, although a single mutation of Trp16 by Ala was found to reduce the binding affinity by only two-fold (ICAM-2/W16 in Table I). This unexpectedly small contribution may be explained by Trp16 being rather bare at the molecular surface. Trp16 is conserved in ICAM-2 but is variant in other ICAMs, CD43 and CD44 (Figure 3B). Regarding the ICAM-2 arginine triplet at the N-terminal region, Arg6 forms a salt bridge (3.5 Å) with Asp252 from the C-terminal end of strand β5C (Figure 2D). Thr9 of the ICAM-2 β-strand forms a hydrogen bond (3.0 Å) to Asn247 of FERM strand β5C. Each substitution of Arg6 or Thr9 with Ala results in a weak reduction in the binding affinity (ICAM-2/R6 and ICAM-2/T9 in Table I).
In conclusion, we suggest a FERM-binding motif, RxxTYxVxxA (x stands for any amino acid residue), for ERM proteins. The key elements in the motif are Y, V and A, which play a central role in hydrophobic interactions with the FERM domain. In FERM-binding peptides from several sources, the Y and V positions are restricted to hydrophobic residues and the A position to small residues (Figure 3B). Two polar residues, R and T, in the motif have some significance in binding by participating in direct contacts with the FERM domain. The RxxTYxVxxA motif of ICAM-2 is followed by the C-terminal basic region, which is important for the strong binding as described in the next section.
C-terminal specificity
The C-terminal region of the ICAM-2 peptide is highly positively charged with five Arg residues (Figure 1C). In our complex structure, this flexible C-terminal region exhibits no specific interactions in terms of structure, but projects toward the highly negatively charged groove between subdomains B and C (Figure 2A). This shallow groove has been postulated as a possible site of interaction with the basic residues of the cytoplasmic tails of the FERM-binding adhesion molecules (Hamada et al., 2000). In fact, deletion of this region reduces the binding affinity to the FERM domain by 9-fold (ICAM-2/s2 in Table I). It is thus conceivable that this region electrostatically interacts with the acidic groove. This interaction provides a rational reason why the FERM binding to ICAM-2 was sensitive to ionic strength (Yonemura et al., 1998).
FERM binding to other adhesion molecules
Recently, ERM-dependent movement of CD43 distal to the T cell/APC (antigen-presenting cell) interaction sites has been shown to be an essential process for formation of the immunological synapse (Allenspach et al., 2001; Delon et al., 2001; Shaw, 2001). We found that the FERM domain binds the CD43 peptide with a Kd value of 104 nM (Table II). The CD43 peptide has Leu and Gly residues at positions corresponding to Tyr10 and Ala15 of the ICAM-2 peptide, respectively. This fact suggests that Tyr and Ala residues at this position are not necessarily a prerequisite for FERM binding and are replaceable by residues with similar properties. Moreover, Thr9 is replaced with an Ala residue. These replacements of the key residues are also seen in the CD44 peptide and may be one reason why the affinities of the CD43 and CD44 peptides are weaker than that of the ICAM-2 peptide. The ICAM-3 peptide exhibits an unexpectedly lower affinity (Kd value of 757 nM). In the ICAM-3 peptide, Ala15 of ICAM-2 is replaced with non-homologous residues and the C-terminal region lacks basic residues (Figure 3B). These possibly reduce the affinity to the FERM domain.
Table II. Binding affinities of the ICAM-2-related peptides for the radixin FERM domain.
The observed binding affinities of ICAMs, CD43 and CD44 are basically consistent with previous results from an in vitro glutathione S-transferase (GST) pull-down binding assay and SPR measurements (Hirao et al., 1996; Heiska et al., 1998; Yonemura et al., 1998). A positive correlation between the isoelectric points of the cyto plasmic tail peptides and their binding affinity to ERM proteins has been pointed out previously (Yonemura et al., 1998). We found no significant binding affinity to ICAM-4 and ICAM-5 (data not shown).
Interestingly, we found that a short cytoplasmic peptide from the vascular cell adhesion molecule 1 (VCAM-1) possesses a region somewhat similar to the FERM-recognition motif of ICAM-2. This VCAM-1 peptide lacks the long C-terminal basic region, while our SPR measurements revealed a significant affinity (149 nM) of this VCAM-1 peptide for the FERM domain. It is likely that the basic residues close to the RxxTYxVxxA motif sequence are more important for the electrostatic interactions than remote basic residues at the C-terminal region. We also found that the neural cell adhesion molecule L1 (NCAM-L1), which shows limited sequence similarity, exhibits a relatively high affinity (61 nM). The affinities of these two newly found adhesion molecules for the radixin FERM domain are stronger than that of ICAM-3. It is noteworthy that recent biochemical studies have shown dynamic interaction of VCAM-1 with ERM proteins in a novel endothelial docking structure for adherent leukocytes (Barreiro et al., 2002) and also functional binding of the axonal NCAM-L1 to ERM proteins (Dickson et al., 2002).
We notice that the FERM-binding motif proposed in this study resembles the juxtamembrane regions of syndecans and neurexins. Syndecans are heparan sulfate proteoglycans expressed on all adherent cells and participate in cell–cell, cell–matrix adhesion and signaling of heparan sulfate growth factors. Neurexins are brain-specific cell surface proteins, which bind neuroligins and neurexophilins, brain-specific adhesion molecules and neuropeptide-like molecules, respectively. We found that the radixin FERM domain binds syndecans and neurexin with relatively low affinities as compared with that of ICAM-2 (Table II). Syndecans and neurexins are known to have PDZ-binding sequences at their C-terminal ends, which bind PDZ-containing adaptor proteins, syntenin and CASK, respectively. In cells, these adaptor proteins may enhance the binding of syndecans and neurexins to ERM proteins. It is also possible that the binding partners of syndecans and neurexins in cells are the other members of ERM proteins or other FERM-containing proteins. We need further experimental tests to answer these questions.
Membrane targeting and unmasking
The N-terminal region of the ICAM-2 peptide contains five basic residues, His1 and 3, and Arg4–6 (Figure 1C). In our structure, this region is flexible. No direct interaction with the FERM domain is observed for these basic residues other than Arg6. This lack of direct interaction seems consistent with substitution of these basic residues with Ala (ICAM-2/RR in Table I) resulting in a weak 2-fold reduction of the binding affinity. A comparison of the FERM–ICAM-2 and FERM–IP3 complex structures reveals that these Arg residues are close to the bound IP3 molecule in the FERM–IP3 complex, but produce no direct contact with the IP3 molecule in the superimposed structures. Recently, several mutations have been introduced to ezrin to analyze the PIP2-binding site (Barret et al., 2000). It has been shown that mutations of basic residues at the membrane-binding surface of the FERM domain impair the PIP2 binding, while these mutations do not impair the in vitro interaction with the cytoplasmic tail of CD44. This result may be interpreted by our crystal structures, in which two binding sites for IP3 and ICAM-2 have no overlap as described above.
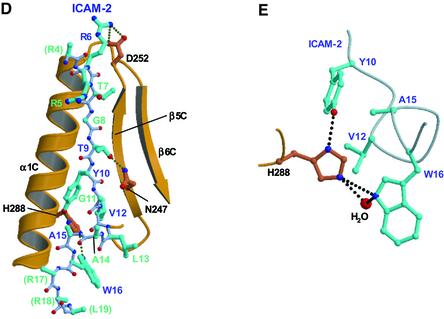
Recent SPR measurements suggest that PIP2 interacts with the ICAM-1 and -2 cytoplasmic tail peptides (Heiska et al., 1998). It seems plausible that in plasma membranes, negatively charged PIP2 molecules colocalize with ICAM-2 and other adhesion molecules having positively charged juxtamembrane regions of the cytoplasmic tails by electrostatic interactions. PIP2 molecules colocalized with adhesion molecules may recruit masked ERM proteins from the cytosol to plasma membranes and unmask ERM proteins for subsequent binding to the cytoplasmic tails of adhesion molecules (Figure 4A).
Fig. 4. Membrane-targeting and unmasking of ERM proteins. (A) Model of ERM proteins bound to both PIP2 in a membrane and the ICAM-2 cytoplasmic tail. The N-terminal FERM, the central helical and the C-terminal tail domains are shown with a triangle in blue, a rectangle in yellow and a block in red, respectively. ERM proteins partition between the cell cortex and cytosol. ICAM-2 in plasma membranes colocalized with PIP2. PIP2 molecules recruit masked ERM proteins from the cytosol to plasma membranes and unmask the masked ERM proteins for subsequent binding to the cytoplasmic tails of adhesion molecules such as ICAM-2 and actin filaments. (B) Comparison of the FERM–ICAM-2 complex with the masked form of moesin. Superposition of the C-terminal tail domain (pink) in the moesin FERM domain–C-tail complex on the radixin FERM domain (gray) bound to the ICAM-2 peptide (blue). (C) Comparison of the β-strand of ICAM-2 (blue green) and strand 1 of the moesin C-terminal tail domain (pink).
The C-terminal tail domain of moesin has been found to mask the acidic groove between subdomains B and C (Pearson et al., 2000), indicating that the C-terminal tail domain in masked full-length ERM proteins blocks the electrostatic interactions between the C-terminal basic region of the ICAM-2 peptide and the acidic groove of the FERM domain. Moreover, the N-terminal region (strand 1) of the C-terminal tail domain has been suggested to bind the groove of subdomain C, the place at which the ICAM-2 peptide was bound in our complex crystal. As shown in Figure 4B, this proposed binding in the masked full-length ERM proteins would directly interfere with binding of the ICAM-2 peptide to the FERM domain, although the sequence homology between the ICAM-2 strand and strand 1 of the C-terminal tail domain is not apparent (Figure 3B). A structure-based comparison indicates Tyr10 of ICAM-2, a key residue for binding, is replaced with Ala492 of the C-terminal tail domain (Figure 4C). In addition to the direct interferences described above, it should be noted that the overall structures of the FERM domains in the ICAM-2-bound and the masked forms show local structural deviations at subdomains B and C, as indicated previously. These deviations cause atom crashes at the interfaces between the C-terminal tail domain and subdomains B and C, when the tail domain is overlaid on the FERM–ICAM-2 complex (Figure 4B).
Comparison with the phosphotyrosine-containing peptide bound to PTB domains
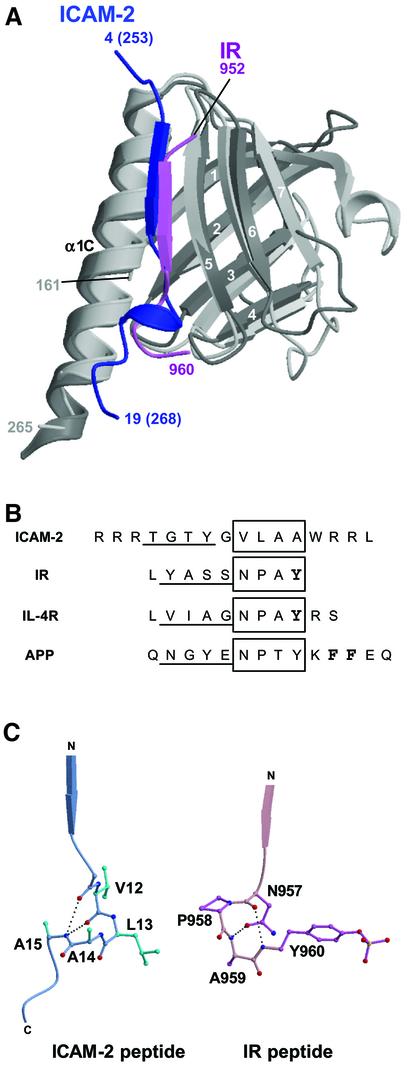
A previous structural comparison (Hamada et al., 2000) has indicated that the fold of subdomain C resembles the PTB domains rather than PH domains. The β-strand of the bound ICAM-2 peptide is superimposable onto that of the phosphotyrosine-containing peptide (pY peptide) from human insulin receptor (IR) bound to the human insulin receptor substrate 1 (IRS-1) PTB domain (Eck et al., 1996) (Figure 5A). The IR and other PTB-binding peptides have NPxY motifs (Figure 5B), which adopt a β-turn conformation at the C-terminal end of the β-strand. The ICAM-2 peptide, however, has no NPxY motif and the region corresponding to this NPxY motif forms a 310 helix followed by the C-terminal region running in a direction different from that of the IR peptide (Figure 5C).
Fig. 5. Comparison of the FERM–ICAM-2 complex with the related PTB–peptide complexes. (A) Superposition of the radixin FERM subdomain C (gray) bound to the ICAM-2 tail (blue) on the IRS-1 PTB domain (light gray) bound to the IR peptide (pink). (B) Peptide alignment based on the superimposed complex structures. The β-strands formed in the complex formations are underlined. The 310 helix in ICAM-2 and the NPxY motif sequences in the PTB domain-binding (IR, IL-4R and APP) peptides are boxed. The phosphotyrosine residues are in bold in the IR and IL-4R peptides. In the APP peptide, two Phe residues anchoring the peptide to the X11 PTB domain are also in bold. (C) Comparison of the 310 helix in the ICAM-2 peptide bound to the radixin FERM domain with the β-turn of the NPxY motif in the IR peptide bound to the IRS-1 PTB domain. The intra-peptide hydrogen bonds are shown as dotted lines.
Compared with the pY peptide binding to PTB domains in a µM range of Kd (Dhe-Paganon et al., 1999), the ICAM-2 binding to the FERM domain is stronger by ∼100-fold. There is, however, no significant difference in their buried accessible surface area at the interfaces, where the peptides have ordered structures. For example, the total buried accessible surface area in the complex between the IR peptide and the IRS-1 PTB domain is ∼1560 Å2 (Eck et al., 1996). The relatively stronger binding of the ICAM-2 peptide to the FERM domain may be attributable to possible electrostatic interactions between the C-terminal basic residues and the highly negatively charged groove between subdomains B and C. Another possible reason for the stronger binding of the ICAM-2 peptide may be attributable to differences in the main-chain–main-chain interactions. Eight main-chain–main-chain hydrogen bonds are formed in the FERM–ICAM-2 complex, whereas five are formed between the IR peptide and the IRS-1 PTB domain. In addition, lack of hydrophobic interactions involving the critical Y residue from the RxxTYxVxxA motif in the IR peptide could also explain the differences in binding affinity.
Interestingly, the neuron-specific X11 PTB domain has been shown to bind a non-pY peptide from β-amyloid precursor protein (APP) (Zhang et al., 1997). Another class of PTB domains, which contains the Numb PTB domain, binds peptides having a GPpY but not NPxpY motif (Li et al., 1998). The current structure provides new evidence that PTB domains have multiple modes of peptide binding with different specificities.
Comparison with β-catenin
It is an interesting question as to what relationship there is between recognition of the cytoplasmic peptides of adhesion molecules for linking the actin cytoskeleton by ERM proteins and other linker proteins. In the β-catenin–E-cadherin complex (Huber and Weis, 2001), the interaction surface involves 100 residues of the E-cadherin peptide, which has an extended structure at the long groove formed by the α-helical armadillo repeats of β-catenin. The cadherin peptide has polar and non-polar interactions with β-catenin primarily mediated by the side chains and raises the total buried accessible surface area of 6100 Å2. No similarity of the peptide conformations and binding modes has been found between our FERM–ICAM-2 and the β-catenin–E-cadherin complexes. Notably, there is no main-chain–main-chain interaction between β-catenin and the E-cadherin peptide. It remains a mystery as to why the ICAM-2 binding affinity to the FERM domain is 14-fold stronger than the binding affinity of cadherin to β-catenin (230 nM), although the total buried accessible surface area in the FERM–ICAM-2 complex is 3.5-fold smaller than that in the β-catenin– E-cadherin complex. A lack of main-chain–main-chain interactions in the peptide binding to β-catenin may be related to the relatively weak binding affinity. In the β-catenin–E-cadherin interactions, phosphorylation of E-cadherin seems to be essential for the affinity of the β-catenin–E-cadherin complex and cell adhesiveness (Lickert et al., 2000).
Implication for other FERM-like domains
FERM-like domains have been found in other cross-linker proteins such as the neurofibromatosis type 2 tumor-suppressor gene product, which is also referred to as merlin/schwannomin (Rouleau et al., 1993; Troffater et al., 1993), talin, which binds integrin-β (Calderwood et al., 1999; Patil et al., 1999), protein tyrosine phosphatases (PTPs) such as FAP-1 (PTP-BAS), which associates with Fas (Sato et al., 1995), focal adhesion kinase (FAK) (Schultz et al., 1998) and membrane-associated unconventional myosins such as myosins VIIa and X (Chishti et al., 1998). Moreover, FERM domains have been found in the Janus kinase (JAK) family non-receptor tyrosine kinases that bind interferon receptors (Girault et al., 1999). Of these proteins, the merlin FERM domain exhibits a relatively high (64%) sequence identity against those of ERM proteins and seems to exhibit a similar specificity in recognition of adhesion molecules. In fact, recently crystallographic studies have revealed the merlin FERM domain structure that closely resembles those of radixin and moesin (Kang et al., 2002; Shimizu et al., 2002). In contrast to the high sequence identity of merlin, the identities of band 4.1 (27%), tailin (24%) and PTPs (∼25–33%) are low, and those of FAK and JAK extremely low. Therefore, it remains unclear whether all of these proteins bind their target receptors/adhesion molecules using a PTB-like subdomain C. The band 4.1 protein binds anion exchanger/band 3 as well as glycophorin C (Jons and Drenckhahn, 1992). Recently, the tyrosine phosphatase PTPD1, which is one of the tyrosine phosphatases possessing a FERM-like domain, has been found to bind a new kinesin-like motor protein, KIF1C (Dorner et al., 1998). Our close inspection of the amino acid sequences at the possible binding regions of these proteins suggests that they may have variants of the FERM-binding motif RxxTYxVxxA for binding to the FERM or FERM-like domains (Figure 3B).
Conclusion
The present work reveals the peptide-binding mode of the radixin FERM domain, which uses the PTB-like subdomain C for recognition of the FERM-binding motif RxxTYxVxxA through a β–β association and most likely the acidic groove between subdomains B and C for electrostatic interactions. Basic residues at the N-terminal flanking region of the motif may not be essential for FERM binding, but may be important for possible interactions with acidic lipid components such as PIP2. The ICAM-2 peptide forms a short 310 helix instead of the β-turn formed by the NPxY motif that is the signature sequence recognized by ‘classical’ PTB domains. Specific contacts have been identified on the complex structure and also been verified using a quantitative binding assay. These contacts, as well as electrostatic interactions involving the C-terminal basic region, whose structure was not specified in this study, collectively achieve high affinity and specificity.
Materials and methods
Protein preparation and binding studies
The FERM domain (1–310 residues) of mouse radixin was expressed as a fusion protein with GST in Escherichia coli and purified by methods described previously (Hamada et al., 2001). For binding studies, SPR measurements were carried out on a BIAcore Biosensor instrument (BIAcore 3000, Pharmacia Biosensor). Biotinylated polypeptides of the juxtamembrane regions of the adhesion molecules, which were synthesized by a Fmoc-based strategy with standard side-chain protecting groups and were purified by reverse phase HPLC, were purchased from Sawady Technology (Tokyo, Japan). The peptide was coupled via the N-terminal biotin moiety to streptavidin-coated sensor chip (sensor chip SA BIAcore AB). The radixin FERM domain (∼0.1–1 µM) was injected into both the peptide-linked and non-linked sensor chips for correction of background signals. All binding experiments were performed at 25°C with a flow rate of 10 µl/min in buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT and 0.05% surfactant P20). The kinetic parameters were evaluated using the BIA evaluation software (Pharmacia). The Kd values were obtained by averaging of at least three independent measurements and are summarized in Table I.
Crystallization and data collection
For crystallization, the purified protein was mixed with the equimolar 28-residue ICAM-2 peptide and concentrated to 20 mg/ml (0.54 mM). The complex crystals were obtained from a 0.27 mM protein/peptide solution with 1:1 stoichiometry containing 50 mM Mes-Na (pH 6.0), 2% polyethylene glycol 6000 (PEG6K), 130 mM NaCl and 0.4 mM DTT at 4°C. MALDI-TOF mass spectroscopy of an aliquot in which crystals were dissolved indicated that the crystals contained the FERM domain and the ICAM-2 peptide (Hamada et al., 2001).
All diffraction data were collected from crystals flash-frozen to 100 K after cryoprotection in a solution containing 7% PEG6K, 35% glycerol and 50 mM Mes-Na buffer. Crystals belonged to space group P3121, which was determined by the following structure analysis, with unit cell parameters, a = b = 100.44 Å, c = 99.49 Å, γ = 120°. A 2.4-Å data set was collected using a charge-coupled area detector (Mar CCD) installed on the beam line BL41XU at the SPring-8 synchrotron facility, Harima, Japan. The wavelength was set to 1.00 Å with a crystal-to-detector distance of 160 mm. The data were collected from three crystals with angular ranges of 180° with step sizes of 1° for an exposure time of 3 s. Intensity data were processed using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997) or DPS-Mosfilm (Rossmann and van Beek, 1999). Two data sets were merged into one set of data for structure determination (Table III).
Table III. Crystallographic analysis of the radixin FERM domain– ICAM-2 complex.
| Intensity data |
|
| Resolution (Å) | 2.40 |
| Reflections | |
| measured | 599708 |
| unique | 23153 |
| Completeness (%)a | 100.0/100.0 |
| Rsymb (%)a | 7.1/13.7 |
| Mean I/σa |
6.0/5.2 |
| Refinement |
|
| Resolution range (Å) | 15.0–2.40 |
| Number of residues | 332 |
| protein | 316 |
| peptide | 16 |
| Atoms included | 2998 |
| protein | 2618 |
| peptide | 116 |
| water | 264 |
| Rcryst factor/Rfree factor (%)c | 22.9/24.0 |
| Mean B factor (Å2) | 49.4 |
| protein | 48.9 |
| peptide | 59.2 |
| R.m.s. deviationsd | 0.008 Å, 1.350°, 1.182° |
aEach pair of values are for overall/outer shell. The resolution ranges of their outer shells are 2.53–2.40 Å.
bRsym = Σ|I – <I>| / ΣI; calculated for all data.
cRcryst and Rfree = Σ||Fo| – |Fc|| / Σ|Fo|, where the free reflections (10% of the total used) were held aside for Rfree throughout refinement.
dThe three values are for bond lengths, bond angles and impropers, respectively.
Structure determination and refinement
The initial phases were calculated by molecular replacement using a search model based on the free state structure of the radixin FERM domain (Hamada et al., 2000). Unambiguous rotation and translation solutions were obtained from several searches using different ranges of intensity data and integration radii with the program AMoRe (Navaza, 1994). After rigid-body refinements of the search model performed with the program CNS (Brünger et al., 1998), the phases were improved by solvent flattening and histogram matching using Solomon (Abrahams and Leslie, 1996). The resultant initial map shows clear electron densities for most of the FERM domain and the peptide. An initial model of the peptide was built into the electron density map using the graphics program O (Jones et al., 1991) as well as rebuilding part of the FERM domain. The model was refined by the method of simulated annealing using CNS (Brünger et al., 1998). After several cycles of rebuilding and refinement, the model was refined to an R value of 22.9% (free R value of 24.0%) for intensity data at a 2.4 Å resolution. The current model includes 316 residues of the protein (six residues arising from the vector), 16 residues of peptide and 264 water molecules. The stereochemical quality of the model was monitored using the program PROCHECK (Laskowski et al., 1993).
In the peptide model, the side chains of Arg4, Arg17, Arg18 and Leu19 were poorly defined and in the current structure are replaced with alanines. No model was built for the three N-terminal and nine C-terminal residues of the peptide, which are poorly defined in the current map. In the protein model, the side chains of Pro2 and Lys3 were poorly defined and in the current structure are replaced with alanines. Asp252 is in disallowed main-chain torsion angle regions.
Ribbon representations of the main-chain folding of the molecule were drawn using the program Molscript (Kraulis, 1991), while molecular surface representations were drawn using the program GRASP (Nicholls et al., 1991).
Acknowledgments
Acknowledgements
We thank Dr S.E.Shoelson (Harvard Medical School, Boston) for providing the coordinates of the IR–IRS-1 PTB domain complex. We also thank Drs K.Okada and M.Kawamoto for their technical assistance in data collection at the SPring-8 Facility, Harima. This work was supported by Grants-in-Aid for Scientific Research to T.H. (12490024, 10359003) and Grants-in-Aid for Scientific Research on Priority Area to S.T. and T.H. (10179103-4) from the MESSC of Japan. K.H. was supported by a research fellowship for Young Scientists from JSPS. T.H. is a member of the TARA project of Tsukuba University. Coordinates for the complex have been deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics under accession No. 1J19.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Allenspach E.J. et al. (2001) ERM-dependent movement of CD43 defines a novel protein complex distal to the immunological synapse. Immunity, 15, 739–750. [DOI] [PubMed] [Google Scholar]
- Amieva M.R. and Furthmayr,H. (1995) Subcellular localization of moesin in dynamic filopodia, retraction fibers and other structures involved in substrate exploration, attachment and cell–cell contacts. Exp. Cell Res., 219, 180–196. [DOI] [PubMed] [Google Scholar]
- Andreoli C., Martin,M., Le Borgne,R., Reggio,H. and Mangeat,P. (1994) Ezrin has properties to self-associate at the plasma membrane. J. Cell Sci., 107, 2509–2521. [DOI] [PubMed] [Google Scholar]
- Arpin M., Algrain,M. and Louvard,D. (1994) Membrane–actin microfilament connections: an increasing diversity of players related to band 4.1. Curr. Opin. Cell Biol., 6, 136–141. [DOI] [PubMed] [Google Scholar]
- Barreiro O., Yánez-Mó,M., Serrador,J.M., Montoya,M.C., Vicente-Manzanares,M., Tejedor,R., Furthmayr,H. and Sánchez-Madrid,F. (2002) Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol., 157, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret C., Roy,C., Montcourrier,P., Mangeat,P. and Niggli,V. (2000) Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP2) binding site in the NH2-terminal domain of ezrin correlates with its altered cellular distribution. J. Cell Biol., 151, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1999) Regulation of cortical structure by the ezrin–radixin–moesin protein family. Curr. Opin. Cell Biol., 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Bretscher A., Chambers,D., Nguyen,R. and Reczek,D. (2000) ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell. Dev. Biol., 16, 113–143. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Zent,R., Grant,R., Rees,D.J., Hynes,R.O. and Ginsberg,M.H. (1999) The Talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem., 274, 28071–28074. [DOI] [PubMed] [Google Scholar]
- Chishti A.H. et al. (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci., 23, 281–282. [DOI] [PubMed] [Google Scholar]
- Delon J., Kaibuchi,K. and Germain,R.N. (2001) Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin. Immunity, 15, 691–701. [DOI] [PubMed] [Google Scholar]
- Dickson T.C., Mintz,C.D., Benson,D.L. and Salton,S.R.J. (2002) Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J. Cell Biol., 157, 1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S., Ottinger,E.A., Nolte,R.T., Eck,M.J. and Shoelson,S.E. (1999) Crystal structure of the pleckstrin homology–phosphotyrosine binding (PH–PTB) targeting region of insulin receptor substrate 1. Proc. Natl Acad. Sci. USA, 96, 8378–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner C., Ciossek,T., Müller,S., Møller,N.P.H., Ullrich,A. and Lammers,R. (1998) Characterization of KIF1C, a new kinesin-like protein involved in vesicle transport from the Golgi apparatus to the endoplasmic reticulum. J. Biol. Chem., 273, 20267–20275. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Dhe-Paganon,S., Trüb,T., Nolte,R.T. and Shoelson,S.E. (1996) Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell, 85, 695–705. [DOI] [PubMed] [Google Scholar]
- Franck Z., Gary,R. and Bretscher,A. (1993) Moesin, like ezrin, colocalizes with actin in the cortical cytoskeleton in cultured cells, but its expression is more variable. J. Cell Sci., 105, 219–231. [DOI] [PubMed] [Google Scholar]
- Gary R. and Bretscher,A. (1995) Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell, 6, 1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault J.A., Labesse.G., Mornon.J.P. and Callebaut,I. (1999) The N-termini of FAK and JAKs contain divergent band 4.1 domains. Trends Biochem. Sci., 24, 54–57. [DOI] [PubMed] [Google Scholar]
- Hamada K., Shimizu,T., Matsui,T., Tsukita,Sh., Tsukita,Sa. and Hakoshima,T. (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J., 19, 4449–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K., Shimizu,T., Matsui,T., Tsukita,Sh., Tsukita,Sa. and Hakoshima,T. (2001) Crystallographic characterization of the radixin FERM domain bound to the cytosolic tail of the adhesion molecule ICAM-2. Acta Crystallogr. D, 57, 891–892. [DOI] [PubMed] [Google Scholar]
- Heiska L., Alfthan,K., Grönholm,M., Vilja,P., Vaheri,A. and Carpén,O. (1998) Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem., 273, 21893–21900. [DOI] [PubMed] [Google Scholar]
- Helander T.S., Carpen,O., Turunen,O., Kovanen,P.E., Vaheri,A. and Timonen,T. (1996) ICAM-2 redistributed by ezrin as a target for killer cells. Nature, 382, 265–268. [DOI] [PubMed] [Google Scholar]
- Hirao M., Sato,N., Kondo,T., Yonemura,S., Monden,M., Sasaki,T., Takai,Y., Tsukita,Sh. and Tsukita,Sa. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol., 135, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A.H. and Weis,W.I. (2001) The structure of the β-catenin/ E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell, 105, 391–402. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jons T. and Drenckhahn,D. (1992) Identification of the binding interface involved in linkage of cytoskeletal protein 4.1 to the erythrocyte anion exchanger. EMBO J., 11, 2863–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.S., Cooper,D.R., Devedjiev,Y., Derewenda,U. and Derewenda,Z.S. (2002) The structure of the FERM domain of merlin, the neurofibromatosis type 2 gene product. Acta Crystallogr. D, 58, 381–391. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Li S.C., Zwahlen,C., Vincent,S.J., McGlade,C.J., Kay,L.E., Pawson,T. and Forman-Kay,J.D. (1998) Structure of a Numb PTB domain–peptide complex suggests a basis for diverse binding specificity. Nat. Struct. Biol., 5, 1075–1083. [DOI] [PubMed] [Google Scholar]
- Lickert H., Bauer,A., Kemler,R. and Stappert,J. (2000) Casein kinase II phosphorylation of E-cadherin increases E-cadherin/β-catenin interaction and strengthens cell–cell adhesion. J. Biol. Chem., 275, 5090–5095. [DOI] [PubMed] [Google Scholar]
- Magendantz M., Henry,M.D., Lander,A. and Solomon,F. (1995) Interdomain interactions of radixin in vitro. J. Biol. Chem., 270, 25324–25327. [DOI] [PubMed] [Google Scholar]
- Mangeat P., Roy,C. and Martin,M. (1999) ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol., 9, 187–192. [DOI] [PubMed] [Google Scholar]
- Nakamura F., Huang,L., Pestonjamasp,K., Luna,E.J. and Furthmayr,H. (1999) Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell, 10, 2669–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Niggli V., Andréoli,C., Roy,C. and Mangeat,P. (1995) Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett., 376, 172–176. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Patil S., Jedsadayanmata,A., Wencel-Drake,J.D., Wang,W., Knezevic,I. and Lam,S.C. (1999) Identification of a talin-binding site in the integrin β3 subunit distinct from the NPLY regulatory motif of post-ligand binding functions. The talin N-terminal head domain interacts with the membrane-proximal region of the β3 cytoplasmic tail. J. Biol. Chem., 274, 28575–28583. [DOI] [PubMed] [Google Scholar]
- Pearson M.A., Reczek,D., Bretscher,A. and Karplus,P.A. (2000) Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell, 101, 259–270. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G. and van Beek,C.G. (1999) Data processing. Acta Crystallogr. D, 55, 1631–1640. [DOI] [PubMed] [Google Scholar]
- Rouleau G.A. et al. (1993) Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature, 363, 515–521. [DOI] [PubMed] [Google Scholar]
- Sato N., Yonemura,S., Obinata,T., Tsukita,Sa. and Tsukita,Sh. (1991) Radixin, a barbed end-capping actin-modulating protein, is concentrated at the cleavage furrow during cytokinesis. J. Cell Biol., 113, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Funayama,N., Nagafuchi,A., Yonemura,S., Tsukita.Sa. and Tsukita,Sh. (1992) A gene family consisting of ezrin, radixin and moesin. Its specific localization at actin filament/plasma membrane association sites. J. Cell Sci., 103, 131–143. [DOI] [PubMed] [Google Scholar]
- Sato T., Irie,S., Kitada,S. and Reed,J.C. (1995) FAP-1: a protein tyrosine phosphatase that associates with Fas. Science, 268, 411–415. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador J.M., Alonso-Lebrero,J.L., del Pozo,M.A., Furthmayr,H., Schwartz-Albiez,R., Calvo,J., Lozano,F. and Sánchez-Madrid,F. (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol., 138, 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador J.M. et al. (1998) CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell–cell contacts. Blood, 91, 4632–4644. [PubMed] [Google Scholar]
- Shaw A.S. (2001) FERMing up the synapse. Immunity, 15, 683–686. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Seto,A., Maita,N., Hamada,K., Tsukita,Sh., Tsukita,Sa. and Hakoshima,T. (2002) Structural basis or the neurofibromatosis type 2: crystal structure of the Merlin FERM domain. J. Biol. Chem., 277, 10332–10336. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Kawashima,A., Nagafuchi,A. and Tsukita,Sh. (1994) Structural diversity of band 4.1 superfamily members. J. Cell Sci., 107, 1921–1928. [DOI] [PubMed] [Google Scholar]
- Trofatter J.A. et al. (1993) A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell, 72, 791–800. [DOI] [PubMed] [Google Scholar]
- Tsukita Sa., Oishi,K., Sato,N., Sagara,J., Kawai,A. and Tsukita,Sh. (1994) ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol., 126, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sa. and Yonemura,S. (1999) Cortical actin organization: lessons from ERM (Ezrin/Radixin/Moesin) proteins. J. Biol. Chem., 274, 34507–34510. [DOI] [PubMed] [Google Scholar]
- Turunen O., Wahlstrom,T. and Vaheri,A. (1994) Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol., 126, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Nagafuchi.A., Sato,N. and Tsukita,Sh. (1993) Concentration of an integral membrane protein, CD43 (leukosialin, sialophorin), in the cleavage furrow through the interaction of its cytoplasmic domain with actin-based cytoskeletons. J. Cell Biol., 120, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Hirao,M., Doi,Y., Takahashi,N., Kondo,T., Tsukita,Sa. and Tsukita,Sh. (1998) Ezrin/Radixin/Moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43 and ICAM-2. J. Cell Biol., 140, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Matsui,T., Tsukita,Sh. and Tsukita,Sa. (2002) Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J. Cell Sci., 115, 2569–2580. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lee,C.H., Mandiyan,V., Borg,J.P., Margolis,B., Schlessinger,J. and Kuriyan,J. (1997) Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J., 16, 6141–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]